Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that can adapt to changing environments and can secrete an exopolysaccharide known as alginate as a protection response, resulting in a colony morphology and phenotype referred to as mucoid. However, how P. aeruginosa senses its environment and activates alginate overproduction is not fully understood. Previously, we showed that Pseudomonas isolation agar supplemented with ammonium metavanadate (PIAAMV) induces P. aeruginosa to overproduce alginate. Vanadate is a phosphate mimic and causes protein misfolding by disruption of disulfide bonds. Here we used PIAAMV to characterize the pathways involved in inducible alginate production and tested the global effects of P. aeruginosa growth on PIAAMV by a mutant library screen, by transcriptomics, and in a murine acute virulence model. The PA14 nonredundant mutant library was screened on PIAAMV to identify new genes that are required for the inducible alginate stress response. A functionally diverse set of genes encoding products involved in cell envelope biogenesis, peptidoglycan remodeling, uptake of phosphate and iron, phenazine biosynthesis, and other processes were identified as positive regulators of the mucoid phenotype on PIAAMV. Transcriptome analysis of P. aeruginosa cultures growing in the presence of vanadate showed differential expression of genes involved in virulence, envelope biogenesis, and cell stress pathways. In this study, it was observed that growth on PIAAMV attenuates P. aeruginosa in a mouse pneumonia model. Induction of alginate overproduction occurs as a stress response to protect P. aeruginosa, but it may be possible to modulate and inhibit these pathways based on the new genes identified in this study.
INTRODUCTION
As a Gram-negative bacterium encounters conditions that threaten its homeostasis, protective responses must be activated in order for the bacterium to survive. Pseudomonas aeruginosa is an opportunistic pathogen that can easily adapt from conditions in the environment to those in the host. Typically, P. aeruginosa is not harmful to healthy humans; however, a compromised immune system or traumatic event such as a wound or a burn can provide an opportunity for P. aeruginosa to cause infection. P. aeruginosa poses a substantial threat to the life span of individuals with the hereditary disease cystic fibrosis (CF). Mutation of the cystic fibrosis transmembrane conductance regulator (CFTR) gene has systemic effects on humans; one of the major causes of morbidity and mortality in these patients is the acquisition of respiratory infections by pathogens such as P. aeruginosa and Burkholderia cepacia complex organisms.
Modern antibiotics have increased the life span of CF patients by controlling bacterial burden in the lungs. Due to intrinsic and acquired drug resistance of P. aeruginosa, the life span of CF patients is still limited. Furthermore, while numerous vaccines for protection against P. aeruginosa have been tested, no vaccine has reached the market (1). Decades of research have shown that P. aeruginosa upregulates mechanisms through both environmental sensing and mutational events to survive in niches such as those found in the CF lung. One particular phenotype has received extensive study over the last 3 decades: alginate overproduction. When P. aeruginosa overproduces the exopolysaccharide known as alginate, a phenotype referred to as mucoid is evident (see Fig. S1 in the supplemental material). CF patients are initially infected with organisms that resemble environmental isolates that do not overproduce alginate (2, 3). Mouse infection studies have also indicated that alginate production is essential for the organism to chronically colonize the lung (4, 5).
P. aeruginosa has at least two well-characterized mechanisms for activating alginate overproduction. The first mechanism is genetic inactivation of the main negative regulator mucA, which encodes an anti-sigma factor. The primary role of MucA is to inhibit the alternative sigma factor σ22 (also known as AlgU or AlgT) by sequestering it to the inner membrane (6). MucA is an inner membrane protein with its C terminus in the periplasm and N terminus in the cytoplasm. When mucA is inactivated (7), σ22 activates the expression of at least 293 genes that are referred to as the σ22 regulon (8). One subset of these genes is the algD alginate biosynthetic operon that encodes the machinery to synthesize and secrete the exopolysaccharide (9). Many P. aeruginosa isolates from CF patients have mutations in the mucA gene (7, 10–13) and are constitutively mucoid.
Recently, a mechanism controlling alginate overproduction, independent of mutations, was characterized and is known as regulated proteolysis of MucA (14). MucA can be proteolytically cleaved by the protease AlgW (8, 15). The AlgW protease can be activated by the specific amino acids of the C terminus of various envelope proteins (15, 16). After AlgW acts upon the C terminus of MucA, a second protease, MucP, further cleaves MucA, likely near the transmembrane-spanning domain (15, 17). At this point, MucA is further processed by the ClpXP protease complex in the cytoplasm, leading to transcriptionally active σ22 (18). This sequence of proteolytic events can be induced by membrane stress. MucB and MucD are also negative regulators of σ22 (19, 20) and are encoded in the same operon as algU and mucA. MucB interacts with the C terminus of MucA and protects it from proteolytic degradation (16). MucD is a serine protease and chaperone (17, 19, 20), which likely performs housekeeping functions in the periplasm by degrading or refolding misfolded proteins. In the absence of MucD, alginate overproduction occurs through MucP proteolysis of MucA (17).
Our recent in vitro work indicates that Pseudomonas isolation agar supplemented with ammonium metavanadate (PIAAMV) induces alginate overproduction by activating the alginate proteolytic cascade (21). Ammonium metavanadate (AMV) is an ammonia salt of vanadate, which is an oxoanion of the transitional metal vanadium. In solution, vanadate resembles phosphate, and therefore, vanadate will likely bind proteins that also bind phosphate (22). Vanadate has been shown to induce modification of lipid A, the anchor of lipopolysaccharide (LPS), which affects cell membrane integrity (21, 23, 24). Based on our previous study which demonstrated that alginate overproduction occurs when P. aeruginosa is grown on PIAAMV, we proposed that vanadate induces envelope stress in this organism (21). We also showed that the three essential components of this medium needed to activate alginate overproduction are triclosan, vanadate, and magnesium chloride (21). However, questions remained as to which genes are required for this effect, how vanadate causes membrane stress, which molecular targets are affected, and, furthermore, how this type of cell membrane stress affects the virulence and transcriptome of P. aeruginosa. Our hypothesis was that growth of P. aeruginosa on PIAAMV affects genes outside the alginate pathway as well as systems that lead to regulation of the mucoid phenotype. To characterize inducible alginate overproduction on PIAAMV, the PA14 nonredundant transposon library was screened on PIAAMV to identify mutants that were not capable of alginate overproduction. By using the lack of alginate overproduction on PIAAMV as a visual reporter, we identified 135 mutants that revealed potentially novel pathways for alleviating cell membrane stress through compensatory pathways. The effect of vanadate on the transcriptome was determined by microarray analysis, which identified large sets of coordinated regulated genes and in various pathways. We observed that growth of P. aeruginosa on PIAAMV attenuated virulence in an acute mouse pneumonia model. Collectively, our data indicate that PIAAMV affects a multitude of gene systems in P. aeruginosa leading to the activation of compensatory pathways linked to the capability of this bacterium to adapt and survive. An understanding of these networks may provide valuable knowledge for developing novel pharmacological agents and strategies against P. aeruginosa.
MATERIALS AND METHODS
Screening of the PA14 nonredundant library of transposon mutants on PIA and PIAAMV.
Pseudomonas isolation agar (PIA) (BD Difco) and PIAAMV (21) were prepared fresh daily in 150-mm petri dishes. The PA14 nonredundant mutant library was obtained from Massachusetts General Hospital and the Broad Institute of MIT and Harvard. As recommended by the PA14 library project (http://ausubellab.mgh.harvard.edu/cgi-bin/pa14/home.cgi), the library was copied according to protocols with the following modifications. Two-milliliter, deep-well, 96-well growth plates containing 500 μl of L-broth supplemented with 15 μg/ml gentamicin were inoculated with a thawed culture from the original plates. Plates were incubated at 37°C overnight with shaking at 60 rpm. One milliliter of L-broth supplemented with glycerol (22.5%, wt/vol) and 15 μg/ml gentamicin was added directly to the 2-ml culture well. One plate required kanamycin for selection, and the protocol was performed as described above, with 200 μg/ml kanamycin in place of gentamicin. The culture grown overnight was used to inoculate the PIA and PIAAMV plates directly. Sterile 200-μl pipette tips were used to pick up approximately 1 μl of the cell suspension to inoculate the PIA and PIAAMV plates. PIA and PIAAMV plates were incubated at 25°C and monitored several times a day to observe the phenotypes of the strains on the two growth media. At 48 h, prospective nonmucoid mutants on PIAAMV were subcultured again on PIA and PIAAMV and incubated at 25°C for 2 to 3 days. To show the phenotypes that were observed, 5-μl suspensions of the strains identified were spotted onto PIAAMV plates and incubated for 3 days at 25°C. Strains confirmed to have a nonmucoid phenotype on PIAAMV were saved and later rechecked by two more independent passages on PIA and PIAAMV; the phenotypes were imaged and visualized under a dissecting microscope at ×10 and ×40 magnifications. Strains that passed these tests were then denoted on the PA14 genome sequence in CLC Workbench version 6, and a genome hit map was generated. The distribution of the PseudoCAP (Pseudomonas Community Annotation Project) classes and the localizations of the mutants identified were generated by extracting annotation information from the PA14 genome (25) with Microsoft Excel 2007. Graphs generated in Excel were then exported and edited in Adobe Illustrator.
Imaging of PA14 mutants.
To visualize the mucoid or nonmucoid phenotypes on PIAAMV, 5-μl cell suspensions were spotted onto PIAAMV and incubated at 25°C for 3 days. The plate was then rested on top of a white light transilluminator. A Canon 450D camera was used to image the plates, with the picture captured at a 45° angle. Images were cropped in Adobe Photoshop CS5 and assembled into a panel in Adobe Illustrator CS5.
Affymetrix GeneChip microarray analysis.
Strain PAO1 was cultured on both PIA and PIAAMV for 18 h. Three separate plates were used for each medium. The cells from each plate were scraped into 4 ml of RNA Protect (Qiagen) and stored immediately at −80°C. RNA isolation, cDNA preparation, labeling, and microarray analysis were performed as previously described (26). Microarray analysis was performed by using DNASTAR Array Star version 5.0. The software was used to normalize the raw expression data and generate gene lists with fold change values. CLC WorkBench version 6 was used to generate the volcano plot comparing the P values to the fold changes of the gene expression data set. Gene lists were compiled with relevant information from the PAO1 annotation (25) in Microsoft Excel 2007. Graphs were exported and annotated in Adobe Illustrator CS5. Microarray data reported previously by Tart et al. (27) were compared to the data set compiled in this study, with a P value cutoff of >0.01. The resulting Venn diagram was further annotated and color coded in Adobe Illustrator CS5.
Murine acute pneumonia model.
P. aeruginosa strain PAO1 was cultured on PIA or PIAAMV (21) for 18 h at 37°C. Cells were swabbed from the agar plates and suspended in phosphate-buffered saline (PBS). Twofold serial dilutions were made, and the optical density at 595 nm (OD595) was measured on a PerkinElmer Victor3 multilabel plate reader (Waltham, MA). Linear regression was used to calculate the volume of the suspension to give the intended dose of 4 × 107 CFU in 20 μl. Doses were prepared for infection, and an aliquot was used to ascertain the absolute CFU value of the dose by serial dilution and plating of 10-μl 10-fold dilutions onto PIA. Eight-week-old female BALB/c mice were obtained from Harlan Laboratories. Mice were anesthetized by intraperitoneal injection of 0.25 ml of ketamine (6.7 mg/ml) and xylazine (1.3 mg/ml) in 0.9% saline. Bacterial doses were administered to the anesthetized mice by pipetting 10 μl directly into each nostril. The infected mice were carefully monitored for the duration of the survival experiments. Four mice in each group were euthanized at 6 h postinfection for the determination of the bacterial burden in the organs. Nasal washes with PBS were used to determine the number of CFU contained in the nares. Lungs of the mice were harvested; one half was used for histology with hematoxylin and eosin staining, and the other was homogenized for plating to determine the CFU loads. For the rechallenge experiments, PAO1 cells were cultured on PIA. The mice were rechallenged with the same dose 1 month after the first challenge, and as a control, a set of naive mice were used for comparison. All mouse experiments were performed according to protocols approved by the University of Virginia Animal Care and Use Committee (protocol number 2844), conforming to AAALAC International accreditation guidelines.
Microarray data accession number.
The microarray data are available on the GEO (Gene Expression Omnibus) website at http://www.ncbi.nlm.nih.gov/projects/geo under accession number GSE48429.
RESULTS AND DISCUSSION
Genes required for the inducible mucoid phenotype of P. aeruginosa were identified by growth on PIAAMV.
In our previous study, we observed that growth on PIAAMV activates proteolysis of MucA, leading to alginate overproduction (21). In that same study, we tested a number of isogenic mutants on PIAAMV to identify predictable genes (e.g., alginate biosynthetic genes and regulators, etc.) that were required for the inducible mucoid phenotype (21). In the current study, the PA14 nonredundant library (28) was screened for other genes required for alginate overproduction on PIAAMV (see Fig. S1A and S1B in the supplemental material). The PA14 mutant library, which includes 5,850 mutants, was replicated on PIA and PIAAMV. It was observed that all strains from the PA14 library were capable of growth on PIA and PIAAMV. The strains were incubated at 25°C, which allowed easier identification of the mucoid phenotype. Nonmucoid strains on PIAAMV were selected and passed three times on PIA and PIAAMV to confirm their lack of alginate overproduction.
From this analysis, four types of genes were identified: those encoding known alginate enzymes and regulators, metabolic genes, genes involved in modulating envelope stress (e.g., alleviating effects), and other genes (see Fig. S1B in the supplemental material). Known genes directly involved in the synthesis, secretion, and regulation of alginate that were inactivated were grouped into the alginate class (see Fig. S1B in the supplemental material). Seventeen percent of the nonmucoid mutants on PIAAMV were of the known alginate class. We also hypothesized that metabolic genes would be required for alginate overproduction. The metabolic class of genes that were required for mucoidy on PIAAMV comprised 24% of the mutants identified. We hypothesize that loss of some genes would alleviate membrane stress caused by PIAAMV. For example, if a gene product was involved in the uptake of vanadate into cells, inactivation of this gene would prohibit the uptake of vanadate, alleviate vanadate-induced stress, and hypothetically decrease the alginate stress response. Forty-four percent of the genes identified in our PA14 screen were grouped into the alleviation class (see Fig. S1B in the supplemental material). In addition to the three main classes of genes required for the mucoid phenotype on PIAAMV, we also identified mutants that could not be classified in the main groups and refer to this group as “others” (15% of the mutants identified). In Table 1, the strains with a nonmucoid phenotype on PIAAMV are indicated and assigned to the four classes and subclasses.
Table 1.
P. aeruginosa genes required for the mucoid phenotype on PIAAMV
| Class and subclass | Locus taga | Gene | Productc | PIAAMV/PIA fold changeb |
|---|---|---|---|---|
| Alginate | PA0762 | algU | Sigma factor AlgU | |
| PA1801 | clpP | ATP-dependent Clp protease proteolytic subunit | ||
| PA1802 | clpX | ATP-dependent Clp protease ATP-binding subunit ClpX | ||
| PA3257 | prc | Periplasmic tail-specific protease | ||
| PA3540 | algD | GDP-mannose 6-dehydrogenase AlgD | 2.14 | |
| PA3542 | alg44 | Alginate biosynthesis protein Alg44 | 2.56 | |
| PA3545 | algG | Alginate-C5-mannuronan epimerase | 2.17 | |
| PA3546* | algX | Alginate biosynthesis protein AlgX | 2.5 | |
| PA3547 | algL | Poly(beta-d-mannuronate) lyase precursor AlgL | 2.5 | |
| PA3548* | algI | Alginate O-acetyltransferase AlgI | 1.84 | |
| PA3550 | algF | Alginate O-acetyltransferase | 2.58 | |
| PA3551* | algA | Mannose-1-phosphate guanylyltransferase/mannose-6-phosphate isomerase | 2.69 | |
| PA3649* | mucP | Putative membrane-associated zinc metalloprotease | ||
| PA4446 | algW | AlgW protein | 1.17 | |
| PA5255 | algQ | Alginate regulatory protein AlgQ | 1.13 | |
| PA5261 | algR | Alginate biosynthesis regulatory protein AlgR | 1.28 | |
| PA5483 | algB | Two-component response regulator AlgB | ||
| Alleviating | ||||
| Cell envelope (lipid A, LPS, and membrane stability) | PA0011 | htrB | Putative 2-OH-lauroyltransferase | |
| PA0337* | ptsP | Phosphoenolpyruvate-protein phosphotransferase | 1.48 | |
| PA0595 | ostA | Organic solvent tolerance protein OstA precursor | 1.24 | |
| PA1180 | phoQ | Two-component sensor PhoQ | ||
| PA2023 | galU | UTP-glucose-1-phosphate uridylyltransferase | ||
| PA2240 | pslJ | Conserved hypothetical protein | 1.54 | |
| PA2525 | ompB | Putative outer membrane protein | ||
| PA2526 | muxC | Putative efflux transporter | 1.42 | |
| PA2527 | muxB | Putative RND efflux transporter | 1.15 | |
| PA2965 | fabF1 | Beta-ketoacyl-acyl carrier protein synthase II | ||
| PA3194 | edd | 6-Phosphogluconate dehydratase | ||
| PA3242 | htrB | Putative lipid A biosynthesis lauroyl/myristoyl acyltransferase | ||
| PA3828 | lptF | Putative LPS permease | 1.45 | |
| PA4293 | pprA | Putative two-component sensor | ||
| PA4370 | icmP | Insulin-cleaving metalloproteinase outer membrane | ||
| PA4404 | Putative peptidase | |||
| PA4406 | lpxC | UDP-3-O-acyl-N-acetylglucosamine deacetylase | ||
| PA4612 | Ankyrin-like protein | |||
| PA5001 | Conserved hypothetical protein; glycosyltransferase | 1.52 | ||
| PA5004 | wabH | Putative glycosyltransferase | ||
| Peptidoglycan | PA0666 | anmK | Putative chaperone | |
| PA0667 | Putative metallopeptidase | |||
| PA1726 | bglX | Periplasmic beta-glucosidase | ||
| PA2621 | clpS | Putative cytoplasmic protease | ||
| PA4001 | sltB1 | Soluble lytic transglycosylase B | 1.31 | |
| PA5124 | ntrB | Two-component sensor NtrB | −1.27 | |
| Phosphate | PA5199* | envZ | Two-component sensor EnvZ | 1.37 |
| PA5235* | glpT | Glycerol-3-phosphate transporter | ||
| PA5361 | phoR | Two-component sensor PhoR | ||
| PA5365 | phoU | Phosphate uptake regulatory protein PhoU | ||
| PA5366 | pstB | Phosphate ABC transporter, ATP-binding protein | ||
| PA5368 | pstC | Phosphate ABC transporter, permease protein | ||
| PA5369 | pstS | Periplasmic phosphate-binding protein | ||
| Chaperones and protein folding | PA0090 | Putative ClpA/B-type chaperone | 3.68 | |
| PA0594 | surA | Peptidyl-prolyl cis-trans isomerase SurA | 1.47 | |
| PA3737 | dsbC | Thiol:disulfide interchange protein DsbC | ||
| PA3821 | secD | Protein export membrane protein SecD | ||
| PA5128 | secB | Secretion protein SecB | 1.19 | |
| PA5568 | yidC | Putative inner membrane protein, 60 kDa | ||
| Phenazine biosynthesis | PA0051 | phzH | Potential phenazine-modifying enzyme | −1.93 |
| PA0652 | vfr | Cyclic AMP receptor-like protein | ||
| PA1003 | mvfR | Transcriptional regulator MvfR | ||
| PA1431 | rsaL | Regulatory protein RsaL | 1.5 | |
| PA1899 | phzA2 | Probable phenazine biosynthesis protein | ||
| PA1905 | phzG2 | Probable pyrodoxamine 5′-phosphate oxidase | ||
| PA2586* | gacA | Response regulator GacA | ||
| PA2770* | Putative phenazine biosynthesis protein, PhzF family | |||
| PA2798 | rssB | Putative two-component response regulator | ||
| PA4209 | phzM | Probable phenazine-specific methyltransferase | ||
| PA4210* | phzA1 | Probable phenazine biosynthesis protein | ||
| PA4211* | phzB1 | Probable phenazine biosynthesis protein | ||
| PA4214 | phzE1 | Phenazine biosynthesis protein PhzE | ||
| Metabolism | ||||
| Metabolism | PA0394 | Putative PLP-dependent enzyme | ||
| PA0558 | Conserved hypothetical protein | −1.68 | ||
| PA0593 | pdxA | Pyridoxal phosphate biosynthetic protein PdxA | 1.43 | |
| PA0597 | Putative nucleotidyltransferase | |||
| PA0793 | Conserved hypothetical protein | |||
| PA0794 | Probable aconitate hydratase | |||
| PA0796 | prpB | Carboxyphosphonoenolpyruvate phosphonomutase | ||
| PA1838 | cysI | Sulfite reductase | −1.25 | |
| PA2068 | Putative MFS transporter | |||
| PA2531 | Putative aminotransferase | −1.82 | ||
| PA2796 | tal | Transaldolase | ||
| PA2804 | Putative phosphohydrolase | |||
| PA3113 | trpF | Phosphoribosyl anthranilate isomerase | 1.57 | |
| PA3772 | Conserved hypothetical protein | |||
| PA4007 | proA | Probable gamma-glutamyl phosphate reductase | ||
| PA4402 | argJ | N-Acetylglutamate synthase | ||
| PA4465 | Putative ATP kinase | |||
| PA4640 | mqo | Malate:quinone oxidoreductase | ||
| PA4758 | carA | Carbamoyl-phosphate synthase small chain | −1.45 | |
| PA5236 | Putative aromatic hydrocarbon reductase | 1.28 | ||
| PA5331 | pyrE | Orotate phosphoribosyltransferase | ||
| Zinc | PA4723 | dksA | Suppressor protein DksA | 1.13 |
| PA5498 | znuA | Periplasmic zinc-binding protein | ||
| Iron | PA0969 | tolQ | TolQ protein | |
| PA2403 | Putative iron-regulated membrane protein | −1.31 | ||
| PA2406 | Conserved hypothetical protein | −1.36 | ||
| PA2407 | Putative adhesion protein | |||
| PA2410 | Putative ABC transporter, periplasmic substrate-binding protein | |||
| Others | ||||
| Motility | PA0411* | pilJ | Type IV pilus methyl-accepting chemotaxis transducer PilJ | |
| PA1086 | flgK | Flagellar hook-associated protein 1, FlgK | ||
| PA1450 | Conserved hypothetical protein | |||
| PA1454 | fleN | Flagellar synthesis regulator FleN | ||
| PA1822 | fimL | Pilin biosynthetic protein | ||
| PA3351* | flgM | Putative negative regulator of flagellin synthesis, FlgM | −1.24 | |
| PA4526 | pilB | Type 4 fimbrial biogenesis protein PilB | ||
| PA4552 | pilW | Type 4 fimbrial biogenesis protein PilW | −2.98 | |
| PA4556 | pilE | Type 4 fimbrial biogenesis protein PilE | −1.58 | |
| Others | PA0124 | Putative plasmid system protein | ||
| PA2423 | Hypothetical | 1.43 | ||
| PA2615 | ftsK | Cell division/stress response protein | 1.18 | |
| PA2780 | Putative transcriptional regulator | |||
| PA3011 | topA | DNA topoisomerase I | ||
| PA4951 | orn | Oligoribonuclease | −1.22 | |
| PA5239* | rho | Transcription termination factor Rho | 1.48 | |
| PA5515 | Conserved hypothetical protein | |||
| PA14_07490 | Hypothetical protein | ND | ||
| PA14_53580 | Hypothetical protein | ND | ||
| PA14_56340 | Intergenic between mntH1 and PA14_56360 | ND |
An asterisk indicates that more than one mutant with a transposon in the same gene was identified.
Shown is the fold change observed for the gene in the PIAAMV/PIA transcriptome analysis. ND, not determined.
RND, resistance nodulation cell division; PLP, pyridoxal 5-phosphate; MFS, major facilitator superfamily.
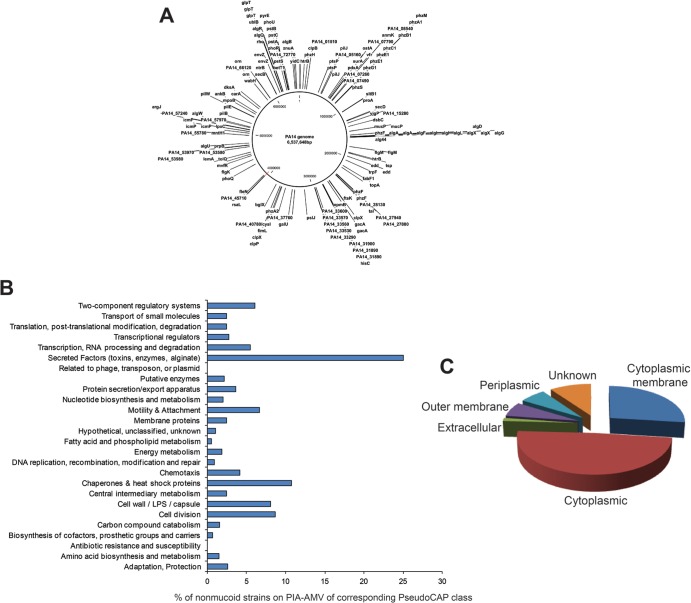
The screen for genes required for the mucoid phenotype on PIAAMV resulted in the identification of 135 nonmucoid PA14 mutants. The PA14 mutant library does have some redundancy, and therefore, of the 135 mutants identified, 117 were of unique genes. Since the PAO1 locus tag number is used more commonly, for clarity, we have displayed the corresponding PAO1 locus tag instead of the PA14 tag, except for genes not present in PAO1 (Table 1). Plotting of the genomic location for each of the mutants identified revealed that genes affecting this phenotype are distributed throughout the P. aeruginosa chromosome (Fig. 1A). When the genes were organized by PseudoCAP classes, as expected, most genes identified were a part of the secreted factor class, including the alginate biosynthesis genes (Fig. 1B). For alginate overproduction on PIAAMV, genes that encode products localizing to all cellular compartments are required (Fig. 1C).
Fig 1.
Genome-wide analysis of genes required for the mucoid phenotype of P. aeruginosa strain PA14 on PIAAMV. Of the 5,850 PA14 mutants screened, 135 mutants with a nonmucoid phenotype on PIAAMV were identified. Of these nonmucoid strains, 117 unique genes were identified as having an effect on the mucoid phenotype of P. aeruginosa on PIAAMV. (A) The PA14 genome with the 135 transposon insertions that abolished the PIAAMV-induced mucoid phenotype of PA14 is shown. (B) Mutants were grouped according to the PseudoCAP class and are shown as a percentage of the total number of genes in the class. (C) The distribution of gene product localization of the nonmucoid mutants on PIAAMV is shown.
Alginate genes are required for the mucoid phenotype on PIAAMV.
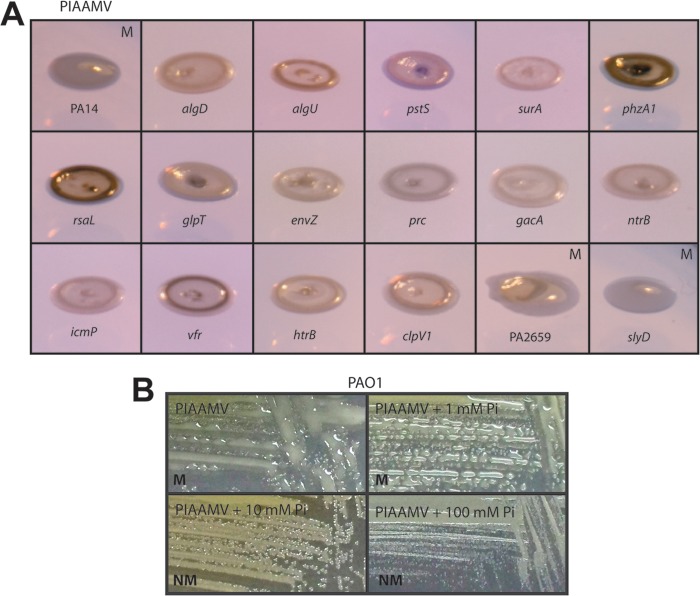
Since PIAAMV causes alginate overproduction by P. aeruginosa (21), we predicted that we would identify mutants with transposon insertions in alginate biosynthesis genes (Table 1). A recent global-scale analysis of regulators of P. aeruginosa phenotypes identified AlgW as a top regulator of transcriptional regulatory networks, since the loss of algW causes a large number of effects on the global genetic network (29). Inactivation of algQ resulted in a visibly nonmucoid phenotype on PIAAMV (Table 1). AlgQ is an anti-sigma factor for the housekeeping sigma factor RpoD (30). It is possible that inactivation of algQ could increase the expression levels of compensatory factors under the control of RpoD that may relieve the stress caused by vanadate and allow P. aeruginosa to be nonmucoid on PIAAMV. Prc is a protease that acts upon mutated MucA proteins (31). We have proposed that Prc may play roles in regulated proteolysis of nonmutated MucA proteins (14), and here we observed that the prc mutant was nonmucoid on PIAAMV, supporting our hypothesis (Fig. 2A).
Fig 2.
Nonmucoid PA14 mutants on PIAAMV and effect of phosphate on phenotype. (A) Five-microliter suspensions of strains were spotted onto PIAAMV and cultured for 72 h at 25°C. The strains were digitally imaged with the plates resting on a white light transilluminator with the camera set at an angle of approximately 45° to show the absence or presence of the mucoid mound caused by alginate overproduction, as demonstrated for strain PA14. Nonmucoid mutants are shown along with several mucoid mutants on PIAAMV: PA2659 and slyD. PA14, slyD, and PA2659 strains are mucoid on PIAAMV and are indicated with an “M,” while all other strains shown are nonmucoid on PIAAMV. (B) PIA contains 0.3 mM phosphate, and PIAAMV contains 0.3 mM phosphate and 0.34 mM vanadate. When the phosphate concentration (with either sodium or potassium phosphate) in PIAAMV was increased to 1.3 mM (0.3 mM PIA supplemented with 1 mM phosphate), the mucoid phenotype was still evident. However, increased phosphate supplementation visibly decreased alginate overproduction to wild-type levels.
Cell envelope genes required for the mucoid phenotype on PIAAMV.
Recent large-scale interaction pathway studies of Escherichia coli indicated that significant cross talk occurs between bacterial envelope gene systems (32). Previously, we showed that PIAAMV, in addition to activating alginate overproduction, also causes palmitoylation of the hydrophobic anchor of a lipopolysaccharide, lipid A (21). This suggested that other cell stress reduction pathways besides alginate are also induced on PIAAMV (14). In this study, we identified two htrB mutants, PA0011 and PA3242, which were nonmucoid on PIAAMV (Table 1). HtrB, a P. aeruginosa orthologue of E. coli LpxL, is an inner membrane acyltransferase responsible for transferring a lauryl group to the lipid A precursor (33); however, P. aeruginosa fatty acylation of lipid A differs from that of E. coli in that the sequence of enzymatic events is not the same (34). A recent study suggests that PA0011 is involved in antibiotic resistance mechanisms (35). In the above-mentioned study, PA0011 was shown to be regulated by quorum sensing (QS), and PA0011 mutants were susceptible to various antibiotics and environmental stresses. It is possible that inactivation of one of the htrB genes may cause compensatory effects or lead to modified lipid A that may alleviate the cell stress caused by PIAAMV.
In the nonmucoid PIAAMV screen, we identified two mutants that function in LPS transport: the ostA (lptD) and lptF (PA3828) mutants (Table 1). LptD plays a role in delivering LPS to the outer leaflet of the outer membrane, whereas LptF is a component of the LPS ABC transporter. Recently, peptide-mimetic antibiotics against P. aeruginosa LptD have been designed and have been shown to have potent antimicrobial activity (36). The inactivation of ostA (lptD) may have effects on the expression of the downstream gene surA, which is also required for the mucoid phenotype on PIAAMV (Table 1) and is discussed below. We observed that the inactivation of lptF (PA3828), which encodes a component of the LPS permease, also caused the nonmucoid phenotype on PIAAMV, and to our surprise, the lptF mutant grew more rapidly than wild-type PA14 on PIAAMV (data not shown). Inactivation of lptF would likely cause cells to have less LPS, and this also suggests that P. aeruginosa has other ways to shuttle LPS, indicating that lptF is not essential or that compensatory factors can make up for the loss of lptF. The amount of O-antigen LPS was measured, and the lptF mutant had significantly less than did wild-type PA14, as determined by Western blotting of isolated LPS detected with LPS serotype-specific antibodies (data not shown). We hypothesize that decreased LPS expression levels may upregulate compensatory factors that change membrane integrity, causing P. aeruginosa to remain nonmucoid on PIAAMV.
Mutation of peptidoglycan genes that turn off the mucoid phenotype on PIAAMV.
Whereas alginate protects the bacterial outer membrane, peptidoglycan (PG) provides structural support to the envelope. Inhibition of PG biosynthesis has been shown to activate regulated proteolysis of MucA and alginate gene expression (8), and, as described below, we observed that many PG genes were upregulated during growth on PIAAMV (see Table S1 in the supplemental material). During bacterial growth, PG is recycled by transport back to the cytoplasm. Transposon mutants of anmK or the next gene in the operon, PA0667, are nonmucoid on PIAAMV (Table 1). AnmK (PA0666) is a kinase that phosphorylates a recycling-intermediate PG sugar (1,6-anhydro-N-acetylmuramic acid), which is then followed by further processing by other enzymes (37). A recent structural analysis has shown that AnmK may also be a hydrolase (38). anmK is transcribed with another gene, PA0667, which is a putative peptidase similar to E. coli YebA. PA0667 and YebA have LytM (lysostaphin) domains, which cleave PG cross-links. Phenotypes of mutants with LytM domains resemble those of amiA, amiB, and amiC mutants, indicating that several sets of amidases may be utilized for PG recycling (39). These data suggest that the blocking of PG recycling stabilizes the membrane during growth on PIAAMV. ClpS is the adaptor protein of the ClpAP protease complex. clpS is in an operon with clpA in P. aeruginosa. The PA14 clpS mutant was nonmucoid on PIAAMV (Table 1). Without clpS, the ClpAP protease would likely recognize different substrates (40). Two recent independent studies have shown that inactivation of clpS increases resistance to β-lactam antibiotics in strain PA14 (41, 42). Based on these data, we hypothesize that the effect of clpS on PIAAMV may be through effects on PG. SltB1 is a membrane-bound transglycosylase involved in PG formation. The sltB1 mutant was nonmucoid on PIAAMV (Table 1). The most probable explanation for this effect is that the inactivation of sltB1 caused increased expression of one of the other PG transglycosylase enzymes.
Genes involved in phosphate uptake and regulation that are required for the mucoid phenotype on PIAAMV, and effect of phosphate supplementation.
phoR, phoU, pstB, and pstS mutants were identified as nonmucoid on PIAAMV (Table 1 and Fig. 2). Based on this observation, we hypothesized that since vanadate is a phosphate mimic, it is possible that vanadate is taken into cells by the pst system of phosphate transport, which has also been speculated by others (43). Phosphates are essential molecules for energy production, nucleic acids, signal transduction, as well as other biological processes. Phosphate uptake effectors are encoded by the pstSCAB-phoU operon and the PhoB/PhoR two-component system (44). PstS is a high-affinity phosphate-binding protein that shuttles phosphate to the PstA, PstB, and PstC phosphate ABC transporters (44). The histidine kinase PhoR senses phosphate concentrations and phosphorylates PhoB (45), which activates the expression of the Pho regulon, leading to the differential expression of 400 proteins (46). PhoU is thought to control the phosphorylation and dephosphorylation of PhoR by interacting with the phosphate transport system (44). Of note, the phosphate transport system is essential for virulence (47).
For P. aeruginosa, it was recently shown that the periplasmic phosphate-binding protein PstS forms large appendage-like extracellular chains (48). Furthermore, a low phosphate level leads to a high PstS expression level (48). We observed that PstS is expressed at the same level on both PIA and PIAAMV (data not shown). PIA is a low-phosphate medium (48), and since no additional phosphate was added to PIAAMV, it is a low-phosphate medium as well. The concentration of phosphate in PIA is approximately 0.3 mM. We previously determined empirically that 0.3 mM AMV added to PIA caused the most robust mucoid phenotype (21). We hypothesized that supplementation of phosphate in PIAAMV would turn off the mucoid phenotype because phosphate would be taken up preferentially by the Pst system instead of vanadate. Increasing the phosphate level to 1 mM with either sodium or potassium phosphate in the growth medium did not turn off the mucoid phenotype (Fig. 2B). However, upon the addition of 10 or 100 mM additional phosphate (sodium or potassium phosphate), both PAO1 (Fig. 2B) and PA14 (data not shown) were nonmucoid on PIAAMV, overcoming the effect of vanadate in the medium. When we put mucA mucoid mutant strain PDO300 on increasing amounts of phosphate, the strain remained mucoid on PIA (data not shown), suggesting that the phosphate suppression of the mucoid phenotype was likely restricted to PIAAMV-induced alginate overproduction.
Chaperones and protein-folding enzymes that are required for mucoidy on PIAAMV.
In our previous study, we observed that the expression levels of multiple stress response chaperones were increased during growth on PIAAMV (21). In the screen for genes required for the mucoid phenotype on PIAAMV, we observed that 12% of all genes encoding chaperones were required (see Fig. S1B in the supplemental material). SurA is a periplasmic protein that is important for proper folding of envelope proteins and participates in chaperoning proteins to the outer membrane. In our current study, surA was required for the mucoid phenotype on PIAAMV (Table 1 and Fig. 2A). We also identified nonmucoid mutants with transposon insertions in all genes of the surA operon (ostA, surA, and pdxA). It is possible that inactivation of either surA or ostA affects pdxA, resulting in the nonmucoid phenotype on PIAAMV. PdxA is involved in the production of pyridoxol phosphate (vitamin B6), which is an enzymatic cofactor for transamination reactions. Vitamin B6 is a cofactor of WbpE, which is an enzyme that is involved in biosynthesis of O-antigen LPS (49). Therefore, it is possible that inactivation of the ostA-surA-pdxA operon may decrease LPS production, as we saw for the lptF LPS permease mutant. The relationship between vitamin B6 biosynthesis and LPS/alginate biosynthesis is under investigation in our laboratory.
We have hypothesized that PIAAMV results in incorrect protein folding in the periplasm (21), which causes regulated proteolysis of MucA and the mucoid phenotype. DsbC is a chaperone with protein disulfide isomerase activity. Interestingly, vanadate causes disulfide bonds to cleave or improperly form (50). Based on this, we would suspect that dsbC is required for growth on PIAAMV; however, the dsbC mutant grew on PIAAMV but was nonmucoid. SecB, SecD, and YidC are proteins of the Sec translocation complex responsible for the translocation of proteins from the cytoplasm to the envelope. Inactivation of any one of these genes would likely decrease the amount of proteins sent to the envelope. Here we observed that the secB, secD, or yidC mutant displayed a nonmucoid phenotype on PIAAMV (Table 1), which suggested that lowered protein content in the periplasm can lower membrane stress during growth under stressful conditions.
Zinc regulation genes are required for the mucoid phenotype on PIAAMV.
ZnuA is a periplasmic protein that binds zinc and participates in its transport to the cytoplasm. DksA (PA4723) is a transcription factor that responds to low zinc levels and interacts directly with RNA polymerase, modulating transcription at various promoters (51). Here we observed that dksA and znuA are required for the mucoid phenotype on PIAAMV (Table 1), suggesting that zinc is important for activating the alginate overproduction stress response.
Iron-regulated genes required for the mucoid phenotype on PIAAMV.
Vanadate has been shown to bind siderophores (52). Iron starvation has been shown to upregulate PA2403 to PA2410, uncharacterized genes that are in a single operon (25). Here, in our study, we identified that PA2403, PA2406, PA2407, and PA2410 were required for the mucoid phenotype on PIAAMV (Table 1). With little information about these genes, it is difficult determine why these mutants are nonmucoid on PIAAMV. However, PA2410 hypothetically encodes a periplasmic solute binding protein, and therefore, we speculate that inactivation of this operon may affect vanadate transport into the cell. Alternatively, the requirement of these iron-regulated genes for the mucoid phenotype on PIAAMV may be indirect through other systems. Others have hypothesized that iron is essential for ATP production by electron transport for biosynthesis of alginate (53). Therefore, these iron-regulated genes may cause metabolic problems that would prohibit the synthesis of alginate.
Relationship between phenazine biosynthesis and the mucoid phenotype on PIAAMV.
P. aeruginosa produces and secretes phenazines, which are redox-active compounds that are toxic to prokaryotes and may play a role in the infectious process (54). P. aeruginosa produces several phenazines, but the best characterized is pyocyanin. Pyocyanin production can be detected in the sputum of cystic fibrosis patients (55), is controlled by QS (54), and acts as a terminal signaling molecule that increases the expression level of the transcription factor SoxR, which in turn activates another subset of genes (56). P. aeruginosa has two sets of pyocyanin gene operons, and mutants of both operons were nonmucoid on PIAAMV (Table 1), suggesting that decreased pyocyanin secretion was sufficient to result in a nonmucoid phenotype on PIAAMV. This is surprising, due to the fact that, since there are two biosynthetic operons, it might be expected that the redundancy would alleviate the effects of inactivating the gene in the other operon. Inactivation of pyocyanin biosynthesis genes caused a black/brown visual phenotype on the colony surface when the strains were grown on PIAAMV (phzA1) (Fig. 2A). We did not notice this black colony phenotype when the strains were grown on PIA (data not shown). Previously, it was noted that siderophores, which bind and sequester iron, also form a complex with vanadium, causing the dark brown phenotype (52). In addition to pyocyanin biosynthesis genes, inactivation of key regulators of pyocyanin such as mvfR, vfr, and gacA (54) also resulted in the nonmucoid phenotype on PIAAMV (Table 1). Vfr is a cyclic AMP receptor protein that has been shown to regulate pyocyanin production (57). GacA is another global transcriptional regulator that affects pyocyanin production (58). mvfR (pqsR) is a transcription factor that regulates QS, which in turn influences pyocyanin production (59); this mutant was also nonmucoid on PIAAMV. We also noticed that the vfr mutant had a dark colony morphology similar to that of the pyocyanin biosynthesis mutants when grown on PIAAMV (Fig. 2A).
The stationary sigma factor RpoS is a negative regulator of pyocyanin production, as inactivation of rpoS leads to increased pyocyanin production (60). RssB is a negative regulator of RpoS (61), and therefore, in the absence of RssB, increased amounts of RpoS would be available in the cell, which would suppress pyocyanin production. We observed here that the rssB mutant was nonmucoid on PIAAMV (Table 1). If mucoidy on PIAAMV is dependent upon pyocyanin production, the mutants that did not produce or that exhibited decreased pyocyanin production may be complemented by the addition of exogenous pyocyanin. We grew the PA14 rsaL mutant, which produces large amounts of pyocyanin (62), and extracted pyocyanin using methods described previously to evaluate the effect of pyocyanin on the transcriptome of P. aeruginosa (56). We then supplied serial dilutions of the crude pyocyanin preparation on PIAAMV plates. Each of the nonmucoid pyocyanin mutants was inoculated onto the plates. The pyocyanin mutants remained nonmucoid on the pyocyanin-supplemented PIAAMV plates (data not shown), indicating that exogenous pyocyanin alone could not complement the pyocyanin mutants to restore the mucoid phenotype on PIAAMV. This finding suggests that the lack of pyocyanin production may affect the efflux of vanadate from the cell or that pyocyanin production itself has other effects on the physiology of the bacterium.
MexGHI-OpmD is a multidrug efflux pump that is also responsible for secretion of pyocyanin (63). A previous study has shown that MexGHI-OpmD is responsible for resistance to and growth in the presence of vanadium (63). Based on this, we hypothesized that turning off pyocyanin production allows more vanadate to be pumped out, which would alleviate vanadate-induced cell stress, and furthermore that the black/brown colony morphology of the pyocyanin mutants is a result of vanadate secretion from the cells. The dark colony morphology has also been observed for environmental Escherichia isolates grown in the presence of vanadium (64). In a study of Shewanella oneidensis, a bacterium that is capable of reducing vanadate into vanadyl ion, it was shown that the brown precipitate is granular vandyl ion (65). Secretion of vanadate from P. aeruginosa would detoxify the cells, which would alleviate the stress that is activating the mucoid phenotype on PIAAMV. We hypothesize that inactivation of pyocyanin genes would allow increased secretion of vanadate from the efflux pumps. RsaL is a negative regulator of pyocyanin and QS (62). Interestingly, we identified that the rsaL mutant was nonmucoid on PIAAMV. This was initially counterintuitive to the hypothesis described above. However, pyocyanin increases the level of expression of the MexGHI-OpmD efflux pump in a SoxR-dependent manner, as determined by microarray analysis (56). These data suggest that vanadate secretion likely alleviates the mucoid phenotype on PIAAMV.
Transcriptome analysis of P. aeruginosa strain PAO1 on PIAAMV.
A large number of studies have examined the global gene expression of various mucoid P. aeruginosa strains (8, 26, 66–71). By compiling multiple studies, a σ22 regulon with at least 293 genes has been established (8). While we know that PIAAMV activates the expression of σ22 genes (21), based on our mutant library screen, we predicted that PIAAMV would also alter the expressions of genes outside the σ22 regulon. We also hypothesized that transcriptomic analysis of P. aeruginosa cells cultured on PIAAMV compared to PIA would facilitate the characterization of new pathways by building upon the genetic screen. The P. aeruginosa Affymetrix GeneChip microarray was designed by using the PAO1 genome sequence (72) rather than the PA14 genome (73); therefore, we used strain PAO1 to observe the effect of PIAAMV on P. aeruginosa. PAO1 was cultured on both PIA and PIAAMV for 18 h at 37°C, and RNA was isolated. P. aeruginosa Affymetrix GeneChips were used to perform microarray analysis in order to characterize the effects on the transcriptome. DNASTAR Array Star version 5.0 was used to normalize the raw data and generate fold change values and statistics.
The P. aeruginosa array contains 5,570 genes, 1,716 of which were statistically differentially (P value of <0.05) expressed on PIAAMV compared to PIA. This corresponded to 31% of the genome, which supports our hypothesis that AMV affects genes outside alginate biosynthesis. The expression levels of 861 genes were increased while the expression levels of 855 genes were decreased during growth on PIAAMV. Previously, we performed shotgun proteomics of PAO1 cells grown on PIA and PIAAMV (21). In that study, 70 proteins were identified, 27 of which were statistically differentially expressed in this microarray (data not shown). The expression of 85% of these genes correlates directly with our previous proteomic analysis.
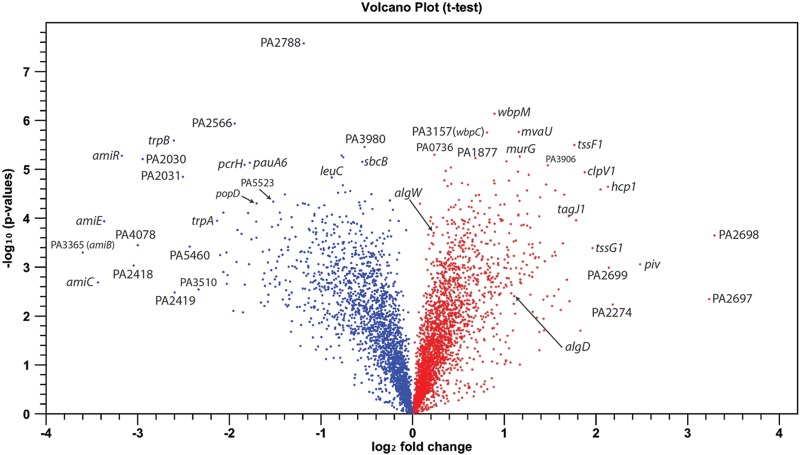
To illustrate the relationship between statistical analysis (P values) and the fold changes observed in our microarray study, a volcano plot was generated (Fig. 3). Red or blue dots indicate genes activated or repressed, respectively, as a result of the growth of strain PAO1 on PIAAMV (Fig. 3). Interestingly, a large number of genes were activated on PIAAMV at higher levels and with lower P values than those for the predicted alginate genes algW and algD (Fig. 3).
Fig 3.
Volcano plot of PAO1 genes dysregulated as a result of ammonium metavanadate. P. aeruginosa strain PAO1 was cultured on both PIA and PIAAMV for 18 h at 37°C. Microarray analysis was performed to determine the effect of AMV on the transcriptome. The log10 P value is plotted on the y axis, and the log2 fold change is plotted on the x axis. Blue or red circles indicate genes repressed or activated, respectively, by AMV, as determined by a comparison of PIAAMV to PIA. Genes highly dysregulated and genes of interest are indicated with their gene names or by PA locus tags. The major P. aeruginosa phenotype caused by PIAAMV, alginate production, is confirmed due to the increased expression levels of genes such as algW and algD. However, this figure illustrates that PIAAMV also has a number of statistically significant effects on many other genes relevant to virulence.
PIAAMV causes differential expression of secreted factors, cell wall, cell division genes; protein secretion apparatus genes; and chaperone, heat shock protein, and adaptation genes.
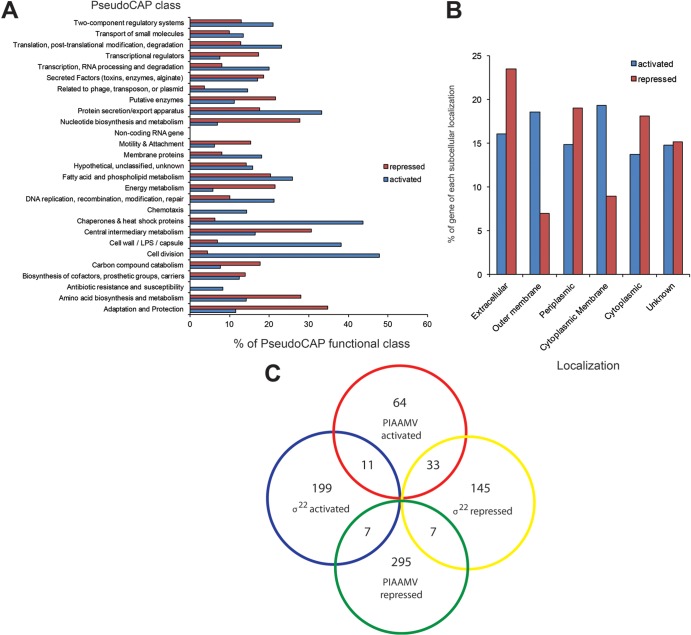
To grasp the distribution of genes affected when P. aeruginosa is cultured on PIAAMV compared to PIA, the PIAAMV list was sorted in relation to the PseudoCAP (Pseudomonas Community Annotation Project) class to which each gene was assigned. These lists were then counted and converted to correspond to the percentages of genes that make up each PseudoCAP class (Fig. 4A). This analysis allows discussion of the relative overall effect on each class of genes. In the class of secreted factors, which includes alginate genes, 20% of genes were activated, and the other 20% were repressed, meaning that 40% of all genes in this class were dysregulated by PIAAMV (Fig. 4A). As expected due to cell membrane stress induced by PIAAMV, a large number of genes corresponding to the cell wall/LPS/capsule class had increased expression levels on PIAAMV (Fig. 4A). Interestingly, half of all genes of the cell division class were activated on PIAAMV (Fig. 4A), while only a few were repressed. Our previous proteomic analysis indicated that several chaperones are upregulated in response to PIAAMV (21); here we noted that 44% of chaperones in the genome were activated in the PIAAMV regulon (Fig. 4A). We would have predicted that adaptation and protection genes would be upregulated on PIAAMV, but we observed that 35% of adaptation and protection genes were surprisingly repressed on PIAAMV (Fig. 4A). Altogether, these data suggested that growth on PIAAMV causes pleiotropic effects on the transcriptome of P. aeruginosa.
Fig 4.
AMV affects expression of an array of genes independent of σ22. PIAAMV activates σ22 and alginate overproduction as a result of cell membrane stress (21). This figure illustrates the distribution of the classes of genes and localizations of gene products affected by PIAAMV. Comparison with known σ22-controlled genes (27) indicates that the effects of AMV are beyond the σ22 regulon. (A) Genes were grouped according to PseudoCAP class (25) and are shown as a percentage of the total number of genes in the class. The percentages of genes of the PseudoCAP class repressed or activated as a result of AMV are indicated by blue and red bars, respectively. (B) Genes significantly dysregulated were also grouped based on their subcellular localizations. These groups are shown as percentages of genes of the corresponding localization, repressed or activated, due to AMV and are indicated by blue and red bars, respectively. (C) A four-way Venn diagram comparing repressed and activated genes of the σ22 regulon to the AMV regulon indicates some overlap, as expected, but clearly indicates that the AMV effect is beyond just the genes controlled by σ22.
Localization of gene products of differentially expressed genes affected by PIAAMV.
Since alginate overproduction due to PIAAMV is likely a result of abrogated envelope proteins leading to the activation of regulated proteolysis of MucA (21), it would seem feasible that most of the effects of PIAAMV would be focused on the envelope. As expected, 18% of genes that encode proteins that localize to the outer membrane were activated (Fig. 4B). However, it is clear that PIAAMV stimulates global effects on P. aeruginosa based on the fact that genes of all subcellular localizations were affected. Strikingly, a large percentage of genes (23%) encoding extracellular gene products were repressed on PIAAMV (Fig. 4B). More genes encoding products that localize to the periplasm were repressed than activated.
The PIAAMV regulon has minimal overlap with the σ22 regulon.
To determine the overlap of the PIAAMV regulon with the σ22 regulon, gene lists from a microarray study that compared a mucoid mucA mutant strain to an isogenic nonmucoid σ22 mutant (AlgU/T) (27) were compared to the PIAAMV regulon gene lists and plotted as a four-way Venn diagram (Fig. 4C) with respect to activated and repressed genes of each gene list. To perform this meta-analysis, we used a PIAAMV regulon gene list generated with cutoffs applied at a P value of <0.01 and a more- or less-than-2-fold change, which mimicked the cutoffs that were used to generate the gene lists in the previous study (27). It should be noted that our analysis was performed with the PAO1 laboratory strain, whereas the study used for comparison used a cystic fibrosis isolate, FRD1 (27). Only a small number of activated or repressed genes were in agreement in this comparison (Fig. 4C), which further suggested that the PIAAMV regulon is composed mainly of genes outside the σ22 regulon.
PIAAMV activates expression of alginate genes.
PIAAMV induces the mucoid phenotype of P. aeruginosa (see Fig. S1A in the supplemental material); therefore, it is predictable that expression of the algD alginate biosynthetic operon would be activated. However, several observations concerning alginate gene expression were noted. The fold changes of the alginate genes were not as high as might have been expected (see Table S1 in the supplemental material) compared to previous arrays that were performed using mucoid strains with inactivated negative regulators of alginate biosynthesis (67, 74). The microarray experiment was performed with PAO1 at early stationary phase (18 h). Since it seems that vanadate is taken up by the phosphate transport system, it is possible that stationary-phase cells are not taking up as much vanadate, which would mean less stress and less alginate gene expression. Another possible explanation for this discrepancy is that even though regulated proteolysis of MucA is occurring, it is likely that more MucA is being expressed. However, the fold changes for algU and mucA were not statistically significant in the context of comparing PIA to PIAAMV, contrary to previously reported observations of mucA mutant strains (67). In a previous study, Wood and Ohman found that the cell membrane disruptor d-cycloserine upregulated algU 5.68-fold in strain PAO1; however, d-cycloserine did not cause alginate overproduction (71). It should be noted that the data set presented here and the above-mentioned data set were analyzed and normalized by using different software packages. Another interesting distinction between our data set and the data set of the d-cycloserine-induced microarray study is that the expression level of the gene encoding the MucA protease AlgW was significantly increased on PIAAMV compared to the level on PIA but not in response to d-cycloserine (71). However, AlgW does play a role in the response to d-cycloserine (8, 71), because regulated proteolysis of MucA occurs when P. aeruginosa is cultured in the presence of d-cycloserine. Although PIAAMV did not cause significant activation of algU, mucA, or mucB, increased expression levels of mucC and mucD were observed (see Table S1 in the supplemental material). In our previous study, we observed increased MucD expression levels on PIAAMV by Western blotting (21), which is confirmed by our analysis here. PIAAMV also induced the expression of algP and algQ (67). The algP gene encodes a histone-like product that may regulate alginate gene expression by either direct or indirect effects (75). AlgQ, as mentioned, is an anti-sigma factor and regulator of sigma D (σ70) (30), similar, as mentioned, to Rsd of E. coli (76). Interestingly, AlgQ is a global regulator that has been characterized as a modulator of QS (77) and pyoverdine production (78). It was proposed previously that algP and algQ were not regulated and are constitutively expressed (79).
PIAAMV activated the expression of O-antigen lipopolysaccharide biosynthesis genes.
In our previous study, we examined the O-antigen LPS chain length and concentration in PAO1 cells cultured on PIA and PIAAMV by Western blotting (21), and no significant differences were detected. However, growth on PIAAMV increased the expression of the entire O-antigen LPS operon (wzz to wbpM [PA3160 to PA3141]), with fold changes similar to those observed for alginate biosynthesis genes, which was noticeable in the volcano plot of the transcriptome (Fig. 3). Recently, large-scale genetic interaction studies indicated that LPS compensates for perturbations introduced by mutations in envelope biosynthesis genes and pathways (32). Cell envelope stress caused by PIAAMV likely activates increased expression levels of LPS genes to alleviate membrane stress. Another possibility that cannot be disregarded is that AMV may directly or indirectly inhibit LPS biosynthesis, and this increased gene expression is an effort to compensate for this inhibition. The differential expression of LPS genes on PIAAMV suggests that this may be a useful medium to evaluate how LPS biosynthetic genes are regulated. It does not seem that phosphate starvation alone increases LPS gene expression levels (80), suggesting that this is likely not the mechanism. In addition to O-antigen genes affected by PIAAMV, other LPS biosynthesis genes were also activated (see Table S1 in the supplemental material).
PIAAMV both activated and repressed the expression of genes involved in biosynthesis and remodeling of peptidoglycan.
PG provides structural support to the envelope of bacteria (81). Under conditions that perturb membrane fluidics, it could be hypothesized PG genes would be differentially expressed to build a sacculus capable of withstanding membrane stress and maintaining homeostasis. We hypothesize that alginate seems to be a last-chance survival method employed by P. aeruginosa to alleviate cell membrane stress. In our previous proteomic analysis of PAO1 cells cultured on PIA and PIAAMV, we observed increased MrcB expression levels (21). MrcB is involved in catalyzing the transglycosylation and transpeptidation of PG precursors. Therefore, we hypothesized that PIAAMV would affect the expression of PG genes. Here we observed that PIAAMV activated the expressions of 35 genes encoding PG synthesis enzymes (see Table S1 in the supplemental material); mrcB expression was upregulated 1.4-fold (see Table S1 in the supplemental material). These data propose several potential effects of PIAAMV on PG. Increasing expression levels of PG factors might compensate for the disruption of proper envelope protein folding, or AMV may be directly inhibiting PG synthesis in a fashion similar to that of d-cycloserine. Most PG genes were upregulated due to PIAAMV; however, the largest fold changes observed in the PIAAMV transcriptome were for the repression of amiE, amiB (PA3365), amiC, amiR, and amiS (PA3362) (Fig. 3; see also Table S1 in the supplemental material), which were repressed by 10-, 12-, 11-, 9-, and 2-fold, respectively (see Table S1 in the supplemental material). These genes encode an amidase (amiE and several regulators) that cleaves the peptide moiety from N-acetylmuramic acid and are important for septation and cell separation during cell division. Inactivation of these genes results in cells with constricted inner membranes and PG, which then leads to abnormally large periplasmic spaces (82). AmiB resembles a ClpB-type chaperone but likely forms an ABC transporter in concert with an integral membrane protein, AmiS (83). AmiC has been shown to be involved in the stability of the cell envelope (84) and is a negative regulator of the amidase operon. AmiR regulates the transcription of the operon likely as an antitermination transcription factor (85). Interestingly, AmiB has been shown to have ATPase activity, and nanomolar concentrations of ammonium vanadate inhibit this activity (83). In addition to the amiEBCRS amidase genes, PIAAMV increased the expression level of one probable amidase, PA2698 (Fig. 3). It is possible that PA2698 encodes an amidase that compensates for the decreased expression levels of the amiEBCRS genes.
Transcriptome data suggest that vanadate directly affects the expression of the amiEBCRS operon, and recently, a study showed that triclosan alone causes decreased expression levels of the amiEBCRS operon (86). In our study, triclosan is in both PIA and PIAAMV. Since vanadate caused decreased amiEBCRS expression levels, we suggest that vanadate may synergistically affect P. aeruginosa with triclosan. Triclosan has been thought to disrupt membranes because it chemically resembles a detergent (87); however, it was shown to directly target and inhibit fatty acid synthesis (88). This would hint that vanadate and triclosan affect similar structures of P. aeruginosa.
PIAAMV activates expression of the T6SS and T2SS.
Growth on PIAAMV activated the expression of type VI secretion system (T6SS) genes as well as type II secretion system (T2SS) genes (Fig. 3; see also Table S1 in the supplemental material). Using a dual-proteomic approach, we previously reported increased amounts of T6SS proteins in a nonmucoid CF isolate compared to an isogenic mucoid isolate (89). The T6SS is regulated by the sensor kinases RetS and GacS (90). Inactivation of retS increased type VI secretion and decreased type III secretion (91). It is possible that vanadate inhibits RetS/GacS pathways, causing increased expression levels of the T6SS and repression of the T3SS. Previous studies have shown that in the absence of MucA repression of σ22 (AlgU), the T3SS is repressed (92), which is an alternative explanation as to why the T3SS is repressed on PIAAMV. A number of genes related to PG synthesis were dysregulated when PAO1 was cultured on PIAAMV, as mentioned above (see Table S1 in the supplemental material). The T6SS delivers two effectors, Tse1 and Tse3, which hydrolyze PG (93). The expression levels of the genes encoding these effectors were significantly increased on PIAAMV (see Table S1 in the supplemental material). These data suggest that the effects on PG may be caused by induction of the T6SS on PIAAMV. ClpV1 (PA0090) is a chaperone that is required for secretion of T6SS effector proteins (94). Inactivation of clpV1 resulted in a nonmucoid phenotype on PIAAMV (Table 1). It is possible that the decreased secretion of T6SS effectors results in cell membrane stress that is lower than the threshold needed to induce alginate overproduction. This interplay between self-imposed cell membrane stress and induction of the mucoid phenotype is interesting and illustrates the dynamic interplay of the various virulence systems of P. aeruginosa.
A number of toxins are secreted by P. aeruginosa via the T2SS. PIAAMV activated the expression of the Xcp type II secretion apparatus genes (95) (see Table S1 in the supplemental material) as well as protease IV (piv). Protease IV degrades surfactant proteins and is important in eye infections but may also contribute to acute lung infection (96). Interestingly, triclosan alone does not increase the gene expression level of either the T6SS or T2SS (86). However, phosphate starvation does increase gene expression levels of both the T6SS and T2SS (80), suggesting that the upregulation of these systems on PIAAMV may be due to the impact of vanadate on phosphate transport.
PIAAMV represses expression of iron uptake genes.
Previous studies indicated that iron-sequestering siderophores may be direct targets of vanadate (52, 63). Vanadate was shown to bind directly to siderophores (52). AMV caused decreased expression levels of many iron uptake genes (see Table S1 in the supplemental material). Since vanadate can bind siderophores in place of iron, it is possible that proteins that regulate gene expression in response to low concentrations of iron can also bind vanadate. Vanadate binding in place of iron could have effects on the entire iron uptake system. This also correlates with the identification of several iron-regulated genes that are required for mucoidy on PIAAMV (Table 1).
PIAAMV activates expression of the genes encoding the MexGHI-OpmD efflux pump.
As mentioned above, the mexGHI-opmD efflux pump has been shown to confer resistance to vanadate (63). Here we observed that the expression levels of the mexGHI-opmD genes were increased in response to PIAAMV (see Table S1 in the supplemental material). Interestingly, the MexGHI-OpmD pump is responsible for secreting vanadium, and it also secretes pyocyanin (63). Accumulation of vanadate in cells likely has many detrimental effects on the cells. We hypothesize that the MexGHI-OpmD efflux pump potentially pumps enough vanadate out of cells to allow the cells to grow in the presence of vanadate. We did not observe globally increased gene expression of all efflux pumps due to growth on vanadate.
Growth on PIAAMV causes attenuated virulence in a murine acute pneumonia model.
P. aeruginosa strains that initially infect the lungs of CF patients are typically similar to wild-type environmental organisms and are nonmucoid. Previous work has shown that during murine lung infection, alginate production is induced (4). Hypermucoid strains, such as PDO300, which has an inactivated mucA gene, are as virulent as wild-type nonmucoid strains in an acute pneumonia model (26). Other studies have shown that early CF isolates are virulent but that later isolates have attenuated virulence in acute infection models (97). Based on these data, we tested whether growth on PIAAMV would have an effect on acute virulence of P. aeruginosa strain PAO1. PAO1 cells were cultured on PIA and PIAAMV for 18 h at 37°C. PAO1 on PIAAMV was visibly mucoid, whereas it was nonmucoid on PIA (data not shown). Doses of 4 × 107 CFU were prepared, and 8-week-old female BALB/c mice were challenged by the intranasal route. Mice were euthanized at 6 h, nasal cavities were washed, and the lungs were harvested to determine bacterial loads and perform histology. Between the two groups of mice, those receiving PAO1 grown on PIA and those receiving PAO1 grown on PIAAMV, no statistical differences in the bacterial loads were observed in either nasal wash specimens (PIA, 2.5 × 107 CFU/ml; PIAAMV, 3.8 × 107 CFU/ml) or the lung (PIA, 4.6 × 107 CFU/lung; PIAAMV, 3.6 × 107 CFU/lung). However, lung histopathology performed with standard hematoxylin and eosin stain revealed that mice infected with PAO1 cultured on PIA had more extensive lung damage, while the lungs from mice infected with PAO1 cultured on PIAAMV more closely resembled the lungs of uninfected mice (data not shown). Based on this, we predicted that growth on PIAAMV would attenuate virulence of P. aeruginosa. To test this hypothesis, we performed subsequent survival experiments with PAO1 cultured on PIA (Fig. 5). PAO1 cultured on PIA caused morbidity of 7 of 8 of the mice challenged (Fig. 5). However, PAO1 cultured on PIAAMV resulted in only 1 death out of 8 mice challenged in two separate independent experiments. These data corroborated our observations at 6 h postinfection and indicated that growth on PIAAMV attenuates P. aeruginosa.
Fig 5.
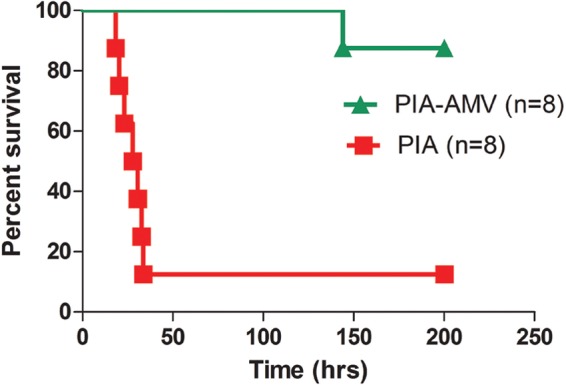
Cultivation of P. aeruginosa strain PAO1 on PIAAMV results in attenuated acute virulence. Strain PAO1 was cultured for 18 h at 37°C on either PIA or PIAAMV, and 4 × 107 CFU were administered to BALB/c mice by the intranasal route. Kaplan-Meier survival curves of BALB/c mice challenged with P. aeruginosa indicate that PIAAMV causes defects in PAO1 acute virulence. Shown are data for two independent experiments (n = 4 in each experiment).
Based on our expression data, it is clear that PIAAMV changes the expressions of many virulence gene systems, but it would have been difficult to predict that virulence would have been affected. Transcriptome analysis indicated that the T6SS was activated while the T3SS was repressed (see Table S1 in the supplemental material). The T3SS is involved in acute virulence, and repression of this system on PIAAMV contributes to the attenuated virulence (98, 99). We also previously observed that PAO1 cells growing on PIAAMV have palmitate on lipid A (21). In human cell lines, synthetic lipid A containing palmitate causes 10- to 100-fold less inflammation (100, 101). It is possible that PAO1 cells cultured on PIAAMV caused less inflammation, but further experiments are needed to test this hypothesis. The iron concentration directly regulates essential virulence factors of P. aeruginosa (102). On PIAAMV, vanadate likely binds siderophores, and we also noticed that the iron uptake system was repressed on PIAAMV (see Table S1 in the supplemental material). It is possible that the effect of vanadate on iron uptake may have contributed to the attenuated virulence. The data presented here indicate that PIAAMV not only activates the general stress response of alginate but also globally affects gene expression and attenuates acute virulence of P. aeruginosa.
Summary.
In this study, we have taken three broad approaches to determine the effect of vanadate-induced stress on P. aeruginosa as a model for inducible alginate overproduction conditions. We used alginate overproduction as a reporter of cell stress and screened the PA14 nonredundant library of mutants on PIAAMV to better understand the genetic elements that contribute to or modulate cell envelope stress. Previously, it was shown that PIAAMV activated MucA proteolysis (21). We hypothesize that most of the mutants identified in the screen have altered MucA proteolysis as a result of their respective mutations, with the exception of genes involved in metabolism and alginate synthesis. Based on this idea, it seems that there may be many systems that modulate regulated proteolysis of MucA. We also determined the effect of P. aeruginosa growth on PIAAMV on the transcriptome. These effects confirmed our hypothesis that PIAAMV affects more than alginate synthesis. We also determined that cultivation of P. aeruginosa on PIAAMV attenuates virulence in an acute pneumonia model. Ultimately, integration of the data generated by these three methods indicates that P. aeruginosa has a vast network of systems that are coordinated for modulating cell stress leading to the mucoid phenotype.
Microarray analysis indicated that growth on PIAAMV resulted in the activation of expression of some virulence factors and the repression of others. PIAAMV induced cell stress, resulting in both activation and repression of genes that correspond to some key known virulence factors. It is difficult to pinpoint which is the overall factor(s) that leads to the attenuated virulence in the acute pneumonia model. Growth on PIAAMV induced expression of the T6SS. CF patients have antibodies against the T6SS, indicating that it is expressed during infection (94). The T3SS is a key factor in acute infection (103), and PIAAMV decreased the expression levels of the T3SS. It is possible that this is one of the major reasons why P. aeruginosa cultured on PIAAMV is attenuated. Since the virulence systems were dysregulated by vanadate, we now wonder if this reprogramming can be used to engineer live attenuated strains for vaccine development. Mice challenged with PAO1 cultured on PIAAMV are protected from subsequent challenge with PAO1 grown on PIA (data not shown). Efforts are currently under way to test whether PIAAMV-cultured P. aeruginosa can be used to protect mice as a vaccine strategy.
Based on our findings presented here, we propose that PIAAMV alters global gene expression and turns on cell stress pathways in P. aeruginosa (Fig. 6). Furthermore, we propose that the genes and pathways identified here can be further studied to understand this general stress response. Vanadate is likely taken up by the phosphate transport system, but P. aeruginosa can also secrete vanadate via efflux pumps (Fig. 6). Vanadate induces the alginate stress response, alters the transcriptome, and attenuates the virulence of P. aeruginosa (Fig. 6). Recently, a systems biology approach has shown that the alginate regulator and protease AlgW is a high-value target for the design of therapeutics because it can be linked to so many other gene systems (104). Our data corroborate the above-mentioned study and suggest that an understanding of the global networks modulating cell stress is necessary and may lead to the development of new strategies to combat multidrug-resistant organisms such as P. aeruginosa.
Fig 6.
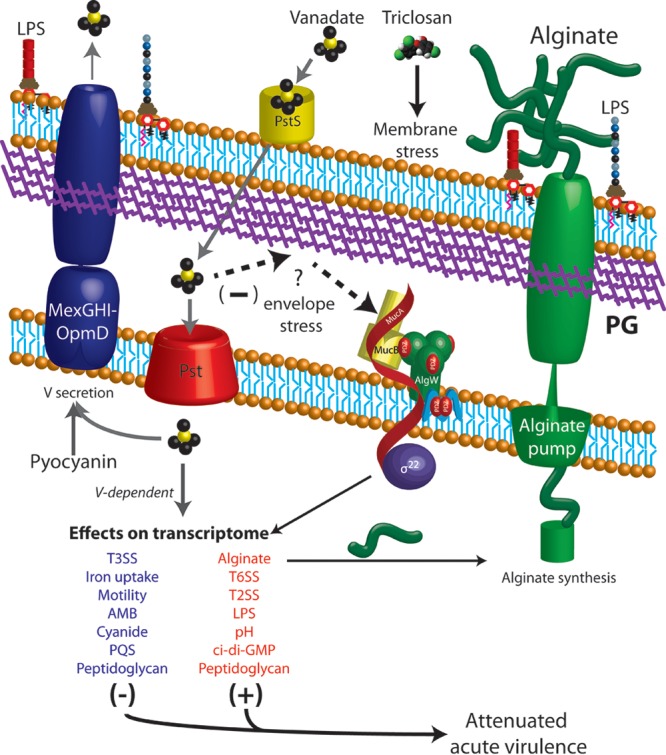
Model of envelope stress caused by PIAAMV leading to global gene expression changes and attenuated virulence in P. aeruginosa. PIAAMV is a medium composed of standard Pseudomonas isolation agar supplemented with 0.34 mM ammonium metavanadate. Vanadate is indicated by the yellow and black structures. Our previous study suggested the P. aeruginosa activates overproduction of the exopolysaccharide known as alginate as a stress response (21). Degradation of the anti-sigma factor MucA during growth on PIAAMV leads to activation of alginate gene expression and secretion of alginate (21). In this study, we screened the PA14 nonredundant library of transposon mutants, which led us to propose the following model. Triclosan from PIA causes membrane stress, as others have described (71). The addition of vanadate to PIAAMV activates alginate overproduction. Mutants of the phosphate transport system were nonmucoid on PIAAMV, which suggested that vanadate may be taken into cells by this system. Due to the identification of several mutants of hydrolytic enzymes involved in peptidoglycan remodeling, we propose that either vanadate may have a direct effect on peptidoglycan or peptidoglycan remodeling might be a way of alleviating the stress caused in the envelope by vanadate. Other mutants suggest that vanadate affects proteins in the cytoplasm. Inactivation of genes involved in pyocyanin biosynthesis suggested that vanadate may be secreted from the MexGHI-OpmD efflux pump, as others have indicated (63). Transport of vanadate is indicated with gray arrows. Vanadate causes both repression and activation of many key virulence systems (see Table S1 in the supplemental material). We note that genes involved in peptidoglycan synthesis and remodeling are both activated and repressed. Furthermore, laboratory strain PAO1 is attenuated when cultured on PIAAMV. Our study suggests that vanadate causes specific effects and envelope stress on P. aeruginosa and may be useful for understanding the various stress response networks. AMB, l-2-amino-4-methoxy-trans-3-butenoic acid; PQS, Pseudomonas quinolone signal.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Jason Papin laboratory and Anna Blazier of the University of Virginia for assisting in the replication of the PA14 mutant library and John Alverdy of the University of Chicago for providing the PstS antibody. We also thank the University of Virginia Research Histology Core and Sandford Feldman of University of Virginia Comparative Medicine for helpful discussions and histology imaging and evaluation.
F.H.D. was supported by a postdoctoral fellowship from the Cystic Fibrosis Foundation (DAMRON10F0). M.J.S. was supported by NIH grant R21 AI1094487. J.B.G. was supported by a grant from the NIH (R01 AI068112).
Footnotes
Published ahead of print 21 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00534-13.
REFERENCES
- 1.Doring G, Pier GB. 2008. Vaccines and immunotherapy against Pseudomonas aeruginosa. Vaccine 26:1011–1024 [DOI] [PubMed] [Google Scholar]
- 2.Doggett RG, Harrison GM, Stillwell RN, Wallis ES. 1966. An atypical Pseudomonas aeruginosa associated with cystic fibrosis of the pancreas. J. Pediatr. 68:215–221 [Google Scholar]
- 3.Hoiby N. 1974. Pseudomonas aeruginosa infection in cystic fibrosis. Relationship between mucoid strains of Pseudomonas aeruginosa and the humoral immune response. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 82:551–558 [PubMed] [Google Scholar]
- 4.Bragonzi A, Worlitzsch D, Pier GB, Timpert P, Ulrich M, Hentzer M, Andersen JB, Givskov M, Conese M, Doring G. 2005. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J. Infect. Dis. 192:410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman FT, Mueschenborn S, Meluleni G, Ray C, Carey VJ, Vargas SO, Cannon CL, Ausubel FM, Pier GB. 2003. Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc. Natl. Acad. Sci. U. S. A. 100:1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowen DW, Deretic V. 2000. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol. Microbiol. 36:314–327 [DOI] [PubMed] [Google Scholar]
- 7.Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 90:8377–8381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wood LF, Leech AJ, Ohman DE. 2006. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of sigma (AlgT) and the AlgW and Prc proteases. Mol. Microbiol. 62:412–426 [DOI] [PubMed] [Google Scholar]
- 9.Franklin MJ, Nivens DE, Weadge JT, Howell PL. 2011. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2:167. 10.3389/fmicb.2011.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boucher JC, Yu H, Mudd MH, Deretic V. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65:3838–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bragonzi A, Wiehlmann L, Klockgether J, Cramer N, Worlitzsch D, Doring G, Tummler B. 2006. Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 152:3261–3269 [DOI] [PubMed] [Google Scholar]
- 12.Ciofu O, Lee B, Johannesson M, Hermansen NO, Meyer P, Hoiby N. 2008. Investigation of the algT operon sequence in mucoid and non-mucoid Pseudomonas aeruginosa isolates from 115 Scandinavian patients with cystic fibrosis and in 88 in vitro non-mucoid revertants. Microbiology 154:103–113 [DOI] [PubMed] [Google Scholar]
- 13.Pulcrano G, Iula DV, Raia V, Rossano F, Catania MR. 2012. Different mutations in mucA gene of Pseudomonas aeruginosa mucoid strains in cystic fibrosis patients and their effect on algU gene expression. New Microbiol. 35:295–305 [PubMed] [Google Scholar]
- 14.Damron FH, Goldberg JB. 2012. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol. Microbiol. 84:595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu D, Eisinger VM, Rowen DW, Yu HD. 2007. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 104:8107–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cezairliyan BO, Sauer RT. 2009. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol. Microbiol. 72:368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damron FH, Yu HD. 2011. Pseudomonas aeruginosa MucD regulates alginate pathway through activation of MucA degradation via MucP proteolytic activity. J. Bacteriol. 193:286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. 2008. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology 154:2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boucher JC, Martinez-Salazar J, Schurr MJ, Mudd MH, Yu H, Deretic V. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 178:511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg JB, Gorman WL, Flynn JL, Ohman DE. 1993. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J. Bacteriol. 175:1303–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damron FH, Davis MR, Jr, Withers TR, Ernst RK, Goldberg JB, Yu G, Yu HD. 2011. Vanadate and triclosan synergistically induce alginate production by Pseudomonas aeruginosa strain PAO1. Mol. Microbiol. 81:554–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crans DC, Smee JJ, Gaidamauskas E, Yang L. 2004. The chemistry and biochemistry of vanadium and the biological activities exerted by vanadium compounds. Chem. Rev. 104:849–902 [DOI] [PubMed] [Google Scholar]
- 23.Tam C, Missiakas D. 2005. Changes in lipopolysaccharide structure induce the sigma(E)-dependent response of Escherichia coli. Mol. Microbiol. 55:1403–1412 [DOI] [PubMed] [Google Scholar]
- 24.Zhou Z, Lin S, Cotter RJ, Raetz CR. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. J. Biol. Chem. 274:18503–18514 [DOI] [PubMed] [Google Scholar]
- 25.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39:D596–D600. 10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damron FH, Owings JP, Okkotsu Y, Varga JJ, Schurr JR, Goldberg JB, Schurr MJ, Yu HD. 2012. Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J. Bacteriol. 194:1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tart AH, Wolfgang MC, Wozniak DJ. 2005. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J. Bacteriol. 187:7955–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galan-Vasquez E, Luna B, Martinez-Antonio A. 2011. The regulatory network of Pseudomonas aeruginosa. Microb. Inform. Exp. 1:3. 10.1186/2042-5783-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dove SL, Hochschild A. 2001. Bacterial two-hybrid analysis of interactions between region 4 of the sigma(70) subunit of RNA polymerase and the transcriptional regulators Rsd from Escherichia coli and AlgQ from Pseudomonas aeruginosa. J. Bacteriol. 183:6413–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reiling SA, Jansen JA, Henley BJ, Singh S, Chattin C, Chandler M, Rowen DW. 2005. Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology 151:2251–2261 [DOI] [PubMed] [Google Scholar]
- 32.Babu M, Diaz-Mejia JJ, Vlasblom J, Gagarinova A, Phanse S, Graham C, Yousif F, Ding H, Xiong X, Nazarians-Armavil A, Alamgir M, Ali M, Pogoutse O, Pe'er A, Arnold R, Michaut M, Parkinson J, Golshani A, Whitfield C, Wodak SJ, Moreno-Hagelsieb G, Greenblatt JF, Emili A. 2011. Genetic interaction maps in Escherichia coli reveal functional crosstalk among cell envelope biogenesis pathways. PLoS Genet. 7:e1002377. 10.1371/journal.pgen.1002377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clementz T, Bednarski JJ, Raetz CR. 1996. Function of the htrB high temperature requirement gene of Escherchia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J. Biol. Chem. 271:12095–12102 [DOI] [PubMed] [Google Scholar]
- 34.Mohan S, Raetz CR. 1994. Endotoxin biosynthesis in Pseudomonas aeruginosa: enzymatic incorporation of laurate before 3-deoxy-D-manno-octulosonate. J. Bacteriol. 176:6944–6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen L, Ma Y, Liang H. 2012. Characterization of a novel gene related to antibiotic susceptibility in Pseudomonas aeruginosa. J. Antibiot. (Tokyo) 65:59–65 [DOI] [PubMed] [Google Scholar]
- 36.Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, Steinmann J, Van der Meijden B, Bernardini F, Lederer A, Dias RL, Misson PE, Henze H, Zumbrunn J, Gombert FO, Obrecht D, Hunziker P, Schauer S, Ziegler U, Kach A, Eberl L, Riedel K, DeMarco SJ, Robinson JA. 2010. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327:1010–1013 [DOI] [PubMed] [Google Scholar]
- 37.Uehara T, Suefuji K, Valbuena N, Meehan B, Donegan M, Park JT. 2005. Recycling of the anhydro-N-acetylmuramic acid derived from cell wall murein involves a two-step conversion to N-acetylglucosamine-phosphate. J. Bacteriol. 187:3643–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bacik JP, Whitworth GE, Stubbs KA, Yadav AK, Martin DR, Bailey-Elkin BA, Vocadlo DJ, Mark BL. 2011. Molecular basis of 1,6-anhydro bond cleavage and phosphoryl transfer by Pseudomonas aeruginosa 1,6-anhydro-N-acetylmuramic acid kinase. J. Biol. Chem. 286:12283–12291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uehara T, Dinh T, Bernhardt TG. 2009. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J. Bacteriol. 191:5094–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dougan DA, Reid BG, Horwich AL, Bukau B. 2002. ClpS, a substrate modulator of the ClpAP machine. Mol. Cell 9:673–683 [DOI] [PubMed] [Google Scholar]
- 41.Alvarez-Ortega C, Wiegand I, Olivares J, Hancock RE, Martinez JL. 2010. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Antimicrob. Agents Chemother. 54:4159–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez L, Breidenstein EB, Song D, Hancock RE. 2012. Role of intracellular proteases in the antibiotic resistance, motility, and biofilm formation of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56:1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies JW, Collins JF. 1970. The induction of Bacillus licheniformis penicillinase by vanadate, molybdate and tungstate anions. Biochim. Biophys. Acta 217:552–554 [DOI] [PubMed] [Google Scholar]
- 44.Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13:198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. 1989. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J. Mol. Biol. 210:551–559 [DOI] [PubMed] [Google Scholar]
- 46.VanBogelen RA, Olson ER, Wanner BL, Neidhardt FC. 1996. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J. Bacteriol. 178:4344–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamarche MG, Wanner BL, Crepin S, Harel J. 2008. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32:461–473 [DOI] [PubMed] [Google Scholar]
- 48.Zaborina O, Holbrook C, Chen Y, Long J, Zaborin A, Morozova I, Fernandez H, Wang Y, Turner JR, Alverdy JC. 2008. Structure-function aspects of PstS in multi-drug-resistant Pseudomonas aeruginosa. PLoS Pathog. 4:e43. 10.1371/journal.ppat.0040043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larkin A, Olivier NB, Imperiali B. 2010. Structural analysis of WbpE from Pseudomonas aeruginosa PAO1: a nucleotide sugar aminotransferase involved in O-antigen assembly. Biochemistry 49:7227–7237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang D, Behrens A, Farahbakhsh M, Gatjens J, Rehder D. 2003. Formation, preservation, and cleavage of the disulfide bond by vanadium. Chemistry 9:1805–1813 [DOI] [PubMed] [Google Scholar]
- 51.Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, de Crecy-Lagard V. 2011. Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol. Microbiol. 79:700–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baysse C, De Vos D, Naudet Y, Vandermonde A, Ochsner U, Meyer JM, Budzikiewicz H, Schafer M, Fuchs R, Cornelis P. 2000. Vanadium interferes with siderophore-mediated iron uptake in Pseudomonas aeruginosa. Microbiology 146:2425–2434 [DOI] [PubMed] [Google Scholar]
- 53.Hassett DJ, Howell ML, Ochsner UA, Vasil ML, Johnson Z, Dean GE. 1997. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J. Bacteriol. 179:1452–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lau GW, Hassett DJ, Ran H, Kong F. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10:599–606 [DOI] [PubMed] [Google Scholar]
- 55.Wilson R, Sykes DA, Watson D, Rutman A, Taylor GW, Cole PJ. 1988. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56:2515–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61:1308–1321 [DOI] [PubMed] [Google Scholar]
- 57.Beatson SA, Whitchurch CB, Sargent JL, Levesque RC, Mattick JS. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 184:3605–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309–319 [DOI] [PubMed] [Google Scholar]
- 59.Farrow JM, III, Sund ZM, Ellison ML, Wade DS, Coleman JP, Pesci EC. 2008. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J. Bacteriol. 190:7043–7051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suh SJ, Silo-Suh L, Woods DE, Hassett DJ, West SE, Ohman DE. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. 1996. The response regulator RssB controls stability of the sigma(S) subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333–1339 [PMC free article] [PubMed] [Google Scholar]
- 62.Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, Venturi V, Zennaro E, Leoni L. 2007. RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 66:1557–1565 [DOI] [PubMed] [Google Scholar]
- 63.Aendekerk S, Ghysels B, Cornelis P, Baysse C. 2002. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 148:2371–2381 [DOI] [PubMed] [Google Scholar]
- 64.Hernandez A, Mellado RP, Martinez JL. 1998. Metal accumulation and vanadium-induced multidrug resistance by environmental isolates of Escherichia hermannii and Enterobacter cloacae. Appl. Environ. Microbiol. 64:4317–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carpentier W, Sandra K, De Smet I, Brige A, De Smet L, Van Beeumen J. 2003. Microbial reduction and precipitation of vanadium by Shewanella oneidensis. Appl. Environ. Microbiol. 69:3636–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Firoved AM, Boucher JC, Deretic V. 2002. Global genomic analysis of AlgU sigma(E)-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J. Bacteriol. 184:1057–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Firoved AM, Deretic V. 2003. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J. Bacteriol. 185:1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Firoved AM, Ornatowski W, Deretic V. 2004. Microarray analysis reveals induction of lipoprotein genes in mucoid Pseudomonas aeruginosa: implications for inflammation in cystic fibrosis. Infect. Immun. 72:5012–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Firoved AM, Wood SR, Ornatowski W, Deretic V, Timmins GS. 2004. Microarray analysis and functional characterization of the nitrosative stress response in nonmucoid and mucoid Pseudomonas aeruginosa. J. Bacteriol. 186:4046–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rao J, DiGiandomenico A, Unger J, Bao Y, Polanowska-Grabowska RK, Goldberg JB. 2008. A novel oxidized low-density lipoprotein-binding protein from Pseudomonas aeruginosa. Microbiology 154:654–665 [DOI] [PubMed] [Google Scholar]
- 71.Wood LF, Ohman DE. 2009. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol. Microbiol. 72:183–201 [DOI] [PubMed] [Google Scholar]
- 72.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 73.He J, Baldini RL, Deziel E, Saucier M, Zhang Q, Liberati NT, Lee D, Urbach J, Goodman HM, Rahme LG. 2004. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. U. S. A. 101:2530–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Damron FH, Qiu D, Yu HD. 2009. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J. Bacteriol. 191:2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deretic V, Konyecsni WM. 1990. A procaryotic regulatory factor with a histone H1-like carboxy-terminal domain: clonal variation of repeats within algP, a gene involved in regulation of mucoidy in Pseudomonas aeruginosa. J. Bacteriol. 172:5544–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jishage M, Ishihama A. 1998. A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 95:4953–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ledgham F, Soscia C, Chakrabarty A, Lazdunski A, Foglino M. 2003. Global regulation in Pseudomonas aeruginosa: the regulatory protein AlgR2 (AlgQ) acts as a modulator of quorum sensing. Res. Microbiol. 154:207–213 [DOI] [PubMed] [Google Scholar]
- 78.Ambrosi C, Tiburzi F, Imperi F, Putignani L, Visca P. 2005. Involvement of AlgQ in transcriptional regulation of pyoverdine genes in Pseudomonas aeruginosa PAO1. J. Bacteriol. 187:5097–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Konyecsni WM, Deretic V. 1990. DNA sequence and expression analysis of algP and algQ, components of the multigene system transcriptionally regulating mucoidy in Pseudomonas aeruginosa: algP contains multiple direct repeats. J. Bacteriol. 172:2511–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bains M, Fernandez L, Hancock RE. 2012. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78:6762–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10:123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, de Pedro MA, Holtje JV. 2001. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167–178 [DOI] [PubMed] [Google Scholar]
- 83.Wilson SA, Williams RJ, Pearl LH, Drew RE. 1995. Identification of two new genes in the Pseudomonas aeruginosa amidase operon, encoding an ATPase (AmiB) and a putative integral membrane protein (AmiS). J. Biol. Chem. 270:18818–18824 [DOI] [PubMed] [Google Scholar]
- 84.Ize B, Stanley NR, Buchanan G, Palmer T. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 48:1183–1193 [DOI] [PubMed] [Google Scholar]
- 85.Lowe N, Rice PM, Drew RE. 1989. Nucleotide sequence of the aliphatic amidase regulator gene (amiR) of Pseudomonas aeruginosa. FEBS Lett. 246:39–43 [DOI] [PubMed] [Google Scholar]
- 86.Chuanchuen R, Schweizer HP. 2012. Global transcriptional responses to triclosan exposure in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 40:114–122 [DOI] [PubMed] [Google Scholar]
- 87.Meincke BE, Kranz RG, Lynch DL. 1980. Effect of irgasan on bacterial growth and its adsorption into the cell wall. Microbios 28:133–147 [PubMed] [Google Scholar]
- 88.Heath RJ, Yu YT, Shapiro MA, Olson E, Rock CO. 1998. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J. Biol. Chem. 273:30316–30320 [DOI] [PubMed] [Google Scholar]
- 89.Rao J, Damron FH, Basler M, Digiandomenico A, Sherman NE, Fox JW, Mekalanos JJ, Goldberg JB. 2011. Comparisons of two proteomic analyses of non-mucoid and mucoid Pseudomonas aeruginosa clinical isolates from a cystic fibrosis patient. Front. Microbiol. 2:162. 10.3389/fmicb.2011.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13:3128–3138 [DOI] [PubMed] [Google Scholar]
- 92.Wu W, Badrane H, Arora S, Baker HV, Jin S. 2004. MucA-mediated coordination of type III secretion and alginate synthesis in Pseudomonas aeruginosa. J. Bacteriol. 186:7575–7585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ball G, Durand E, Lazdunski A, Filloux A. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43:475–485 [DOI] [PubMed] [Google Scholar]
- 96.Malloy JL, Veldhuizen RA, Thibodeaux BA, O'Callaghan RJ, Wright JR. 2005. Pseudomonas aeruginosa protease IV degrades surfactant proteins and inhibits surfactant host defense and biophysical functions. Am. J. Physiol. Lung Cell. Mol. Physiol. 288:L409–L18. 10.1152/ajplung.00322.2004 [DOI] [PubMed] [Google Scholar]
- 97.Lore NI, Cigana C, De Fino I, Riva C, Juhas M, Schwager S, Eberl L, Bragonzi A. 2012. Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PLoS One 7:e35648. 10.1371/journal.pone.0035648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roy-Burman A, Savel RH, Racine S, Swanson BL, Revadigar NS, Fujimoto J, Sawa T, Frank DW, Wiener-Kronish JP. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767–1774 [DOI] [PubMed] [Google Scholar]
- 99.Vance RE, Rietsch A, Mekalanos JJ. 2005. Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect. Immun. 73:1706–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feist W, Ulmer AJ, Musehold J, Brade H, Kusumoto S, Flad HD. 1989. Induction of tumor necrosis factor-alpha release by lipopolysaccharide and defined lipopolysaccharide partial structures. Immunobiology 179:293–307 [DOI] [PubMed] [Google Scholar]
- 101.Loppnow H, Brade L, Brade H, Rietschel ET, Kusumoto S, Shiba T, Flad HD. 1986. Induction of human interleukin 1 by bacterial and synthetic lipid A. Eur. J. Immunol. 16:1263–1267 [DOI] [PubMed] [Google Scholar]
- 102.Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 99:7072–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frank DW. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26:621–629 [DOI] [PubMed] [Google Scholar]
- 104.Sigurdsson G, Fleming RM, Heinken A, Thiele I. 2012. A systems biology approach to drug targets in Pseudomonas aeruginosa biofilm. PLoS One 7:e34337. 10.1371/journal.pone.0034337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.