Abstract
All bacterial quorum sensing (QS) systems are based on the production, secretion, and detection of small signaling molecules. Gram-positive bacteria typically use small peptides as QS effectors, and each QS circuit generally requires the interaction of a single signaling molecule with a single receptor protein. The recently described Rgg2 and Rgg3 (Rgg2/3) regulatory circuit of Streptococcus pyogenes (group A streptococcus [GAS]) is one of only a few QS circuits known to utilize multiple signaling peptides. In this system, two distinct, endogenously produced peptide pheromones (SHP2 and SHP3) both function to activate the QS circuit. The aim of this study was to further define the roles of SHP2 and SHP3 in activation of the Rgg2/3 QS system, specifically with regard to shp gene identity and dosage. Results from our studies using transcriptional reporters and isogenic GAS mutants demonstrate that shp gene dosage does contribute to Rgg2/3 system induction, as decreased gene dosage results in decreased or absent induction. Beyond this, however, data indicate that the shp genes possess distinct potentials for supporting system activation, with shp3 more readily able to support system activation than shp2. Studies using synthetic peptides and shp gene mutants indicate that the disparate activities of endogenous SHPs are due to production, rather than signaling, differences and are conferred by the N-terminal regions rather than the C-terminal signaling regions of the peptides. These data provide evidence that the N-terminal, noneffector sequences of SHP pheromones influence their production efficiencies and thereby the relative activation potentials of endogenous SHPs.
INTRODUCTION
Bacteria commonly benefit from a prevalent form of bacterial communication known as quorum sensing (QS), a process which enables the coordination of responses across a population. This process occurs via the production, secretion, and detection of small, secreted signaling molecules which are often referred to as pheromones or autoinducers. The latter term references the common phenomenon wherein the signaling molecule positively regulates its own expression. Gram-positive bacteria often utilize processed oligopeptides as QS molecules, and these are genetically encoded and ribosomally generated within the cell. Either the general secretory (Sec) system or specialized transporters belonging to the ATP-binding cassette (ABC) family are responsible for active transport of these peptides out of the cell due to their inability to permeate the cell membrane (1, 2). At points between translation, export, and detection, peptides are subject to various modification events, including processing and/or cyclization (for a review, see reference 3). Following signal accumulation in the extracellular milieu, detection of the peptide signal can occur either at the surface of the cell via interaction with a sensor kinase or intracellularly via direct interaction with a cytoplasmic regulatory protein.
There are two groups of receptor proteins now known to function in intracellular detection of QS peptides: the RNPP family, named for its four prototypical members (Rap, NprR, and PlcR, each found in several Bacillus species, and PrgX of Enterococcus faecalis) (4, 5), and the Rgg family of transcriptional regulators widespread among the Firmicutes (6–12). Although the majority of these QS systems function with only a single peptide pheromone and single cognate receptor protein, several rely on the coordinated actions of multiple signaling peptides and/or receptors. The best-studied example is the PrgX system of E. faecalis. The transcriptional repressor PrgX interacts with both the chromosomally encoded activator peptide cCF10 and the plasmid-encoded inhibitor peptide iCF10 (13–15). In simple terms, cCF10 induces expression of plasmid conjugation genes through disruption of intermolecular PrgX interactions, whereas iCF10 promotes repression by stabilizing PrgX tetramer formation. Protein crystallization and modeling studies have revealed that both peptides bind at the same site of PrgX and that the disparity in sequence between these two peptides promotes their antagonistic activities (for a review, see reference 16).
The Rgg2 and Rgg3 (Rgg2/3) regulatory circuit of Streptococcus pyogenes that was recently described by our laboratory is another QS system that utilizes two distinct endogenously produced peptide pheromones in a single QS circuit (6). This system utilizes two Rgg-family regulators (Rgg2 and Rgg3) that respond to adjacently encoded signaling peptides (SHP2 and SHP3) to control gene expression and biofilm formation. Rgg2 is a transcriptional activator of target genes, including the shp genes themselves, whereas Rgg3 represses expression of these genes. The C-terminal eight amino acids of the SHP peptides are identical at seven of eight positions and comprise the active signaling molecules (Fig. 1A), and both mature peptides function intracellularly to induce system activation through derepression of Rgg3 and activation of Rgg2 (6, 10). Interestingly, this is the only Rgg-mediated QS system thus far identified to involve the coordinated action of multiple Rgg proteins and natively produced signaling peptides rather than a single receptor-peptide pair.
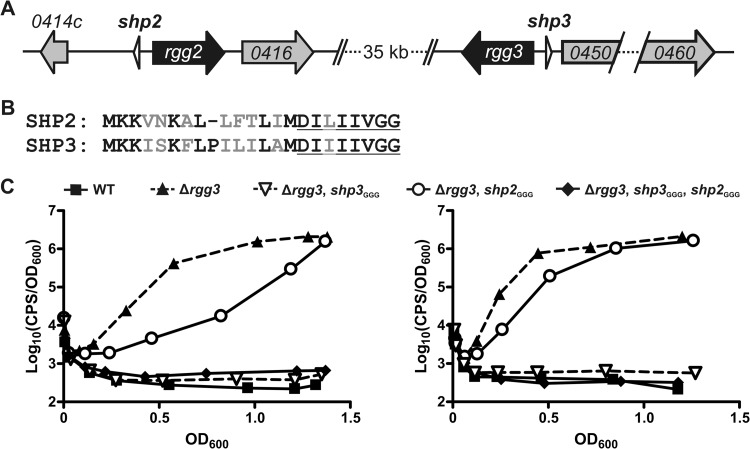
Fig 1.
Inactivation of shp genes differentially affects Rgg2/3 system autoinduction in the Δrgg3 background. (A) Organization of the rgg2 and rgg3 loci in the NZ131 genome. (B) Alignment of full-length prepeptides SHP2 and SHP3. Nonidentical amino acids are in gray font, and the C-terminal eight residues comprising the mature, active peptides are underlined. (C) Luciferase expression from Pshp2 reporter integrated into WT (BNL148), Δrgg3 (BNL149), Δrgg3 shp3GGG (BNL203), Δrgg3 shp2GGG (BNL176), and Δrgg3 shp3GGG shp2GGG (BNL199) isogenic mutant strains. Panels show results obtained with biological replicates using the same strains on separate occasions, and expression trends are representative of at least three independent experiments.
Despite both utilizing two endogenously produced peptide pheromones, the PrgX and Rgg2/3 systems are distinct in several ways. Unlike the case in E. faecalis, where only a single peptide-responsive regulator, PrgX, governs system activation, the Rgg2 and Rgg3 regulators of S. pyogenes both appear to respond to peptide signals. Unlike the PrgX system, where the two peptides have opposite activities, both SHP2 and SHP3 function to activate the Rgg2/3 system. Finally, unlike cCF10 and iCF10, which share less than 50% identity, the C-terminal eight amino acids that comprise the active portions of SHP2 and SHP3 differ by only a single residue (possessing leucine and isoleucine, respectively).
The aim of this study was to further define the precise roles of SHP2 and SHP3 in activation of the Rgg2/3 QS system. More specifically, we sought to determine if the SHP peptides possess distinct activities and/or if shp gene dosage is a factor for system activation. As expected, shp gene dosage does contribute to system activation. Unexpectedly, however, the data reveal that the shp2 and shp3 genes possess distinct potentials for supporting system activation in a manner independent of their C-terminal active regions, with shp3 possessing enhanced activity compared to shp2. Comparable activities of exogenously added synthetic peptides indicate that the disparate activities of endogenous SHPs result from differential production and/or secretion, providing evidence that the N-terminal, noneffector sequences of SHP pheromones influence their production efficiencies and thereby the relative activation potentials of endogenous SHPs.
MATERIALS AND METHODS
Strains, plasmids, and culture media.
All strains used in this study are listed in Table 1, and construction of mutant strains is described in detail below. All plasmids and primers used in this study are listed in Tables 2 and 3, respectively. Group A streptococcus (GAS) was routinely grown in Todd-Hewitt medium (TH; BD Biosciences) supplemented with 2% (wt/vol) yeast extract (Y; Amresco). For all reporter expression studies, GAS was grown in chemically defined medium (CDM) (6) made no more than 24 h prior to the experiment and containing 1% (wt/vol) glucose at 37°C. When necessary, antibiotics were included at the following concentrations: chloramphenicol (Cm), 3 μg ml−1; erythromycin (Erm), 0.5 μg ml−1; and spectinomycin (Spec), 100 μg ml−1. Escherichia coli strains DH10β (Invitrogen) and BH10C (17) were used for cloning and were grown in Luria-Bertani (LB) broth or on LB agar with antibiotics at the following concentrations: chloramphenicol, 10 μg ml−1; erythromycin, 500 μg ml−1; and spectinomycin, 100 μg ml−1.
Table 1.
Bacterial strains used in this study
| Strain | Description or genotype; phenotypea | Reference or source |
|---|---|---|
| NZ131 | Wild-type M49 S. pyogenes isolate | 18, 31 |
| BNL148 | NZ131 with integrated pBL111 Pshp2 reporter; Ermr | 6 |
| BNL149 | JCC131 with integrated pBL111 Pshp2 reporter; Cmr Ermr | 6 |
| BNL169 | NZ131 Δrgg3::cat shp2GGG; Cmr | This study |
| BNL170 | NZ131 shp2GGG shp3GGG; unmarked | 19 |
| BNL176 | BNL169 with integrated pBL111 Pshp2 reporter; Cmr Ermr | This study |
| BNL177 | BNL170 with integrated pBL111 Pshp2 reporter; Ermr | 19 |
| BNL186 | NZ131 Δrgg3::cat shp2leu17ile; Cmr | This study |
| BNL187 | NZ131 Δrgg3::cat Δshp2→shp3; Cmr | This study |
| BNL192 | BNL187 with integrated pBL111 Pshp2 reporter; Cmr Ermr | This study |
| BNL193 | NZ131 Δrgg3::cat shp2GGG shp3GGG; Cmr | This study |
| BNL196 | BNL186 with integrated pBL111 Pshp2 reporter; Cmr Ermr | This study |
| BNL198 | NZ131 Δrgg3::cat Δshp3→shp2; Cmr | This study |
| BNL199 | BNL193 with integrated pBL111 Pshp2 reporter; Cmr Ermr | This study |
| BNL202 | BNL198 with integrated pBL111 Pshp2 reporter; Cmr Ermr | This study |
| BNL203 | JCC177 with integrated pBL111 Pshp2 reporter; Cmr Ermr | This study |
| BNL206 | BNL170 with integrated pJC219 Pshp3 reporter; Cmr Ermr | This study |
| BNL208 | NZ131 Δrgg3::cat shp3GGG Δshp2→shp3; Cmr | This study |
| BNL210 | NZ131 Δrgg3::cat shp2GGG Δshp3→shp2; Cmr | This study |
| BNL212 | NZ131 Δrgg3::cat Δshp3→shp2 Δshp2→shp3; Cmr | This study |
| BNL216 | BNL208 with integrated pBL111 Pshp2 reporter; Cmr Ermr | This study |
| BNL220 | BNL210 with integrated pBL111 Pshp2 reporter; Cmr Ermr | This study |
| BNL222 | BNL212 with integrated pBL111 Pshp2 reporter; Cmr Ermr | This study |
| BNL225 | NZ131 Δrgg3::cat shp3ile18leu; Cmr | This study |
| BNL227 | BNL225 with integrated pBL111 Pshp2 reporter; Cmr Ermr | This study |
| JCC131 | NZ131 Δrgg3::cat; Cmr | 6 |
| JCC177 | NZ131 Δrgg3::cat shp3GGG; Cmr | This study |
Cm, chloramphenicol; Erm, erythromycin.
Table 2.
Plasmids used in this study
| Plasmid | Description; phenotypea | Reference or source |
|---|---|---|
| p7INT | Shuttle-suicide vector that integrates at streptococcal bacteriophage T12 attB site; Ermr | 23 |
| pBL111 | DNA fragment containing the shp2 promoter (500 bp) fused to luxAB and cloned into p7INT; Ermr | 6 |
| pBL119 | pFED760 containing DNA fragment encompassing shp2 and flanking DNA; Ermr | 19 |
| pBL120 | pFED760-based vector for mutation of shp2 start codon to GGG; Ermr | 19 |
| pBL121 | pBL119 with shp2 leu17 codon mutated to code for ile; Ermr | This study |
| pBL122 | pFED760-based vector for replacement of shp2 with shp3; Ermr | This study |
| pBL123 | pFED760-based vector for simultaneous rgg3::cat and shp3→shp2 mutations; Cmr Ermr | This study |
| pBL124 | pJC175 with shp3 ile18 codon mutated to code for leu; Cmr Ermr | This study |
| pFED760 | Shuttle vector pGh9-ISS1 deleted for ISS1 element; temp-sensitive; Ermr | 11, 20 |
| pJC175 | pFED760-based vector for replacement of rgg3 with cat cassette; Cmr Ermr | 6 |
| pJC180 | pFED760-based vector for mutation of shp3 start codon to GGG; Ermr | 19 |
| pJC219 | DNA fragment containing the shp3 promoter (384 bp) fused to luxAB and cloned into p7INT; Ermr | This study |
Cm, chloramphenicol; Erm, erythromycin.
Table 3.
Primers used in this study
| Constructed plasmid | Primer | Sequencea | Description |
|---|---|---|---|
| pBL121 | BL84 | CTTTTTACATTGATCATGGATATCATCATTATCGTTGGTGGATAACC | Inverse PCR S primer to mutate shp2 leu17 codon with nucleotide changes underlined |
| BL85 | GGTTATCCACCAACGATAATGATGATATCCATGATCAATGTAAAAAG | Inverse PCR AS primer to mutate shp2 leu17 codon with nucleotide changes underlined | |
| pBL122 | BL80 | GCGTGCCCGGGCCACTTTAAATAAATAATCTGTGAG | S primer for shp2 and flanking regions; XmaI |
| BL81 | GCGTGCCCGGGATCGGTTTATCTGTAATATTTTTTTGACAAAGGTT | AS primer for shp2 and flanking regions; XmaI | |
| BL86 | ATTTTCTTCATGACTGTCTCCTTTCTGATTTTCTATTTTGTACT | AS primer for shp2 upstream flanking region with complementary sequence to shp3 underlined | |
| BL87 | TTGTTGGTGGGTAACCTTTCGCTTAAAAACAATCCACTTTG | S primer for shp2 downstream flanking region with complementary sequence to shp3 underlined | |
| BL88 | AGGAGACAGTCATGAAGAAAATTTCAAAATTTTTGCCGATTTTAA | S primer for shp3 with complementary sequence to shp2 upstream region underlined | |
| BL89 | GCGAAAGGTTACCCACCAACAATAATGATAATATCCATTGCTAAA | AS primer for shp3 with complementary sequence to shp2 downstream region underlined | |
| pBL123 | BL90 | CATGGCGGCCGCAAAGCGAAAAAAATGACACATTTATTC | S primer for shp3 and flanking regions; NotI |
| BL91 | CATGGCGGCCGCGAAAAAAAATATTTACCATCGTTAA | AS primer for shp3 and flanking regions; NotI | |
| BL92 | AACTTTTTTCATAAGTGGACTTCCTTTCAGTTTTTATTTATTGTA | AS primer for shp3 upstream flanking region with complementary sequence to shp2 underlined | |
| BL93 | ATCGTTGGTGGATAAGTTACTTAGGTGTATACCTAAGTCTTTAGATACTCTA | S primer for shp3 downstream flanking region with complementary sequence to shp2 underlined | |
| BL94 | GAAGTCCACTTATGAAAAAAGTTAATAAAGCTTTGCTTTTTACA | S primer for shp2 with complementary sequence to shp3 upstream region underlined | |
| BL95 | TAAGTAACTTATCCACCAACGATAATCAGGATATCCATGAT | AS primer for shp2 with complementary sequence to shp3 downstream region underlined | |
| pBL124 | BL96 | ATTTTAGCAATGGATATTCTGATTATTGTTGGTGGG | Inverse PCR S primer to mutate shp3 ile18 codon with nucleotide changes underlined |
| BL97 | CCCACCAACAATAATCAGAATATCCATTGCTAAAAT | Inverse PCR AS primer to mutate shp3 ile18 codon with nucleotide changes underlined | |
| pJC219 | BL27 | GCGTGGAATTCGCCTTTAATTTTATTATGGT | AS primer for luxAB; EcoRI |
| JC156 | ATGAAGTTTGGAAATATTTGTTTTTCG | S primer for luxAB | |
| JC238 | CATGGGATCCGTGTCGGAAAGTAAACATGC | S primer for shp3 promoter; BamHI | |
| JC239 | CAAATATTTCCAAACTTCATAAGTGGACTTCCTTTCAG | AS primer for shp3 promoter with complementary sequence to luxAB underlined |
Restriction sites are in bold.
Construction of mutant GAS strains.
All S. pyogenes strains used in this study were derived from the serotype M49 strain NZ131 (18). The plasmids used to delete rgg3 (pJC175) and to mutate the translational start codons of shp2 (pBL120) and shp3 (pJC180) were described previously (6, 19).
Vectors used for the allelic replacement of shp2 were generated as follows. To construct a plasmid in which the shp2 codon encoding leucine at amino acid position 17 was mutated to encode isoleucine, pBL119 was used as the template in an inverse PCR with primer pair BL84 and BL85, resulting in plasmid pBL121. For the vector used in allelic replacement of shp2 with shp3, upstream and downstream regions flanking the shp2 gene were amplified by PCR with primer pair BL80 and BL86 and primer pair BL87 and BL81 using NZ131 DNA as the template, with the antisense and sense primers for the upstream and downstream regions, respectively, containing sequence complementary to the shp3 gene. A separate PCR was used to amplify shp3 with primers BL88 and BL89, each of which contained complementary sequence to the upstream or downstream flanking region of shp2. The upstream region and shp3 PCR products were fused together by overlap extension PCR using the primers BL80 and BL89. The product of this fusion was then used together with the downstream region PCR fragment in a second overlap extension PCR using the primers BL80 and BL81. This final fusion product was inserted at the XmaI site of the temperature-sensitive vector pFED760 (11, 20) to generate pBL122.
Vectors used for the allelic replacement of shp3 simultaneously with rgg3::cat were generated as follows. For the vector used in allelic replacement of shp3 with shp2, upstream and downstream regions flanking the shp3 gene were amplified by PCR with primer pair BL90 and BL92 and primer pair BL91 and BL93 using JCC131 DNA as the template, with the antisense and sense primers for the upstream and downstream regions, respectively, containing sequence complementary to the shp2 gene. A separate PCR was used to amplify shp2 with primers BL94 and BL95, each of which contained complementary sequence to the upstream or downstream flanking region of shp3. The upstream region and shp2 PCR products were fused together by overlap extension PCR using the primers BL90 and BL95, the product of which was then used together with the downstream region PCR fragment in a second overlap extension PCR using primers BL90 and BL91. This final fusion product was inserted at the NotI site of pFED760 to generate pBL123. To construct a plasmid in which the shp3 codon encoding isoleucine at amino acid position 18 was mutated to encode leucine, pJC175 was used as the template in an inverse PCR with primer pair BL96 and BL97, resulting in plasmid pBL124.
All deletion vectors were electroporated into NZ131, and a two-step temperature-dependent selection process was used to isolate mutants of interest (21). Briefly, cells containing each deletion construct were grown at the permissive temperature (30°C) and then shifted to 37°C and plated on the appropriate antibiotic to select for bacteria in which the plasmid had integrated at one of the flanking regions. Cells were then grown at the permissive temperature to allow the plasmid to recombine out of the chromosome, and loss of Erm resistance was used to identify a successful second crossover event and loss of the deletion vector. Genotypes were confirmed by PCR and sequencing. This process was repeated to construct double and triple mutants.
Construction of luciferase transcriptional reporters.
Construction of Pshp2-lux reporter plasmid pBL111 was reported previously (6). For the Pshp3-lux reporter plasmid, a 384-bp region directly upstream of shp3, but not including the shp3 gene itself, was amplified from NZ131 genomic DNA using primers JC238 and JC239, and Vibrio fischeri luxAB genes were amplified from pCN58 (22) using primers JC156 and BL27. The promoter product was fused to luxAB in an overlap extension PCR using JC238 and BL27, and this product was inserted into EcoRI and BamHI sites of p7INT (23) to generate pJC219. All reporter plasmids were electroporated into GAS, and site-specific integration of the plasmids at a tRNA(Ser) gene was confirmed by PCR.
Luciferase transcriptional reporter assays.
For luciferase assays monitoring autoinduction, cells from overnight cultures grown in THY at 30°C were diluted 160-fold into CDM and incubated at 37°C. At each time point, 50 μl of each culture was removed to an opaque 96-well plate, samples were exposed to decyl aldehyde (Sigma) fumes for 30 s, and luminescence was quantified (in counts per second [cps]) using a Turner Biosystems Veritas Microplate Luminometer. The optical density of the culture at 600 nm (OD600) was also measured at each time point using a Spectronic 20D spectrophotometer (Milton Roy). Relative luminescence was calculated by normalizing cps to OD.
Luminescence experiments with synthetic peptides were performed in two different manners. For Fig. 2A, reporter strains were grown in CDM to an OD of 0.3 to 0.5 and then diluted 1:11 into fresh CDM containing peptide of interest, and OD and luminescence were monitored as described above. For Fig. 2B and C, reporter strains were grown in CDM to an OD of 0.3 to 0.5 and then diluted 1:11 into fresh CDM. Aliquots of the dilute culture were then distributed into the wells of a clear-bottom 96-well plate containing peptides of interest. Volatile decyl aldehyde was provided as a 1% solution in mineral oil between the wells of the microplate as previously described (24), and the plate was lidded and parafilmed to ensure entrapment of the volatile content. Cells were grown at 37°C in a Synergy 2 plate reader (BioTek) with continuous fast shaking to deter cell sedimentation and facilitate accurate evaluation of the culture density, and OD600 and cps were measured every 10 min throughout growth. The maximum cps/OD600 value was determined for each well and plotted against peptide concentration to determine peptide-specific activities.
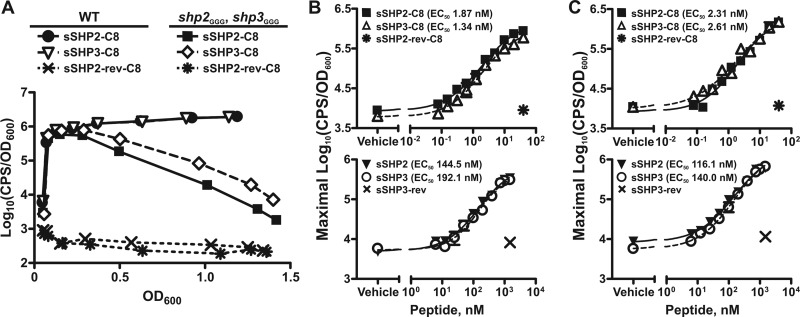
Fig 2.
Synthetic SHP peptides are equivalently able to independently support Rgg2/3 activation. (A) Luciferase expression from Pshp2 reporter integrated into WT (BNL148) and shp3GGG shp2GGG (BNL177) strains grown in the presence of 5 nM sSHP2-C8, sSHP3-C8, or control peptide sSHP2-rev-C8. Data shown are representative of at least three independent experiments. (B and C) Dose-response curves for maximal relative luminescence from Pshp2 (B) and Pshp3 (C) reporters integrated into the shp3GGG shp2GGG strain (BNL177 and BNL206, respectively) in response to synthetic C8 (top panels) or full-length (bottom panels) SHP peptides. Single concentrations of reverse peptides were included as controls. Data shown represent means and standard errors of the means (error bars) of the results of three independent experiments, with EC50s calculated from dose-response curves (GraphPad Prism, version 5.0).
Synthetic peptides.
Crude preparations of synthetic peptides used in luciferase assays were purchased from Neo-Peptide (Cambridge, MA), with purities ranging from 31% to 51%. Synthetic peptides were reconstituted as 2 mM stocks in dimethyl sulfoxide (DMSO) and stored at −80°C.
RESULTS
Disruption of shp2 and disruption of shp3 have differential impacts on system autoinduction in the absence of rgg3.
It was previously shown that in the absence of rgg3, the Rgg2/3 system is activated in an Rgg2-dependent manner and results in high-level expression from the shp promoters (6). It was also shown that simultaneous deletion of shp3 and rgg3 abolished system activation (6), indicating that removal of rgg3 was not sufficient and that SHPs, or at least SHP3, were required for Rgg2-mediated system activation. We questioned if the abolition of autoinduction in the Δrgg3-shp3 mutant occurred because both shp genes are required due to peptide dosage or if only shp3 is required because SHP3 possesses distinct activity that cannot be compensated for by SHP2. To assess these possibilities, we systematically generated Δrgg3 mutants that also harbored mutations, changing the translation initiation codons of shp2 or shp3 or both shp genes to GGG (shp2GGG, shp3GGG) and then observed luminescence production from the Pshp2 reporter in the various mutant strains. Mutation of the start codon was anticipated to abolish translation of the peptide, while use of the Δrgg3 background allowed us to observe endogenous system activation without adding synthetic peptide, since the lack of a repressor in this strain allows for natural autoinduction of the system (6). In agreement with previous results, deletion of rgg3 (Δrgg3; BNL149) resulted in high-level luminescence production whereas there was no detectable reporter induction in the wild-type (WT) (BNL148) reporter strain. Also in agreement with previous results, mutation of shp3 abolished reporter induction in the Δrgg3 strain (Δrgg3, shp3GGG; BNL203) (Fig. 1B). It should be noted that the ability of the start codon mutation to abolish reporter induction in a manner equivalent to that seen when the entire shp3 gene was deleted (6) indirectly confirms that the start codon mutation is sufficient to prevent SHP production or at least reduces SHP production to levels below that required for detection by the system. Unexpectedly, mutation of shp2 in the Δrgg3 strain (Δrgg3, shp2GGG; BNL176) did not abolish reporter induction; however, there was a difference in the timings of reporter induction, with luminescence induction in the Δrgg3 shp2GGG mutant reproducibly slower than that in the Δrgg3 strain. A reporter strain lacking rgg3 and carrying initiation codon mutations in both shp genes (Δrgg3, shp2GGG, shp3GGG; BNL199) produced no detectable luminescence, akin to the profiles of WT and Δrgg3 Δshp3GGG reporter strains (Fig. 1B). Expression profiles determined using a Pshp3 reporter were comparable to those seen with the Pshp2 reporter in the various strains (data not shown). Overall, these data suggest that SHP3, but not SHP2, is required for system activation in the absence of rgg3 under these growth conditions. They also indicate that although shp2 alone is not sufficient for system activation, SHP2 does contribute to system induction, as indicated by the timing difference between activation in the Δrgg3 and activation in the Δrgg3 shp2GGG reporter strains (Fig. 1B).
As can be noted in the replicate panels in Fig. 1B, we found that although the luminescence trend of each strain was consistent, the exact timing of the reporter induction varied slightly throughout our experiments, especially in shp mutant strains. We have found that the timing of full induction correlates, at least in part, with the age of the CDM, such that the older the CDM, the later the luminescence plateau is reached in strains that autoinduce successfully (data not shown). Unfortunately, despite efforts to optimize medium preparation and culture conditions to maximize reproducibility in our experiments, there remained variability in the timing of induction, although we were able to achieve values correlating to relative light production consistently plateauing between OD600 of 0.5 and 1.0 in the Δrgg3 mutant. Therefore, in order to make conclusions regarding system induction differences in the various strains, we included both WT and Δrgg3 reporter strains as controls in all experiments to provide reference curves against which to compare the mutant expression patterns. As such, all conclusions regarding the timing of system induction should be regarded as qualitative rather than quantitative.
sSHP2-C8 and sSHP3-C8 can independently induce the Rgg2/3 circuit with comparable specific activities.
The lack of autoinduction in the Δrgg3 shp3GGG strain but presence of autoinduction in the Δrgg3 shp2GGG strain (Fig. 1B) indicated that the two pheromones possess distinct abilities to activate the Rgg2/3 system, with SHP3 being required for system induction but not SHP2. This appeared to conflict with previous studies in which both synthetic SHP2 and SHP3 C8 peptides (sSHP2-C8 and sSHP3-C8) were able to activate the Rgg2/Rgg3 QS system in a wild-type (WT) background (6). However, in the WT background, endogenous production of both SHPs resulting from positive autoregulation of the shp genes in response to synthetic SHPs precluded the determination of independent SHP activities. Thus, we sought to examine if synthetic SHPs could independently support system activation using transcriptional reporter strains in which the translation initiation codons of shp2 and shp3 were mutated from ATG to GGG (shp2GGG, shp3GGG; BNL177 and BNL206). We reasoned that if SHP3, but not SHP2, were indeed required for activation of the system, addition of sSHP3 to a shp-null strain should result in system activation whereas addition of sSHP2 should not.
It was previously demonstrated that the C-terminal 8 amino acids of the SHP peptides comprise the active signaling molecules. Addition of either sSHP2-C8 or sSHP3-C8 to the shp-null strain carrying the Pshp2 reporter (BNL177) at a concentration of 5 nM resulted in robust luminescence expression, whereas a control peptide with the reverse sequence of SHP2-C8 (sSHP2-rev-C8) had no inducing activity (Fig. 2A). These data demonstrate that the active portions of both SHP2 and SHP3 can in fact activate the Rgg2/3 system independently and that SHP3 is not required for system induction when SHP2-C8 is provided exogenously. Of note, when the sSHP-induced luminescence profiles of the shp2GGG shp3GGG mutant were compared to those of a WT strain carrying the same reporter (BNL148), it was evident that an intact positive-feedback loop contributed to overall system activation in the WT strain. Following addition of synthetic peptide, luminescence was highly induced in both strains but declined over time in the double shp mutant whereas it remained high throughout growth in the WT reporter assay (Fig. 2A). These results confirm the importance of endogenous peptide production for sustained induction following initial system activation. Comparable luminescence expression patterns were observed in WT and shp2GGG shp3GGG strains carrying a comparably constructed Pshp3 transcriptional reporter (JCC181 and BNL206, respectively), indicating that peptide-mediated system activation is not promoter specific in nature under these conditions (data not shown).
A possible reason that the shp genes have disparate abilities to independently support autoinduction but are equivalently able to induce the system when added exogenously is that the synthetic peptide concentrations being used were greater than those natively produced by the bacteria. This could potentially mask differential peptide activities detectable only at lower peptide concentrations. Thus, we performed synthetic peptide titrations using both full-length and C8 peptides to determine if the specific induction activities of SHP2 and SHP3 were indeed different. We performed these studies using both the Pshp2 and Pshp3 reporters in the shp2GGG shp3GGG background (BNL177 and BNL206, respectively) to simultaneously determine if there were peptide-specific and/or promoter-specific differences in induction potential at the lower peptide concentrations. In agreement with our earlier synthetic peptide studies, there were no statistical differences between the effective peptide concentrations of sSHP2-C8 and sSHP3-C8 that elicited half-maximal relative luminescence induction (50% effective concentrations [EC50s]) using either reporter (Fig. 2B and C, top panels). Likewise, full-length sSHP2 and sSHP3 also had comparable EC50s with both reporters (Fig. 2B and C, bottom panels). These results further confirm that SHP2 and SHP3 are functionally redundant for system activation but fail to explain why the shp2 gene alone is incapable of supporting autoinduction in the Δrgg3 shp3GGG mutant.
Differential induction potentials of the shp genes are not due to differences in promoter strength.
During the completion of the experiments described above, we noticed that basal and maximal luminescence levels from the Pshp3 reporter were slightly but reproducibly higher than those seen with the Pshp2 reporter. This led us to investigate if differences in promoter strength could account for the differences in induction potential of the shp2 and shp3 genes. If the shp3 promoter was indeed stronger than the shp2 promoter, it could be that more SHP3 than SHP2 is produced by cells, resulting in nonequivalent abilities of the two genes to support autoinduction. To test this, we generated additional Δrgg3 strains in which one shp gene harbors the GGG start codon mutation and the second shp gene is replaced in the chromosome with the open reading frame of the first shp gene (Δrgg3, shp3GGG, Δshp2→shp3, BNL208; Δrgg3, shp2GGG, Δshp3→shp2, BNL210). These strains still only have one functional shp gene, but it is expressed from the promoter of the other shp gene. We then compared Pshp2 reporter expression in these new strains to that in the parent Δrgg3 strain and in strains with a single shp gene under the control of its native promoter. When we did this, we found that a single copy of shp2 was unable to support any detectable autoinduction regardless of whether the gene was expressed from its own promoter (Δrgg3, shp3GGG, BNL203) or the shp3 promoter (Δrgg3, shp2GGG, Δshp3→shp2, BNL220) (Fig. 3A). In contrast, a single shp3 gene successfully induced luminescence when expressed from either the native shp3 promoter (Δrgg3, shp2GGG, BNL176) or from the shp2 promoter (Δrgg3, shp3GGG, Δshp2→shp3, BNL216), both being delayed for induction compared to the Δrgg3 strain (Fig. 3A). It should be noted that shp3 was better able to induce autoinduction when expressed from its native promoter, which again indicates some low-level differences in promoter strength, in agreement with the basal luminescence levels detected from our reporters. However, whatever this difference may be does not account for the differential induction capacities of shp2 and shp3, given that shp3 is sufficient to support detectable autoinduction whereas shp2 is not, regardless of the promoter driving its expression.
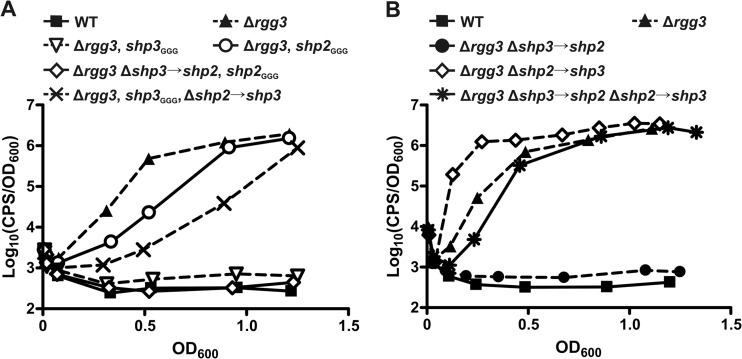
Fig 3.
The enhanced system induction potential of shp3 compared to shp2 is not a consequence of differential promoter strengths. (A) Luciferase expression from a Pshp2 reporter integrated into WT (BNL148), Δrgg3 (BNL149), Δrgg3 shp3GGG (BNL203), Δrgg3 shp2GGG (BNL176), Δrgg3 Δshp3→shp2 shp2GGG (BNL220), and Δrgg3 Δshp2→shp3 shp3GGG (BNL216) strains. (B) Luciferase expression from Pshp2 reporter integrated into WT (BNL148), Δrgg3 (BNL149), Δrgg3 Δshp3→shp2 (BNL202), Δrgg3 Δshp2→shp3 (BNL192), and Δrgg3 Δshp3→shp2 Δshp2→shp3 (BNL222) strains. Expression trends are representative of at least three independent experiments.
To further confirm that promoter strength disparities could not fully account for the disparate induction potentials of the shp genes, we generated additional strains in the Δrgg3 background in which we replaced one shp gene with a second copy of the other shp gene (Δrgg3, Δshp2→shp3, BNL187; Δrgg3, Δshp3→shp2, BNL198). We also generated a strain in which the shp genes are swapped (Δrgg3, Δshp3→shp2, Δshp2→shp3, BNL212) such that there was still one copy of each shp gene but they were each under the control of the opposite gene's promoter. We then compared reporter expression in these shp-swap strains to that in the Δrgg3 parent strain. When shp2 was replaced by shp3, resulting in a strain carrying two copies of shp3 (Δrgg3, Δshp2→shp3, BNL192), the Pshp2 reporter was induced earlier than in the parent Δrgg3 strain (BNL149) (Fig. 3B). Surprisingly, a strain carrying two copies of shp2 was unable to support autoinduction (Δrgg3, Δshp3→shp2, BNL202), further pointing to an inherent difference in the induction potentials of endogenous SHP2 and SHP3. Notably, the strain in which both shp genes were swapped (Δrgg3, Δshp3→shp2, Δshp2→shp3, BNL222) had a luminescence profile similar to that of the parent Δrgg3 strain (Fig. 3B). These data confirm that promoter strength does not account for the differential abilities of shp2 and shp3 to support autoinduction: shp2 cannot support autoinduction even when present in two copies and expressed from the shp3 promoter whereas shp3 is better able to induce the system than shp2 even when expressed from the shp2 promoter. These data also confirm that shp gene dosage contributes to system induction, at least for shp3, as two copies of shp3 resulted in premature activation of the reporter (Fig. 3B) whereas one copy of the gene resulted in delayed activation compared to the Δrgg3 strain results (Fig. 3A).
Differential induction potentials of the shp genes reside in the N termini of the peptides.
The disparate ability of shp3 but not shp2 to individually support system activation in the Δrgg3 background (Fig. 1B and 3A), and the premature reporter induction seen when shp2 was replaced by shp3 in the Δrgg3 strain versus the lack of reporter induction in a strain carrying only two copies of shp2 (Fig. 3B), conflicted with the ability of both sSHP2 and sSHP3 to induce system activation when added exogenously (Fig. 2) and led us to question if the SHP pro peptides possess distinct activities apart from the activities of the active portion of the peptides. As mentioned previously, the C-terminal active portions of SHP2 and SHP3 are highly similar whereas the N-terminal regions are much more disparate (Fig. 1A), and thus we postulated that the N-terminal regions of these peptides may have been contributing to the differences we were observing.
To examine this possibility, we generated strains in which only the C8 region of the shp gene was swapped for that of the other shp gene. Since the C8 peptides differ by only a single residue (leucine and isoleucine), this was achieved by site-directed mutagenesis of a single codon within each shp gene. The codon encoding leucine at position 17 of SHP2 was changed to encode isoleucine (shp2leu17ile), and the codon encoding isoleucine at position 18 of the SHP3 peptide was altered to encode leucine (shp3ile18leu). A strain carrying the shp2leu17ile allele will in effect produce WT SHP3 peptide from the shp3 locus and a SHP2Nterm-SHP3C8 hybrid peptide from the shp2 locus. Similarly, a strain with the shp3ile18leu allele will produce WT SHP2 from the shp2 locus and a SHP3Nterm-SHP2-C8 hybrid peptide from the shp3 locus. We again engineered these mutations in the Δrgg3 background (BNL186 and BNL225) and then introduced the Pshp2 reporter and assessed the timing of system autoinduction compared to that seen with other reporter strains.
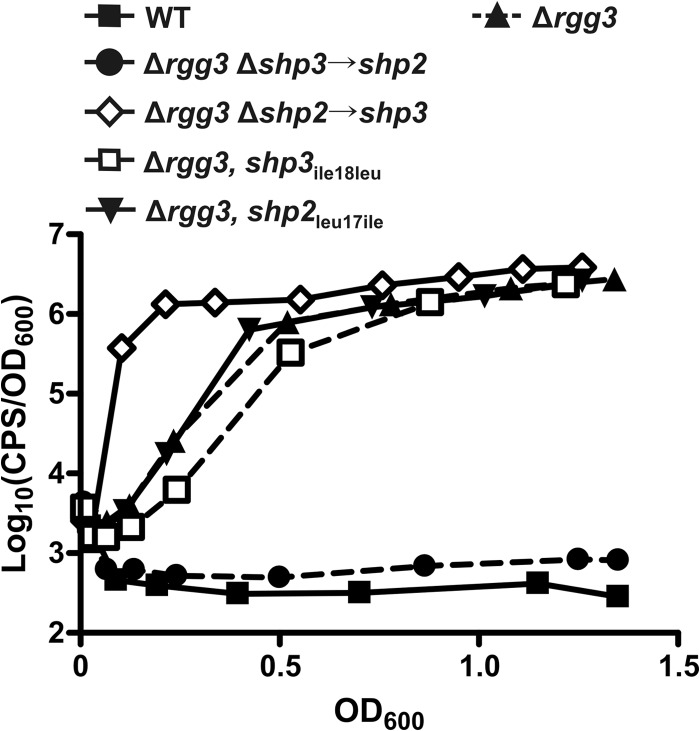
Unlike the strain carrying two copies of shp3 (Δrgg3 Δshp2→shp3, BNL192), swapping of only the C-terminal isoleucine of SHP3 (Δrgg3, shp2leu17ile, BNL196) did not result in early reporter induction (Fig. 4). It is evident that the hybrid SHP2Nterm-SHP3C8 peptide is produced, however, given that the timing of system induction in the Δrgg3 shp2leu17ile strain mirrors that of the Δrgg3 strain (Fig. 4) and is not delayed as in the Δrgg3 shp2GGG strain (Fig. 1B). Likewise, replacement of the C-terminal isoleucine of SHP3 with leucine (Δrgg3, shp3ile18leu, BNL227) failed to abolish reporter induction, unlike the case when the entire shp3 gene was replaced with shp2 (Δrgg3 Δshp3→shp2, BNL202) (Fig. 4). The cause of the slight delay in the Δrgg3 shp3ile18leu strain compared to the Δrgg3 parent strain remains unclear. Overall, these data indicate that the N-terminal portions of the SHPs are responsible for the differential induction activities of shp2 and shp3, as swapping of only the active C-terminal region does not reproduce the luminescence profiles seen when the entire shp gene is swapped. These results are the first evidence that the N-terminal, nonactive portions of the endogenously produced SHP peptides factor into their native signaling potentials.
Fig 4.
The N-terminal regions of the peptides confer the differential system induction potentials of shp2 and shp3. Data represent luciferase expression from a Pshp2 reporter integrated into WT (BNL148), Δrgg3 (BNL149), Δrgg3 Δshp3→shp2 (BNL202), Δrgg3 Δshp2→shp3 (BNL192), Δrgg3 shp3ile18leu (BNL227), and Δrgg3 shp2leu17ile (BNL196) strains. Expression trends are representative of at least three independent experiments.
DISCUSSION
It has become evident that peptide-mediated QS systems have diverse circuit architectures, with most systems utilizing only a single peptide pheromone but others requiring multiple signals that interact with a single or multiple receptor proteins. Clear examples of the latter are PrgX, which interacts with peptides cCF10 and iCF10 (13–15), and the more recently discovered Rgg2 and Rgg3 regulators that each interact with both SHP2 and SHP3 (6, 10). The previous evidence of redundancy in SHP stimulatory activities (6, 10) and the conservation of both shp genes in all sequenced GAS genomes led us to investigate the roles of SHP peptide specificity and signal dosage in the Rgg2/3 QS system. Our genetic studies have led to three general conclusions. The mature SHP2 and SHP3 peptides are redundant in their ability to activate the Rgg2/3 system (Fig. 2). Separately, shp gene dosage contributes to the timing of Rgg2/3 system induction, with a reduced gene copy number resulting in delayed system induction (Fig. 1B and 3). Lastly, and perhaps most intriguingly, full-length endogenous SHP2 and SHP3 possess distinct potentials for supporting system activation independently of their highly similar C-terminal active regions (Fig. 4). Overall, these data support our hypothesis that both SHP identity and signal dosage contribute to activation of the Rgg2/3 system.
The data presented herein represent the first evidence that the noneffector, N-terminal regions of SHP2 and SHP3 influence their overall system activation potential in GAS. As mentioned above, the N-terminal regions of SHP2 and SHP3 are much more divergent than the nearly identical C-terminal region which contains the active signaling portion of the molecule (Fig. 1A). Although not directly involved in activation of their cognate receptors, the N-terminal regions of signaling peptides could be imagined to indirectly affect signaling potential at several steps of the circuit, namely, SHP maturation and/or export, and we hypothesize that the differential activation potentials of shp2 and shp3 indeed result from one or both of these steps. The possibility cannot be ruled out, however, that the production differences between the SHPs occur at the mRNA level rather than the peptide level, perhaps due to disparate translation efficiencies of the N termini or differential interactions of small RNAs with the shp mRNAs. Additional studies will be needed to address these possibilities.
Very little is known regarding processing of SHP peptides. Evidence from our laboratory supports the idea of a prominent role for the membrane-associated metalloprotease Eep in the Rgg2/3 system; however, Eep was not essential for activation of the system when SHP3 was overexpressed, indicating that additional enzymes can participate in processing of at least SHP3 under certain conditions (6). Redundancy in peptide processing may be a conserved trend in intracellular peptide signaling systems, as is the case in B. subtilis, where three proteases (subtilisin, Epr, and Vpr) have been implicated in the maturation of Phr peptides (25). In E. faecalis, Eep has been demonstrated to function in the maturation of both cCF10 and iCF10 from their precursor molecules (26, 27), but cCF10 production is additionally believed to involve signal II peptidase and a yet-to-be-identified carboxy peptidase which are required to fully separate the mature signaling peptide from its lipoprotein precursor (27). Given that Eep recognizes determinants in E. faecalis in the N-terminal, nonsignal portion of signal precursor molecules (27), it is plausible that the disparate N termini of SHP2 and SHP3 are differentially recognized by Eep, resulting in superior processing of SHP3 compared to SHP2. However, the possibility cannot be excluded that native processing of SHP2 and that of SHP3 are performed by different and/or multiple proteolytic enzymes. Genetic and biochemical experiments are under way to examine native SHP processing, assess potential SHP substrate specificity corresponding to Eep, and identify the potential secondary pathway that facilitates processing of SHP3 when overexpressed in the absence of Eep.
Identification of the mechanism(s) by which cognate peptides of Rgg regulators are exported from the cell also remains elusive. The amino acid sequences of SHP2 and SHP3 (Fig. 1A) resemble only vaguely the general signal peptides used by Gram-positive bacteria to target proteins for export via Sec (28), and online prediction algorithms designed to identify Sec-targeting sequences disagree as to whether SHP2 and/or SHP3 are Sec substrates, depending on the program being used (data not shown). Regardless of the identity of the export system, be it Sec or a designated ABC transporter, it is possible that the N-terminal regions of the SHP peptides result in unequal degrees of recognition by the systems, thereby resulting in preferential export of one peptide, presumably SHP3, over the other. It will be interesting to determine how SHP peptides are exported from the cell, the sequence determinants that facilitate recognition by the export system, and if export is coupled to peptide maturation or if these are temporally and/or spatially distinct steps in the circuit.
With regard to the functional redundancy of SHP2 and SHP3 in system activation, it should be noted that the EC50s for the full-length synthetic peptides were 50 to 140 times less than those for their C8 counterparts when tested for induction activity in a shp-null background. Additionally, the maximum induction by C8 peptides was always higher than that for the full-length peptides (Fig. 2B and C). Given that GAS secretes numerous proteases (29), it is possible that the activation by exogenously added, full-length synthetic SHPs results from some degree of nonspecific but productive peptide cleavage by proteases not normally involved in SHP peptide maturation. Such nonspecific cleavage would likely also result in the nonproductive degradation of a percentage of the added sSHPs, thereby reducing the effective peptide concentration available for system induction and resulting in higher effective EC50s. The C8 peptides, which presumably do not require any additional processing steps to be active, may be less-optimal substrates for these proteases or may be imported into the cell more efficiently, resulting in overall lower EC50s. Hopefully, further elucidation of SHP maturation will help address such uncertainties.
On a final note, we find it interesting that GAS has evolved to utilize two Rgg-SHP pairs in a single circuit when other systems appear to be fully functional with just one. It is appealing to speculate that having two peptide-encoding genes with disparate activation potentials confers some type of advantage for the bacteria, especially given that both pairs are conserved among all sequenced GAS strains. One possibility is that it is energetically costly to have the system in an “on” state, and requiring two peptides to be sufficiently produced prior to robust induction may help prevent premature system activation. Another possibility is that having multiple peptides driven by separate promoters could allow more regulatory inputs into the system than if just one shp gene were involved. Although the conserved Rgg binding site is highly conserved in the two shp promoters, the remainder of the intergenic regions between each shp and rgg gene is highly divergent (10) and could support differential regulation by other regulatory proteins. There is evidence supporting this idea, at least in part, as the metal-dependent regulator MtsR is known to regulate the shp3 promoter but is not known to have any role in shp2 regulation (30). Taking this idea farther, if regulatory inputs are indeed distinct at the two shp promoters, it may be that shp2 is in fact sufficient to support autoinduction under specific, yet-to-be determined conditions despite not having sufficient activity under the growth conditions used for these studies. Differential regulation of processing enzymes and/or export machinery in response to cellular and/or environmental stimuli may also be a factor in determining if and when the Rgg2/3 system is activated, further expanding the levels at which other signals could influence this system.
ACKNOWLEDGMENTS
Support for this work was provided by the NIH, grant AI091779, and the Burroughs Wellcome Fund.
Footnotes
Published ahead of print 19 July 2013
REFERENCES
- 1.Stephenson S, Mueller C, Jiang M, Perego M. 2003. Molecular analysis of Phr peptide processing in Bacillus subtilis. J. Bacteriol. 185:4861–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, Gray L, Novick RP, Ji G. 2002. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus. J. Biol. Chem. 277:34736–34742 [DOI] [PubMed] [Google Scholar]
- 3.Thoendel M, Horswill AR. 2010. Biosynthesis of peptide signals in gram-positive bacteria. Adv. Appl. Microbiol. 71:91–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Declerck N, Bouillaut L, Chaix D, Rugani N, Slamti L, Hoh F, Lereclus D, Arold ST. 2007. Structure of PlcR: Insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria. Proc. Natl. Acad. Sci. U. S. A. 104:18490–18495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocha-Estrada J, Aceves-Diez AE, Guarneros G, de la Torre M. 2010. The RNPP family of quorum-sensing proteins in Gram-positive bacteria. Appl. Microbiol. Biotechnol. 87:913–923 [DOI] [PubMed] [Google Scholar]
- 6.Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. 2011. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog. 7:e1002190. 10.1371/journal.ppat.1002190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleuchot B, Gitton C, Guillot A, Vidic J, Nicolas P, Besset C, Fontaine L, Hols P, Leblond-Bourget N, Monnet V, Gardan R. 2011. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol. Microbiol. 80:1102–1119 [DOI] [PubMed] [Google Scholar]
- 8.Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192:1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim M, Guillot A, Wessner F, Algaron F, Besset C, Courtin P, Gardan R, Monnet V. 2007. Control of the transcription of a short gene encoding a cyclic peptide in Streptococcus thermophilus: a new quorum-sensing system? J. Bacteriol. 189:8844–8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lasarre B, Aggarwal C, Federle MJ. 2013. Antagonistic Rgg regulators mediate quorum sensing via competitive DNA binding in Streptococcus pyogenes. mBio 3:e00333–12. 10.1128/mBio.00333-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78:589–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mashburn-Warren L, Morrison DA, Federle MJ. 2012. The cryptic competence pathway in Streptococcus pyogenes is controlled by a peptide pheromone. J. Bacteriol. 194:4589–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori M, Sakagami Y, Ishii Y, Isogai A, Kitada C, Fujino M, Adsit JC, Dunny GM, Suzuki A. 1988. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J. Biol. Chem. 263:14574–14578 [PubMed] [Google Scholar]
- 14.Nakayama J, Ruhfel RE, Dunny GM, Isogai A, Suzuki A. 1994. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J. Bacteriol. 176:7405–7408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi K, Brown CK, Gu ZY, Kozlowicz BK, Dunny GM, Ohlendorf DH, Earhart CA. 2005. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc. Natl. Acad. Sci. U. S. A. 102:18596–18601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunny GM, Johnson CM. 2011. Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Curr. Opin. Microbiol. 14:174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howell-Adams B, Seifert HS. 2000. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol. Microbiol. 37:1146–1158 [DOI] [PubMed] [Google Scholar]
- 18.McShan WM, Ferretti JJ, Karasawa T, Suvorov AN, Lin S, Qin B, Jia H, Kenton S, Najar F, Wu H, Scott J, Roe BA, Savic DJ. 2008. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J. Bacteriol. 190:7773–7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cook LC, LaSarre B, Federle MJ. 2013. Interspecies communication among commensal and pathogenic streptococci. mBio 4():e00382-13. 10.1128/mBio.00382-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Degnan BA, Fontaine MC, Doebereiner AH, Lee JJ, Mastroeni P, Dougan G, Goodacre JA, Kehoe MA. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68:2441–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 70:6076–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McShan WM, McLaughlin RE, Nordstrand A, Ferretti JJ. 1998. Vectors containing streptococcal bacteriophage integrases for site-specific gene insertion. Methods Cell Sci. 20:51–57 [Google Scholar]
- 24.Bachmann H, Kleerebezem M, van Hylckama Vlieg JE. 2008. High-throughput identification and validation of in situ-expressed genes of Lactococcus lactis. Appl. Environ. Microbiol. 74:4727–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanigan-Gerdes S, Dooley AN, Faull KF, Lazazzera BA. 2007. Identification of subtilisin, Epr and Vpr as enzymes that produce CSF, an extracellular signalling peptide of Bacillus subtilis. Mol. Microbiol. 65:1321–1333 [DOI] [PubMed] [Google Scholar]
- 26.An FY, Sulavik MC, Clewell DB. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 181:5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandler JR, Dunny GM. 2008. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J. Bacteriol. 190:1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Wely KH, Swaving J, Freudl R, Driessen AJ. 2001. Translocation of proteins across the cell envelope of Gram-positive bacteria. FEMS Microbiol. Rev. 25:437–454 [DOI] [PubMed] [Google Scholar]
- 29.Cole JN, Barnett TC, Nizet V, Walker MJ. 2011. Molecular insight into invasive group A streptococcal disease. Nat. Rev. Microbiol. 9:724–736 [DOI] [PubMed] [Google Scholar]
- 30.Toukoki C, Gold KM, McIver KS, Eichenbaum Z. 2010. MtsR is a dual regulator that controls virulence genes and metabolic functions in addition to metal homeostasis in the group A streptococcus. Mol. Microbiol. 76:971–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon D, Ferretti JJ. 1991. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol. Lett. 66:219–224 [DOI] [PubMed] [Google Scholar]