Abstract
The obligatory aerobic acetic acid bacterium Gluconobacter oxydans oxidizes a variety of substrates in the periplasm by membrane-bound dehydrogenases, which transfer the reducing equivalents to ubiquinone. Two quinol oxidases, cytochrome bo3 and cytochrome bd, then catalyze transfer of the electrons from ubiquinol to molecular oxygen. In this study, mutants lacking either of these terminal oxidases were characterized. Deletion of the cydAB genes for cytochrome bd had no obvious influence on growth, whereas the lack of the cyoBACD genes for cytochrome bo3 severely reduced the growth rate and the cell yield. Using a respiration activity monitoring system and adjusting different levels of oxygen availability, hints of a low-oxygen affinity of cytochrome bd oxidase were obtained, which were supported by measurements of oxygen consumption in a respirometer. The H+/O ratio of the ΔcyoBACD mutant with mannitol as the substrate was 0.56 ± 0.11 and more than 50% lower than that of the reference strain (1.26 ± 0.06) and the ΔcydAB mutant (1.31 ± 0.16), indicating that cytochrome bo3 oxidase is the main component for proton extrusion via the respiratory chain. Plasmid-based overexpression of cyoBACD led to increased growth rates and growth yields, both in the wild type and the ΔcyoBACD mutant, suggesting that cytochrome bo3 might be a rate-limiting factor of the respiratory chain.
INTRODUCTION
Gluconobacter oxydans is a Gram-negative, obligate aerobic, rod-shaped alphaproteobacterium belonging to the family of acetic acid bacteria (1). Due to its many membrane-bound dehydrogenases that incompletely oxidize sugars, sugar alcohols, and other compounds stereo- and regioselectively in the periplasm, G. oxydans has been used for decades in industrial biotechnology (2). Important applications are in the production of vitamin C and of 6-amino-l-sorbose, an intermediate in the synthesis of the antidiabetes drug miglitol (3). Only small fractions of the carbon and energy sources are metabolized in the cytoplasm, either via the pentose phosphate pathway or via the Entner-Doudoroff pathway, and it was demonstrated that the former one is of major importance for growth on mannitol and glucose (4, 5). Moreover, it was recently shown that the pentose phosphate pathway has partially cyclic operation during growth on glucose (6).
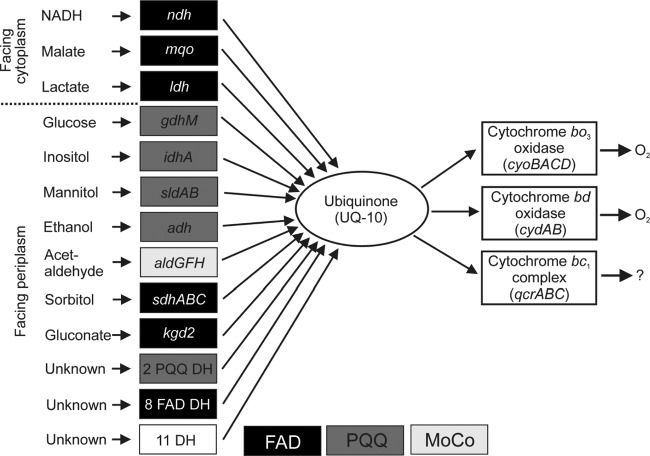
G. oxydans possesses 32 membrane-bound dehydrogenases, 11 with known and 21 with unknown substrate specificity (Fig. 1) (7, 8). Electrons are transferred to ubiquinone from the various dehydrogenases via pyrroloquinoline quinone (PQQ), PQQ plus cytochrome c, or flavin adenine dinucleotide (FAD). Electron transfer from ubiquinol to oxygen is catalyzed by two terminal oxidases, cytochrome bd and cytochrome bo3 (9) (Fig. 1). Interestingly, the genome sequence of G. oxydans 621H revealed the presence of the qcrABC genes for a cytochrome bc1 complex and of a cycA gene for a soluble cytochrome c, whereas genes for a cytochrome c oxidase were absent (8). Furthermore, genome sequencing revealed that G. oxydans possesses only a non-proton-pumping single subunit NADH dehydrogenase encoded by the ndh gene. Whereas genes for succinate:quinone oxidoreductase were absent, an mqo gene for malate:quinone oxidoreductase was present.
Fig 1.
Overview of the components of the respiratory chain of G. oxydans. DH, dehydrogenase; FAD, flavin adenine dinucleotide; PQQ, pyrroloquinoline quinone; MoCo, molybdopterin.
The cytochrome bo3 quinol oxidase of Gluconobacter suboxydans IFO 12528 (now G. oxydans) has been purified and characterized (10). It belongs to the superfamily of heme-copper oxygen reductases and contains heme b, heme o, and a copper ion as prosthetic groups. The reaction catalyzed by cytochrome bo3 quinol oxidase contributes to the generation of proton motive force in two ways. On the one hand, ubiquinol is oxidized at the periplasmic side of the membrane, with the protons released in the periplasm and the electrons transferred to the binuclear heme o3-Cu center. As the protons for water formation stem from the cytoplasm, an electrochemical proton gradient is established. On the other hand, heme-copper oxygen reductases use the free energy associated with electron transfer from ubiquinol to dioxygen to pump protons across the membrane, further contributing to the establishment of proton motive force (11). In Escherichia coli the bo3 oxidase is synthesized under conditions of high aeration.
Cytochrome bd oxidase of G. oxydans was characterized as a cyanide-insensitive oxidase (9). Biochemical and spectroscopic studies of overexpression strains revealed the presence of heme b558, b595, and d (12, 13). Cytochrome bd oxidases do not pump protons but can generate proton motive force by oxidizing ubiquinol at the periplasmic side of the membrane and taking up protons for water formation from the cytoplasm (14, 15). In Escherichia coli maximal synthesis of cytochrome bd occurs under microaerobic conditions (16).
The use of G. oxydans in biotechnological production processes, which are often whole-cell biotransformations, is limited due to its poor cell yield, which results in a cost-intensive formation of biomass. G. oxydans IFO3293 and G. oxydans 621H cells cultivated with glucose as the carbon source in a bioreactor reach a cell yield of 0.09 g of cells (dry weight) [gcdw]/g of glucose (4, 17). In comparison to these values, E. coli reaches a value of 0.49 gcdw/g of glucose, and Bacillus subtilis reaches a yield of 0.32 gcdw/g of glucose (18, 19). It has been shown for several respiring bacteria that the cell yield correlates with the H+/O ratio measured by the oxygen pulse method (20, 21). Acetobacter pasteurianus for example has a H+/O ratio of 1.9 ± 0.1 and a cell yield of 13.1 gcdw/mol of ethanol, whereas Paracoccus pantotrophus shows a H+/O ratio of 4.9 ± 0.3 and reaches a cell yield of 25.2 gcdw/mol of ethanol (21).
To gain further insights into the respiratory energy metabolism of G. oxydans, in-frame deletion mutants of the terminal oxidases were constructed and characterized with respect to growth, respiration activity, and H+/O ratio. The results show that cytochrome bo3 oxidase is of major importance for growth of G. oxydans, whereas cytochrome bd oxidase is dispensable under the tested conditions.
MATERIALS AND METHODS
Materials.
Chemicals were obtained from Sigma-Aldrich (Taufkirchen, Germany), Qiagen (Hilden, Germany), Merck (Darmstadt, Germany), and Roche Diagnostics (Mannheim, Germany).
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were cultivated in lysogeny broth (LB) medium (22, 23) or on LB agar plates at 37°C. When required, kanamycin was added to a final concentration of 50 μg ml−1. G. oxydans ATCC 621H Δupp (ATCC 621H is identical to DSM 2343) strains were cultivated on mannitol medium containing 220 mM (4%, wt/vol) mannitol, 5 g liter−1 yeast extract, 2.5 g liter−1 MgSO4 · 7 H2O, 1 g liter−1 (NH4)2SO4, and 1 g liter−1 KH2PO4. The initial pH value of the medium was 6.0. G. oxydans possesses a natural resistance against cefoxitin. As a precaution to prevent bacterial contaminations, 50 μg ml−1 cefoxitin was added to the preculture medium. When required, kanamycin was added (50 μg ml−1).
Table 1.
Bacterial strains, plasmids, and oligonucleotides used in this work
| Strain, plasmid, or oligonucleotide | Characteristic(s) or primer sequencea | Source and/or reference |
|---|---|---|
| Strains | ||
| E. coli DH5αF′ | F′ ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 relA1 | 25 |
| E. coli S17-1 | ΔrecA endA1 hsdR17 supE44 thi-1 tra+ | 51 |
| G. oxydans Δupp | G. oxydans 621H derivative with a deletion of GOX0327 coding for uracil phosphoribosyltransferase; reference strain in this study | 26 |
| G. oyxdans Δupp ΔcydAB | G. oxydans Δupp derivative with a deletion of the genes cydA (GOX0278) and cydB (GOX0279) coding for cytochrome bd oxidase | This work |
| G. oxydans Δupp ΔcyoBACD | G. oxydans Δupp derivative with a deletion of the genes cyoBACD (GOX1911 to GOX1914) coding for cytochrome bo3 oxidase | This work |
| Plasmids | ||
| pAJ63a | Kanr; pK18mobGII derivative; lacZ mob oriV; contains GOX0327 for uracil phosphoribosyltransferase including its promoter region | 26 |
| pAJ-cydAB | Kanr; pAJ63a derivative used for marker-free deletion of the cydAB genes; contains an overlap extension PCR product covering the 20-bp upstream region of edd and the 36-bp downstream region of eda | This work |
| pAJ-cyoBACD | Kanr; pAJ63a derivative used for marker-free deletion of the cyoBACD genes; contains an overlap extension PCR product covering 20-bp upstream region and the 67-bp downstream region of gnd | This work |
| pBBR1p384 | Kanr; pBBR1MCS-2 derivative containing the promoter region of gox0384 for target gene expression | 52; U. Deppenmeier, University of Bonn, Germany |
| pBBR1p384-cyoBACD | Kanr; pBBR1p384 derivative expressing cyoBACD | This work |
| pBBR1p384-cydAB | Kanr; pBBR1p384 derivative expressing cydAB | This work |
| pBBR1p384-cydABCD | Kanr; pBBR1p384 derivative expressing cydABCD | This work |
| Oligonucleotides | ||
| cydAB_forw_FlA | TCTCCGGACAACCGGATCAC | |
| cydAB_rev_FlA | GTAGTGATGTCCGGCCATGTCGATTGCCTTCTGGGTAGATGGCGAAACGC | |
| cydAB_forw_FlB | AAGGCAATCGACATGGCCGGACATCACTACCACTGAGAACAGGGAGGCCG | |
| cydAB_rev_FlB | CTGTTCGACAGTCTGCATCGC | |
| cydAB_mut_forw | ATCGCTCTCACAGCATCG | |
| cydAB_mut_rev | GGGCATGTGTCGTATGTC | |
| cydAB_screen_forw | TGAACACGCTGGAATACC | |
| cydAB_screen_rev | CATCGTGTCGTGCAGTTC | |
| cyoBACD_forw_FlA | GGGTTCGCTCAGATAACAAG | |
| cyoBACD_rev_FlA | GGACATCATGTTGATCATCCGTTCCGGCTTGCAGTAGTCGCCGGGAAACG | |
| cyoBACD_forw_FlB | AAGCCGGAACGGATGATCAACATGATGTCCCGCTAAGGTTTCTTTCGAAA | |
| cyoBACD_rev_FlB | ACGGATTGCGTGGAAGTC | |
| cyoBACD_mut_forw | GTCTATGATGCGCTGACG | |
| cyoBACD_mut_rev | CGATCGGAGACGTCAATG | |
| cyoBACD_forw | CTGGCGAAGAACAAGAAG | |
| cyoBACD_rev | GGTGCTGGTGTTCATGTG | |
| cydAB_compl_forw_XhoI | AGTTCGCTCGAGCTACCCAGAAGGCAATCGACb | |
| cydAB_compl_rev_KpnI | TGACTTGGTACCCAGCCCAGATCGCCACGAACc | |
| cydCD_compl_forw_EcoRI | GGCACGGAATTCGTCGGGCTTTTGCGACCTATd | |
| cydCD_compl_rev_XhoI | GCGTAGCTCGAGCAGGATAGAGAGGTGTGAAGb | |
| cyoBACD_compl_forw_XhoI | TGCGATCTCGAGACTACTGCAAGCCGGAACGGb | |
| cyoBACD_compl_rev_KpnI | GTACTGGGTACCAAGGGCTGGCAGGATTTCTCc |
The sequence is given 5′ to 3′. Underlined sequences indicate the regions used for overlap extension PCR or restriction enzyme sites.
Contains an XhoI site.
Contains a KpnI site.
Contains an EcoRI site.
Cloning and DNA techniques.
For DNA manipulation standard, methods were used as described by Sambrook and Russell (24). For PCRs, genomic DNA isolated from G. oxydans 621H Δupp was used as the template. Competent cells of E. coli were prepared with CaCl2 and transformed as described by Hanahan et al. (25). DNA sequencing was performed by Agowa (Berlin, Germany) and Eurofins MWG Operon (Ebersberg, Germany). Oligonucleotides were synthesized by Biolegio (Nijmegen, Netherlands) and are listed in Table 1.
Construction of in-frame deletion mutants.
Construction of G. oxydans Δupp in-frame deletion mutants of the genes cydAB and cyoBACD was performed using a recently described method in which fluorouracil is used to select for the second homologous recombination event leading to chromosomal excision of the plasmid vector (26). For the deletion of cyoBACD, plasmid pAJ-cyoBACD was constructed as follows. A total of 500 bp of the upstream region of cyoA and 500 bp of the downstream region of cyoD were amplified using the oligonucleotide pairs cyoBACD_forw_FlA/cyoBACD_rev_FlA and cyoBACD_forw_FlB/cyoBACD_rev_FlB and Phusion DNA polymerase (Fisher Scientific, Schwerte, Germany). The two PCR products were fused by overlap extension PCR with Platinum Pfx DNA polymerase (Invitrogen, Darmstadt, Germany) using the oligonucleotides cyoBACD_forw_FlA/cyoBACD_rev_FlB. The resulting 1-kb PCR product was cloned into the SmaI-restricted plasmid pAJ63a that is nonreplicative in G. oxydans. After conjugative transfer of pAJ-cyoBACD from E. coli S17-1 to G. oxydans Δupp, colonies were screened by PCR for chromosomal integration of pAJ-cyoBACD via a single homologous crossover event using the oligonucleotides cyoBACD_mut_forw and cyoBACD_mut_rev. Positive clones were inoculated overnight in 20 ml of mannitol medium containing cefoxitin and kanamycin and used as a preculture for fluorouracil-enforced vector excision, as described previously (5). Fluorouracil-resistant colonies were screened by PCR for the chromosomal deletion of cyoBACD using the oligonucleotide pair cyoBACD_mut_forw/cyoBACD_mut_rev and cyoBACD_forw/cyoBACD_rev. The up- and downstream regions of the deleted genes were sequenced, and the corresponding strain was named G. oxydans Δupp ΔcyoBACD. Deletion of the cydAB genes was performed analogously with the help of plasmid pAJ-cydAB, which contains the upstream region of cydA fused to the downstream region of cydB. The desired strain obtained after allelic exchange was called G. oxydans Δupp ΔcydAB.
Complementation studies.
For complementation of the deletion mutants, the broad-host-range plasmid pBBR1p384 was used to construct three expression plasmids. The cyoBACD region (GOX1911 to GOX1914) including 20 bp upstream of the predicted start codon of cyoB was amplified using Phusion DNA polymerase (Fisher Scientific, Schwerte, Germany) with the oligonucleotides cyoBACD_compl_forw_XhoI and cyoBACD_compl_rev_KpnI. The cydAB region (GOX0278 to GOX0279) including 20 bp upstream of the predicted start codon of cydA was amplified with the oligonucleotides cydAB_compl_forw_XhoI and cydAB_compl_rev_KpnI. The PCR products were digested with XhoI and KpnI and cloned into pBBR1p384 restricted with the same enzymes, resulting in plasmids pBBR1p384-cyoBACD and pBBR1p384-cydAB. The cydCD region (GOX2409 to GOX2410) including 31 bp upstream of the predicted start codon was amplified using Phusion DNA polymerase (Fisher Scientific, Schwerte, Germany) with the oligonucleotides cydCD_compl_forw_EcoRI and cydCD_compl_rev_XhoI. The PCR product was digested with EcoRI and XhoI and cloned into pBBR1p384-cydAB restricted with the same enzymes, resulting in plasmid pBBR1p384-cydABCD. Using E. coli S17-1 as host, these plasmids and as a control pBBR1p384 vector were transferred into the desired G. oxydans strains by conjugation.
For growth experiments with the plasmid-carrying strains, 20 ml of mannitol medium in a 100-ml baffled shake flask (three baffles, 1 by 2.5 cm each) was inoculated with cells from agar plates and incubated for 24 h at 30°C and 140 rpm. A second preculture (80 ml of mannitol medium in a 500-ml baffled shake flask with three baffles, 1.5 by 5 cm each) was inoculated with 1% (vol/vol) of the first preculture and incubated for 16 h at 30°C and 140 rpm. This preculture was then used for inoculation of the main culture (100 ml of mannitol medium in a 500-ml baffled shake flask) to an optical density at 600 nm (OD600) of 0.3. At selected time points, samples for the measurement of the optical density and for high-performance liquid chromatography (HPLC) analysis were taken. All cultivations were carried out in a Minitron incubation shaker (Infors HT, Basel, Switzerland).
Characterization of respiration activity and growth.
Cultivations of G. oxydans 621H Δupp and deletion mutants were performed using a self-made respiration activity monitoring system, RAMOS, which enables the online measurement of the oxygen transfer rate (OTR), carbon dioxide transfer rate (CTR), and respiratory quotient (RQ) in eight parallel flasks (27). The OTR describes the total molar flow of oxygen per time from the gas phase to the liquid volume. Since the microorganisms in the liquid phase consume this oxygen, OTR is equal to oxygen uptake rate. To determine the OTR, modified 250-ml shake flasks were used, each with four additional ports in the headspace. These ports are necessary for supplying gas, purging exhaust gas, inoculating the culture, and connecting an electrochemical oxygen sensor located in the headspace of the flasks. The aeration of 10 ml min−1 per flask is adjusted by a thermal mass flow controller to guarantee similar aeration conditions as in normal shake flasks with a cotton plug (28, 29). To determine the OTR, the valves for gas inlet and outlet for each flask were periodically closed (5 min closed and 15 min open). Due to the interruption of the air supply and the continued respiration activity of the microorganisms, the oxygen partial pressure in the headspace of the flasks decreases while the carbon dioxide partial pressure decreases. The partial pressures are monitored by an oxygen sensor and a total pressure sensor. Based on the slope of the decrease of the oxygen partial pressure over time, the OTR is calculated. CTR can be calculated by the difference of the total pressure and the oxygen partial pressure, and RQ is calculated as the ratio of CTR/OTR (27). During the experiments the temperature was kept constant at 30°C by placing the RAMOS device in an incubation hood. Commercial versions of RAMOS are available from Kuhner (Birsfelden, Switzerland) or Hitech Zang (Herzogenrath, Germany). This device has already been intensively in other investigations to study and characterize microbial systems (30–32). Off-line sampling was conducted with the help of parallel 250-ml unbaffled shake flasks running simultaneously under identical conditions (30°C; 50-mm shake diameter) as the RAMOS cultivations in a ISF-4-W Kuhner shaker (Birsfelden, Switzerland). Each sample flask was used for only one sampling in order to avoid a change of the flask filling volume. For the preculture, mannitol medium was inoculated with G. oxydans cells from a glycerol stock and incubated overnight at 30°C. The main culture was inoculated with the preculture to an OD600 of 0.1. Nonlimiting oxygen supply was achieved by using 10 ml of mannitol medium and a shaking frequency of 350 rpm. For oxygen-limiting conditions, different volumes (20 to 60 ml) of mannitol medium were used with a shaking frequency of 200 rpm. Samples of the cultures grown in parallel to the RAMOS cultures were taken at selected time points and analyzed regarding pH, optical density, and sugar concentrations.
H+/O measurements.
For determination of H+/O ratios, 50 ml of mannitol medium (G. oxydans) or LB medium (E. coli) was inoculated to an OD600 of 0.3. Cells were grown to an OD600 of 1.5 at 30°C (G. oxydans) or 37°C (E. coli) at 140 rpm in baffled 500-ml shake flasks (three baffles, 1.5 by 5 cm each) in a Minitron incubation shaker (Infors HT, Basel, Switzerland). Cells were harvested by centrifugation at 10,414 × g for 3 min at 4°C and washed twice with 25 ml of 200 mM KCl each. Cells were resuspended in 1 ml of 200 mM KCl and centrifuged for 5 min at 16,100 × g and 4°C. The supernatant was discarded, and the cells were resuspended in 200 μl of 200 mM KCl. Fifty microliters of the cell suspension was added into a closed reaction vessel containing 2.5 ml of an anaerobic buffer composed of 1 mM morpholineethanesulfonic acid (MES), pH 6.0, 200 mM KCl, 50 mM potassium thiocyanate (KSCN), 10 mM mannitol, and 24 μg ml−1 valinomycin, leading to a final OD600 of about 7 (33). The vessel was thermostatted at 30°C and kept anaerobic by a flow of argon. The cell suspension in the vessel was continuously mixed with a magnetic stirrer. The pH of the medium was recorded using a micro-pH electrode (Mettler Toledo, Gießen, Germany) in combination with a Seven Easy pH meter (Mettler Toledo, Gießen, Germany) and the software LabX direct pH (Mettler Toledo, Gießen, Germany). After the pH remained constant, defined volumes (12.5 μl to 50 μl) of air-saturated 150 mM KCl solution were added. The solubility of O2 in 150 mM KCl at 30°C was 223 μM (corresponding to 0.223 nmol O2/μl or 0.446 ng-atoms of O/μl). Calibration of the pH electrode was performed using 2.5 μl, 5 μl, and 10 μl of an anaerobic 5 mM HCl solution (5 μl of 5 mM HCl is equivalent to 25 ng-ions of H+). The maximal acidification observed after addition of air-saturated KCl was used for calculation of the H+/O ratio without correction for the proton influx into the cells that occurred during the acidification (34). The determined H+/O ratios therefore represent minimal values. Doubling of the concentrations of valinomycin or thiocyanate did not significantly affect the measured H+/O values, indicating that proton translocation was not limited by insufficient charge compensation.
Respirometry.
A respirometer with an electrochemical oxygen electrode was used to characterize the oxygen affinity of the reference strain and the deletion mutants. Two milliliters of an overnight culture was centrifuged (5 min at 10,000 × g at room temperature), and the supernatant was removed. The cell pellet was resuspended in fresh mannitol medium to obtain a cell suspension with an OD600 of 15. To calibrate the respirometer, 4.9 ml of mannitol medium was added to the measuring chamber, which was constantly mixed by a magnetic stirrer. Air saturation of the medium was achieved via surface aeration until a constant value was reached. Then, 100 μl of the concentrated cell suspension was transferred to the measuring chamber, and the chamber was immediately closed. Bubbles inside the measuring chamber had to be avoided since they would act as an oxygen source during the measurement. The voltage corresponding to the dissolved oxygen tension (DOT) was recorded continuously by a computer. The oxygen solubility of the mannitol medium at 22.5°C was 244 μM, calculated as described previously (35–37).
Determination of substrate and product concentrations by HPLC analysis.
One milliliter of culture was centrifuged for 5 min at 10,000 × g, and the supernatant was filtered through a 0.2-μm-pore-size polyvinylidene difluoride (PVDF) filter (Carl Roth, Karlsruhe, Germany) prior to HPLC analysis. Mannitol and fructose were quantified using a Carbohydrate Pb2+ column (300 by 8 mm) including a precolumn (CS-Chromatographie Service, Langerwehe, Germany) at 80°C in the experiments accompanying RAMOS cultivations. H2O was used as eluent at a flow rate of 0.6 ml min−1. Retention times for fructose and mannitol were 20 and 30 min, respectively. 5-Ketofructose as well as fructose and mannitol (complementation experiments) were quantified with a Rezex RCM-Monosaccharide 300- by 7.8-mm column (Phenomenex, Aschaffenburg, Germany) at 60°C using H2O as the eluent at a flow rate of 0.6 ml min−1. The above-mentioned metabolites were detected by a refractive index detector. Retention times for 5-ketofructose, fructose, and mannitol were 13.47, 15.23, and 19.95 min, respectively. Calibration curves were made using a series of standards ranging from 0 to 10 g liter−1 mannitol, 0 to 15 g liter−1 fructose, and from 0 to 5 g liter−1 for 5-ketofructose.
DNA microarrays.
For DNA microarray analysis of strain G. oxydans Δupp ΔcyoBACD versus the reference strain G. oxydans Δupp, cells were cultivated in baffled shake flasks with mannitol as the carbon source and harvested at an OD600 of 1.5. RNA extraction, cDNA labeling and DNA microarray analysis were performed as described previously (38).
Microarray data accession number.
The DNA microarray data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO series accession number GSE47782.
RESULTS
Construction of G. oxydans mutants lacking terminal oxidases.
In order to investigate the role of the two terminal oxidases in G. oxydans, deletion mutants lacking either the cydAB genes for cytochrome bd oxidase or the cyoBACD genes for cytochrome bo3 oxidase were constructed. To obtain the cyd deletion mutant, 12 fluorouracil-resistant colonies obtained after the second homologous recombination event were analyzed by PCR, and three of them were found to have the cydAB deletion. G. oxydans Δupp ΔcydAB clone 1 was used for further experiments. To identify a strain lacking cytochrome bo3 oxidase, more than 500 fluorouracil-resistant colonies had to be analyzed after the second homologous recombination event, and only one of them contained the desired cyoBACD deletion. The strain was named G. oxydans Δupp ΔcyoBACD. The 100-fold difference in the appearance of cydAB and cyoBACD mutants already indicates that cytochrome bo3 oxidase is important for growth.
Growth and respiration activity of terminal oxidase mutants.
Growth experiments on agar plates containing mannitol medium revealed a severe growth defect of strain G. oxydans Δupp ΔcyoBACD as the colonies grew much more slowly and reached a much smaller size than those of the reference strain. In contrast, strain G. oxydans Δupp ΔcydAB behaved like the reference strain (Fig. 2). Growth and respiration activity of the terminal oxidase deletion mutants were analyzed in more detail using RAMOS and shake flask cultivations, both under oxygen-excess and oxygen-limited conditions (Fig. 3 and 4 and Table 2). The results obtained confirmed the findings obtained on agar plates. The final OD600 values reached by G. oxydans Δupp ΔcyoBACD were 39 to 43% lower than those of the wild type, whereas the Δupp ΔcydAB strain reached 100 to 103% of the wild-type level.
Fig 2.
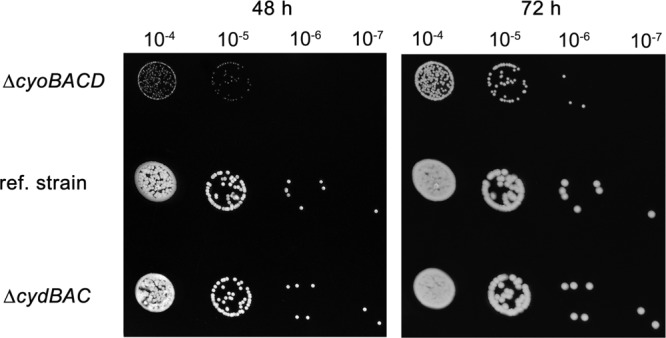
Growth of the indicated G. oxydans strains on agar plates containing mannitol medium with 1.5% agar. The plates were incubated for 48 h or 72 h at 30°C. A preculture was grown in 20 ml of mannitol medium for 24 h. After adjusting the OD600 of all precultures to 1, the suspension was diluted 104- to 107-fold in mannitol medium. Ten microliters of each dilution was dropped on the agar plates. Three biological replicates were performed with comparable results, and one representative experiment is shown here. ref, reference.
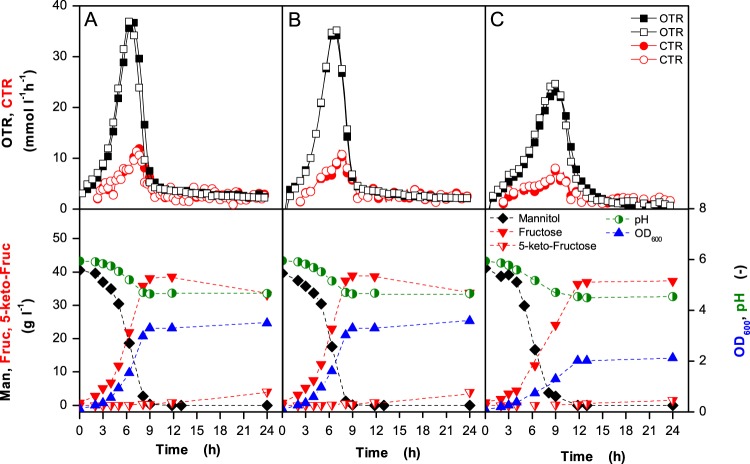
Fig 3.
Growth of G. oxydans Δupp (A), G. oxydans Δupp ΔcydAB (B), and G. oxydans Δupp ΔcyoBACD (C) in mannitol medium at 30°C under oxygen excess. The 250-ml flasks contained 10 ml of medium and were shaken at 350 rpm with a shaking diameter of 50 mm. In the upper panels, the oxygen transfer rates (OTRs) and the carbon dioxide transfer rates (CTRs) of two independent cultures are shown. In the lower panels, the OD600, the pH of the medium, and the concentrations of mannitol (Man), fructose (Fruc), and 5-keto-fructose (5-keto-Fruc) are shown. RAMOS experiments were conducted as duplicates and showed excellent reproducibility.
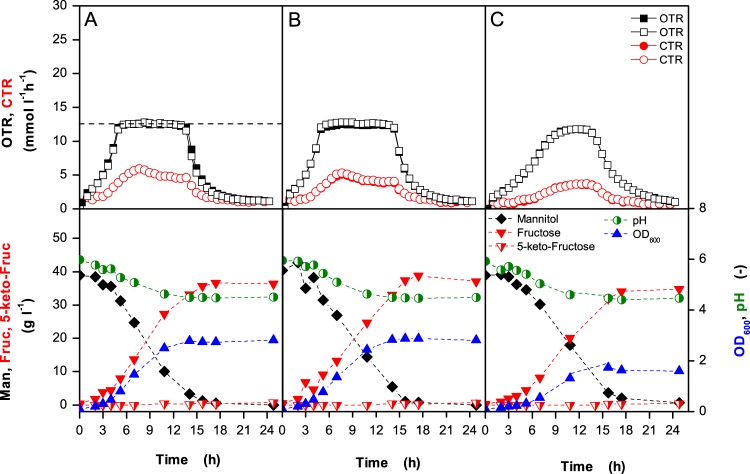
Fig 4.
Growth of G. oxydans Δupp (A), G. oxydans Δupp ΔcydAB (B), and G. oxydans Δupp ΔcyoBACD (C) in mannitol medium at 30°C under oxygen limitation. The 250-ml flasks contained 40 ml of medium and were shaken at 200 rpm with a shaking diameter of 50 mm. In the upper panels, the oxygen transfer rates (OTRs) and the carbon dioxide transfer rates (CTRs) of two independent cultures are shown. In the lower panels, the OD600, the pH of the medium, and the concentrations of mannitol (Man), fructose (Fruc), and 5-keto-fructose (5-keto-Fruc) are shown. RAMOS experiments were conducted as duplicates and showed an excellent reproducibility. In panel A the maximal oxygen transfer capacity is illustrated as an example by a dashed horizontal line.
Table 2.
Growth-related parameters and respiration activities obtained during cultivation of the indicated G. oxydans strains under a nonlimiting oxygen supply and under oxygen limitationa
| Condition and G. oxydans strain | μmax (h−1) | OD600 | OTRmax (mM h−1) | Specific maximal O2 consumption rate (μmol min−1 mg protein−1)b | Total O2 consumption (mM)c | CTRmax (mM h−1) | Specific maximal CO2 production rate (μmol min−1 mg protein−1)b | Total CO2 formed (mM)c |
|---|---|---|---|---|---|---|---|---|
| Nonlimiting oxygen supply | ||||||||
| Δupp strain | 0.40 | 3.49 | 37 | 2.08 (100) | 194 | 12 | 0.62 (100) | 100 |
| Δupp ΔcydAB strain | 0.41 | 3.58 | 36 | 2.05 (99) | 183 | 11 | 0.57 (92) | 91 |
| Δupp ΔcyoBACD strain | 0.30 | 2.12 | 24 | 1.56 (75) | 154 | 8 | 0.52 (84) | 66 |
| Oxygen limitation | ||||||||
| Δupp strain | 0.38 | 2.81 | 13 | 0.93 (100) | 162 | 6 | 0.38 (100) | 66 |
| Δupp ΔcydAB strain | 0.37 | 2.81 | 13 | 0.93 (100) | 171 | 5.5 | 0.35 (92) | 65 |
| Δupp ΔcyoBACD strain | 0.23 | 1.59 | 12 | 0.76 (82) | 142 | 3.5 | 0.22 (58) | 44 |
Dry cell weight (DCW) per liter was obtained using following empirical correlation: DCW = 0.1675 ln (OD600) + 0.4684 (R2 = 0.9593); protein was calculated as 50% of DCW. Mean values of two independent cultures are shown which deviated by less than 5%. max, maximum.
Calculated using the OD600 values at the time of maximal OTR and CTR, respectively. Values in parentheses are percentages for consumption and production relative to the corresponding values for the Δupp strain, set at 100%.
Under oxygen excess, G. oxydans Δupp and G. oxydans Δupp ΔcydAB reached their maximal OTRs of about 37 mmol liter−1 h−1 after 7 h. The time points of maximal OTR correlated with the time points at which more than 60% of mannitol had been consumed, which is predominantly oxidized in the periplasm to fructose (5). G. oxydans Δupp ΔcyoBACD reached a maximal OTR of 24 mmol liter−1 h−1 after 9 h and had a 27% reduced growth rate. The specific maximal oxygen consumption rate of the cyo deletion mutant was reduced by 25% compared to that of the reference strain, whereas that of the cyd deletion mutant was almost identical to that of the reference strain. The specific maximal CO2 production rate of the cyo deletion mutant was decreased by 16% compared to the reference strain, whereas that of the cyd deletion mutant was decreased by 8%. After complete consumption of mannitol, the OTRs of all three strains decreased to 2 to 5 mmol liter−1 h−1, showing that only a small fraction of fructose was either oxidized in the periplasm to ketofructose or in the cytoplasm to CO2 and acetate. During the cultivations, the pH dropped from 6 to 4.5, presumably due to acetate formation (5).
The results of the oxygen-limited cultivations of the G. oxydans Δupp, Δupp ΔcydAB, and Δupp ΔcyoBACD strains are shown in Fig. 4 and Table 2. In the case of the reference strain and the cyd deletion mutant, the OTR initially increased and then abruptly reached a plateau when oxygen became limiting. The height of this plateau, known as maximum oxygen transfer capacity, represents the maximum OTR provided by the bioreactor under these operating conditions. As soon as the maximum oxygen transfer capacity is reached, the microorganisms are oxygen limited. For the G. oxydans Δupp and Δupp ΔcydAB strains, this plateau remained constant over several hours until mannitol was completely oxidized, and then the OTR decreased again. In the case of the cyo deletion mutant, the OTR kinetics was different as the OTR was diminished long before the cells reached the maximum oxygen transfer capacity. As a consequence, there was no abrupt transition from oxygen excess to oxygen limitation, and the plateau phase was much shorter. To further investigate this phenomenon, RAMOS experiments were conducted with the reference strain and the cyo deletion mutant using various medium volumes (20 to 60 ml) to achieve different degrees of oxygen limitation (Fig. 5). In all cases, the OTR kinetics of the reference strain and the cyo deletion mutant were comparable to those shown in Fig. 4. Thus, the reference strain and the cyd deletion mutant are capable of high respiration rates also when oxygen becomes limiting, whereas the cyo deletion mutant lacks this ability. This suggests that cytochrome bo3 oxidase has a high-oxygen affinity, whereas cytochrome bd oxidase has a low-oxygen affinity. Similar results as shown here for mannitol-grown cells were also observed for glucose-grown cells (data not shown), confirming that they are not dependent on the carbon source.
Fig 5.
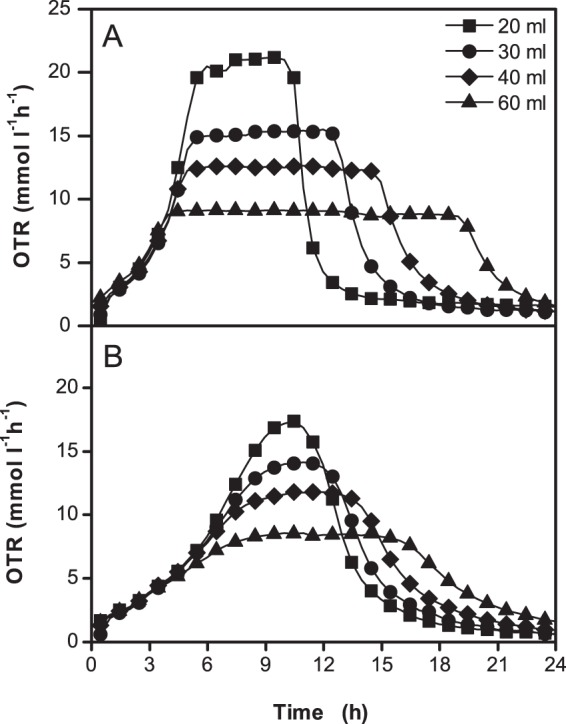
Comparison of OTRs of strains G. oxydans Δupp (A) and G. oxydans Δupp ΔcyoBACD (B) in mannitol medium at 30°C under oxygen-limited conditions. The 250-ml unbaffled flasks contained 20 ml, 30 ml, 40 ml, or 60 ml of medium and were shaken at 200 rpm with a shaking diameter of 50 mm. The maximal OTR values obtained for the reference strain were 21.0, 15.0, 12.5, and 9.0 mmol liter−1 h−1, and those for the cyo deletion mutant were 18.5, 14.5, 11.5, 8.5 mmol liter−1 h−1, respectively.
Respirometry.
To further analyze the respiration activities of the reference strain and the terminal oxidase deletion mutants, oxygen consumption was measured in a respirometer (Fig. 6). Oxygen consumption by the strain lacking cytochrome bo3 oxidase occurred significantly more slowly than that of the reference strain and the bd oxidase deletion mutant. Comparison of the slope of DOT curves, which is equivalent to the oxygen transfer rates, demonstrated a 30% lower value for the cyo deletion mutant (2.3 ± 0.06 mM h−1 for the Δupp ΔcyoBACD strain, 3.55 ± 0.11 mM h−1 for the Δupp ΔcydAB strain, and 3.26 ± 0.03 mM h−1 for the Δupp strain). The oxygen consumption rates of the reference strain and the cyd deletion mutant decreased when DOT ran below ∼10%, whereas oxygen consumption rate of the cyo deletion mutant had already decreased when the DOT ran below 25%. Based on the data shown in Fig. 6, half-maximum oxygen consumption rates were estimated at 7 μM oxygen for the Δupp and Δupp ΔcydAB strains and at 21 μM oxygen for the Δupp ΔcyoBACD strain. These values are roughly comparable to the KM values for oxygen of purified bo3 oxidase (2 to 3 μM) and purified bd oxidase (21 μM) as determined from the deoxygenation kinetics of oxymyoglobin (13).
Fig 6.
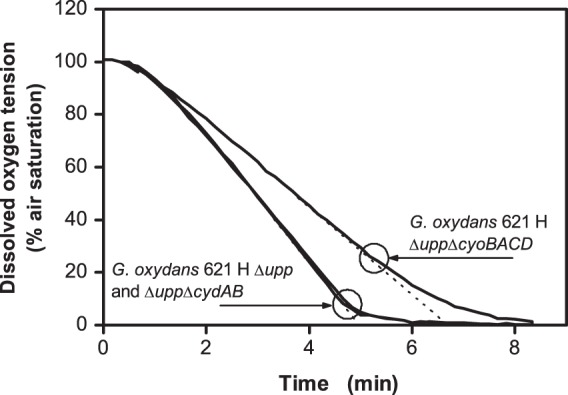
Oxygen consumption of G. oxydans Δupp, G. oxydans Δupp ΔcyoBACD, and G. oxydans Δupp ΔcydAB in a respirometer with an electrochemical oxygen electrode. Experimental conditions were as follows: complex medium with 40 g liter−1 mannitol; room temperature. The OD600 was adjusted to 0.3 for all strains. The circles show the dissolved oxygen tension where the oxygen consumption rate started to decrease. The experiments were conducted in triplicate with comparable results, and one experiment for each strain is shown here.
H+/O ratio measurements.
The deletion mutants constructed in this work allowed us to determine the H+/O ratios in the presence of only one terminal oxidase. The experiments were performed with mannitol as an energy source, which is oxidized in the periplasm by the major polyol dehydrogenase SldAB to fructose, with concomitant reduction of ubiquinone. The average H+/O ratio of the reference strain G. oxydans Δupp containing both terminal oxidases determined from 10 independent experiments was 1.26 ± 0.06 (Table 3). Control experiments under identical conditions (30°C, pH 6) using E. coli DH5αF′ showed an average H+/O ratio of 3.04 ± 0.11, indicating that the low values measured for G. oxydans were not an artifact of the experimental setup. The G. oxydans Δupp ΔcydAB strain lacking cytochrome bd oxidase showed a H+/O ratio (1.31 ± 0.16) comparable to that of the reference strain, which agrees with the unimpaired growth of the mutant under the tested conditions. The Δupp ΔcyoBACD mutant, on the other hand, showed a 56% reduction of the H+/O ratio (0.56 ± 0.11) compared to the reference strain, indicating that cytochrome bo3 oxidase is predominantly responsible for respiratory proton translocation in G. oxydans. The low H+/O ratio of the cyo deletion mutant correlates with its poor growth on agar medium and in shake flask experiments.
Table 3.
H+/O ratio of three G. oxydans strains and E. coli as a reference with mannitol as the carbon source
| Strain | H+/O ratio at the indicated vol of air-saturated 150 mM KCl solution (no. of expts)a |
Avg H+/O ratio | ||
|---|---|---|---|---|
| 12.5 μl | 25 μl | 50 μl | ||
| G. oxydans strains | ||||
| Δupp strain | 1.19 ± 0.11 (10) | 1.34 ± 0.10 (10) | 1.24 ± 0.10 (10) | 1.26 ± 0.10 |
| Δupp ΔcydAB strain | ND | 1.31 ± 0.16 (8) | ND | 1.31 ± 0.16 |
| Δupp ΔcyoBACD strain | ND | 0.56 ± 0.11 (8) | ND | 0.56 ± 0.11 |
| E. coli DH5αF′ | 2.89 ± 0.23 (10) | 3.16 ± 0.21 (10) | 3.05 ± 0.17 (10) | 3.04 ± 0.11 |
The H+/O ratio measured by the oxygen pulse method. ND, not determined.
E. coli was used as a control strain for the experimental setup.
Complementation of terminal oxidases.
For complementation studies, the genes cyoBACD, cydAB, and cydABCD with their native ribosomal binding sites were cloned into the expression vector pBBR1p384, resulting in plasmids pBBRp384-cyoBACD, pBBRp384-cydAB, and pBBRp384-cydABCD, respectively. After transfer of the plasmids into the desired G. oxydans strains, growth of the recombinants in mannitol medium was tested. Plasmid-based expression of either cydAB or cydABCD in the reference Δupp strain and in the Δupp ΔcydAB mutant had no influence on the growth properties (data not shown). In contrast, plasmid-based expression of the cyoBACD genes led to improved growth (Fig. 7). The Δupp strain carrying pBBRp384-cyoBACD showed an 8% higher growth rate than the Δupp strain carrying pBBRp384 (from 0.39 ± 0.01 h−1 to 0.42 ± 0.00 h−1), and the final OD600 was increased by 20% (from 4.05 ± 0.13 to 4.88 ± 0.09). In the Δupp ΔcyoBACD mutant strain carrying pBBRp384-cyoBACD, the growth rate was increased by 60% compared to the strain carrying pBBRp384 (from 0.27 ± 0.03 h−1 to 0.43 ± 0.00 h−1), and the final OD600 increased by 69% (from 2.92 ± 0.05 to 4.93 ± 0.05). The delay observed in Fig. 7 for mannitol oxidation and fructose formation by strain G. oxydans Δupp(pBBRp384-cyoBACD) compared to the Δupp ΔcyoBACD(pBBRp384-cyoBACD) strain is probably due to a higher initial mannitol concentration of the former culture.
Fig 7.
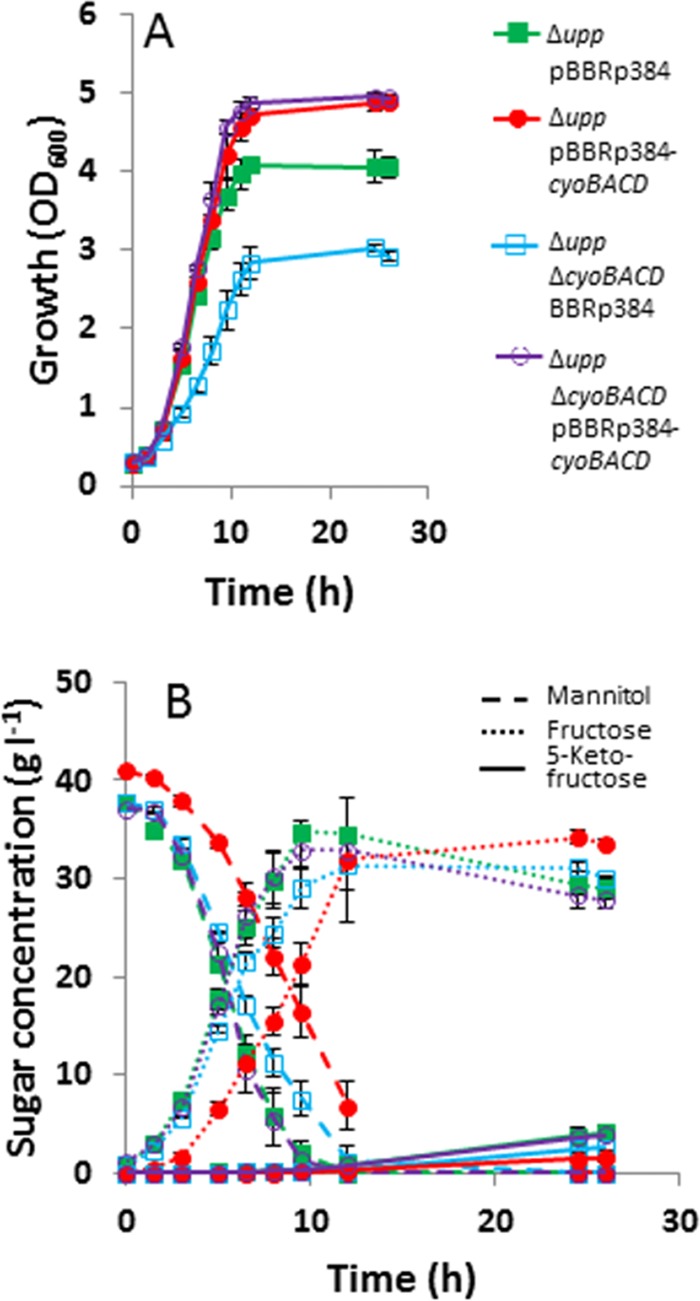
Growth (A), substrate consumption and product formation (B) of the G. oxydans Δupp(pBBRp384), Δupp(pBBRp384-cyoBACD), Δupp ΔcyoBACD(pBBRp384), and Δupp ΔcyoBACD(pBBRp384-cyoBACD) strains. The strains were cultivated in 100-ml baffled shake flasks with mannitol medium at 30°C and 140 rpm. Mean values and standard deviations of three independent cultures are shown.
DISCUSSION
In this study, the roles of the two terminal oxidases of G. oxydans 621H, cytochrome bd oxidase and cytochrome bo3 oxidase, were investigated. Deletion of the corresponding genes revealed that neither of these oxidases is essential for growth in a mannitol-yeast extract medium but that the bo3 oxidase is of major importance for the build-up of proton motive force and biomass formation.
Shake flask and RAMOS cultivations demonstrated that the absence of cytochrome bd oxidase had no influence on growth and respiration activity under either oxygen excess or oxygen limitation conditions. The reference strain and the bd oxidase deletion mutant consumed comparable quantities of oxygen (oxygen excess condition, 194 and 182 mM; oxygen limitation condition, 162 and 171 mM) (Table 2). For complete oxidation of 220 mM mannitol to fructose, 110 mM O2 is required, and the rest is used for oxidation of mannitol or fructose to CO2 and acetate in the cytoplasm, for partial oxidation of fructose to ketofructose in the periplasm, and for oxidation of components of the yeast extract (Table 2). The strain lacking cytochrome bo3 oxidase showed reduced oxygen consumption (oxygen excess condition, 154 mM; oxygen limitation condition, 142 mM), which is in agreement with the reduced formation of CO2, 5-ketofructose, and biomass.
The oxygen consumption rates of the cyo deletion mutant in the RAMOS and respirometer studies was about 25 to 30% lower than those of the reference strain and the cyd deletion mutant. This relatively small reduction is presumably due to an increased synthesis of cytochrome bd oxidase in the cyo deletion mutant, which was indicated by an increased peak at 630 nm in reduced-minus-oxidized difference spectra of purified membranes of this mutant and by a 3.6- to 5.4-fold increased mRNA level of the cydAB genes (GOX0278-GOX0279) in DNA microarray experiments comparing the mRNA levels of the Δupp ΔcyoBACD and Δupp strains (see Table S1 and Fig. S1 in the supplemental material). This suggests that synthesis of the bd oxidase is increased in the cyo deletion mutant and that G. oxydans is capable of sensing and responding to a deficiency in its respiratory capacity.
When the RAMOS cultivations were performed under conditions leading to oxygen limitation during growth, the OTR of the cyo deletion mutant hardly reached the horizontal plateau that was observed for the reference strain and the cyd deletion mutant (Fig. 5). Instead, the OTR of the cyo deletion mutant slowly approached the maximal value possible under the chosen conditions. This behavior might be caused by a low-oxygen affinity of cytochrome bd, the only remaining terminal oxidase in the cyo deletion mutant. Support for this assumption was obtained by the respirometer measurements of the cyo deletion mutant, in which the initially linear oxygen consumption rate had already decreased when the dissolved oxygen tension (DOT) fell below 25%, whereas in case of the wild type and the cyd deletion mutant, it did not decrease until the DOT fell below 10% (Fig. 6).
Our evidence that cytochrome bd oxidase of G. oxydans has a significantly lower oxygen affinity than cytochrome bo3 oxidase is in agreement with recent biochemical data. Based on the cyanide insensitivity of G. oxydans cytochrome bd, Matsushita and coworkers designated this enzyme cyanide-insensitive oxidase (CIO) and the corresponding genes cioA and cioB instead of cydA and cydB (12). In a recent study, they purified and characterized the terminal oxidases of G. oxydans and, for comparison, cytochrome bd of E. coli (13). Their results indicated significant differences between CIO and E. coli cytochrome bd: besides a much lower sensitivity to cyanide (50% inhibitory concentration [IC50] of 118 mM versus 4.2 mM for E. coli bd), the CIO enzyme has a higher sensitivity to azide (IC50 of 0.8 mM versus 4.8 mM), an exceptionally high turnover number, and a KM value for oxygen (21 μM) which is 10-fold higher than the one of E. coli cytochrome bd (2 μM) and about 7-fold higher than the one measured for G. oxydans cytochrome bo3 (3 μM). A phylogenetic analysis supported the idea that the cytochrome bd oxidase superfamily includes a specific CIO clade with members from the alpha, beta-, and gammaproteobacteria (13). Based on these data, the use of a different designation for members of the CIO clade appears reasonable and will be used by us in future studies.
In agreement with the growth properties of the G. oxydans cyd and cyo deletion mutants, the absence of cytochrome bd oxidase did not affect respiration-driven proton extrusion (H+/O ratio of 1.3), whereas the lack of cytochrome bo3 oxidase caused a reduction of more than 50% (H+/O ratio of 0.56). That the absence of the non-proton-pumping cytochrome bd oxidase did not improve the H+/O ratio might be due to a much lower content of the bd oxidase than of the bo3 enzyme and to the proposed lower oxygen affinity of the bd oxidase, which should favor oxygen reduction by the higher-affinity bo3 oxidase under the experimental conditions of an oxygen pulse. In previous studies, H+/O ratios of 1.7 to 2.2 were reported for G. oxydans cells supplied with glycerol, glucose, lactate, or ethanol at pH 6 (39). These values are higher than those determined by us, which could be due to the use of other substrates or differences in the experimental conditions.
The H+/O ratio of 1.3 measured for G. oxydans is more than 2-fold lower than the ratio of 3.0 determined for E. coli using a nonoptimized system. In the literature, H+/O ratios reported for E. coli vary between 3.4 and 4.9 (20, 34, 40). A major difference between the respiratory chains of the two species is the lack of the multisubunit proton-pumping NADH dehydrogenase I (NDH-I) in G. oxydans. However, NDH-I is preferentially synthesized during anaerobic growth in the presence of alternate electron acceptors, whereas during aerobic growth the non-proton-pumping single-subunit NDH-II is the dominant NADH:Q oxidoreductase (41). Therefore, it is unclear to what extent NDH-I (H+/e− stoichiometry of 2; [42]) contributes to the H+/O ratios measured for E. coli. For cytochrome bo3 oxidase, a H+/e stoichiometry of 2, and therefore a H+/O ratio of 4, can be assumed (11), and for cytochrome bd oxidase a H+/e stoichiometry of 1 and a H+/O ratio of 2 are assumed (43, 44). As the bd oxidase was not relevant for growth of G. oxydans and proton translocation under the conditions used in our study, a H+/O ratio approaching 4 might be expected for G. oxydans. However, both our results as well as those presented by Matsushita et al. (39) are much lower than a value of 4. There are a number of very speculative hypotheses to explain the low H+/O ratios determined for G. oxydans. (i) The cytoplasmic membrane of G. oxydans might be leakier for protons than the one of E. coli, causing a non-energy-conserving backflow of protons into the cells (45). (ii) The respiratory chain could involve a reverse electron transfer coupled to an influx of protons (46). (iii) Cytochrome bo3 oxidase might not function as a primary proton pump but as a Na+ pump, as has been described for cytochrome bo3 oxidase of Vitreoscilla sp. (47, 48). In this context it is interesting that G. oxydans possesses two different F1Fo-type ATP synthases (49), one of which might use Na+ as coupling ion (8, 38). The comparative DNA microarray analysis of the cyo deletion mutant versus the reference strain showed an upregulation of genes (GOX2167 to GOX2175) coding for the putative Na+-dependent F1Fo-ATPase (see Table S1 in the supplemental material).
Surprisingly, complementation of the G. oxydans cyo deletion mutant with the expression plasmid pBBR1p384-cyoBACD resulted not only in a reversal to wild-type-like growth but also to an increased growth rate and an increased final OD600. A comparable growth behavior was observed when the Δupp reference strain was transformed with this plasmid (Fig. 7). An increased cell yield was also observed for an E. coli mutant deficient in the bo3 and the bd-I oxidase when the bo3 oxidase gene was overexpressed by a plasmid (20). The possibility that an increased level of bo3 oxidase in G. oxydans causes a shift of the electron flux from the non-proton-pumping bd oxidase to the proton-pumping bo3 oxidase appears unlikely as the G. oxydans cyd deletion mutant did not show improved growth although only the bo3 oxidase remained as a terminal oxidase. Another explanation for the improved growth is the assumption that the oxidative capacity of G. oxydans under the experimental conditions used is limited by the activity of the bo3 oxidase and can be increased by its overproduction. A limitation of oxygen consumption by the activity of the terminal oxidases was recently reported for membranes of a G. oxydans strain with an increased membrane-bound glucose dehydrogenase activity (50). Although the major polyol dehydrogenase SldAB responsible for mannitol oxidation to fructose was not overproduced in our strain, its activity might be high enough to satisfy an increased capacity for oxygen reduction due an elevated level of bo3 oxidase. In addition, the oxidation of components of the yeast extract can also contribute to the electron flux to the bo3 oxidase. An increased rate of oxygen reduction by the bo3 oxidase is coupled to an increased rate of proton motive force generation and oxidative phosphorylation, which can explain the increased growth rate and maybe also the increased cell yield. Although additional studies are required to clarify the positive effect of bo3 overproduction, the result shows that respiratory chain components are a promising starting point for further optimization of G. oxydans for its use in biotechnological applications. For the development of strains with a higher biomass yield, studies are planned that are directed at the repair of defective metabolic pathways by genomic integration of heterologous genes, such as succinate dehydrogenase, and at an increased cytoplasmic rather than periplasmic metabolism of the carbon source by deleting suitable membrane-bound dehydrogenases. If these approaches are successful, the possibilities for using G. oxydans as a broadly applicable host for oxidative biotransformations in industry will be considerably improved.
Supplementary Material
ACKNOWLEDGMENTS
We are most grateful to Armin Ehrenreich and Wolfgang Liebl (Technical University of Munich, Germany) for providing the strains and protocols used for generating the G. oxydans deletion mutants and to Uwe Deppenmeier (University of Bonn, Germany) for providing plasmid pBBR1p384. We thank Jayne Louise Wilson (The University of Sheffield, United Kingdom) for advising us on the method of H+/O measurement. We thank Dietmar Laudert, Günter Pappenberger, and Hans-Peter Hohmann (DSM Nutritional Products) for their scientific input and their continued disposition for discussion.
We also thank DSM Nutritional Products (Kaiseraugst, Switzerland) for financial support. This work was funded by the German Ministry of Education and Research (BMBF) within the GenoMik-Transfer program (grant 0315632D and B).
Footnotes
Published ahead of print 12 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00470-13.
REFERENCES
- 1.Kersters K, Lisdiyanti P, Komagata K, Swings J. 2006. The family Acetobacteriaceae: the genera Acetobacter, Acidomonas, Asaia, Gluconacetobacter, Gluconobacter and Kozakia, p 163–200 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes, vol. 5, 3rd ed. Springer-Verlag GmbH, Heidelberg, Germany [Google Scholar]
- 2.De Ley J, Gillis M, Swings J. 1984. The genus Gluconobacter, p 267–278 In Krieg NR, Holt JG. (ed), Bergey's manual of systematic bacteriology, vol. 1 Williams and Wilkins, Baltimore, MD [Google Scholar]
- 3.Raspor PP, Goranovič D. 2008. Biotechnological applications of acetic acid bacteria. Crit. Rev. Biotechnol. 28:101–124 [DOI] [PubMed] [Google Scholar]
- 4.Richhardt J, Bringer S, Bott M. 2013. Role of the pentose phosphate pathway and the Entner-Doudoroff pathway in glucose metabolism of Gluconobacter oxydans 621H. Appl. Microbiol. Biotechnol. 97:4315–4323 [DOI] [PubMed] [Google Scholar]
- 5.Richhardt J, Bringer S, Bott M. 2012. Mutational analysis of the pentose phosphate and Entner-Doudoroff pathways in Gluconobacter oxydans reveals improved growth of a Δedd Δeda mutant on mannitol. Appl. Environ. Microbiol. 78:6975–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanke T, Nöh K, Noack S, Polen T, Bringer S, Sahm H, Wiechert W, Bott M. 2013. Combined fluxomics and transcriptomics analysis of glucose catabolism via a partially cyclic pentose phosphate pathway in Gluconobacter oxydans 621H. Appl. Environ. Microbiol. 79:2336–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deppenmeier U, Hoffmeister M, Prust C. 2002. Biochemistry and biotechnological applications of Gluconobacter strains. Appl. Microbiol. Biotechnol. 60:233–242 [DOI] [PubMed] [Google Scholar]
- 8.Prust C, Hoffmeister M, Liesegang H, Wiezer A, Fricke WF, Ehrenreich A, Gottschalk G, Deppenmeier U. 2005. Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat. Biotechnol. 23:195–200 [DOI] [PubMed] [Google Scholar]
- 9.Ameyama M, Matsushita K, Shinagawa E, Adachi O. 1987. Sugar-oxidizing respiratory chain of Gluconobacter suboxydans. Evidence for a branched respiratory chain and characterization of respiratory chain-linked cytochromes. Agic. Biol. Chem. 51:2943–2950 [Google Scholar]
- 10.Matsushita K, Shinagawa E, Adachi O, Ameyama M. 1987. Purification, characterization and reconstitution of cytochrome o-type oxidase from Gluconobacter suboxydans. Biochim. Biophys. Acta 894:304–312 [Google Scholar]
- 11.Puustinen A, Finel M, Virkki M, Wikström M. 1989. Cytochrome o (bo) is a proton pump in Paracoccus denitrificans and Escherichia coli. FEBS Lett. 249:163–167 [DOI] [PubMed] [Google Scholar]
- 12.Mogi T, Ano Y, Nakatsuka T, Toyama H, Muroi A, Miyoshi H, Migita CT, Ui H, Shiomi K, Omura S, Kita K, Matsushita K. 2009. Biochemical and spectroscopic properties of cyanide-insensitive quinol oxidase from Gluconobacter oxydans. J. Biochem. 146:263–271 [DOI] [PubMed] [Google Scholar]
- 13.Miura H, Mogi T, Ano Y, Migita CT, Matsutani M, Yakushi T, Kita K, Matsushita K. 2013. Cyanide-insensitive quinol oxidase (CIO) from Gluconobacter oxydans is a unique terminal oxidase subfamily of cytochrome bd. J. Biochem. 153:535–545 [DOI] [PubMed] [Google Scholar]
- 14.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 1807:1398–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller MJ, Gennis RB. 1985. The cytochrome d complex is a coupling site in the aerobic respiratory chain of Escherichia coli. J. Biol. Chem. 260:14003–14008 [PubMed] [Google Scholar]
- 16.Fu HA, Iuchi S, Lin EC. 1991. The requirement of ArcA and Fnr for peak expression of the cyd operon in Escherichia coli under microaerobic conditions. Mol. Gen. Genet. 226:209–213 [DOI] [PubMed] [Google Scholar]
- 17.Krajewski V, Simić P, Mouncey NJ, Bringer S, Sahm H, Bott M. 2010. Metabolic engineering of Gluconobacter oxydans for improved growth rate and growth yield on glucose by elimination of gluconate formation. Appl. Environ. Microbiol. 76:4369–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dauner M, Sonderegger M, Hochuli M, Szyperski T, Wüthrich K, Hohmann HP, Sauer U, Bailey JE. 2002. Intracellular carbon fluxes in riboflavin-producing Bacillus subtilis during growth on two-carbon substrate mixtures. Appl. Environ. Microbiol. 68:1760–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soini J, Ukkonen K, Neubauer P. 2008. High cell density media for Escherichia coli are generally designed for aerobic cultivations—consequences for large-scale bioprocesses and shake flask cultures. Microb. Cell Fact. 7:26. 10.1186/1475-2859-7-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minohara S, Sakamoto J, Sone N. 2002. Improved H+/O ratio and cell yield of Escherichia coli with genetically altered terminal quinol oxidases. J. Biosci. Bioeng. 93:464–469 [DOI] [PubMed] [Google Scholar]
- 21.Luttik M, Van Spanning R, Schipper D, Van Dijken JP, Pronk JT. 1997. The low biomass yields of the acetic acid bacterium Acetobacter pasteurianus are due to a low stoichiometry of respiration-coupled proton translocation. Appl. Environ. Microbiol. 63:3345–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25.Hanahan D, Jessee J, Bloom FR. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63–113 [DOI] [PubMed] [Google Scholar]
- 26.Peters B, Junker A, Brauer K, Mühlthaler B, Kostner D, Mientus M, Liebl W, Ehrenreich A. 2013. Deletion of pyruvate decarboxylase by a new method for efficient markerless gene deletions in Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 97:2521–2530 [DOI] [PubMed] [Google Scholar]
- 27.Anderlei T, Zang W, Papaspyrou M, Büchs J. 2004. Online respiration activity measurement (OTR, CTR, RQ) in shake flasks. Biochem. Eng. J. 17:187–194 [Google Scholar]
- 28.Anderlei T, Büchs J. 2001. Device for sterile online measurement of the oxygen transfer rate in shaking flasks. Biochem. Eng. J. 7:157–162 [DOI] [PubMed] [Google Scholar]
- 29.Mrotzek C, Anderlei T, Henzler H, Büchs J. 2001. Mass transfer resistance of sterile plugs in shaking bioreactors. Biochem. Eng. J. 7:107–112 [DOI] [PubMed] [Google Scholar]
- 30.Peña C, Galindo E, Büchs J. 2011. The viscosifying power, degree of acetylation and molecular mass of the alginate produced by Azotobacter vinelandii in shake flasks are determined by the oxygen transfer rate. Proc. Biochem. 46:290–297 [Google Scholar]
- 31.Seletzky JM, Noak U, Fricke J, Welk E, Eberhard W, Knocke C, Büchs J. 2007. Scale-up from shake flasks to fermenters in batch and continuous mode with Corynebacterium glutamicum on lactic acid based on oxygen transfer and pH. Biotechnol. Bioeng. 98:800–811 [DOI] [PubMed] [Google Scholar]
- 32.Silberbach M, Maier B, Zimmermann M, Büchs J. 2003. Glucose oxidation by Gluconobacter oxydans: characterization in shaking-flasks, scale-up and optimization of the pH profile. Appl. Microbiol. Biotechnol. 62:92–98 [DOI] [PubMed] [Google Scholar]
- 33.Bott M, Thauer RK. 1989. Proton translocation coupled to the oxidation of carbon monoxide to CO2 and H2 in Methanosarcina barkeri. Eur. J. Biochem. 179:469–472 [DOI] [PubMed] [Google Scholar]
- 34.Lawford HG, Haddock BA. 1973. Respiration-driven proton translocation in Escherichia coli. Biochem. J. 136:217–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumpe A, Deckwer W-D. 1979. Estimation of O2 and CO2 solubilities in fermentation media. Biotechnol. Bioeng. 21:1075–1078 [Google Scholar]
- 36.Schumpe A, Quicker G, Deckwer W-D. 1982. Gas solubilities in microbial culture media. Adv. Biochem. Eng. 24:1–38 [Google Scholar]
- 37.Rischbieter E, Schumpe A. 1996. Gas solubilities in aqueous solutions of organic substances. J. Chem. Eng. Data 41:809–812 [Google Scholar]
- 38.Hanke T, Richhardt J, Polen T, Sahm H, Bringer S, Bott M. 2012. Influence of oxygen limitation, absence of the cytochrome bc1 complex and low pH on global gene expression in Gluconobacter oxydans 621H using DNA microarray technology. J. Biotechnol. 157:359–372 [DOI] [PubMed] [Google Scholar]
- 39.Matsushita K, Nagatani Y, Shinagawa E, Adachi O, Ameyama M. 1989. Effect of extracellular pH on the respiratory chain and energetics of Gluconobacter suboxydans. Agric. Biol. Chem. 53:2895–2902 [Google Scholar]
- 40.Brice JM, Law JF, Meyer DJ, Jones CW. 1974. Energy conservation in Escherichia coli and Klebsiella pneumoniae. Biochem. Soc. Trans. 2:523–526 [Google Scholar]
- 41.Tran QH, Bongaerts J, Vlad D, Unden G. 1997. Requirement for the proton-pumping NADH dehydrogenase I of Escherichia coli in respiration of NADH to fumarate and its bioenergetic implications. Eur. J. Biochem. 244:155–160 [DOI] [PubMed] [Google Scholar]
- 42.Bogachev AV, Murtazina RA, Skulachev VP. 1996. H+/e− stoichiometry for NADH dehydrogenase I and dimethyl sulfoxide reductase in anaerobically grown Escherichia coli cells. J. Bacteriol. 178:6233–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puustinen A, Finel M, Haltia T, Gennis RB, Wikström M. 1991. Properties of the two terminal oxidases of Escherichia coli. Biochemistry 30:3936–3942 [DOI] [PubMed] [Google Scholar]
- 44.Jasaitis A, Borisov VB, Belevich NP, Morgan JE, Konstantinov AA, Verkhovsky MI. 2000. Electrogenic reactions of cytochrome bd. Biochemistry 39:13800–13809 [DOI] [PubMed] [Google Scholar]
- 45.Brown GC. 1992. The leaks and slips of bioenergetic membranes. FASEB J. 6:2961–2965 [DOI] [PubMed] [Google Scholar]
- 46.van der Oost J, Schepper M, Stouthamer AH, Westerhoff HV, van Spanning RJ, de Gier JW. 1995. Reversed electron transfer through the bc1 complex enables a cytochrome c oxidase mutant (delta aa3/cbb3) of Paracoccus denitrificans to grow on methylamine. FEBS Lett. 371:267–270 [DOI] [PubMed] [Google Scholar]
- 47.Kim SK, Stark BC, Webster DA. 2005. Evidence that Na+-pumping occurs through the D-channel in Vitreoscilla cytochrome bo. Biochem. Biophys. Res. Commun. 332:332–338 [DOI] [PubMed] [Google Scholar]
- 48.Park C, Moon JY, Cokic P, Webster DA. 1996. Na+-translocating cytochrome bo terminal oxidase from Vitreoscilla: some parameters of its Na+ pumping and orientation in synthetic vesicles. Biochemistry 35:11895–11900 [DOI] [PubMed] [Google Scholar]
- 49.Dibrova DV, Galperin M, Mulkidjanian A. 2010. Characterization of the N-ATPase, a distinct, laterally transferred Na+-translocating form of the bacterial F-type membrane ATPase. Bioinformatics 26:1473–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyer M, Schweiger P, Deppenmeier U. 2013. Effects of membrane-bound glucose dehydrogenase overproduction on the respiratory chain of Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 97:3457–3466 [DOI] [PubMed] [Google Scholar]
- 51.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic-engineering-transposon mutagenesis in Gram-negative bacteria. Nat. Biotehnol. 1:784–791 [Google Scholar]
- 52.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.