Abstract
Ribonucleotide reductases (RNRs) are essential enzymes for DNA synthesis because they are responsible for the production of the four deoxyribonucleotides (dNTPs) from their corresponding ribonucleotides. Escherichia coli contains two classes of aerobic RNRs, encoded by the nrdAB (class Ia) and nrdHIEF (class Ib) operons, and a third RNR class, which is functional under anaerobic conditions and is encoded by the nrdDG (class III) operon. Because cellular imbalances in the amounts of the four dNTPs cause an increase in the rate of mutagenesis, the activity and the expression of RNRs must be tightly regulated during bacterial chromosome replication. The transcriptional regulation of these genes requires several transcription factors (including DnaA, IciA, FIS [factor for inversion stimulation], Fnr, Fur, and NrdR), depending on the RNR class; however, the factors that dictate the expression of some RNR genes in response to different environmental conditions are not known. We show that H-NS modulates the expression of the nrdAB and nrdDG operons. H-NS represses expression both in aerobically and in anaerobically growing cells. Under aerobic conditions, repression occurs at the exponential phase of growth as well as at the transition from the exponential to the stationary phase, a period when no dNTPs are needed. Under anoxic conditions, repression occurs mainly in exponentially growing cells. Electrophoretic mobility assays performed with two DNA fragments from the regulatory region of the nrdAB operon demonstrated the direct interaction of H-NS with these sequences.
INTRODUCTION
Ribonucleotide reductases (RNRs) are essential enzymes, for both prokaryotes and eukaryotes, that are responsible for the production of the four deoxyribonucleotides (dNTPs) from their corresponding ribonucleotides. Three classes of RNRs have been described to date, differing in the mechanism used for radical generation, in structure, and in oxygen dependence (1). The class I RNRs consist of two homodimeric proteins: a large subunit (α) that contains the catalytic site and the binding site for allosteric effectors and a smaller subunit (β) harboring the metallo radical cofactor that stabilizes the free tyrosyl radical linked to a diiron-oxo center that initiates cysteine activation for enzymatic function. Class I enzymes function only under aerobic conditions and are found in all eukaryotes and some prokaryotes. Class I is further subdivided into classes Ia and Ib, which are encoded by the nrdAB and nrdHIEF genes, respectively. A third subclass, class Ic, was established because some RNRs, such as those from Chlamydia trachomatis, lack the tyrosyl radical. The class II enzymes require S-adenosylcobalamin (AdoCob) as a radical generator and do not depend on oxygen for activity, thereby functioning in aerobic or anaerobic environments. Class II RNRs consist predominantly of homodimers encoded by the nrdJ gene. This RNR class is found in prokaryotes, archaebacteria, and some lower eukaryotes. Lastly, the class III RNR reductases, encoded by the nrdDG operon, use S-adenosylmethionine (SAM) and an iron-sulfur cluster to generate a glycyl radical that is extremely sensitive to oxygen. Therefore, enzymes belonging to this class are strictly anaerobic and are found only in prokaryotes able to grow under anaerobic conditions (1, 2).
An unresolved question is why several RNR classes are encoded in the genome of a single organism. Whereas almost all eukaryotes encode only class Ia RNRs, prokaryotes contain genes encoding single copies of different RNR classes (Ia or Ib; II or III) or encoding combinations of different RNR classes (Ia+Ib, Ib+II, or Ia+Ib+III). In some instances, all classes (Ia+II+III) are present simultaneously (3). The simultaneous presence of different RNR classes in some microorganisms raises the necessity for a fine-tuning of RNR activity. It is well known that the activity of RNRs is tightly regulated by the cell cycle and environmental triggers to generate and maintain proper dNTP pools that ensure the fidelity of DNA synthesis and repair (2, 4, 5).
Escherichia coli is a good example of a microorganism expressing several RNRs. Enzymes belonging to classes Ia, Ib, and III are expressed from the nrdAB, nrdDG, and nrdHIEF operons, respectively (6–8). In this bacterium, the expression of RNR genes can be positively or negatively regulated at the transcriptional level; nrdAB expression is repressed by the NrdR protein, whereas the DnaA, IciA, and FIS (factor for inversion stimulation) transcriptional regulators induce the expression of this operon (for a review, see reference 4). Other cis-acting sites and an AT-rich region located in the promoter region are important for coupling the transcription of the gene to the cell cycle (9, 10). Recently, we have discovered a new transcriptional factor (NrdR) that is a repressor of the three different nrd genes in aerobically growing E. coli cells (6).
The nucleoid-associated protein H-NS is a global transcriptional repressor that controls the expression of several environmentally regulated genes. This protein is widespread in Gram-negative bacteria (11, 12) and has been best characterized in Escherichia coli and related genera. H-NS plays a dual role, both as an architectural protein that contributes to nucleoid structure and as a global modulator of gene expression (see references 13 and 14 for reviews). The E. coli hns gene encodes a 137-amino-acid protein with a molecular mass of 15.4 kDa. H-NS consists of an N-terminal dimerization domain and a C-terminal DNA-binding domain that are separated by a linker domain. H-NS is a highly abundant protein that binds to DNA in a non-sequence-specific manner but has a preference for intrinsically curved AT-rich regions. The H-NS protein is capable of interacting with itself and other proteins in addition to DNA. Indeed, the generation of homodimers and -oligomers appears to be a key process that allows H-NS to modulate gene expression (13, 15, 16). The mechanism of transcriptional repression by H-NS involves binding to high-affinity DNA sequences to initiate the oligomerization of H-NS along the DNA, resulting in a higher-order nucleoprotein complex. This nucleoprotein structure leads to the repression of transcription either by occluding the binding of RNA polymerase or by trapping RNA polymerase (17, 18).
In this study, we provide evidence indicating that H-NS modulates the expression of the E. coli class Ia (nrdAB) and class III (nrdDG) RNRs in the transition from the exponential to the stationary growth phase and under anaerobic conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and plasmids used in this study are described in Table 1. To test the effect of either temperature or osmolarity on nrdA expression, cells were grown either in Luria-Bertani (LB) broth at 25°C or 37°C or in modified LB broth containing NaCl at final concentrations of 170 or 500 mM. The cells were grown with continuous shaking at 150 rpm. When required, LB broth was supplemented with 10 μg ml−1 gentamicin, 50 μg ml−1 kanamycin, 50 μg ml−1 ampicillin, 30 μg ml−1 chloramphenicol, and 17 μg ml−1 tetracycline. Anaerobic growth was carried out in Hungate tubes, as described previously (26).
Table 1.
Strains, plasmids, and bacteriophages used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Plasmids | ||
| pGEM-T easy | A/T cloning vector, Apr | Promega |
| pLG338-30 | Low-copy-no. vector; oripSC101, Apr | 19 |
| pETS130-GFP | Green fluorescent protein gene cloned into a high-copy-no. cloning vector; Gmr | 20 |
| pUJ8 | Promoterless vector for transcriptional lacZ fusions, Apr | 21 |
| pUTminiTn5Cm | Mini-Tn5 Cm in plasmid pUT; Apr Cmr | 21 |
| pLGHNSEC | hns cloned into the BamHI site of pLG338-30 | 22 |
| pETS150 | nrdA promoter cloned into the BamHI site of pETS130-GFP; Gmr | 8 |
| pETS152 | nrdD promoter cloned into the BamHI site of pETS130-GFP; Gmr | 8 |
| pETS151 | nrdH promoter cloned into the BamHI site of pETS130-GFP; Gmr | 8 |
| pETS163 | nrdA promoter cloned into the BamHI and SmaI site of pUJ8; Apr | This work |
| pETS166 | PnrdA::lacZ from pUJ8 cloned into the NotI site of pUTminiTn5Cm; Cmr | This work |
| pETS168 | PnrdA::lacZ (ΔHNS binding site) from pUJ8 cloned into the NotI site of pUTminiTn5Cm; Cmr | This work |
| Strains | ||
| DH5α | recA1 endA1 hsdR17 supE44 thi-1 relA1 Δ(lacZYA-argF)U169 deoR ϕ80dlacZM15 | Laboratory stock |
| MG1655 | F− ilvG rph1; wild type | Laboratory stock |
| MG1655Δhns | MG1655 trp::Tn10 Δhns | This work |
| AAG1 | MG1655 ΔlacZ | 23 |
| AAG1Δhns | AAG1 trp::Tn10 Δhns | Marta Gibert |
| S17λpir | Tpr Smr recA thi pro hsdR−M+ RP4: 2-Tc::Mu Km Tn7 λpir | Laboratory strain |
| ETS114 | MG1655trp::Tn10Δhns | 24 |
| ETS115 | MG1655 fis::Km | 25; this work |
| ETS116 | MG1655Δhns fis::Km | 25; this work |
| ETS117 | MG1655trp::Tn10 ΔstpA60::Km | 24; this work |
| ETS118 | MG1655Δhns trp::Tn10 stpA60::Km | 24; this work |
| ETS119 | AAG1 with PnrdA::lacZ | This work |
| ETS121 | AAG1 with PnrdA::lacZ(ΔHNS binding site) | This work |
| ETS122 | AAG1Δhns with PnrdA::lacZ | This work |
| ETS123 | MG1655 iciA::Km | 25; this work |
| ETS124 | MG1655Δhns iciA::Km | 25; this work |
| Phage | ||
| P1vir | Laboratory stock |
To construct the strains AAG1Δhns, ETS114, ETS115, ETS116, ETS117, and ETS118, P1vir-mediated transduction was performed as described previously (27) using lysates obtained from the strains BSN27 (24), JW3229 (25), and BSN28 (24).
DNA manipulation.
Restriction endonucleases, T4 DNA ligase, alkaline phosphatase, DNA polymerase (Klenow fragment) and DNA-modifying enzymes were purchased from Fermentas and used according to the manufacturer's instructions. Plasmid DNA was isolated using the QIAprep spin miniprep kit (Qiagen), as described by the manufacturer.
PCR amplifications were performed using high-fidelity polymerase (Fermentas); 2× PCR master mix was used for screening assays, according to the manufacturer's specifications (Fermentas), with the primers described in Table 2. The other molecular assays and manipulations were performed using standard procedures (28).
Table 2.
Primers used in this study
| Name | Sequence (5′→3′) | Application |
|---|---|---|
| Gfp-mut3-rev | GAATTGGGACAACTCCAGTG | Cloning assessment |
| PnrdABamHI-up | ACCCGGGATCATTTTCTATAAGACGG | Cloning, probe amplification |
| PnrdASmaI-lw | AGGATCCAGCAGATTCTGATTCATG | Cloning |
| PA-450 ext-lw | CGCATGCATTGCACCGGGTTGAACAAT | Cloning |
| PA-400 extpho-up | GTACGCTTAAAGTCATGAATAATTTTCTTATAATATAAGG | Cloning |
| PnrdA-50pb-up | ATCCACAAAGTTATGCACTTGC | Probe amplification |
| PnrdA-50pb-lw | TAACTCAGGAAGGAAAAAGTGG | Probe amplification |
| PnrdA-300pb-lw | TTGTTGATGGCGAATGGTTGTT | Probe amplification |
| PnrdA-400pb-up | TTAAAGTCATGAATAATTTTCTTATAATATAAG | Probe amplification |
| PnrdA-120pb-lw | AGAGAAAAATTTGTTAAAAATAACTGTTCG | Probe amplification |
| PnrdA-172pb-up | AGGTTAGATAAATTGATATAGATGGC | Probe amplification |
| PnrdA-235pb-up | TAGATCAATTTTTGCAATCATTAGCAA | Probe amplification |
| StpA-up | GATCGCTTACACTACGCGACG | Mutation assessment |
| StpA-lw | CAGCGACATCCGGCCTC | Mutation assessment |
| E. coli fis-up | GCGTAAATTCTGACGTACTGAC | Mutation assessment |
| E. coli fis-lw | TTTACGCAGCGTACCACGG | Mutation assessment |
| PBR-E | ATTATCATGACATTAACC | Cloning assessment |
| HNS-3 | CCACCCCAATATAAGTTTGAG | Mutation assessment |
| HNSBDist | CCGGATCCTAAAAAATCCCGC | Mutation assessment |
| M13-dir | GTTTTCCCAGTCACGAC | Cloning assessment |
| M13-rev | CAGGAAACAGCTATGAC | Cloning assessment |
| kT | CGGCCACAGTCGATGAATCC | Cloning assessment |
| k1 | CAGTCATAGCCGAATAGCCT | Cloning assessment |
| k2 | CGGTGCCCTGAATGAACTGC | Cloning assessment |
Expression analysis of green fluorescent protein (GFP).
Overnight cultures of E. coli wild-type MG1655 and its isogenic hns mutant derivative transformed with pETS130, pETS150, pETS151, or pETS152 were adjusted to an OD550 of 0.05 in 10 ml LB broth and incubated at 37°C. At different points of the growth curve, 1 ml of bacterial cells was collected by centrifugation and resuspended in 1 ml of fixing solution (1× PBS and 4% formaldehyde) for 10 min on ice. The fixed cultures were subsequently washed twice with 1× PBS. Samples in triplicate were plated in 96-well solid polystyrene plates (Fisher Scientific), and the fluorescence was measured using an FLx800 fluorescence microplate reader (BioTek).
Construction of lacZ transcriptional fusions.
Transcriptional fusions were constructed by inserting the promoter fragments into plasmid pUJ8 to generate transcriptional lacZ fusions (21). To construct the PnrdA-lacZ fusion, a 702-bp fragment encompassing the nrdA promoter region was amplified by PCR using primers PnrdABamHI-up and PnrdASmaI-lw (Table 2) and cloned into plasmid pUJ8 using the BamHI-SmaI sites to generate the pETS163 plasmid. This plasmid DNA was sequenced to ensure that the fragments were inserted in the correct orientation and that no mutation had been introduced during amplification and cloning.
pETS168 (PnrdA-lacZΔH-NS binding site) was constructed by amplification of the nrdA promoter region from nucleotide position −575 to −450 and from −400 to +127 using primers PnrdABamHI-up and PA-450ext-lw for the upstream region and PA-400extpho-up and PnrdASmaI-lw for the more proximal region. Both amplified regions were extracted from 2% agarose gels and ligated. PCR amplification using the PnrdABamHI-up and PnrdASmaI-lw primers generated an nrdA promoter region band (652 bp) without the putative H-NS binding site of approximately 50 bp located at a position centered at −425 nt. The amplified region was cloned into pUJ8 using the BamHI-SmaI sites.
To transfer the lacZ fusions into the E. coli chromosome as a single copy, the recombinant plasmids pETS163 and pETS165 were digested with NotI, and the fragments containing the promoter fusion were subcloned into the suicide plasmid pUTminiTn5Cm (21), generating pETS166 and pETS168, respectively. The recombinant plasmids were transferred by conjugation using S17.1 (λ-pir) to a Rifr derivative of E. coli MG1655, and kanamycin-resistant transconjugants were screened for the loss-of-vector-mediated ampicillin resistance, as described previously (21). The presence of the desired fusion (ETS119 and ETS121) in the chromosome of several independent exconjugants was established by PCR analysis and β-galactosidase production (data not shown).
β-Galactosidase assay.
Overnight cultures were subcultured in fresh LB medium with the appropriate antibiotics and grown with shaking. Samples were collected at different stages of growth specified for each individual experiment, and the β-galactosidase activities were determined independently at least three times, as described previously (27). Unless otherwise indicated, the standard deviations were less than 10%.
Band shift assays.
To investigate the binding of regulatory proteins to the three nrd promoters, electrophoretic band shift assays were performed, as described previously (29). DNA fragments of different lengths and comprising different sites were synthetized and purified (the primers are listed in Table 2). H-NS-DNA binding was performed in 20 μl of 40 mM Tris-HCl (pH 8.0), 100 mM KCl, 1 mM dithiothreitol (DTT), 5 mM potassium phosphate, 5% glycerol, and 50 mM EDTA, with increasing concentrations of purified H-NS (29) (0.5 to 4 μM) and 30 ng of DNA. After incubation for 20 min at 25°C, the protein-DNA complexes were subjected to electrophoretic separation at 100 V through 0.8% agarose gels in 0.5× TBE buffer. The DNA was visualized by ethidium bromide staining.
Western blotting.
Crude E. coli protein extracts were prepared using the BugBuster extraction reagent (Novagen). The protein extracts were loaded onto 7.5% minigels (5 μg of protein per lane) and separated by electrophoresis using a Bio-Rad minigel apparatus (Bio-Rad). The proteins were electroblotted onto polyvinylidene difluoride (PVDF) membranes (Immun-Blot; Bio-Rad) using a semidry transfer system (Bio-Rad), and the membranes were blocked overnight at 4°C in 5% skim milk in phosphate-buffered saline (PBS). The membranes were probed with a 1:10,000 dilution of rabbit anti-Pseudomonas aeruginosa NrdA (20). The detection of primary antibodies was performed using donkey anti-rabbit (Bio-Rad) horseradish peroxidase-conjugated secondary antibodies at a 1/50,000 dilution and visualized using the Amersham ECL Prime Western blotting reagent (GE Healthcare) according the manufacturer's protocol. Imaging was performed using an ImageQuant LAS 4000 mini-imager (GE Healthcare) with the chemiluminescence high-sensitivity setting. The protein concentrations were determined using the Bio-Rad Bradford assay.
RESULTS
H-NS represses expression of class Ia and III RNRs.
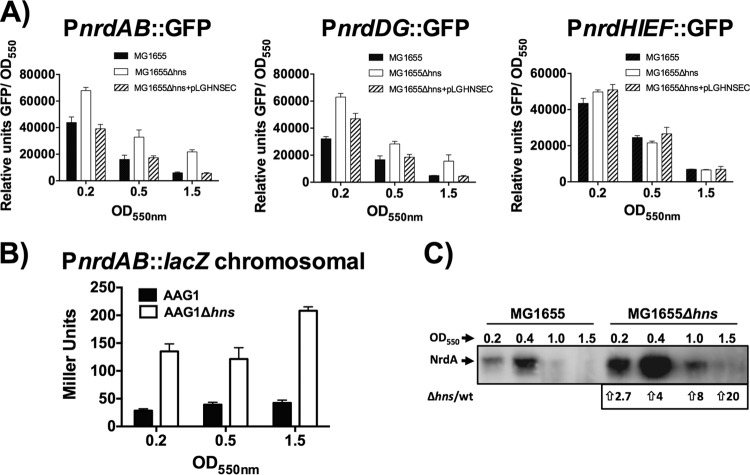
Several genes involved in modifying DNA architecture (e.g., those encoding FIS and DnaA) have been shown to be involved in the transcriptional regulation of E. coli nrdAB genes (4). These factors appear to respond to nutritional changes (FIS) (30) or the DnaA concentration itself (31). Taking into account the facts that nrd genes are also environmentally regulated and that H-NS influences DNA architecture and modulates gene expression in response to environmental factors (32–34), we hypothesized that H-NS might participate in the modulation of the expression of some of the nrd operons. To test this hypothesis, we measured the expression of the different nrd promoters in strains MG1655 and MG1655Δhns using plasmids carrying transcriptional fusions of each nrd promoter region with the green fluorescent protein (GFP), as previously described (8). In exponentially growing cells, GFP expression from the plasmid pETS150 (PnrdA::GFP) was twice as high in the Δhns cells as in the wild-type cells. The difference in GFP expression increased to 3.5-fold in cells entering the stationary phase (Fig. 1A). The same effect of the Δhns allele was observed in cells carrying the plasmid pETS152 (PnrdD::GFP) (Fig. 1A). Complementation with an hns+ gene cloned into the plasmid pLGHNSEC (Table 1) returned the expression of GFP protein to the level of the wild-type cells. No change in expression was found when the cells carrying the PnrdHIEF promoter region (pETS151) were depleted of the H-NS protein (Fig. 1A). These results suggest that class Ia (nrdAB) and III (nrdDG) but not Ib (nrdHIEF) RNR genes could be modulated by H-NS.
Fig 1.
H-NS modulates the expression of class I and III RNRs. (A) Fluorescence was measured in strains MG1655 and MG1655Δhns harboring plasmids pETS150 (PnrdAB-GFP), pETS152 (PnrdDG-GFP), and pETS151 (PnrdHIEF-GFP). The hns gene cloned into plasmid pLGHNSEC was used to complement hns deficiency in strain MG1655Δhns. The measurements at different points of growth at 37°C in LB were background subtracted using the normalized fluorescence from a culture of MG1655 at the points indicated (A550 = 0.2, 0.5, and 1.5). The fluorescence was normalized to the optical density at 550 nm (OD550) and is given in relative units. The points represent the mean of three independent experiments, and the error bars denote the standard deviations. (B) Expression of the PnrdAB::lacZ transcriptional fusion inserted as a single chromosomal copy (ETS119 and ETS122). β-Galactosidase activity was determined in strains ETS119 and ETS122 harboring the chromosomal fusion. Samples were collected at different stages of growth in LB medium at 37°C. The values are in Miller units. The points represent the means from three independent experiments, and the error bars denote the standard deviations. (C) Immunodetection of NrdA protein in aerobic cultures of strains MG1655 and MG1655Δhns.
We then focused our study on the aerobic RNRs (class Ia) and constructed a transcriptional fusion of the nrdAB promoter region to β-galactosidase as a reporter gene (see Materials and Methods). The use of the transcriptional β-galactosidase fusion inserted as a single chromosomal copy provided results comparable to those obtained using GFP fusions (Fig. 1B). When strains AAG1 (ETS119) and AAG1Δhns (ETS122) were compared, the transcriptional PnrdA::lacZ chromosomal fusion showed 4.2- and 3.1-fold induction, respectively, in the exponential phase of growth and a somewhat higher induction (6.6-fold) at the onset of the stationary phase (Fig. 1B).
To correlate the transcriptional data with protein expression, we measured the levels of NrdA protein in wild-type MG1655 cells and its isogenic Δhns mutant by Western blotting (Fig. 1C). NrdA was expressed at higher levels in the hns mutant than in the wild type, in both the exponential and stationary growth phases. HNS modulation of NrdA is FIS and IciA independent and is not influenced by StpA.
Considering that FIS and IciA modulate nrdAB expression (30, 35), we decided to test whether H-NS modulation of these genes was FIS or IciA dependent. To this end, NrdA levels were detected by Western blotting in the wild-type MG1655 strain and its Δhns, Δfis, and Δfis Δhns or ΔiciA Δhns derivatives. The results indicated that FIS (Fig. 2A) and IciA (Fig. 2B) do not influence the H-NS regulation of NrdA.
Fig 2.
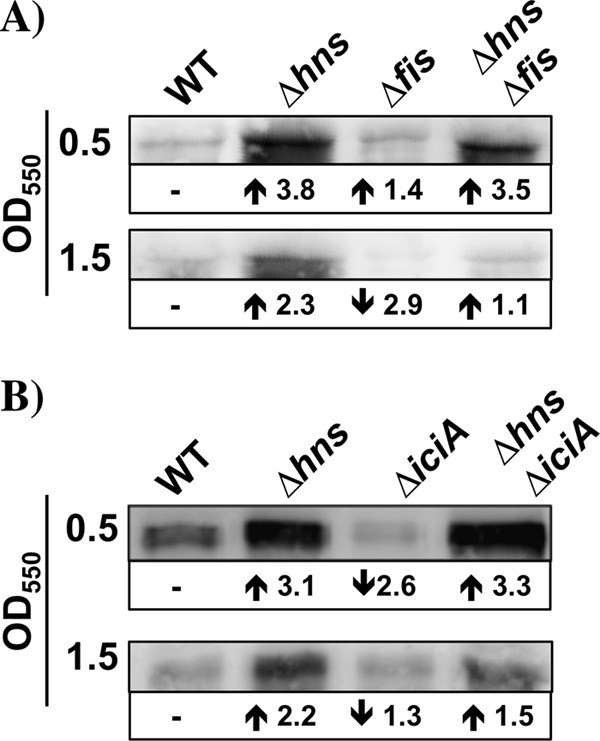
Modulation of NrdA by H-NS is not influenced by FIS or IciA. Immunodetection of the NrdA protein in aerobically growing cells of the wild-type (MG1655) strain and its hns (MG1655Δhns), fis (ETS115), and hns fis (ETS116) derivatives (A) and wild-type (MG1655) strain and its hns (MG1655Δhns), iciA (ETS123), and hns iciA (ETS124) derivatives (B).
It has been reported that the overexpression of the StpA paralogue in hns mutants partially compensates for the loss of H-NS (36). We also tested whether NrdA expression was further deregulated in a Δhns ΔstpA genetic background. Western blotting detection of NrdA in wild-type and Δhns, ΔstpA, and Δhns ΔstpA mutant cells showed that StpA does not attenuate NrdA expression in either exponential- or stationary-growth-phase cells (Fig. 3).
Fig 3.
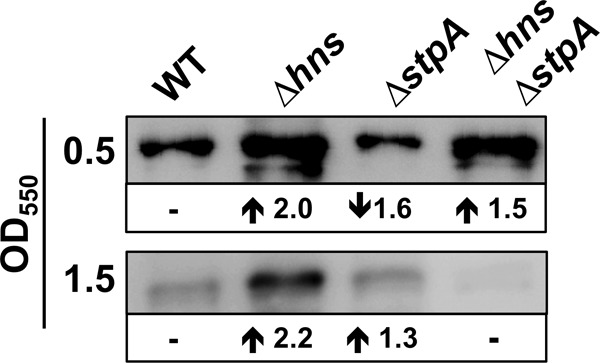
Specific H-NS repression of NrdA is not influenced by StpA. Immunodetection of the NrdA protein in aerobically growing cells of the wild-type strain MG12655 and its corresponding hns (MG1655Δhns), stpA (ETS117), and hns stpA (ETS118) mutant derivatives.
H-NS protein binds specifically to two DNA regions in the promoter and coding nrdA regions.
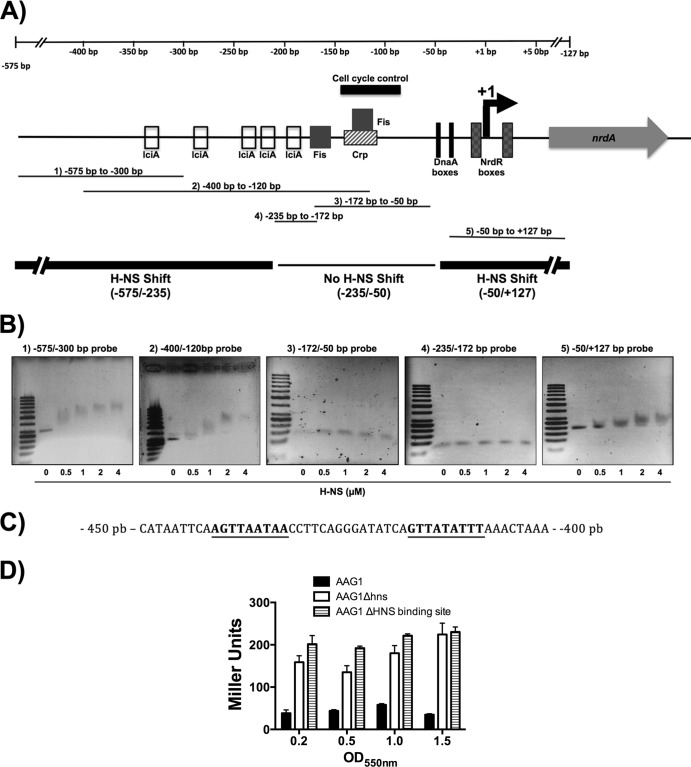
The finding that H-NS is able to repress the expression of nrdAB and nrdDG in vivo prompted us to investigate its interaction with the nrdA, nrdH, and nrdD promoters. To this end, DNA fragments corresponding to the entire nrdA, nrdD, and nrdH promoters (703 bp, nucleotide positions −575 to +127; 261 bp, −244 to +17; and 441 bp, −419 to + 22, respectively) were used for band shift assays in the presence of purified H-NS protein. As observed in Fig. 4, when the PnrdA and PnrdD DNA fragments were used in the presence of increasing H-NS concentrations, protein-DNA complexes exhibiting decreased mobility were detected, thus suggesting the progressive occupancy of multiple binding sites by H-NS. However, low-mobility protein-DNA complexes were not observed when a fragment corresponding to the PnrdH promoter was used, thus indicating that this fragment contains no binding site for H-NS. We next performed fine mapping of the putative H-NS binding sites in the PnrdA operon by using several overlapping DNA fragments from the promoter region and from the nrdA coding sequence. H-NS preferentially binds to two DNA regions, one of which includes sequences that map upstream of the known binding sites for the transcription factors that regulate the expression of nrdA (FIS and IciA) (Fig. 5B). The 3′ end of the fragment to which H-NS binds preferentially is located 235 bp upstream of the nrdA transcriptional start site (4). The deletion analysis of this fragments shows that the putative H-NS binding site of approximately 50 bp is centered at position −425. This sequence is AT rich (68.4%), a feature typically found in other well-characterized H-NS binding regions. In addition, this region contains two sequences (AGTTAATAA and GTTATATTT) that are highly similar to the H-NS consensus sequence described by Lang et al. (41). The second region includes sequences within the nrdA structural gene, 127 bp downstream of the nrdA transcriptional start site (Fig. 5B). Its AT content is also high (58.6%), but it does not contain either of the two H-NS consensus sequences.
Fig 4.
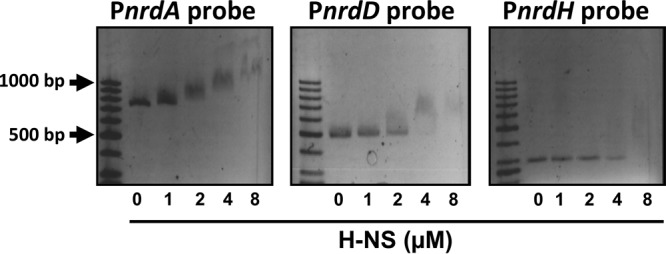
The H-NS protein binds to class Ia and III DNA promoter-regulatory regions. EMSAs performed with purified H-NS protein and promoter-regulatory regions of different RNR class genes present on the E. coli genome (PnrdA, class Ia; PnrdD, class III; and PnrdH, class Ib). H-NS protein was added at the concentrations indicated below the gels.
Fig 5.
The H-NS protein binds to two DNA regions in the promoter and coding nrdA regions of the gene. (A) Schematic representation of the E. coli nrdAB promoter region (PnrdA). PnrdA contains multiple transcription factor binding sites; the locations and scale are based both on the complete description available in the EcoCyc database (www.ecocyc.org) (50) and on available experimental data (30, 35). The positions indicated are based on the +1 start site of PnrdA transcription, 110 bp upstream of the ATG codon of nrdA (51). Short DNA fragments of the PnrdA region (bands 1 to 5) were used for the EMSA analysis (B). (B) EMSAs with purified H-NS protein and different short DNA fragments of the PnrdA promoter region, as indicated in panel A. H-NS was added at the concentration indicated at the bottom. (C) Putative H-NS 50 bp binding sequence in the nrdAB promoter region. Bold underlined type shows the putative DNA sequences identical to the H-NS consensus sequence region described previously (41). (D) Effect of the deletion of the putative PnrdA H-NS binding site on nrdA expression. β-Galactosidase activity was determined in strains ETS119 (AAG1), ETS122 (AAG1Δhns), and ETS121 (PnrdA::lacZ, ΔH-NS binding site). Cells were grown aerobically in LB medium at 37°C, and samples were collected at different points of growth. The β-galactosidase values are in Miller units. The points represent the means from three independent experiments, and the error bars denote standard deviations.
To verify that the identified upstream H-NS binding region is functional in vivo, we constructed a transcriptional PnrdA::lacZ fusion with a deletion in the putative H-NS binding region (see Materials and Methods). The expression of the chromosomal PnrdA::lacZ fusion in strain ETS121 (ΔHNS binding site) was similar to that of the corresponding Δhns mutant (Fig. 5C), indicating the relevance of these sequences to the ability of H-NS to modulate the expression of the PnrdAB operon.
Osmolarity, temperature, and anaerobiosis: the H-NS modulatory role for the nrdAB operon.
Considering that H-NS modulates gene expression in response to environmental factors, we next analyzed whether this protein modulates the expression of the nrdAB operon in response to specific environmental stimuli. For these studies, we evaluated temperature, osmolarity, and anaerobiosis. We tested first whether nrdA expression is sensitive to osmolarity and temperature using strains ETS119 and ETS122 (nrdAB::lacZ). To assess the effect of osmolarity, we evaluated the β-galactosidase activity in cells growing in LB broth containing either 0, 170, or 500 mM NaCl. The values of nrdA expression in cells grown under low-osmolarity (0 M NaCl) and high-osmolarity (500 mM NaCl) conditions were related to those obtained from cells grown in standard LB broth (170 mM), which was taken as the reference (Fig. 6A). Osmolarity did not influence nrdA expression; accordingly, the modulatory role of H-NS with regard to nrdA was not affected by the osmolarity of the growth medium. With respect to temperature, the evaluation of β-galactosidase showed that nrdA expression is upregulated when cells are grown at a low temperature (Fig. 6B). H-NS does not participate in the temperature-dependent regulation of nrdA, as shown by evaluation of β-galactosidase in strain ETS119 at high and low temperatures.
Fig 6.
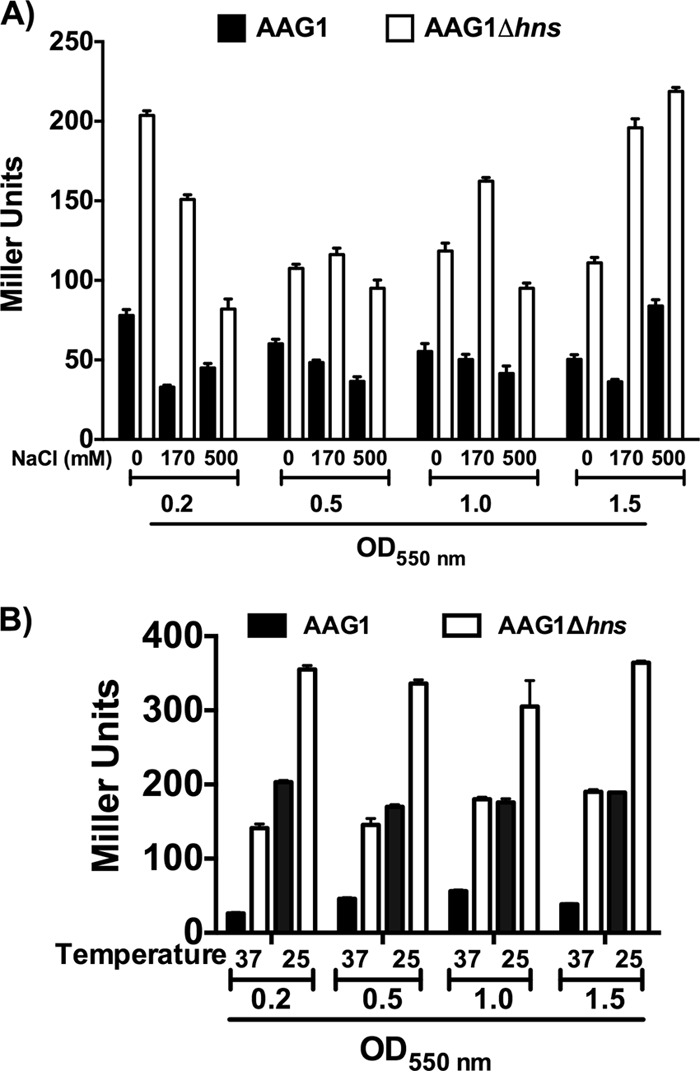
Effects of osmolarity and temperature on nrdAB expression. The effects of osmolarity (A) and temperature (B) on the expression of the PnrdAB::lacZ transcriptional fusion with β-galactosidase were measured in strains ETS119 and ETS122. Samples at different NaCl concentrations were collected at different stages of growth in LB medium at 37°C. For temperature-dependent expression, cells were grown at 25°C and 37°C. The values are in Miller units. The points represent the means from three independent experiments, and the error bars denote standard deviations.
As indicated in the introduction, class Ia RNRs are not active under anaerobic conditions, although other RNRs (class III RNRs) can function under these conditions. Hence, it was interesting to evaluate whether H-NS plays a role in nrdAB repression when E. coli cells are growing anaerobically. Therefore, an expression analysis was performed with strains ETS119, ETS121, and ETS122 (Fig. 7A) at different stages of growth (optical densities at 550 nm [OD550] of 0.2, 0.5, and 1) according to the growth curve shown in Fig. 7B. Compared with the wild-type strain, the expression of nrd in the Δhns mutant was increased in both exponentially growing cells (6.4 and 2.9 times) and stationary-phase cells (2.2 times). To corroborate the transcriptional data, we measured the anaerobic levels of the NrdA protein in wild-type MG1655 and its isogenic Δhns mutant by Western blotting (Fig. 7C). NrdA was expressed at higher levels in the hns mutant than in the wild type in both the exponential and early stationary growth phases.
Fig 7.
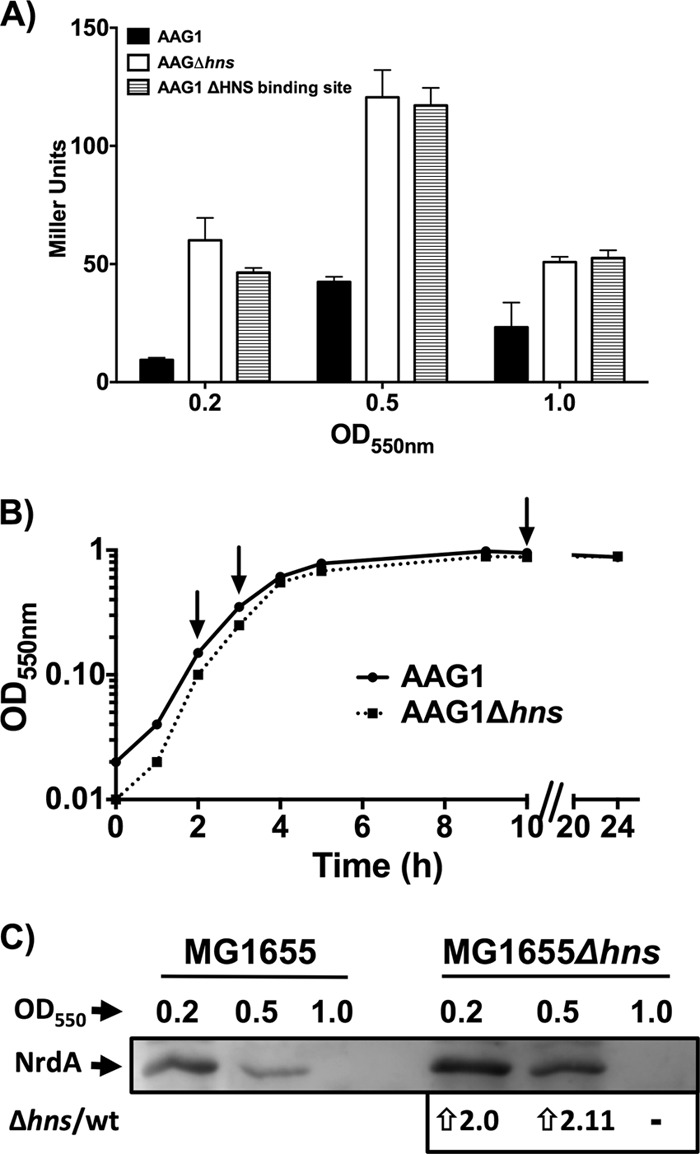
Class Ia RNR is repressed anaerobically by H-NS. (A) β-Galactosidase was measured in strains harboring chromosomal PnrdAB::lacZ transcriptional fusions. β-Galactosidase activity was determined in strains ETS119 (AAG1), ETS122 (AAG1Δhns), and ETS121 (PnrdA::lacZ, ΔH-NS binding site). Cells were grown anaerobically in LB medium at 37°C, and samples were collected at different points of the growth curve. The points represent the means from three independent experiments, and the error bars denote standard deviations. (B) Anaerobic growth curves of strains ETS119 and ETS122 indicating the points of sample collection and corresponding assay for β-galactosidase (A) and for NrdA immunodetection (C). (C) Immunodetection of NrdA protein in anaerobically growing cells of strains MG1655 and (MG1655Δhns).
DISCUSSION
Oxygen availability is an important signal for many pathogenic bacteria to be able to colonize their hosts and cause intestinal and extraintestinal infections in human and animals (37). For this reason, several bacterial genes associated with colonization and infection need to respond and adapt to a shift from an aerobic to an anaerobic environment. Oxygen availability also influences the expression of several housekeeping functions that are essential for adaptation to anaerobic niches: RNRs are one example. The Escherichia coli genome encodes two aerobic class I RNRs (class I, encoded by nrdAB, and class Ib, encoded by nrdHIEF) and one class III RNR (nrdDG). Remarkably, class Ia RNR expression is repressed in anaerobic conditions; enzyme activity is also repressed under these conditions (26). In contrast, class III RNR enzyme expression is induced and fully active only under anaerobic conditions (26, 38).
RNR expression is affected by other physiological stimuli in addition to oxygen availability, and several factors have been shown to control the transcriptional regulation of the different RNR genes (2, 4). The nrdAB operon is transcriptionally activated by FIS, IciA, and DnaA and is cell cycle regulated; furthermore, its expression is not dependent on Arc or Fnr (38). However, the activation of Fnr-dependent transcriptional regulation triggers the expression of nrdDG during anaerobic growth (38, 39). The expression of the nrdHIEF operon is activated by Fur, but it is not sensitive to the regulatory mechanisms reported to influence nrdAB expression. It was recently reported that this operon can substitute for the enzymatic activity of class Ia RNRs when Mn ions are present instead of Fe (40).
In spite of the available information, the factors that dictate the expression of some RNR genes in response to certain specific environmental conditions remain to be elucidated. Examples are the repression of nrdAB operon expression under anaerobic conditions and the downregulation of nrdAB expression at the transition from the exponential to the stationary phase. At the onset of the stationary phase, dNTPs are not required because DNA replication is inhibited.
In this study, we identified some aspects of the regulatory mechanisms modulating the expression of E. coli nrd genes. We assigned a role to the H-NS protein as a modulator of the expression of the nrdAB and nrdDG genes. The expression of the nrdAB and nrdDG operons is upregulated in an hns mutant under several growth conditions. Furthermore, using the nrdAB operon as a model, we demonstrated the specific interaction of H-NS with two DNA fragments of the operon, one of which maps upstream of the promoter and the other of which is located within the nrdA coding sequence. The DNA sequences of both fragments are AT rich, and in addition, the upstream fragment includes DNA sequences identical to the H-NS consensus sequence region described previously (41). H-NS binding to two target sequences located at a distance apart has been reported for most H-NS-modulated operons (17, 42), and it is not uncommon that one of the binding sites maps downstream of the promoter, as is the case for the bgl operon (43). In vivo assays of nrdAB expression in cells lacking the upstream H-NS binding site confirm the role of these sequences as H-NS targets. Our findings prompted us to search previous global transcriptomic studies to assess if these approaches provided evidence that the nrdAB operon is targeted by H-NS both in E. coli and in Salmonella. This is the case (44, 45).
In aerobically growing E. coli cells, H-NS represses nrdAB expression under all conditions tested. Osmolarity does not influence expression of this operon and, accordingly, this environmental factor did not modify the effect of the hns allele on nrdAB expression. With respect to temperature, it should be noted that the temperature-dependent modulation of gene expression by H-NS usually occurs in genes or operons that are downregulated at low temperature, which is not the case for the nrdAB operon, which is upregulated at low temperature. Hence, it is not surprising that H-NS does not play a role in the temperature-dependent regulation of this operon. It should be pointed out here that H-NS has also been associated to chromosome replication. It has been suggested that H-NS, directly or indirectly, facilitates the initiation of chromosome replication (46). The H-NS concentration appears to play a significant role in chromosome replication (47). Therefore, H-NS may extend its role in chromosome replication by modulating the nrdAB operon. The intracellular amount of free H-NS protein (not bound to other target sequences) may be critical both in influencing chromosome replication and in dictating nrdAB transcription.
The role of H-NS-mediated repression of gene expression when cells grow anaerobically is well documented (48, 49). Considering that NrdAB activity is not required when cells grow anaerobically, we hypothesized that NrdAB expression could be repressed in anoxically growing cells and that H-NS might play a relevant role silencing nrdAB expression under these conditions. In fact, nrdAB expression is increased in strain AGG1Δhns growing anaerobically. Nevertheless, the extent of repression is similar to that observed in aerobically growing cells. Hence, a specific role for H-NS repressing nrdAB in response to anaerobiosis cannot be drawn from the data obtained.
The results presented in this work provide evidence that the nrdAB operon is sensitive to H-NS-mediated silencing under a wide range of growth conditions.
ACKNOWLEDGMENTS
This work was supported by the Ministerio de Economia y Competitividad with grants BFU2011-24066 and ERA-NET PathoGenoMics (BIO2008-04362-E) to E.T., BFU2010-21836-C02-02 to C.M., BFU2010-21836-C02-01 to A.J. and CSD2008-00013 grant to A.J., C.M., and E.T. This work was also supported by the Generalitat de Catalunya 2009SGR66. E.T. was supported by the Ramón y Cajal and I3 program from the Ministerio de Ciencia e Innovación.
Footnotes
Published ahead of print 19 July 2013
REFERENCES
- 1.Nordlund P, Reichard P. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75:681–706 [DOI] [PubMed] [Google Scholar]
- 2.Torrents E, Sahlin M, Sjöberg B-M. 2008. The ribonucleotide reductase family—genetics and genomics, p 17–77 In Andersson KK. (ed), Ribonucleotide reductases. Nova Science Publishers, New York, NY [Google Scholar]
- 3.Lundin D, Torrents E, Poole AM, Sjöberg B-M. 2009. RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genomics 10:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrick J, Sclavi B. 2007. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol. Microbiol. 63:22–34 [DOI] [PubMed] [Google Scholar]
- 5.Mathews CK. 2006. DNA precursor metabolism and genomic stability. FASEB J. 20:1300–1314 [DOI] [PubMed] [Google Scholar]
- 6.Torrents E, Grinberg I, Gorovitz-Harris B, Lundstrom H, Borovok I, Aharonowitz Y, Sjöberg BM, Cohen G. 2007. NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. J. Bacteriol. 189:5012–5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 8.Cendra MDM, Juarez A, Torrents E. 2012. Biofilm modifies expression of ribonucleotide reductase genes in Escherichia coli. PLoS One 7:e46350. 10.1371/journal.pone.0046350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobson BA, Fuchs JA. 1998. Multiple cis-acting sites positively regulate Escherichia coli nrd expression. Mol. Microbiol. 28:1315–1322 [DOI] [PubMed] [Google Scholar]
- 10.Jacobson BA, Fuchs JA. 1998. A 45 bp inverted repeat is required for cell cycle regulation of the Escherichia coli nrd operon. Mol. Microbiol. 28:1307–1314 [DOI] [PubMed] [Google Scholar]
- 11.Bertin P, Benhabiles N, Krin E, Laurent-Winter C, Tendeng C, Turlin E, Thomas A, Danchin A, Brasseur R. 1999. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in gram-negative bacteria. Mol. Microbiol. 31:319–329 [DOI] [PubMed] [Google Scholar]
- 12.Tendeng C, Bertin PN. 2003. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol. 11:511–518 [DOI] [PubMed] [Google Scholar]
- 13.Dorman CJ. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391–400 [DOI] [PubMed] [Google Scholar]
- 14.Rimsky S. 2004. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 7:109–114 [DOI] [PubMed] [Google Scholar]
- 15.Rimsky S, Zuber F, Buckle M, Buc H. 2001. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 42:1311–1323 [DOI] [PubMed] [Google Scholar]
- 16.Dorman CJ, Deighan P. 2003. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 13:179–184 [DOI] [PubMed] [Google Scholar]
- 17.Dame RT, Wyman C, Wurm R, Wagner R, Goosen N. 2002. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 277:2146–2150 [DOI] [PubMed] [Google Scholar]
- 18.Esposito D, Petrovic A, Harris R, Ono S, Eccleston JF, Mbabaali A, Haq I, Higgins CF, Hinton JC, Driscoll PC, Ladbury JE. 2002. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J. Mol. Biol. 324:841–850 [DOI] [PubMed] [Google Scholar]
- 19.Stoker NG, Fairweather NF, Spratt BG. 1982. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene 18:335–341 [DOI] [PubMed] [Google Scholar]
- 20.Sjöberg BM, Torrents E. 2011. Shift in ribonucleotide reductase gene expression in Pseudomonas aeruginosa during Infection. Infect. Immun. 79:2663–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Lorenzo V, Herrero M, Jakubzik U, Timmis KN. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia J, Madrid C, Cendra M, Juarez A, Pons M. 2009. N9L and L9N mutations toggle Hha binding and hemolysin regulation by Escherichia coli and Vibrio cholerae H-NS. FEBS Lett. 583:2911–2916 [DOI] [PubMed] [Google Scholar]
- 23.Aberg A, Shingler V, Balsalobre C. 2008. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 67:1223–1241 [DOI] [PubMed] [Google Scholar]
- 24.Johansson J, Dagberg B, Richet E, Uhlin BE. 1998. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J. Bacteriol. 180:6117–6125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garriga X, Eliasson R, Torrents E, Jordan A, Barbe J, Gibert I, Reichard P. 1996. nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem. Biophys. Res. Commun. 229:189–192 [DOI] [PubMed] [Google Scholar]
- 27.Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29.Madrid C, Nieto JM, Paytubi S, Falconi M, Gualerzi CO, Juarez A. 2002. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 184:5058–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Augustin LB, Jacobson BA, Fuchs JA. 1994. Escherichia coli Fis and DnaA proteins bind specifically to the nrd promoter region and affect expression of an nrd-lac fusion. J. Bacteriol. 176:378–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olliver A, Saggioro C, Herrick J, Sclavi B. 2010. DnaA-ATP acts as a molecular switch to control levels of ribonucleotide reductase expression in Escherichia coli. Mol. Microbiol. 76:1555–1571 [DOI] [PubMed] [Google Scholar]
- 32.White-Ziegler CA, Davis TR. 2009. Genome-wide identification of H-NS-controlled, temperature-regulated genes in Escherichia coli K-12. J. Bacteriol. 191:1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stella S, Falconi M, Lammi M, Gualerzi CO, Pon CL. 2006. Environmental control of the in vivo oligomerization of nucleoid protein H-NS. J. Mol. Biol. 355:169–174 [DOI] [PubMed] [Google Scholar]
- 34.Forns N, Juarez A, Madrid C. 2005. Osmoregulation of the HtrA (DegP) protease of Escherichia coli: an Hha-H-NS complex represses HtrA expression at low osmolarity. FEMS Microbiol. Lett. 251:75–80 [DOI] [PubMed] [Google Scholar]
- 35.Han JS, Kwon HS, Yim JB, Hwang DS. 1998. Effect of IciA protein on the expression of the nrd gene encoding ribonucleoside diphosphate reductase in E. coli. Mol. Gen. Genet. 259:610–614 [DOI] [PubMed] [Google Scholar]
- 36.Sonden B, Uhlin BE. 1996. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 15:4970–4980 [PMC free article] [PubMed] [Google Scholar]
- 37.Jones SA, Gibson T, Maltby RC, Chowdhury FZ, Stewart V, Cohen PS, Conway T. 2011. Anaerobic respiration of Escherichia coli in the mouse intestine. Infect. Immun. 79:4218–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boston T, Atlung T. 2003. FNR-mediated oxygen-responsive regulation of the nrdDG operon of Escherichia coli. J. Bacteriol. 185:5310–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roca I, Ballana E, Panosa A, Torrents E, Gibert I. 2008. Fumarate and nitrate reduction (FNR) dependent activation of the Escherichia coli anaerobic ribonucleotide reductase nrdDG promoter. Int. Microbiol. 11:49–56 [PubMed] [Google Scholar]
- 40.Martin JE, Imlay JA. 2011. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol. Microbiol. 80:319–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon CL, Rimsky S, Stella S, Babu MM, Travers A. 2007. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 35:6330–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang FC, Rimsky S. 2008. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 11:113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dole S, Nagarajavel V, Schnetz K. 2004. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 52:589–600 [DOI] [PubMed] [Google Scholar]
- 44.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238 [DOI] [PubMed] [Google Scholar]
- 45.Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM. 2011. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 39:2073–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katayama T, Takata M, Sekimizu K. 1996. The nucleoid protein H-NS facilitates chromosome DNA replication in Escherichia coli dnaA mutants. J. Bacteriol. 178:5790–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atlung T, Hansen FG. 2002. Effect of different concentrations of H-NS protein on chromosome replication and the cell cycle in Escherichia coli. J. Bacteriol. 184:1843–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krishnan HH, Ghosh A, Paul K, Chowdhury R. 2004. Effect of anaerobiosis on expression of virulence factors in Vibrio cholerae. Infect. Immun. 72:3961–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim MJ, Lim S, Ryu S. 2008. Molecular analysis of the Salmonella typhimurium tdc operon regulation. J. Microbiol. Biotechnol. 18:1024–1032 [PubMed] [Google Scholar]
- 50.Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muniz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP, Karp PD. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 39:D583–D590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuggle CK, Fuchs JA. 1986. Regulation of the operon encoding ribonucleotide reductase in Escherichia coli: evidence for both positive and negative control. EMBO J. 5:1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]