Abstract
The emergence of Acinetobacter baumannii as an increasingly multidrug-resistant nosocomial pathogen largely relies on acquisition of resistance genes via horizontal gene transfer. Here, we demonstrate that many clinical isolates of A. baumannii take up DNA while they move along wet surfaces. We show that both motility and DNA uptake are abolished after inactivation of pilT, which putatively encodes the type 4 pilus (T4P) retraction ATPase, and comEC, which putatively encodes the DNA uptake channel, respectively. Inactivation of pilT correlates with an increase in the number and length of pili with an average diameter of 7.2 nm. In the Galleria mellonella infection model, the comEC mutant is significantly attenuated, whereas the pilT mutant is not, dissecting biologically distinct roles of T4P and the DNA uptake channel. Collectively, these findings promote our understanding of the mechanisms of DNA uptake and resistance development in A. baumannii, which may also apply to other important pathogens.
INTRODUCTION
The capability of Acinetobacter baumannii to undergo horizontal gene transfer (HGT) events considerably contributes to the alarming resistance development of this emerging pathogen (1–6). However, while the low-pathogenic relative Acinetobacter baylyi ADP1 (BD4) is a model organism to study DNA uptake from the environment (7–12), to date only a single isolate of A. baumannii has been shown to be naturally competent for transformation (13).
It is long known that members of the genus Acinetobacter, though lacking flagella, can move along wet surfaces in an intermittent and jerky way termed twitching motility (14–16). Henrichsen and Blom were the first to propose that Acinetobacter twitching motility was related to the expression of polar fimbriae (16, 17). Since that time, twitching motility has been intensively studied in many genera, including Myxococcus, Neisseria, Pseudomonas, and Haemophilus, firmly establishing that this form of movement is powered by depolymerization of type 4 pili (T4P) (18–21). Only very recently was Acinetobacter motility further elucidated, providing evidence that at least in part it is driven by means of T4P in A. baumannii (22–26). Specifically, inactivation of pilT, putatively encoding an ATPase responsible for T4P retraction, reduced surface motility by roughly 50% (24). Residual activity observed with this pilT mutant could be due to the pilT paralogue pilU, known to be present in many representatives of A. baumannii (23). Alternatively, another mode of surface-associated motility could be active under the same experimental conditions. Actually, forms of motility seemingly different from twitching have been described for Acinetobacter. Barker and Maxted (27) found that when Acinetobacter strains were stab inoculated into semisolid media, some showed surface motility called “swarming,” while others exhibited spreading at the bottom of the petri dish beneath the medium or both forms in parallel. In addition, spreading at the surface sometimes was found to be accompanied by the formation of ditches in the agar surface, and no signs of jerking movements were found by these authors under the conditions tested (27). Even though phenotypically distinct, all forms of motility described for A. baumannii have been shown to depend on synthesis of the polyamine 1,3-diaminopropane (26). Surface motility of A. baumannii was further shown to be controlled by blue light sensing (28) and quorum sensing (24, 29) and to depend on iron availability (30, 31).
Besides conveying twitching motility, T4P also permit DNA uptake in a number of Gram-negative bacteria (19), and in Neisseria gonorrhoeae, for example, T4P are involved in both motility and DNA uptake (32). The mechanistic role that T4P play in DNA uptake is not clearly defined but requires the secretin PilQ for DNA passage through the outer membrane (33, 34). Transport via the inner membrane is mediated by a ComA/ComEC membrane channel (12, 33, 35). While A. baylyi harbors a comA-like transporter gene that has been shown to be required for natural transformation (36), A. baumannii harbors a comEC-like gene (37) for which no functional characterization is published to date and which exhibits only about 50% identity to A. baylyi ComA on the protein level.
Sequencing of A. baumannii genomes is steadily revealing that members of this species are in extensive genetic exchange with related species and also across the genus, family, and order barriers, suggesting that natural competence could contribute to this continuous DNA uptake (1, 4, 37, 38). Although an apparently complete set of genes required for natural transformation competence seems to be present in A. baumannii (23, 25, 37), to date, only a single isolate has been described to be naturally competent (13). Given the potential role of T4P in surface-associated motility of A. baumannii (17, 23–26) and their established contribution to DNA uptake in various species (12, 19, 35), we speculated that A. baumannii might develop competence for DNA uptake while moving along wet surfaces in a T4P-dependent manner.
MATERIALS AND METHODS
Motility and transformation.
Motility plates were composed of 0.5% agarose, 5 g/liter of tryptone, and 2.5 g/liter of NaCl as described previously (26). The inoculum was stabbed into the semisolid medium to enable spread of bacteria at both the surface of the medium and the interphase between the bottom of the petri dish and the medium. Two alternative transformation procedures were performed. The transforming DNA (30 μg per plate) could be added to the molten medium immediately before pouring the plates. The plates were then inoculated by stabbing with a pipette tip. A single colony from a blood agar plate stored in the refrigerator for no longer than 2 weeks was touched with the pipette tip, which was then stabbed into the DNA-doped motility plate seven times. Alternatively, the DNA can be mixed with the inoculum of bacteria and can then be stabbed into the motility medium (seven times, pipetting 2 μl of the mixture with each stabbing). To this end, a suspension of bacteria (generated from a single colony resuspended in 20 μl of sterile phosphate-buffered saline [PBS]) is produced and mixed with equal volumes of the transforming DNA (400 ng/μl). The precise optical density (OD) of the bacterial suspension had no significant effect on the transformation efficiency. The latter procedure yielded higher transformation rates than did the standard procedure in which the transforming DNA (30 μg per plate) was mixed into the medium prior to pouring into petri dishes, albeit at the expense of somewhat increased variance. The method using mixtures of bacteria and DNA was also used to determine the transformation rates given in Table 1. After inoculation, the transformation plates were sealed with Parafilm to prevent desiccation, which proved detrimental to both motility and transformation efficiency. The plates were incubated for 18 h at 37°C. The bacteria were then flushed off the motility medium with 1 ml of sterile PBS. The suspension was adjusted to 10 optical densities (so that the 10-fold dilution yielded an OD at 600 nm [OD600] of 1.0), and 100 μl was plated on the appropriate selective agar (typically 30 μg/ml of kanamycin). Tenfold dilution series were performed from the OD-adjusted PBS suspension to determine the number of CFU for calculation of transformation rates (number of transformants divided by total CFU). Chromosomal DNA for transformation experiments was purified with the MasterPure DNA purification kit (Epicentre Biotechnologies). Sterility of transforming DNA was controlled by plating. Effective transformation with DNA from ATCC 17978 mutants 27 and 179 was confirmed by PCR on selected colonies after subculturing of these colonies. Direct colony PCR from the selection plate is not recommended, since the background of transforming DNA as well as the background of DNA from killed bacteria can lead to ambiguous results. Subsequently, DNA sequencing was performed to confirm homologous recombination events. Phenotypic features such as motility morphotypes were used as additional controls. DNase I treatment of the mixture of transforming DNA and bacterial inoculum significantly reduced the transformation rates, while treatment of the bacteria flushed off the motility plates with DNase I did not interfere with the transformation rates.
Table 1.
Transformation rates of 10 naturally competent A. baumannii isolatesa
| Strain | Mean transformation rate (SD) |
||
|---|---|---|---|
| Mutant 27 DNAb | Mutant 179 DNAc | Plasmid pWH1266::Kmd | |
| 07-028 | 5.85E−08 (6,78E−08) | 1.29E−06 (8.27E−07) | 0 (0) |
| 07-099 | 0 (0) | 6.44E−08 (1.04E−07) | 0 (0) |
| 07-095 | 4.09E−06 (1.06E−06) | 1.13E−04 (2.56E−05) | 4.53E−06 (1.61E−06) |
| 07-101 | 4.34E−07(2.56E−07) | 5.39E−05 (3.51E−05) | 3.91E−08 (3.91E−08) |
| 07-102 | 5.82E−05 (5.25E−06) | 2.62E−03 (7.12E−04) | 0 (0) |
| 07-105 | 5.90E−08 (4.82E−08) | 2.41E−06 (1.17E−06) | 5.16E−08 (5.16E−08) |
| 07-111 | 1.87E−07 (8.50E−09) | 6.37E−06 (4.09E−06) | 0 (0) |
| 10-096 | 7.75E−06 (3.21E−06) | 5.72E−04 (2.76E−04) | 9.28E−07 (1.03E−07) |
| DSM 30011 | 2.59E−06 (8.98E−07) | 1.99E−04 (1.26E−04) | 0 (0) |
| BMBF 320 | 2.94E−06 (1.50E−06) | 1.07E−05 (3.09E−06) | 1.36E−06 (5.22E−07) |
To obtain chromosomal DNA for transformation experiments, Acinetobacter baumannii ATCC 17978 was mutagenized with transposon EZ-Tn5 <KAN-2> (Epicentre Biotechnologies) as previously described (26). From resulting mutants 27 and 179, harboring transposon insertions in genes A1S_2167 (encoding cytochrome o ubiquinol oxidase subunit I) and A1S_2846 (encoding sulfite reductase), respectively, chromosomal DNA was purified. Plasmid transformation was studied with a derivative of pWH1266 (39), designated pWH1266::Km. Transformation experiments were performed as described in Materials and Methods.
Three independent experiments.
Four independent experiments.
Two independent experiments.
Plasmid transformation was studied with a derivative of pWH1266 (39), designated pWH1266::Km, which was isolated from Escherichia coli DH5α. Plasmid pWH1266 confers resistance to ampicillin and tetracycline. Since all 10 naturally competent isolates are sensitive to kanamycin but not all are sensitive to either ampicillin or tetracycline, we mutagenized the plasmid with transposon EZ-Tn5 <KAN-2> (Epicentre Biotechnologies) to obtain pWH1266::Km. Transposon insertion after nucleotide position 207 (39) as verified by DNA sequencing did not interfere with plasmid stability or copy number. Effective transformation with plasmid pWH1266::Km was confirmed by isolation of the plasmid from a number of colonies and detection of the Kmr cassette in the pWH1266 background by PCR. To this end, forward primer FP3 5′-GAGTTGAAGGATCAGATCACGC-3′ binding inside EZ-Tn5 <KAN-2> and reverse primer pWH1266-rev1 5′-GCCTAGAACGTCATAGGAAGCG-3′ binding inside pWH1266 were combined, resulting in a PCR product of approximately 1,250 bp.
A. baumannii mutants.
Transforming DNA was obtained from transposon mutant derivatives of A. baumannii ATCC 17978 mutagenized with the EZ-Tn5 <KAN-2> transposon (Epicentre Biotechnologies). Screening of a transposon mutant library of A. baumannii ATCC 17978 for motility phenotypes revealed a motility-deficient mutant with a transposon insertion in A1S_2610, encoding a homologue of the ComEC competence protein family. Since ATCC 17978 is unable to move at the interphase between the medium and the bottom of the petri dish (26), we used the chromosomal DNA of this comEC::Km mutant to transform naturally competent isolates 07-095, 07-102, and DSM 30011 exhibiting motility at the interphase. Chromosomal DNA of A. baumannii M2 pilT::Km (24) was obtained from Philip N. Rather.
Electron microscopy studies.
Appropriate strains were stab inoculated seven times into motility agarose (0.5% agarose) and incubated at 37°C for approximately 18 h. Colonies formed on the agarose surface were gently resuspended in 0.9 ml of HEPES buffer (mixture of 0.85 ml of H2O plus 0.05 ml of 1 M HEPES [pH 7.2]), and the cells were subsequently fixed by addition of 0.1 ml of paraformaldehyde (20%). The agarose layer was then removed from the petri dishes, and the bacteria sticking to the polystyrene petri dishes (“interphase”) were gently resuspended in HEPES buffer and fixed as described above. Due to the poor growth of the pilT and comEC mutants at the interphase, these strains were stab inoculated 10 times on each plate, and material obtained from three plates was pooled in 1 ml of buffer to yield enough bacteria. Actually, these mutants exhibited no spread at the interphase but formed colonies at the sites of stab inoculation.
Negative-staining electron microscopy was conducted as described by Laue and Bannert (40). Briefly, suspensions of fixed bacteria were applied onto sample supports (drop-on-grid procedure) that had been pretreated with alcian blue or by glow discharge. After brief washes on distilled water, adsorbed bacteria were stained with uranyl acetate (0.5% in water). Samples were inspected with a transmission electron microscope (Tecnai12; FEI Corp.) operated at 120 kV. Images were taken using a 1k slow-scan CCD-camera (MegaviewIII; Olympus Soft Imaging Solutions). Measurements at high resolution were calibrated by using a precise calibration standard (Magical; Technoorg-Linda Ltd.).
Galleria mellonella infection model.
Infection of waxmoth larvae was performed as described recently (26).
RESULTS
Do A. baumannii isolates take up DNA while they move?
To determine whether A. baumannii isolates take up DNA while they move, we selected 28 clinical isolates of A. baumannii from our collection sensitive to the antibiotic kanamycin. We performed transformation experiments using chromosomal DNA of Km-resistant transposon mutant derivatives of A. baumannii strain ATCC 17978. We doped a semisolid medium facilitating surface-associated motility with the transforming DNA and subsequently inoculated A. baumannii isolates to allow them to move along the wet surface. Figure 1 illustrates the morphotypic variance among the isolates under these conditions. After 18 h, the bacteria were rinsed off and plated on kanamycin plates to select for transformants (see Materials and Methods) (Table 1). We identified 10 out of 28 isolates (36%) that were competent for the uptake of the naked DNA. Transformation rates varied depending on isolates and on the locus of homologous recombination, with rates ranging from 3 × 10−3 to 6 × 10−8 for the most efficiently transforming DNA (Table 1). Only 5 of the 10 naturally competent isolates could be transformed with the plasmid tested, a derivative of pWH1266 (39) harboring an insertion of transposon EZ-Tn5 Kan2 (Table 1). The transformation competence and efficiency appeared to be unpredictable from the motility phenotypes and did not correlate with the velocity of motility.
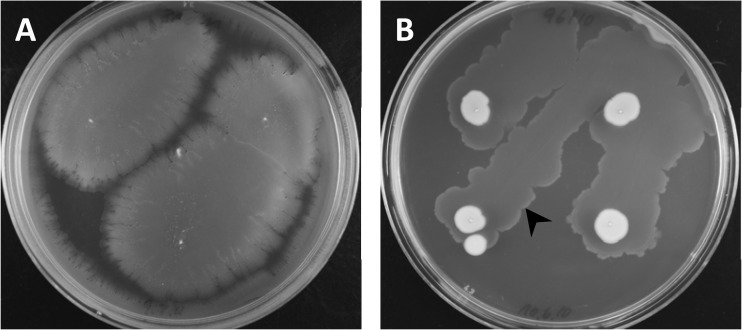
Fig 1.
Transformation of A. baumannii on motility medium. Semisolid medium facilitating surface motility (26) was doped with transforming DNA and inoculated with A. baumannii (the medium was stabbed four times with DSM 30011 [A] and 10-096 [B]). The plates were incubated overnight at 37°C, and the bacteria were floated off the medium the next day and plated on selective medium. The arrowhead indicates the frontline of growth at the interphase (between medium and bottom of petri dish).
In contrast to A. baylyi BD413 (9) and A. baumannii A118 (13), planktonic cells of our isolates were not naturally competent. While competence of A. baylyi BD413 depends on the growth phase and reaches its maximum during early logarithmic growth (41), we could not observe transformation of A. baumannii isolates under any condition other than in association with motility. In effect, when we spread the bacteria on DNA-doped solid medium which did not permit movement of the bacteria and which differed only in the concentration of agarose (1.5% instead of 0.5%) from transformation-permissive conditions, not a single transformation event was detectable with any of our strains. Also, addition of 3 to 5 μg of transforming DNA (chromosomal DNA of ATCC 17978 transposon mutants or plasmid pWH1266::Km) to 3 ml of logarithmic LB cultures (cultures with an OD600 of 0.5, 1, or 2 were tested) followed by 1 h of incubation at 37°C before plating on selective agar did not yield a single transformant. Further, addition of transforming DNA (3 to 5 μg) to pellicle-forming cultures (3 ml incubated at 20 and 37°C) produced no transformants. Collectively, the 10 naturally competent isolates described here appeared transformable only while moving on semisolid media.
Impact of pilT inactivation on motility and natural competence.
Our discovery of a direct coupling of motility and DNA uptake suggests the involvement of T4P and competence proteins mediating DNA import. To challenge this hypothesis, we first made use of a recently described pilT mutant of A. baumannii M2 (24). Also illustrating the methodological impact of our finding, we used chromosomal DNA of this mutant to generate pilT mutant derivatives of our naturally competent isolates 07-095 and 07-102 (Fig. 2). The pilT disruption abolished spread of the mutant bacteria at the boundary between the semisolid medium and the bottom of the petri dish (“interphase” motility) but had comparably little influence on motility along the air-medium boundary (“surface”). Surface motility of mutant 07-102 pilT::Km was slightly elevated compared to that of its parental strain (Fig. 2A), while surface motility of 07-095 pilT::Km was unaffected (Fig. 2B). Taken together, these findings may suggest that motility at the interphase is indeed driven by T4P and therefore may represent twitching motility as recently claimed by others (23, 25). Moreover, we could demonstrate that pilT inactivation annihilated natural transformation competence in both isolates (Fig. 3).
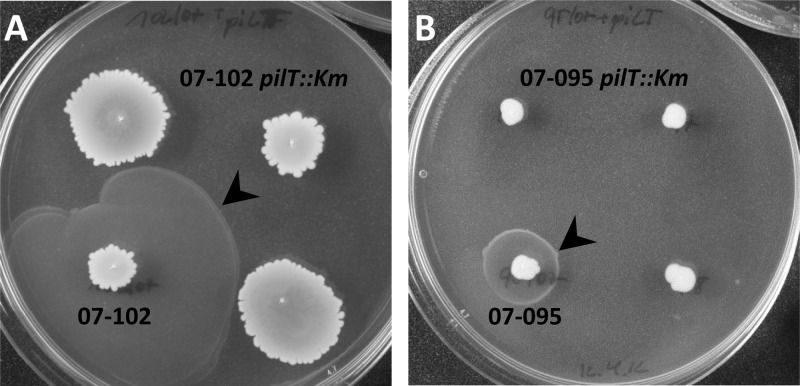
Fig 2.
Inactivation of pilT in A. baumannii abolishes twitching-like motility. A. baumannii isolates 07-095 and 07-102 were transformed on motility plates as described in the text using chromosomal DNA derived from A. baumannii M2 pilT::Km to generate pilT mutants 07-095 pilT::Km and 07-102 pilT::Km, respectively. Of the mutants, three independent colonies were inoculated each on a motility plate together with the respective parental strain. The photos shown were taken after incubation for 18 h at 37°C and subsequent incubation for 24 h at 20°C. The latter incubation was solely to intensify the biofilm formed at the interphase (arrowheads) to facilitate photography.
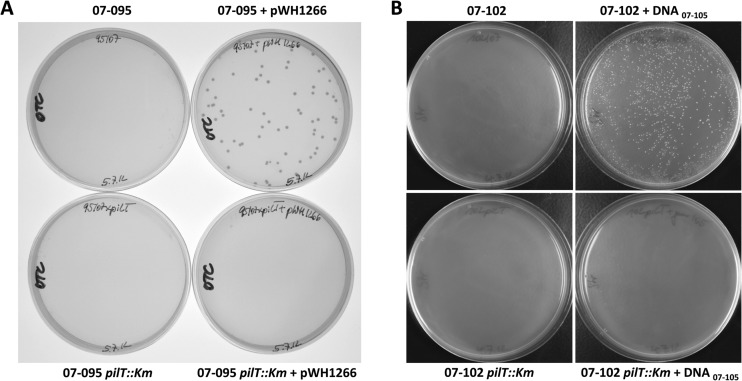
Fig 3.
Inactivation of pilT annihilates natural transformation competence of A. baumannii. Mutant strain 07-095 pilT::Km and its parental strain were incubated on motility plates with or without plasmid pWH1266 (39) conferring resistance to ampicillin and tetracycline. The bacteria were then floated off the motility plates, and after adjustment of optical densities, the bacteria were plated on selective LB agar plates containing 20 μg/ml of oxytetracycline to select for transformants (A). While the parental strain 07-095 was transformed, its 07-095 pilT::Km mutant derivative was not. (B) Isolate 07-102, which is unable to take up plasmid pWH1266 by natural competence (Table 1), and its mutant 07-102 pilT::Km were incubated on motility plates doped with or without chromosomal DNA derived from the streptomycin-resistant isolate 07-105 and subsequently plated on selective LB agar with streptomycin (20 μg/ml) (B). The 07-102 pilT::Km mutant was not transformable, in contrast to its parental strain.
Impact of comEC inactivation on motility and natural competence.
Next, to further characterize the mechanistic coupling of motility and DNA uptake, we studied the impact of comEC inactivation on motility and transformation properties. Orthologues of comEC are required for DNA uptake in different bacteria (12, 33). A comEC::Km transposon mutant derivative of A. baumannii ATCC 17978 was recently identified in a screen for mutations affecting motility (unpublished results). Since ATCC 17978 was not naturally competent in our hands, we used the chromosomal DNA of the ATCC 17978 comEC::Km mutant to transform naturally competent isolates 07-095 and 07-102. We found that inactivation of comEC abolished both twitching motility at the interphase and natural transformation competence (Fig. 4). Motility at the surface was also significantly reduced, in line with the identification of the ATCC 17978 comEC::Km mutant in the course of a screening for motility defects.
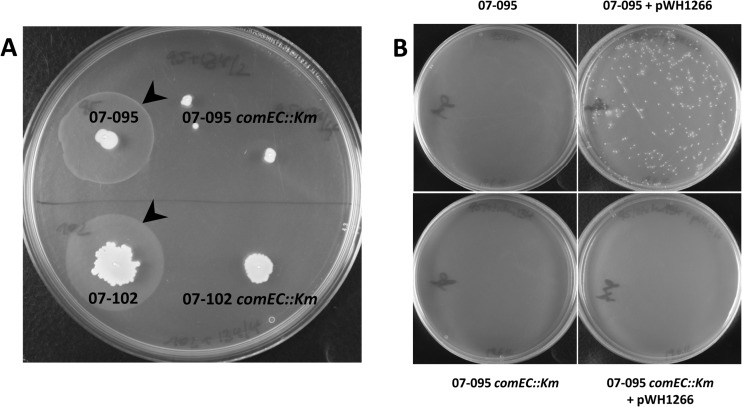
Fig 4.
Inactivation of comEC in A. baumannii abolishes twitching-like motility and natural transformation competence. A. baumannii isolates 07-095 and 07-102 were transformed on motility plates as described in the text using chromosomal DNA derived from A. baumannii ATCC 17978 comEC::Km to generate comEC mutants 07-095 comEC::Km and 07-102 comEC::Km, respectively. (A) Subsequently, both mutants and their respective parental strains were inoculated into motility medium as described in the text. Motility at the interphase was observed with the parental strains (arrowheads) but not with the mutant derivatives. (B) To prove an involvement of comEC in natural competence, 07-095 comEC::Km and its parental strain were incubated on motility plates with or without plasmid pWH1266 conferring resistance to ampicillin and tetracycline. The bacteria were then floated off the motility plates, and after adjustment of optical densities, the bacteria were plated on LB agar plates containing 100 μg/ml of ampicillin to select for transformants. While strain 07-095 was readily transformable, its comEC-inactivated derivative was not.
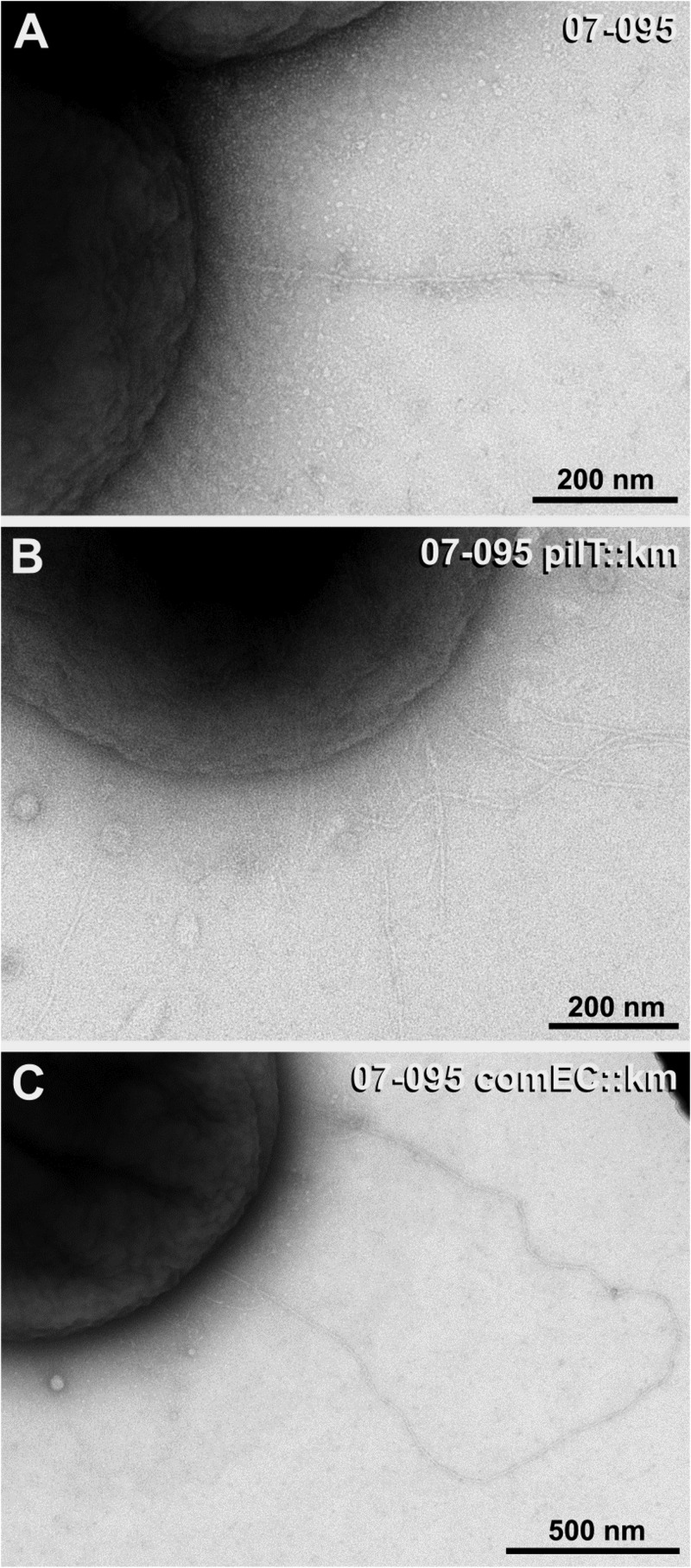
Electron microscopy studies reveal a hyperpiliation phenotype of pilT mutants.
We then applied transmission electron microscopy (TEM) to identify T4P in A. baumannii and to determine the influence of pilT and comEC inactivation on the piliation state. To this end, naturally competent isolates 07-095 and 07-102 and their pilT and comEC mutant derivatives were stab inoculated into motility agarose and the bacteria were collected from the surface and the interphase. In accordance with published work on Acinetobacter pili (17, 42–44), thin (∼ 4 nm wide) and thick (∼ 7 nm wide) pili could be observed. In both parental strains, the thick pili were only rarely found in surface-grown bacteria (approximately 1 pilus per 25 to 50 cells with a typical length of up to 2 μm) and even more sporadic in the interphase-derived preparations (Table 2). In contrast, with both pilT mutants, on average, more than one thick pilus was found per cell in surface-derived preparations and the length of the pili was significantly increased compared to that in the parental strains (typically between 2 and 6 μm) (Fig. 5A and B). Even more pili were found in the pilT mutant preparations derived from the interphase (more than 3 to 5 pili per cell). With regard to the comEC mutant phenotypes, the strains differed. While the comEC mutant of 07-095 was similar to the pilT mutant (Fig. 5C), thick pili were only sporadically found in 07-102 comEC::Km. Taken together, our data demonstrate a hyperpiliation phenotype of the pilT mutants regarding the thick pili, suggesting that these represent indeed T4P. These supposed T4P have an average diameter of 7.2 nm (standard deviation ± 1 nm) as determined from 109 individual measurements on 20 pili.
Table 2.
Evaluation of TEM negative staining of A. baumannii obtained from motility plates
| Strain and sample site | 7-nm pili | Relative frequency of 7-nm pilia |
|---|---|---|
| 07-095 surface | Rarely, but regularly (∼1 pilus per 50 cells); length, ≤2 μm | + |
| 07-095 pilT::Km surface | >1 per cell; length, 2–6 μm | ++ |
| 07-095 comEC::Km surface | ∼1 per cell; length, ≥2 μm | ++ |
| 07-102 surface | Rarely, but regularly (∼1 pilus per 25-50 cells); length, ≤2 μm | + |
| 07-102 pilT::Km surface | ≥1 per cell; length, 2–6 μm | ++ |
| 07-102 comEC::Km surface | A single sporadic pilus detected | − |
| 07-095 interphase | Sporadic | + |
| 07-095 pilT::Km interphase | >3 per cell on avg; length, short and long (≥2 μm) | +++ |
| 07-095 comEC::Km interphase | ∼1 per cell; length, ≥2 μm | ++ |
| 102/07 interphase | Sporadic; amt and length not determinable | + |
| 102/07 pilT::Km interphase | >5 per cell on avg; length, short and long (≥2 μm) | +++ |
| 102/07 comEC::Km interphase | No pili detected | − |
For 7-nm pili, symbols are as follows: −, no pili or single detection; +, sporadic or up to 1 pilus per 25 to 50 cells; ++, ∼1 pilus per cell; +++, ≥3 pili per cell on average; unbiased estimation of the pili distribution on the cells was not possible, because bacteria formed cluster on the sample supports.
Fig 5.
Transmission electron microscopy reveals a hyperpiliation phenotype of pilT mutants. Images show representative cells of naturally competent A. baumannii 07-095 (A) and its pilT (B) and comEC (C) mutant derivatives. In the pilT::Km (B) and comEC::Km (C) mutants, number of pili and length are increased in comparison to those of the wild type (A) (compare also Table 2).
Dissection of independent functions of pilT and comEC in the Galleria mellonella infection model.
Finally, we additionally generated pilT and comEC mutants of naturally competent strains DSM 30011 and 10-096 to study the applicability of natural competence for rapid generation of mutants and to compare the mutants in the Galleria mellonella caterpillar infection model (45). While we were able to introduce pilT::Km into strain 10-096 by natural transformation with chromosomal DNA derived from A. baumannii M2 pilT::Km (24), we were unsuccessful in generating 10-096 comEC::Km using donor DNA from ATCC 17978 comEC::Km, 07-095 comEC::Km, and 07-102 comEC::Km, although we had confirmed the presence of the comEC locus in strain 10-096. Conversely, we successfully generated DSM 30011 comEC::Km using ATCC 17978 comEC::Km donor DNA, while we failed to generate a pilT mutant despite confirmed presence of the pilT gene in DSM 30011. Detailed sequence analyses of donor and acceptor sites may pave the way to identification of determinants that restrict uptake and recombination events in these strains.
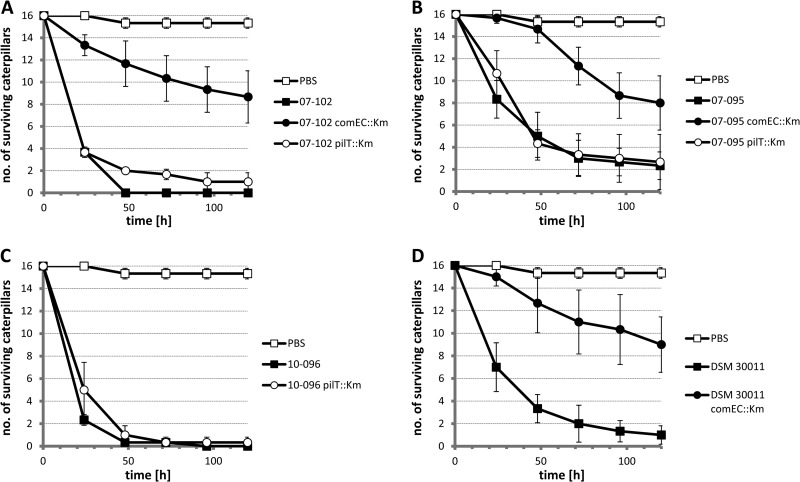
The pilT and comEC derivatives of the naturally competent isolates 07-095, 07-102, 10-096, and DSM 30011 were then characterized in the Galleria mellonella infection model in comparison to their parental strains (Fig. 6). Consistently, these experiments revealed a significant attenuation of the comEC mutants in all strains tested (Fig. 6A, B, and D), whereas pilT mutants were not significantly attenuated (Fig. 6B and C) compared to their parental strains or were only marginally attenuated as in the case of 07-102 comEC::Km (Fig. 6A; compare parental strain and mutant 48 h and 72 h postinfection). Collectively, these data demonstrate that comEC fulfills an important function during infection and that PilT-driven T4P retraction is dispensable under these conditions.
Fig 6.
The comEC locus is important for virulence in the Galleria mellonella infection model, while pilT is not. Galleria mellonella caterpillars were infected with A. baumannii strains as indicated or mock infected with PBS. The numbers of bacteria used for infection (CFU) were ∼106 for isolates 07-095 and DSM 30011 as well as their mutant derivatives and ∼5 × 105 for isolates 07-102 and 10-096 and respective derivatives. The average of three independent replicates (groups of 16 larvae each) is plotted, with error bars representing ±1 standard deviation.
DISCUSSION
A. baumannii genomes are significantly formed by HGT events (1–4). This is particularly true with respect to genetic determinants conferring antibiotic resistance which have been presumably acquired in part from distinctly related species belonging to the Enterobacteriaceae and Pseudomonas (1). The apparent formation of so-called genetic exchange communities (46) is further illustrated by the recent finding that a potent resistance determinant, the New Delhi metallo-β-lactamase 1 (NDM-1) first discovered in Klebsiella pneumoniae and Escherichia coli (47), probably originated in Acinetobacter (48) and can be transferred among A. baumannii isolates via natural transformation competence (49). Mechanistically, conjugative transfer can only partially explain the multitude of HGT events in Acinetobacter, given that tra and mob genes required for conjugative transfer are missing on most sequenced Acinetobacter plasmids (38). Recently, another possible HGT pathway was identified in A. baumannii showing that outer membrane vesicles can mediate transfer of resistance genes (50). Hitherto, only a single isolate of A. baumannii was known to be competent for DNA uptake (13). Here, we add to the understanding of HGT in A. baumannii, demonstrating natural competence in 10 out of 28 (36%) antibiotic-sensitive clinical isolates. Next, we will investigate if natural competence is prevalent among multidrug-resistant isolates, as this may indicate that it contributes to the acquisition of novel resistance genes. Owing to their multidrug resistance, it is technically difficult and problematic from an ethical point of view to transform these isolates with other resistance genes. Therefore, we need to develop alternative methods for the phenotypic display of transformation events.
So far, the only representatives of the genus Acinetobacter known to be naturally competent were A. baylyi ADP1 (BD4) (7–12) and A. baumannii A118 (13). Both are transformable when grown in liquid cultures, with ADP1 known to reach highest competence during early logarithmic growth (41). However, we failed to transform any of our competent isolates under comparable conditions, suggesting significant regulatory and/or mechanistic differences. Interestingly, the ComA DNA uptake channel known to be involved in competence of A. baylyi ADP1 (36) is only about 50% identical to ComEC of A. baumannii. It remains to be determined whether A. baumannii A118 harbors an uptake channel of the ComA or the ComEC type to further estimate whether different uptake channels could contribute to the mechanistic differences. Another significant difference in the endowment with competence genes between A. baylyi and A. baumannii as figured out by Smith et al. (37) refers to A. baylyi comP, which encodes a pilin-like protein involved in DNA uptake but obviously not involved in pilus formation (44).
Inactivation of pilT has been studied in a number of bacteria exhibiting twitching motility. In Neisseria gonorrhoeae, inactivation of pilT abolished both natural transformation and twitching motility even though the amount and length of T4P were found to be unaffected (32). Similarly, T4P-driven motility was abolished in the pilT mutant of Myxococcus xanthus, while piliation was apparently unaffected (51). In contrast and similar to our observations, pilT inactivation in Pseudomonas aeruginosa resulted in a hyperpiliation phenotype (52, 53), and the same was also found in Synechocystis sp. strain PCC6803 (54).
Mechanistically, our data suggest that in A. baumannii, T4P are required for motility at the interphase, as this form of motility was abolished upon pilT inactivation. Thus, as already suggested by others (17, 23, 25), this form of motility can be well termed twitching motility now. The finding that pilT inactivation can interfere with but not abolish surface motility as demonstrated here and as described by Clemmer et al. (24) suggests that T4P are expressed under these conditions, as has been demonstrated here, but are not the (only) driving force of surface motility. Our finding that T4P are expressed both at the surface and at the interphase is further compatible with our observation that transformants could be obtained by flushing off bacteria from only the surface or the interphase. To control whether transformation rates were different at the surface and at the interphase, we mixed the DNA with the agarose medium prior to casting the plates to produce a medium with a constant DNA concentration. After stab inoculation, the bacteria were then separately recovered from surface and interphase, and no significant difference in the transformation rates at both sites could be observed (data not shown). Collectively, these findings show that transformation occurs at both sites of motility and correlates with the presence of T4P.
It will be interesting to learn whether the unprecedented direct mechanistic coupling of motility and DNA uptake applies to other bacteria. A number of pathogens harboring T4P, including Pseudomonas aeruginosa and enterohemorrhagic E. coli (EHEC), are highly suspicious of being competent given the excessive HGT documented for their genomes, but to date they have not been shown to undergo transformation naturally (55–57).
Our finding that the comEC mutants are attenuated in the Galleria mellonella infection model while the pilT mutants are not is unexpected. To our knowledge, this is the first time that DNA uptake channels of the ComA/ComEC type have been implicated in virulence. This could point to a role of the channel independent of DNA uptake and T4P-dependent motility. Alternatively, it is tempting to speculate that DNA uptake could become important during infection as a way to open up DNA as a nutrient source. However, the fact that pilT inactivation abolished DNA uptake on motility plates but had little to no effect on virulence argues against this speculation. The contribution of DNA uptake channels to virulence should now be tested in other pathogens and other infection models. Targeting DNA uptake systems might become an interesting strategy to suppress virulence and resistance development of pathogens in the hospital environment.
ACKNOWLEDGMENTS
We thank Philip N. Rather for providing chromosomal DNA of A. baumannii M2 pilT::Km and Paul G. Higgins and Christine Heider for critical reading of a previous version of the manuscript.
G.W. conceived of the study. G.W., J.P., M.L., and E.S. performed experiments and analyzed and interpreted the data. G.W. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Published ahead of print 12 July 2013
REFERENCES
- 1.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mussi MA, Limansky AS, Relling V, Ravasi P, Arakaki A, Actis LA, Viale AM. 2011. Horizontal gene transfer and assortative recombination within the Acinetobacter baumannii clinical population provide genetic diversity at the single carO gene, encoding a major outer membrane protein channel. J. Bacteriol. 193:4736–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahl JW, Johnson JK, Harris AD, Phillippy AM, Hsiao WW, Thom KA, Rasko DA. 2011. Genomic comparison of multi-drug resistant invasive and colonizing Acinetobacter baumannii isolated from diverse human body sites reveals genomic plasticity. BMC Genomics 12:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snitkin ES, Zelazny AM, Montero CI, Stock F, Mijares L, Program NCS, Murray PR, Segre JA. 2011. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc. Natl. Acad. Sci. U. S. A. 108:13758–13763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G. 2012. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337:1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juni E, Janik A. 1969. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J. Bacteriol. 98:281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmen R, Vosman B, Buijsman P, Breek CK, Hellingwerf KJ. 1993. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J. Gen. Microbiol. 139:295–305 [DOI] [PubMed] [Google Scholar]
- 9.Lorenz MG, Wackernagel W. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metzgar D, Bacher JM, Pezo V, Reader J, Doring V, Schimmel P, Marliere P, de Crecy-Lagard V. 2004. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 32:5780–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott KT, Neidle EL. 2011. Acinetobacter baylyi ADP1: transforming the choice of model organism. IUBMB Life 63:1075–1080 [DOI] [PubMed] [Google Scholar]
- 12.Averhoff B, Friedrich A. 2003. Type IV pili-related natural transformation systems: DNA transport in mesophilic and thermophilic bacteria. Arch. Microbiol. 180:385–393 [DOI] [PubMed] [Google Scholar]
- 13.Ramirez MS, Don M, Merkier AK, Bistue AJ, Zorreguieta A, Centron D, Tolmasky ME. 2010. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J. Clin. Microbiol. 48:1488–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lautrop H. 1961. Bacterium anitratum transferred to the genus Cytophaga. Int. Bull. Bacteriol. Nomenclature Taxonomy 11:107–108 [Google Scholar]
- 15.Lautrop H. 1965. Gliding motility in bacteria as a taxonomic criterion. Publ. Fac. Sci. Univ. JE Purkyne, ser K. 35:322–327 [Google Scholar]
- 16.Henrichsen J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrichsen J, Blom J. 1975. Correlation between twitching motility and possession of polar fimbriae in Acinetobacter calcoaceticus. Acta Pathol. Microbiol. Scand B 83:103–115 [DOI] [PubMed] [Google Scholar]
- 18.Merz AJ, So M, Sheetz MP. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98–102 [DOI] [PubMed] [Google Scholar]
- 19.Pelicic V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827–837 [DOI] [PubMed] [Google Scholar]
- 20.Burrows LL. 2005. Weapons of mass retraction. Mol. Microbiol. 57:878–888 [DOI] [PubMed] [Google Scholar]
- 21.Mattick JS. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289–314 [DOI] [PubMed] [Google Scholar]
- 22.Henrichsen J. 1983. Twitching motility. Annu. Rev. Microbiol. 37:81–93 [DOI] [PubMed] [Google Scholar]
- 23.Antunes LC, Imperi F, Carattoli A, Visca P. 2011. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6:e22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clemmer KM, Bonomo RA, Rather PN. 2011. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157:2534–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eijkelkamp BA, Stroeher UH, Hassan KA, Papadimitrious MS, Paulsen IT, Brown MH. 2011. Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol. Lett. 323:44–51 [DOI] [PubMed] [Google Scholar]
- 26.Skiebe E, de Berardinis V, Morczinek P, Kerrinnes T, Faber F, Lepka D, Hammer B, Zimmermann O, Ziesing S, Wichelhaus TA, Hunfeld KP, Borgmann S, Grobner S, Higgins PG, Seifert H, Busse HJ, Witte W, Pfeifer Y, Wilharm G. 2012. Surface-associated motility, a common trait of clinical isolates of Acinetobacter baumannii, depends on 1,3-diaminopropane. Int. J. Med. Microbiol. 302:117–128 [DOI] [PubMed] [Google Scholar]
- 27.Barker J, Maxted H. 1975. Observations on the growth and movement of Acinetobacter on semi-solid media. J. Med. Microbiol. 8:443–446 [DOI] [PubMed] [Google Scholar]
- 28.Mussi MA, Gaddy JA, Cabruja M, Arivett BA, Viale AM, Rasia R, Actis LA. 2010. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J. Bacteriol. 192:6336–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stacy DM, Welsh MA, Rather PN, Blackwell HE. 2012. Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N-acyl homoserine lactones. ACS Chem. Biol. 7:1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. 2011. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 12:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McQueary CN, Kirkup BC, Si Y, Barlow M, Actis LA, Craft DW, Zurawski DV. 2012. Extracellular stress and lipopolysaccharide modulate Acinetobacter baumannii surface-associated motility. J. Microbiol. 50:434–443 [DOI] [PubMed] [Google Scholar]
- 32.Wolfgang M, Lauer P, Park HS, Brossay L, Hebert J, Koomey M. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321–330 [DOI] [PubMed] [Google Scholar]
- 33.Krüger N-J, Stingl K. 2011. Two steps away from novelty—principles of bacterial DNA uptake. Mol. Microbiol. 80:860–867 [DOI] [PubMed] [Google Scholar]
- 34.Assalkhou R, Balasingham S, Collins RF, Frye SA, Davidsen T, Benam AV, Bjoras M, Derrick JP, Tonjum T. 2007. The outer membrane secretin PilQ from Neisseria meningitidis binds DNA. Microbiology 153:1593–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claverys JP, Martin B. 2003. Bacterial “competence” genes: signatures of active transformation, or only remnants? Trends Microbiol. 11:161–165 [DOI] [PubMed] [Google Scholar]
- 36.Friedrich A, Hartsch T, Averhoff B. 2001. Natural transformation in mesophilic and thermophilic bacteria: identification and characterization of novel, closely related competence genes in Acinetobacter sp. strain BD413 and Thermus thermophilus HB27. Appl. Environ. Microbiol. 67:3140–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fondi M, Bacci G, Brilli M, Papaleo MC, Mengoni A, Vaneechoutte M, Dijkshoorn L, Fani R. 2010. Exploring the evolutionary dynamics of plasmids: the Acinetobacter pan-plasmidome. BMC Evol. Biol. 10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunger M, Schmucker R, Kishan V, Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51 [DOI] [PubMed] [Google Scholar]
- 40.Laue M, Bannert N. 2010. Detection limit of negative staining electron microscopy for the diagnosis of bioterrorism-related micro-organisms. J. Appl. Microbiol. 109:1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmen R, Vosman B, Kok R, van der Zee JR, Hellingwerf KJ. 1992. Characterization of transformation-deficient mutants of Acinetobacter calcoaceticus. Mol. Microbiol. 6:1747–1754 [DOI] [PubMed] [Google Scholar]
- 42.Gohl O, Friedrich A, Hoppert M, Averhoff B. 2006. The thin pili of Acinetobacter sp. strain BD413 mediate adhesion to biotic and abiotic surfaces. Appl. Environ. Microbiol. 72:1394–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grzanka A, Gospodarek E, Domaniewski J. 1996. A comparison of three staining methods in estimation of structures on the cell wall surface of Acinetobacter junii by using electron microscope. Acta Microbiol. Pol. 45:233–239 [PubMed] [Google Scholar]
- 44.Porstendörfer D, Gohl O, Mayer F, Averhoff B. 2000. ComP, a pilin-like protein essential for natural competence in Acinetobacter sp. strain BD413: regulation, modification, and cellular localization. J. Bacteriol. 182:3673–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC, Jr, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob. Agents Chemother. 53:2605–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skippington E, Ragan MA. 2011. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol. Rev. 35:707–735 [DOI] [PubMed] [Google Scholar]
- 47.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toleman MA, Spencer J, Jones L, Walsh TR. 2012. blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:2773–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Gottig S, Hunfeld KP, Seifert H, Witte W, Higgins PG. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 66:1998–2001 [DOI] [PubMed] [Google Scholar]
- 50.Rumbo C, Fernandez-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G. 2011. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:3084–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu SS, Wu J, Kaiser D. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109–121 [DOI] [PubMed] [Google Scholar]
- 52.Bradley DE. 1974. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology 58:149–163 [DOI] [PubMed] [Google Scholar]
- 53.Whitchurch CB, Hobbs M, Livingston SP, Krishnapillai V, Mattick JS. 1991. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene 101:33–44 [DOI] [PubMed] [Google Scholar]
- 54.Bhaya D, Bianco NR, Bryant D, Grossman A. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37:941–951 [DOI] [PubMed] [Google Scholar]
- 55.Sinha S, Redfield RJ. 2012. Natural DNA uptake by Escherichia coli. PLoS One 7:e35620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Schaik EJ, Giltner CL, Audette GF, Keizer DW, Bautista DL, Slupsky CM, Sykes BD, Irvin RT. 2005. DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J. Bacteriol. 187:1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xicohtencatl-Cortes J, Monteiro-Neto V, Saldana Z, Ledesma MA, Puente JL, Giron JA. 2009. The type 4 pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J. Bacteriol. 191:411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]