Abstract
The purple sulfur bacterium Allochromatium vinosum DSM 180T is one of the best-studied sulfur-oxidizing anoxygenic phototrophic bacteria, and it has been developed into a model organism for laboratory-based studies of oxidative sulfur metabolism. Here, we took advantage of the organism's high metabolic versatility and performed whole-genome transcriptional profiling to investigate the response of A. vinosum cells upon exposure to sulfide, thiosulfate, elemental sulfur, or sulfite compared to photoorganoheterotrophic growth on malate. Differential expression of 1,178 genes was observed, corresponding to 30% of the A. vinosum genome. Relative transcription of 551 genes increased significantly during growth on one of the different sulfur sources, while the relative transcript abundance of 627 genes decreased. A significant number of genes that revealed strongly enhanced relative transcription levels have documented sulfur metabolism-related functions. Among these are the dsr genes, including dsrAB for dissimilatory sulfite reductase, and the sgp genes for the proteins of the sulfur globule envelope, thus confirming former results. In addition, we identified new genes encoding proteins with appropriate subcellular localization and properties to participate in oxidative dissimilatory sulfur metabolism. Those four genes for hypothetical proteins that exhibited the strongest increases of mRNA levels on sulfide and elemental sulfur, respectively, were chosen for inactivation and phenotypic analyses of the respective mutant strains. This approach verified the importance of the encoded proteins for sulfur globule formation during the oxidation of sulfide and thiosulfate and thereby also documented the suitability of comparative transcriptomics for the identification of new sulfur-related genes in anoxygenic phototrophic sulfur bacteria.
INTRODUCTION
Anoxygenic purple sulfur bacteria like the gammaproteobacterium Allochromatium vinosum, a member of the family Chromatiaceae, are capable of growth as photolithoautotrophs. Sunlight is the primary energy source, electrons are obtained from reduced sulfur compounds, i.e., an inorganic source, and cellular carbon is obtained via reductive carbon dioxide fixation. An understanding of the biological processes involved in sulfur oxidation is of major interest, since purple sulfur bacteria flourish wherever light reaches sulfidic water layers or sediments and often occur as dense accumulations in conspicuous blooms in freshwater as well as in marine aquatic ecosystems. Here, we have focused on their role as major players in the reoxidation of sulfide produced by sulfate-reducing bacteria in deeper anoxic layers.
Most of our knowledge about the oxidation of reduced sulfur compounds in anoxygenic purple sulfur bacteria comes from enzyme assays and sequence analysis of specific gene clusters in Allochromatium vinosum DSM 180T. The organism is relatively easy to handle and has become a laboratory model organism for the investigation of sulfur oxidation pathways. A. vinosum is genetically accessible (1, 2), its complete genomic sequence is known (3; see also http://www.ncbi.nlm.nih.gov/), and a system for complementation of mutations is available (4–6). Characterization of mutant strains carrying sulfur-related gene insertions or deletions has allowed development of hypotheses for the sulfur oxidation pathways, not only in this organism but also in many other sulfur-oxidizing bacteria (7).
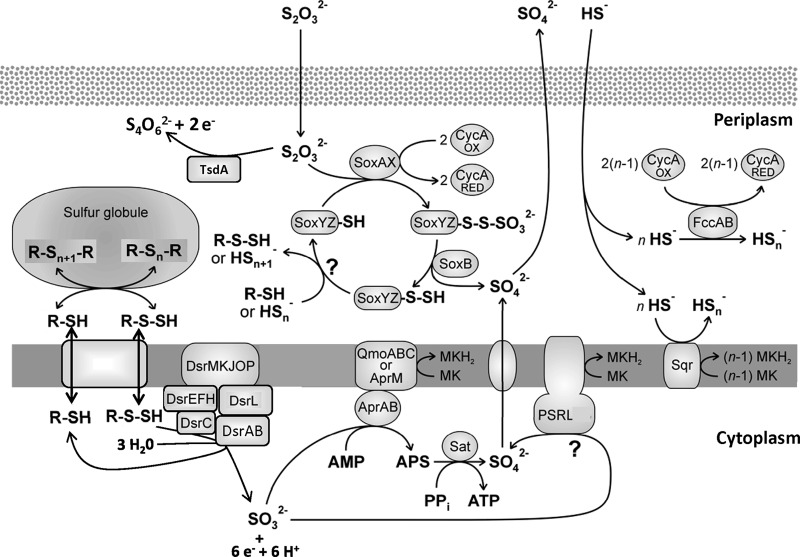
The substrates used by purple sulfur bacteria include primarily sulfide, polysulfides, and elemental sulfur (8). A. vinosum is an especially metabolically versatile purple sulfur bacterium that can also utilize thiosulfate and sulfite. In addition, the organism can also grow independently of reduced sulfur compounds. During photoorganoheterotrophic growth, organic acids like malate are suitable substrates. Growth on sugars is not possible (3, 8). Detailed biochemical characterization of enzymes and proteins in combination with molecular genetics and mutation analyses have demonstrated that all sulfane sulfur regardless of whether it stems originally from sulfide, polysulfides, elemental sulfur, or thiosulfate enters the sulfur oxidation pathway via the formation of sulfur globules (Fig. 1) (7). These reside in the bacterial periplasm and are enclosed by an envelope consisting of at least three different hydrophobic proteins, SgpA, SgpB, and SgpC (9). A. vinosum possesses the genetic capacity to form at least three different enzymes that participate in the oxidation of sulfide, namely, the periplasmic flavocytochrome c and the membrane-bound sulfide:quinone-oxidoreductases SqrD and SqrF, which are both predicted to be oriented toward the periplasm (3, 10, 11). Notably, SqrD is present in purple sulfur bacteria that produce sulfur globules in the periplasm, while it appears to be absent in those species that exclusively produce extracellular sulfur globules. In addition, SqrF has been reported to be important for growth of the green sulfur bacterium Chlorobaculum tepidum at high sulfide concentrations (11). For the oxidation of thiosulfate to sulfate, A. vinosum utilizes the Sox proteins SoxYZ, SoxB, SoxXAK, and SoxL (6, 12). Under slightly acidic conditions, thiosulfate can also be oxidized to tetrathionate via the diheme cytochrome c thiosulfate dehydrogenase (4, 6). For further oxidation of stored sulfur, the Dsr system, including dissimilatory sulfite reductase (DsrAB), is of essential importance, as evidenced by several A. vinosum dsr mutants that are unable to degrade sulfur globules (13–16). Detection of all enzymes necessary for the oxidation of sulfite, the product of the Dsr pathway, to sulfate has still not been achieved. Indirect oxidation via adenosine-5′-phosphosulfate (APS) catalyzed by APS reductase and ATP sulfurylase is a well-described pathway; however, it is neither generally present nor essential (2, 7, 17).
Fig 1.
Current model of sulfur oxidation in purple sulfur bacteria. The figure is modified from that of Frigaard and Dahl (7) and used here with permission of the publisher.
Besides these comparatively well-described sulfur oxidation systems, A. vinosum contains the genetic information for several rhodaneses, sulfur relay proteins, and polysulfide reductase-like proteins with unknown function, and approximately 30% of the A. vinosum genes encode hypothetical proteins with unknown function (3). In addition, uptake and oxidation of externally supplied elemental sulfur by A. vinosum and other purple sulfur bacteria as well as by green sulfur bacteria are still unresolved (18). Hence, it is obvious that many questions remain open, not only concerning oxidative sulfur metabolism but also with regard to general adaptations to various environmental conditions by purple sulfur bacteria.
We now seek to provide a more comprehensive and coherent picture of sulfur oxidation and bioenergetic processes in A. vinosum. To this end, we set out to apply DNA microarray technology and provide whole-genome transcriptional profiling of the response of A. vinosum to the presence of four different reduced sulfur compounds, i.e., sulfide, elemental sulfur, thiosulfate, and sulfite. This allowed us not only to confirm former results but also to identify new genes encoding proteins with appropriate subcellular localization and properties to participate in oxidative dissimilatory sulfur metabolism. The four genes that exhibited the strongest increases of relative mRNA levels on sulfide and elemental sulfur were chosen for inactivation and phenotypic analyses, respectively, of the corresponding mutant strains. This approach verified the importance of the encoded proteins for the formation of sulfur globules during the oxidation of sulfide and thiosulfate and thereby also documented the suitability of comparative transcriptomics for the identification of new sulfur-related genes in anaerobic phototrophic sulfur bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table S1 of the supplemental material. Allochromatium vinosum DSM 180T wild-type and mutant cells were cultivated photolithoautotrophically in batch culture at 30°C under anaerobic conditions and with continuous illumination either in completely filled screw-cap culture bottles or in thermostatted glass fermentors (culture volume, 1 liter) containing modified Pfennig's medium (referred to here as 0 medium, without sulfide) (6). The concentration of ammonium chloride was set to 1.2 g liter−1. Sulfide (4 mM, 6 mM, or 8 mM), thiosulfate (10 mM), sulfite (5 mM), or sulfur (50 mM) was added to the cultures as the sulfur source. For photoorganoheterotrophic growth on malate with sulfate as the sole sulfur source, MgCl2 was replaced by MgSO4. To maintain a pH of 7.0 during growth in fermentors, sterile HCl (0.5 M) and Na2CO3 (0.5 M) solutions were added automatically. Cells of A. vinosum, grown photoorganoheterotrophically on malate (RCV medium [19]) for 3 days, were used as the inoculum for growth experiments. The culture volume of the precultures was 500 ml. Inoculum cells were harvested by centrifugation (10 min, 2,680 × g) and washed once in 0 medium. For DNA microarray experiments, cells were cultivated in a volume of 100 ml RCV or 0 medium supplemented with the respective sulfur source. These cultures were inoculated with stationary-phase cells taken from 100 ml of preculture. The culture volume for phenotypic analyses of wild-type and mutant strains was 250 ml. In these experiments, for phenotypic characterization, the starting optical density at 690 mm (OD690) was set to 0.8 to 0.9. Escherichia coli strains were cultured in LB medium (20). Antibiotics were used at the following concentrations (in μg ml−1): for E. coli, ampicillin at 100, kanamycin at 50; for A. vinosum, ampicillin at 20, kanamycin at 10, rifampin at 50.
Recombinant DNA techniques.
Standard molecular techniques were used (20, 21). Cloning experiments were carried out in E. coli DH5α (see Table S1 in the supplemental material). Chromosomal DNA of A. vinosum was obtained by Sarkosyl lysis (22) and purified by phenol-chloroform extraction and dialysis against water. Southern hybridizations were performed overnight at 68°C as described previously (23). PCR with Taq DNA polymerase was done as described previously (2), and PCR with mutagen primers and using Pfu DNA polymerase to insert restriction sites was performed according to the protocol supplied by Stratagene (La Jolla, CA). DNA probes for Southern hybridizations were digoxigenin labeled by PCR (24).
Construction and analysis of A. vinosum mutant strains.
For the replacement of Alvin_1196 and _1197, Alvin_1468, Alvin_2093, and Alvin_3072, respectively, in the genome of A. vinosum with a kanamycin cassette, splicing by overlap extension-PCR fragments were constructed using primers Avin1196-97Del1 to -4, Del-1468_1 to _4, Del-2093_1 to _4, and Del-3072_1 to _4, respectively (see Table S1 in the supplemental material). Each fragment was inserted into the mobilizable plasmid pSUP301 (25) via HindIII restriction sites, resulting in plasmids pSUP301-Del1196/97, pSUP301-Del1468, pSUP301-Del2093, and pSUP-Del3072. The kanamycin cassette of plasmid pHP45ΩKm was integrated into these plasmids via EcoRI restriction sites. The final mobilizable constructs pSUP301-Del1196/97Km, pSUP301-Del1468Km, pSUP301-Del2093Km, and pSUP301-Del3072Km were transferred from E. coli S17.1 to A. vinosum Rif50 by conjugation (1). Transconjugants were selected on RCV plates containing the appropriate antibiotics under anoxic conditions in the light. Double-crossover recombinants lost the vector-carried ampicillin resistance. Finally, the genotypes of double-crossover recombinants were verified in Southern hybridization experiments.
Analysis of sulfur compounds.
For determination of elemental sulfur according to the methods described by Bartlett and Skoog (26), a cell pellet containing up to 200 nmol sulfur was resuspended in 200 μl distilled water (dH2O), and 100 μl of a 0.2 M sodium cyanide solution was added. The mixture was incubated for 10 min at 100°C, followed by the addition of 650 μl dH2O and 50 μl ferric nitrate reagent [30 g Fe(NO3)3 · 9H2O, 40 ml 55% HNO3, and 100 ml dH2O]. The mixture was centrifuged (16,000 × g, 2 min), and the absorption at 460 nm was measured against that of a reagent blank. A calibration curve was recorded for sodium thiocyanate. Thiosulfate and tetrathionate were measured via cyanolysis, and sulfite was measured by the fuchsin method as described by Dahl (2). Determination of sulfate was done according to the method described by Sörbo (27).
DNA microarrays.
Custom DNA microarrays were obtained from Agilent Technologies (Waldbronn, Germany). Agilent's eArray platform was used to design oligonucleotide probes and assemble the custom 4-by-44,000 60-mer microarray. For genome-wide gene expression analysis of A. vinosum, the NCBI cDNA sequences from the annotation of the genome (NC_013851), as well as plasmid pALVIN01 (NC_013852) and plasmid pALVIN02 (NC_013862), listing the protein-coding genes and the structural RNA-coding genes, were used as input in an eArray to design one 60-mer probe for each gene and using the best probe methodology. For A. vinsoum, 3,340 specific oligonucleotide probes for the annotated genes were designed by using eArray. The custom array design also included specific oligonucleotide probes from other bacterial genomes not relevant to this study and also Agilent's control spots.
RNA preparation and cDNA synthesis.
Total RNA of A. vinosum strain Rif50 (16) was isolated from cells grown photoorganoheterotrophically on 22 mM malate or photolithoautotrophically on 50 mM sulfur, 4 mM sulfide, 10 mM thiosulfate, or 5 mM sulfite in 100-ml culture bottles, based on a procedure described for Gram-negative bacteria (21). Briefly, cells of two culture aliquots (35 ml each) were harvested (12,800 × g, 10 min, 4°C), and each pellet was resuspended in 350 μl RLT buffer (Qiagen, Hilden, Germany) containing 10 mM dithiothreitol (DTT) and disrupted by vortexing (Silamat S 6; Ivoclar Vivadent AG, Schaan, Lichtenstein) by using 0.1-mm zirconia/silica beads (Roth, Karlsruhe, Germany). Cell debris and glass beads were sedimented (15,000 × g, 2 min, 4°C), and the supernatant was mixed with 500 μl phenol (1 min), followed by the addition of 500 μl chloroform and a further mixing step. After centrifugation (10,000 × g, 10 min, 4°C), RNA of the supernatant was precipitated by the addition of a 1/10 volume of 3 M sodium acetate (pH 6) and two volumes of ice-cold ethanol. Precipitated RNA was collected by centrifugation (10,000 × g, 10 min, 4°C), and the pellets generated from 70-ml cultures were resuspended in 100 μl RNase-free water. The RNA was further purified using the RNAeasy total RNA kit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. The RNA concentration was measured photometrically with an Analytic Jena Specord 210 spectrophotometer (20).
For synthesis of fluorescently labeled cDNA with the fluorescent nucleotide analogues Cy3-dCTP and Cy5-dCTP (GE Healthcare, Freiburg, Germany), 25 μg of RNA and 500 ng of random hexamer primers were used. The reaction mixture (30 μl) contained 3 μl 1 mM Cy3-dCTP or Cy5-dCTP, 3 μl 0.1 M DTT, 6 μl 5× first-strand buffer (Invitrogen, Karlsruhe, Germany), 0.6 μl deoxynucleoside triphosphate mix (25 mM [each] dATP, dUTP, and dGTP and 10 mM dCTP), and 2 μl SuperScript II reverse transcriptase (Invitrogen, Darmstadt, Germany). The mixture was incubated for 10 min at room temperature, 110 min at 42°C, then stopped by the addition of 10 μl 0.1 N NaOH, incubated for 10 min at 70°C, and then neutralized by the addition of 10 μl 0.1 N HCl. cDNA samples were purified by washing three times with water on a Microcon column (YM-30; Millipore).
Hybridization of DNA microarrays and data analysis.
Purified cDNA samples to be compared were pooled, and the prepared two-color samples were hybridized at 65°C while rotating for 17 h using Agilent's gene expression hybridization kit, hybridization oven, and hybridization chamber. After hybridization, the arrays were washed using Agilent's wash buffer kit according to the instructions of the manufacturer. Fluorescence of hybridized DNA microarrays was determined at 532 nm (Cy3) and 635 nm (Cy5) at 5-μm resolution with a GenePix 4000B laser scanner and GenePix Pro 6.0 software (Molecular Devices, Sunnyvale, CA). Fluorescence images were saved to raw data TIFF files (GenePix Pro 6.0). Quantitative TIFF image analysis was carried out using GenePix image analysis software, and results were saved as GPR files (GenePix Pro 6.0). For background correction of spot intensities, ratio calculations, and ratio normalizations, GPR files were processed using the BioConductor R-packages limma and marray (http://www.bioconductor.org). For further analysis, the processed and loess-normalized data as well as detailed experimental information according to MIAME (28) were stored in the in-house DNA microarray database of the institute IBG-1: Biotechnology of the Forschungszentrum Jülich GmbH (29) and were also deposited in NCBI's Gene Expression Omnibus database. The DNA microarray analysis was repeated independently at least three times with biological replicates. To search the data for differentially expressed genes based on the normalized Cy5/Cy3 ratio, which reflects the relative RNA level change, the criteria flags of a normalized Cy5/Cy3 ratio of ≥0 (via GenePix Pro 6.0) and signal/noise ratio of ≥3 for Cy5 (F635 [median]/B635 [median], where F635 represents the fluorescence at 635 nm and B635 represents the background at 635 nm) or Cy3 (F532 [median]/B532 [median]) were used. If both signal/noise ratios were <3, then signals were considered too weak to conclude that the RNA level changed. Furthermore, P values were calculated by using a paired Student t test (Microsoft Excel) to compare the relative RNA levels of a gene in the replicates to the relative RNA levels of all other genes in the replicates.
The genome of A. vinosum contains 3,366 genes, all of which were represented on the DNA microarray chips used for the experiments. After normalization and filtering of signal/noise ratios of ≥3 in the elemental sulfur experiment 3,027 gene-representing spots could be evaluated at least three times in four replicates. In the sulfide, thiosulfate, and sulfite experiments, 2,644, 2,713, and 3,185 gene-representing spots, respectively, could be evaluated at least twice in three replicates. The interactive tool VENNY for comparing lists with Venn diagrams (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was applied to identify specific groups of genes, using an RNA level change threshold of 2-fold or more.
Expression studies based on qRT-PCR.
For quantitative real-time PCR (qRT-PCR) experiments, total RNA was isolated from A. vinosum cells grown in thermostatted glass fermentors using the same procedure described above. RNA samples of 100 ng were used for qRT-PCR analysis via the QuantiTect SYBR green RT-PCR kit (Qiagen, Hilden, Germany) and the iCycler iQ real-time detection system (Bio-Rad, Munich, Germany) according to the instructions of the manufacturers. Control reactions with reverse transcriptase omitted were performed for each RNA sample. Approximately 100-bp fragments of Alvin_1196 and Alvin_1197, which are nearly identical on the nucleotide level, and approximately 150-bp fragments of Alvin_0258, Alvin_2600, Alvin_2601, Alvin_3028, and the reference locus, Alvin_0486, encoding a uroporphyrinogen decarboxylase (Uro-D), were amplified (see Table S1 in the supplemental material) with an annealing temperature of 58°C. The qRT-PCR conditions were as follows: 30 min at 50°C (reverse transcription using random nonamer primers), 15 min at 95°C (inactivation of the reverse transcriptase and activation of the polymerase), 40 cycles of 15 s at 94°C, 30 s at 58°C, and 30 s at 72°C, followed by melting curve analysis, in which the temperature was increased every 10 s by 1°C, from a start at 35°C to 95°C. The samples were automatically quantified with the iCycler iQ software (Bio-Rad, Munich, Germany).
Microarray data accession number.
The DNA microarray data were deposited in NCBI's Gene Expression Omnibus database under accession number GSE44042.
RESULTS AND DISCUSSION
Transcript profiling using DNA microarrays.
In this study, the relative genomic expression profiles of A. vinosum DSM 180T growing photolithoautotrophically on different sulfur compounds were determined and compared to those of cells grown photoorganoheterotrophically on malate at exactly the same light intensity. The malate-containing medium was supplied with 0.815 mM sulfate in order to satisfy the sulfur requirement for biosynthesis of sulfur-containing cell constituents. Three independent cultures each, grown on sulfide, thiosulfate, or sulfite, were harvested 1 h, 2 h, or 7 h after inoculation, respectively. When elemental sulfur was the substrate, four independent cultures were harvested 3 h after inoculation. The incubation periods chosen corresponded to those after which A. vinosum exhibits maximum stable oxidation rates for the respective provided sulfur source (2, 6, 13, 18). Doubling times for A. vinosum wild type are in the range of 7 to 10 h in the exponential phase of photoorganoheterotrophic and photolithoautotrophic growth in batch culture (2, 30) (see Table S4 in the supplemental material). Accordingly, appreciable growth of the cells had not occurred in any of the cultures at the time of transcript analysis. We thus ensured similar physiological states for all cultures compared and minimized effects that could be exerted by different growth rates.
Relative mRNA levels of 206 genes on sulfide, 104 on thiosulfate, 233 on elemental sulfur, and 253 on sulfite increased at least 2-fold. On the other hand, relative mRNA levels of 166 genes on sulfide, 125 on thiosulfate, 353 on elemental sulfur, and 171 on sulfite decreased at least 2-fold (see Fig. S1 in the supplemental material). Relative mRNA levels of about 100 genes (for each substrate) were specifically enhanced above the threshold on sulfide, elemental sulfur, or sulfite. In contrast, relative mRNA levels of only 20 genes were specifically increased when thiosulfate was the substrate (see Fig. S1b). We defined a strict threshold for especially strongly affected genes and compiled all the genes for which relative mRNA levels changed 5-fold or more on at least one of the sulfur compounds or a minimum of 2-fold on all four tested sulfur compounds (Tables 1 and 2). Results for genes known to be involved in dissimilatory sulfur metabolism are outlined in Table 3.
Table 1.
Differentially expressed genes in A. vinosum after growth on different reduced sulfur compounds in comparison with growth on malatea
| Locus tag | Gene | Annotation | Strand | Avg fold change after growth onb: |
|||
|---|---|---|---|---|---|---|---|
| Sulfide | Thiosulfate | Sulfur | Sulfite | ||||
| Alvin_0040 | atpB | ATP synthase F0, A-subunit | + | 0.93 | 0.74 | 0.17 | 0.35 |
| Alvin_0041 | atpE | ATP synthase F0, C-subunit | + | 1.17 | 0.79 | 0.17 | 0.50 |
| Alvin_0042 | atpF | ATP synthase F0, B-subunit | + | 1.13 | 0.87 | 0.20 | 0.44 |
| Alvin_0043 | atpH | ATP synthase F1, δ-subunit | + | 1.15 | 1.18 | 0.12 | 0.44 |
| Alvin_0044 | atpA | ATP synthase F1, α-subunit | + | 1.30 | 1.14 | 0.20 | 0.61 |
| Alvin_0629 | α/β-Hydrolase fold protein | − | 4.02 | 0.61 | 0.15 | 1.01 | |
| Alvin_0741 | sufD | FeS assembly protein SufD | + | 0.20 | 0.47 | 4.12 | 1.30 |
| Alvin_0742 | FeS assembly SUF system protein | + | 0.20 | 0.51 | 4.12 | 1.46 | |
| Alvin_0852 | TonB-dependent vitamin B12 receptor | − | 3.18 | 0.61 | 0.19 | 0.65 | |
| Alvin_0913 | gcvP | Glycine dehydrogenase (decarboxylating) | − | 0.43 | 0.35 | 0.40 | 0.44 |
| Alvin_0982 | rpmI | Ribosomal protein L35 | + | 1.06 | 0.51 | 0.19 | 0.49 |
| Alvin_1093 | fccA | Cytochrome c class I | − | 0.13 | 1.99 | 0.56 | 1.39 |
| Alvin_1095 | NapC/NirT cytochrome c domain protein | − | 0.08 | 1.27 | 0.80 | 0.90 | |
| Alvin_1196 | Hypothetical protein Alvin_1196 | + | 0.03 | 1.42 | 98.00 | 3.95 | |
| Alvin_1197 | Hypothetical protein Alvin_1197 | + | 0.03 | 1.37 | 116.63 | 3.88 | |
| Alvin_1420 | Transcriptional regulator, BadM/Rrf2 family | + | 0.07 | 0.99 | 8.70 | 1.86 | |
| Alvin_1436 | Cold shock DNA-binding domain protein | + | 2.29 | 0.71 | 0.10 | 0.29 | |
| Alvin_1525 | feoB | Ferrous iron transport protein B | − | 0.20 | 4.31 | 4.07 | 0.73 |
| Alvin_1527 | feoA | FeoA family protein | − | 0.20 | 2.37 | 3.92 | 0.52 |
| Alvin_1848 | aceA | Isocitrate lyase | − | 0.02 | 1.01 | 15.40 | 2.92 |
| Alvin_2012 | Hypothetical protein Alvin_2012 | + | 4.48 | 0.78 | 0.20 | 1.36 | |
| Alvin_2245 | hemP | Hemin uptake protein HemP | + | 0.17 | 1.78 | 2.71 | 1.06 |
| Alvin_2262 | metE | 5-Methyltetrahydropteroyltriglutamate/homocysteine S-methyltransferase | + | 0.39 | 0.59 | 0.08 | 0.44 |
| Alvin_2337 | rplQ | Ribosomal protein L17 | − | 1.10 | 0.90 | 0.18 | 0.61 |
| Alvin_2338 | rpoA | DNA-directed RNA polymerase, α-subunit | − | 0.94 | 0.55 | 0.17 | 0.63 |
| Alvin_2339 | rpsD | Ribosomal protein S4 | − | 1.04 | 0.58 | 0.16 | 0.60 |
| Alvin_2340 | rpsK | 30S ribosomal protein S11 | − | 1.26 | 0.70 | 0.18 | 0.54 |
| Alvin_2341 | rpsM | 30S ribosomal protein S13 | − | 1.12 | 0.57 | 0.20 | 0.68 |
| Alvin_2342 | rpmJ | Ribosomal protein L36 | − | 1.27 | 1.03 | 0.18 | 0.70 |
| Alvin_2343 | secY | Preprotein translocase, SecY subunit | − | 0.64 | 0.64 | 0.14 | 0.57 |
| Alvin_2357 | rpsC | Ribosomal protein S3 | − | 1.02 | 0.76 | 0.15 | 0.55 |
| Alvin_2358 | rplV | Ribosomal protein L22 | − | 0.92 | 0.73 | 0.14 | 0.46 |
| Alvin_2359 | rpsS | Ribosomal protein S19 | − | 0.91 | 0.79 | 0.12 | 0.48 |
| Alvin_2360 | rplB | Ribosomal protein L2 | − | 0.95 | 0.71 | 0.13 | 0.37 |
| Alvin_2361 | Ribosomal protein L25/L23 | − | 0.85 | 0.63 | 0.15 | 0.55 | |
| Alvin_2362 | Ribosomal protein L4/L1e | − | 0.92 | 0.69 | 0.14 | 0.41 | |
| Alvin_2363 | rplC | 50S ribosomal protein L3 | − | 0.73 | 0.48 | 0.16 | 0.53 |
| Alvin_2364 | rpsJ | Ribosomal protein S10 | − | 0.80 | 0.67 | 0.17 | 0.70 |
| Alvin_2371 | Ribosomal protein L7/L12 | − | 1.47 | 0.91 | 0.12 | 0.59 | |
| Alvin_2372 | rplJ | Ribosomal protein L10 | − | 1.07 | 0.61 | 0.10 | 0.43 |
| Alvin_2373 | rplA | Ribosomal protein L1 | − | 0.85 | 0.61 | 0.17 | 0.62 |
| Alvin_2374 | rplK | Ribosomal protein L11 | − | 0.64 | 0.91 | 0.17 | 0.80 |
| Alvin_2440 | cysB | Transcriptional regulator, LysR family | − | 0.06 | 0.07 | 0.61 | 0.27 |
| Alvin_2441 | cysA | Sulfate ABC transporter, ATPase subunit, CysA | − | 0.17 | 0.22 | 0.70 | 0.40 |
| Alvin_2442 | cysW | Sulfate ABC transporter, inner membrane subunit, CysW | − | 0.09 | 0.11 | 0.65 | 0.35 |
| Alvin_2443 | cysT | Sulfate ABC transporter, inner membrane subunit, CysT | − | 0.06 | 0.07 | 0.66 | 0.25 |
| Alvin_2444 | cysP | Sulfate ABC transporter, periplasmic sulfate-binding protein, CysP | + | 0.04 | 0.28 | 1.35 | 0.35 |
| Alvin_2446 | cysI | Nitrite and sulfite reductase 4Fe-4S region, CysI | + | 0.01 | 0.02 | 0.48 | 0.16 |
| Alvin_2447 | cysH | Adenylylsulfate reductase, thioredoxin dependent, CysH | + | 0.01 | 0.01 | 0.46 | 0.14 |
| Alvin_2448 | cysD | Sulfate adenylyltransferase, small subunit, CysD | + | 0.01 | 0.02 | 0.53 | 0.19 |
| Alvin_2449 | cysN | Sulfate adenylyltransferase, large subunit, CysN | + | 0.02 | 0.04 | 0.76 | 0.19 |
| Alvin_2451 | Molybdopterin oxidoreductase 4Fe-4S region | + | 0.15 | 0.66 | 1.17 | 3.36 | |
| Alvin_2767 | DEAD/DEAH box helicase domain protein | + | 0.95 | 0.76 | 0.16 | 0.33 | |
Data are compiled for genes for which transcription levels decreased at least 5-fold on one sulfur source or at least 2-fold on all tested sulfur sources; the average fold change for the corresponding microarray ratio(s) is highlighted in bold. Genes involved in light harvesting are excluded here; they are summarized in Table S2 in the supplemental material.
The complete microarray data sets can be found in Table S3 in the supplemental material and in the GEO database (see Materials and Methods).
Table 2.
Differentially expressed genes in A. vinosum after growth on different reduced sulfur compounds in comparison with growth on malatea
| Locus tag | Gene | Annotation | Strand | Avg fold change after growth onb: |
|||
|---|---|---|---|---|---|---|---|
| Sulfide | Thiosulfate | Sulfur | Sulfite | ||||
| Alvin_0055 | SOUL heme-binding protein | + | 0.51 | 0.88 | 5.80 | 1.73 | |
| Alvin_0082 | msrA | Peptide methionine sulfoxide reductase | − | 0.33 | 0.75 | 7.54 | |
| Alvin_0283 | RNP-1 like RNA-binding protein | + | 17.56 | 8.39 | 2.96 | 4.55 | |
| Alvin_0314 | pgk | Phosphoglycerate kinase | − | 5.07 | 1.81 | 0.87 | 0.74 |
| Alvin_0316 | tkt | Transketolase | − | 9.86 | 3.37 | 0.98 | 1.26 |
| Alvin_0345 | Sulfur relay protein, TusE/DsrC/DsvC family | − | 0.23 | 1.25 | 24.74 | 3.59 | |
| Alvin_0476 | Hypothetical protein Alvin_0476 | + | 21.29 | 1.00 | 0.65 | ||
| Alvin_0492 | Hypothetical protein Alvin_0492 | − | 0.75 | 1.38 | 5.92 | 3.33 | |
| Alvin_0562 | prk | Phosphoribulokinase | − | 8.37 | 3.05 | 1.16 | 0.83 |
| Alvin_0680 | Protein of unknown function, DUF1271 | + | 0.36 | 1.07 | 6.57 | 1.57 | |
| Alvin_0750 | Two-component transcriptional regulator, LuxR family | − | 5.95 | 1.00 | 1.87 | 1.01 | |
| Alvin_0804 | aceF | Pyruvate dehydrogenase complex dihydrolipoamide acetyltransferase | − | 5.36 | 1.42 | 0.66 | 0.88 |
| Alvin_0805 | aceE | 2-oxo-Acid dehydrogenase E1 subunit, homodimeric type | − | 5.03 | 1.08 | 0.78 | 1.08 |
| Alvin_0961 | Hypothetical protein Alvin_0961 | − | 8.25 | 0.82 | 0.89 | 0.76 | |
| Alvin_0962 | Ankyrin | − | 16.04 | 1.18 | 1.31 | 1.96 | |
| Alvin_1188 | pilE | Fimbrial protein precursor PilE (MS11 antigen) | + | 6.76 | 13.26 | 14.40 | 5.76 |
| Alvin_1196 | Hypothetical protein Alvin_1196 | + | 0.03 | 1.42 | 98.00 | 3.95 | |
| Alvin_1197 | Hypothetical protein Alvin_1197 | + | 0.03 | 1.37 | 116.63 | 3.88 | |
| Alvin_1248 | cas6 | CRISPR-associated protein, Cas6-related protein | − | 0.73 | 1.34 | 9.19 | |
| Alvin_1250 | Hypothetical protein Alvin_1250 | − | 0.60 | 1.23 | 10.60 | 2.67 | |
| Alvin_1305 | Protein of unknown function, DUF2092, periplasmic | + | 4.82 | 4.59 | 2.80 | 2.36 | |
| Alvin_1306 | Hypothetical protein Alvin_1306 | + | 5.10 | 6.24 | 3.81 | 2.93 | |
| Alvin_1317 | sreA | Molybdopterin oxidoreductase 4Fe-4S region, cytoplasm | + | 14.58 | 3.06 | 0.25 | 2.38 |
| Alvin_1318 | sreB | 4Fe-4S ferredoxin iron-sulfur-binding domain protein | + | 5.12 | 1.68 | 0.57 | 1.41 |
| Alvin_1324 | garB | Redoxin domain protein | − | 0.50 | 1.82 | 14.68 | 4.47 |
| Alvin_1365 | rbcA | Ribulose-bisphosphate carboxylase | + | 47.14 | 4.22 | 0.41 | 1.23 |
| Alvin_1366 | rbcB | Ribulose-bisphosphate carboxylase | + | 22.90 | 2.24 | 0.69 | 1.09 |
| Alvin_1367 | CbbQ/NirQ/NorQ domain protein | + | 29.07 | 2.89 | 0.69 | 0.98 | |
| Alvin_1368 | von Willebrand factor type A | + | 20.61 | 1.62 | 0.70 | 1.06 | |
| Alvin_1379 | leuA | 2-Isopropylmalate synthase | + | 7.97 | 3.56 | 1.08 | |
| Alvin_1381 | Hypothetical protein Alvin_1381 | − | 5.47 | 4.55 | 6.21 | 11.76 | |
| Alvin_1394 | Cytochrome b561 | − | 8.66 | 2.00 | 0.54 | 2.36 | |
| Alvin_1395 | Cytochrome c family protein | − | 27.44 | 1.99 | 0.35 | 4.24 | |
| Alvin_1398 | Hypothetical protein Alvin_1398 | − | 5.55 | 0.68 | 0.30 | 0.89 | |
| Alvin_1420 | Transcriptional regulator, BadM/Rrf2 family | + | 0.07 | 0.99 | 8.70 | 1.86 | |
| Alvin_1468 | YceI family protein | − | 0.43 | 1.58 | 27.04 | 5.07 | |
| Alvin_1508 | Sulfur relay protein, TusE/DsrC/DsvC family | − | 5.86 | 3.58 | 1.58 | 0.65 | |
| Alvin_1713 | Isochorismatase hydrolase | + | 6.34 | 2.08 | 1.30 | 1.32 | |
| Alvin_1737 | Dinitrogenase iron-molybdenum cofactor biosynthesis protein | − | 6.59 | 0.78 | 1.22 | 0.53 | |
| Alvin_1740 | Dinitrogenase iron-molybdenum cofactor biosynthesis protein | − | 16.72 | 0.66 | |||
| Alvin_1741 | Hypothetical protein Alvin_1741 | − | 17.94 | 0.59 | |||
| Alvin_1848 | aceA | Isocitrate lyase | − | 0.02 | 1.01 | 15.40 | 2.92 |
| Alvin_1918 | Glutaredoxin-like protein | − | 0.48 | 1.04 | 5.62 | 2.12 | |
| Alvin_1920 | sodB | Superoxide dismutase | + | 0.38 | 1.63 | 9.65 | 2.90 |
| Alvin_2001 | Putative transcriptional regulator, Crp/Fnr family | + | 9.41 | 6.98 | 1.91 | 1.52 | |
| Alvin_2010 | Hypothetical protein Alvin_2010 | + | 14.22 | 2.14 | 0.76 | 4.49 | |
| Alvin_2011 | Hypothetical protein Alvin_2011 | + | 6.10 | 0.78 | 0.49 | 2.24 | |
| Alvin_2032 | Peroxiredoxin | − | 0.28 | 1.04 | 13.88 | 2.68 | |
| Alvin_2033 | Oxidoreductase FAD/NAD(P)-binding domain protein | + | 0.29 | 0.90 | 5.99 | 1.57 | |
| Alvin_2036 | hynL | Nickel-dependent hydrogenase, large subunit | − | 9.86 | 1.89 | ||
| Alvin_2037 | isp1 | Protein of unknown function, DUF224, cysteine-rich region domain protein, DsrM-like | − | 10.81 | 1.26 | 0.69 | 1.32 |
| Alvin_2038 | isp2 | Hypothetical protein Alvin_2038, DsrK-like | - | 16.10 | 1.87 | 0.65 | 2.15 |
| Alvin_2039 | hynS | Hydrogenase (NiFe), small subunit | − | 8.19 | 1.97 | 0.71 | 0.86 |
| Alvin_2040 | hupH | HupH hydrogenase expression protein | − | 19.44 | 2.36 | 0.37 | 0.87 |
| Alvin_2093 | Protein of unknown function, DUF2189, transmembrane | − | 0.40 | 2.62 | 25.62 | 7.04 | |
| Alvin_2100 | Hypothetical protein Alvin_2100 | + | 0.39 | 3.59 | 8.13 | 1.44 | |
| Alvin_2107 | Hypothetical protein Alvin_2107 | − | 11.36 | 6.00 | 1.53 | 0.82 | |
| Alvin_2136 | Hypothetical protein Alvin_2136 | + | 49.59 | 1.05 | 1.39 | 0.78 | |
| Alvin_2497 | Hypothetical protein Alvin_2497 | + | 16.01 | 0.70 | 0.60 | 0.84 | |
| Alvin_2498 | Nitrogen fixation-related protein | + | 29.76 | 0.78 | 0.55 | 0.85 | |
| Alvin_2499 | cydA | Cytochrome bd ubiquinol oxidase, subunit I | + | 6.55 | 1.73 | 0.49 | 2.76 |
| Alvin_2500 | cydB | Cytochrome d ubiquinol oxidase, subunit II | + | 5.71 | 0.92 | 0.82 | 2.76 |
| Alvin_2501 | Cyd operon protein YbgT | + | 5.29 | 1.39 | 1.07 | 2.74 | |
| Alvin_2507 | Host attachment protein | + | 2.53 | 2.81 | 4.61 | 4.99 | |
| Alvin_2515 | Hypothetical protein Alvin_2515 | + | 27.80 | 5.91 | 1.66 | 0.86 | |
| Alvin_2572 | rpoH | RNA polymerase, s32 subunit, RpoH | − | 0.47 | 1.46 | 21.69 | 3.58 |
| Alvin_2600 | SirA family protein, TusA | + | 6.84 | 3.79 | 0.87 | 1.02 | |
| Alvin_2601 | Hypothetical protein Alvin_2601, DsrE-like | + | 5.74 | 3.06 | 1.34 | 1.29 | |
| Alvin_2651 | Protein of unknown function, DUF323 | + | 2.36 | 2.82 | 2.96 | 2.30 | |
| Alvin_2661 | Protein of unknown function, DUF323 | − | 0.56 | 1.00 | 7.87 | 1.67 | |
| Alvin_2667 | Iron-sulfur cluster assembly accessory protein | + | 0.34 | 0.91 | 8.79 | 1.82 | |
| Alvin_2705 | Hemerythrin-like metal-binding protein | + | 1.69 | 2.84 | 4.82 | 6.77 | |
| Alvin_2962 | Putative sodium symporter protein | + | 5.25 | 2.58 | 1.50 | 1.04 | |
| Alvin_2965 | TPR repeat-containing protein | + | 0.72 | 3.42 | 27.11 | 2.96 | |
| Alvin_2980 | Hypothetical protein Alvin_2980 | − | 0.35 | 0.70 | 7.01 | 2.02 | |
| Alvin_3032 | Hypothetical protein Alvin_3032 | + | 15.32 | 6.44 | 1.55 | 0.99 | |
| Alvin_3072 | Hypothetical protein Alvin_3072 | + | 75.81 | 1.25 | 1.81 | 1.55 | |
| Alvin_3073 | C4-dicarboxylate transporter/malic acid transport protein | + | 14.23 | 1.27 | 1.30 | 1.51 | |
| Alvin_3136 | Hypothetical protein Alvin_3136 | + | 5.37 | 2.08 | 1.64 | 3.68 | |
| Alvin_3140 | CRISPR-associated protein, NE0113 family | + | 3.51 | 2.12 | 1.96 | 5.51 | |
| Alvin_3141 | Hypothetical protein Alvin_3141 | + | 4.48 | 2.55 | 2.05 | 4.99 | |
| Alvin_3194 | Hypothetical protein Alvin_3194 | + | 2.64 | 3.42 | 4.86 | 8.72 | |
| Alvin_R0001 | rrs | 16S rRNA | + | 1.38 | 1.44 | 9.57 | |
| Alvin_R0004 | rrl | 23S rRNA | + | 1.00 | 1.65 | 1.34 | 7.16 |
| Alvin_R0008 | Asn tRNA | + | 2.09 | 2.02 | 3.36 | 5.35 | |
| Alvin_R0011 | Leu tRNA | + | 1.44 | 2.17 | 2.99 | 5.52 | |
| Alvin_R0013 | Glu tRNA | − | 2.41 | 2.65 | 3.36 | 10.12 | |
| Alvin_R0015 | Val tRNA | + | 1.64 | 1.63 | 2.30 | 5.16 | |
| Alvin_R0021 | Asp tRNA | + | 2.12 | 2.16 | 3.59 | 4.51 | |
| Alvin_R0023 | Gly tRNA | − | 8.12 | 22.64 | |||
| Alvin_R0030 | rrl | 23S rRNA | − | 0.91 | 1.49 | 1.30 | 8.27 |
| Alvin_R0037 | Phe tRNA | + | 1.96 | 4.56 | 7.15 | ||
| Alvin_R0049 | Gly tRNA | − | 1.02 | 5.88 | |||
| Alvin_R0051 | Gln tRNA | − | 3.86 | 2.98 | 7.45 | ||
| Alvin_R0052 | Lys tRNA | + | 1.89 | 1.82 | 3.68 | 10.55 | |
| Alvin_R0063 | Glu tRNA | + | 2.58 | 2.03 | 5.39 | 7.89 | |
Data are compiled for genes for which transcription levels increased at least 5-fold on one sulfur source or at least 2-fold on all tested sulfur sources; the average fold change for the corresponding microarray ratio(s) is highlighted in bold. Genes known to be involved in dissimilatory sulfur metabolism and light-harvesting genes are excluded here; data for those genes are compiled in Table 3 and in Table S2 in the supplemental material, respectively.
The complete microarray data sets can be found in Table S3 in the supplemental material and in the GEO database (see Materials and Methods).
Table 3.
Different transcription levels for A. vinosum genes encoding proteins with established functions in oxidative sulfur metabolisma
| Locus tag | Gene | Annotation | Strand | Avg fold change after growth onb: |
Reference(s) | |||
|---|---|---|---|---|---|---|---|---|
| Sulfide | Thiosulfate | Sulfur | Sulfite | |||||
| Alvin_0091 | tsdA | Thiosulfate dehydrogenase | + | 1.11 | 0.66 | 1.83 | 1.72 | 4 |
| Alvin_0358 | sgpB | Sulfur globule protein | + | 62.95 | 5.73 | 3.07 | 3.29 | 44 |
| Alvin_1092 | fccB | Flavocytochrome c sulfide dehydrogenase flavin–binding protein | − | 1.10 | 1.42 | 10 | ||
| Alvin_1093 | fccA | Cytochrome c class I | – | 0.13 | 1.99 | 0.56 | 2.28 | 10 |
| Alvin_1118 | sat | Sulfate adenylyltransferase | + | 7.60 | 1.842 | 0.73 | 1.72 | |
| Alvin_1119 | aprM | Hypothetical protein | + | 9.66 | 1.90 | 0.89 | 2.42 | |
| Alvin_1120 | aprB | Adenylylsulfate reductase, β–subunit | + | 8.96 | 1.98 | 1.04 | 2.37 | 66 |
| Alvin_1121 | aprA | Adenylylsulfate reductase, α-subunit | + | 7.72 | 2.27 | 1.10 | 1.78 | 66 |
| Alvin_1195 | sqrF | Sulfide:quinone oxidoreductase | + | 1.90 | 1.35 | 1.54 | 1.90 | 11 |
| Alvin_1251 | dsrA | DsrA | + | 97.62 | 54.54 | 23.64 | 3.20 | 13 |
| Alvin_1252 | dsrB | DsrB | + | 84.74 | 41.34 | 14.10 | 2.23 | 13 |
| Alvin_1253 | dsrE | DsrE | + | 90.13 | 30.57 | 8.40 | 1.52 | 13, 51 |
| Alvin_1254 | dsrF | DsrF | + | 35.78 | 16.58 | 5.33 | 0.98 | 13, 51 |
| Alvin_1255 | dsrH | DsrH | + | 25.28 | 8.40 | 3.29 | 0.96 | 13, 51 |
| Alvin_1256 | dsrC | DsrC | + | 14.24 | 3.97 | 2.25 | 1.18 | 13, 51 |
| Alvin_1257 | dsrM | DsrM | + | 19.68 | 6.23 | 2.64 | 1.04 | 13, 35 |
| Alvin_1258 | dsrK | DsrK | + | 15.02 | 6.16 | 2.82 | 0.97 | 13, 35 |
| Alvin_1259 | dsrL | DsrL | + | 19.22 | 5.90 | 3.89 | 1.35 | 14, 16 |
| Alvin_1260 | dsrJ | DsrJ | + | 22.96 | 5.87 | 3.37 | 1.67 | 5, 14 |
| Alvin_1261 | dsrO | DsrO | + | 9.70 | 5.94 | 2.40 | 1.03 | 14, 35 |
| Alvin_1262 | dsrP | DsrP | + | 17.47 | 6.67 | 2.47 | 1.00 | 14, 35 |
| Alvin_1263 | dsrN | DsrN | + | 2.60 | 2.23 | 0.55 | 14, 16 | |
| Alvin_1264 | dsrR | DsrR | + | 1.74 | 1.30 | 1.37 | 1.11 | 14 |
| Alvin_1265 | dsrS | DsrS | + | 1.29 | 1.29 | 1.10 | 1.18 | 14 |
| Alvin_1325 | sgpC | Sulfur globule protein | + | 11.61 | 1.94 | 0.74 | 5.17 | 44 |
| Alvin_1905 | sgpA | Sulfur globule protein | + | 2.26 | 2.54 | 2.50 | 1.29 | 44 |
| Alvin_2111 | soxY | SoxY | + | 0.32 | 1.08 | 1.58 | 1.44 | 6 |
| Alvin_2112 | soxZ | SoxZ | + | 0.45 | 0.93 | 1.71 | 3.71 | 6 |
| Alvin_2145 | sqrD | Sulfide:quinone oxidoreductase | + | 1.31 | 1.54 | 1.59 | 3.28 | 11 |
| Alvin_2167 | soxB | SoxB | − | 0.85 | 1.15 | 1.72 | 3.45 | 6 |
| Alvin_2168 | soxX | SoxX | + | 1.02 | 2.02 | 5.81 | 3.06 | 6 |
| Alvin_2169 | soxA | SoxA | + | 0.90 | 1.84 | 4.87 | 2.36 | 6 |
| Alvin_2170 | soxK | SoxXA-binding protein | + | 0.83 | 1.50 | 4.25 | 1.04 | 6 |
| Alvin_2171 | soxL | Sulfur transferase, periplasm | + | 0.80 | 1.20 | 3.53 | 6, 12 | |
Transcriptional changes of more than 5-fold on at least one sulfur source or of more than 2-fold on all tested sulfur compounds tested are highlighted in bold.
The complete microarray data sets can be found in Table S3 in the supplemental material and in the GEO database (see Materials and Methods).
It is very important to note that the switch from photoorganoheterotrophic growth on malate to photolithoautotrophic growth on reduced sulfur sources alters not only the electron donor (malate versus a sulfur compound) but also the carbon source (malate versus carbon dioxide) for the cells. This imparts effects on the transcriptomic profiles that need to be carefully dissected. In the following paragraphs, we therefore first briefly describe and discuss the most important general transcriptomic responses observed, before we turn specifically to the effects on sulfur metabolism and finally to the transcriptome-aided identification of new sulfur-related genes.
Transcriptomic responses on carbon metabolism.
The transcriptomes of A. vinosum cells grown on reduced sulfur compounds showed elevated mRNA levels for components of the CO2 fixation machinery compared to cells grown with malate (Table 2). The transcript for the genes in cluster Alvin_1365 to _1368, encoding the major RuBisCO species RbcAB under standard photolithoautotrophic growth conditions (31), showed significantly increased levels during growth on sulfide and thiosulfate, with values up to 50-fold (Table 2; see Table S3 in the supplemental material). The genes for the second RuBisCO, RbcSL (Alvin_2749 and _2750), and some carboxysome-related genes were positively affected by the presence of elemental sulfur, albeit to a lesser extent (see Table S3). Interestingly, Alvin_2545, encoding a RuBisCO-like protein that has been speculated to be involved in the sulfur metabolism of the green sulfur bacterium Chlorobaculum tepidum (32), also revealed significantly increased relative mRNA levels on sulfide (3.5-fold higher). We also found highly elevated relative mRNA levels on sulfide for Alvin_0314, Alvin_0316, and Alvin_0562, which encode further key enzymes of the Calvin cycle, namely, phosphoglycerate kinase, transketolase, and phosphoribulokinase, respectively (Table 2). Some A. vinosum genes for enzymes of the glyoxylate cycle (Alvin_3052 [citrate synthase] and Alvin_1848 [isocitrate lyase]) (Table 2; see also Table S3 in the supplemental material) exhibited higher relative mRNA levels under autotrophic growth conditions, matching previous results for this organism (33).
Transcriptomic responses on the photosynthetic machinery and on hydrogen metabolism.
Transcriptomic effects concerning the photosynthetic machinery are compiled and are briefly discussed in Table S2 in the supplemental material. A. vinosum possesses four hydrogenase gene clusters (3). The hyn cluster (Alvin_2036 to _2040) revealed increased relative mRNA levels, with values of up to 20-fold higher during growth on sulfide (Table 2). Interestingly, Alvin_2037 and Alvin_2038 encode proteins resembling two components of the DsrMKJOP transmembrane complex, which is encoded in the dsr operon, namely, DsrM and DsrK (34). DsrM is a membrane-spanning b-type cytochrome, DsrK is an iron-sulfur protein resembling heterodisulfide reductase from methanogenic archaea (35). The Alvin_2037 and Alvin_2038 proteins are not copurifed with the A. vinosum Hyn [NiFe] hydrogenase (36). The observed transcriptional response of the complete hyn cluster to the presence of sulfide may point to a so-far-underestimated direct connection of hydrogen and sulfur metabolism in purple sulfur bacteria.
Further transcriptomic responses of genes with assigned functions.
Specifically on sulfide alone, relative mRNA levels for several genes of a large cluster encoding the subunits of proton-translocating NADH:quinone oxidoreductase (complex I; Alvin_2407 to _2432) were enhanced. In A. vinosum, electrons originating from sulfide are fed into the quinone pool via sulfide:quinone oxidoreductase (7, 10). Complex I can then catalyze the reverse, proton motive force-consuming electron flow from quinol to NAD+ to provide reduction equivalents for CO2 fixation (37). This interpretation is supported by the enhanced relative mRNA levels of NAD/NADP transhydrogenase genes (Alvin_0834 to _0836) on sulfide. NADPH2 is the immediate electron donor for the reducing enzymes of the Calvin cycle.
When A. vinosum was grown with elemental sulfur, relative mRNA levels for a number of genes for proteins involved in iron trafficking (Alvin_1525 to _1527 and Alvin_1856), components of iron-sulfur cluster assembly systems (Alvin_0110, Alvin_0739 to _0742, Alvin_2667 and _2668), a ferric uptake regulator (Alvin_2243), hemin uptake (Alvin_2245). and a protein of the iron-storage ferritin family (Alvin_2742) were found to be significantly enhanced, pointing to an especially high demand of the cells for iron-containing and/or iron-sulfur cluster-containing proteins.
The relative mRNA levels of several flagellum-related genes (Alvin_0660 to Alvin_0675, Alvin_1951 to _1954, and Alvin_2913 to _2946) were significantly increased on elemental sulfur and/or sulfite but not on sulfide or thiosulfate (see Table S3 in the supplemental material). While a possible connection of flagellation and sulfite was not apparent, flagellar proteins have recently been speculated to be involved in direct physical contact with insoluble elemental sulfur for oxidation in Aquifex aeolicus (38). A close physical contact between the cell surface and elemental sulfur is also necessary for uptake of this water-insoluble substance in A. vinosum (18). Enhanced relative mRNA was furthermore noted on sulfur and/or sulfite for different cell surface structure-related genes: Alvin_1188 (fimbrial protein precursor PilE), the type IV pilus assembly protein PilZ (Alvin_0408, _1569, _2127, and _2226), Alvin_3021, encoding another copy of PilE, and Alvin_3058, encoding a type IV pilus secretin, PilQ. Although pilus formation in purple sulfur bacteria has so far not been systematically assessed and to the best of our knowledge never been described for A. vinosum, we cannot exclude that pili may play a role in electron transfer or in establishing physical contact between cells and elemental sulfur. It is interesting that spinae, surface appendages of some green sulfur bacterial strains (39, 40), have been suggested to play a role in adhesion of sulfur globules to the cells (39).
Transcriptomic responses of assimilatory sulfate reduction genes.
Major decreases in mRNA abundance were observed for the cys genes for assimilatory sulfate reduction (Table 1; see also Fig. S2 in the supplemental material), a pathway needed for synthesis of sulfur-containing cell constituents during photoorganoheterotrophic growth in the absence of reduced inorganic sulfur (41). The tight regulation of the assimilatory sulfate reduction pathway, which is dependent on the availability of reduced sulfur, has been well described for a vast number of Gram-negative bacteria, including E. coli (42).
Transcriptional response of genes for established sulfur-oxidizing proteins.
The results for those 34 genes encoding proteins for which an involvement in oxidative sulfur metabolism has already been proven are compiled in Table 3. For three of these genes (dsrA, dsrB, and sgpB) significantly elevated relative mRNA levels were found on all of the tested sulfur compounds. Among these, a 97.6-fold increase for the dsrA transcript and a 63-fold increase for the sgpB mRNA levels on sulfide versus malate were the strongest responses (Table 3). Oxidation of externally added sulfite does not require the upstream reaction steps, i.e., formation of a sulfur globule envelope and oxidation of stored sulfur via the Dsr system (Fig. 1). In accordance, we observed prominent transcriptional responses for 11 of the corresponding genes (dsrEFHCMKLJOP and sgpA) in cultures grown on sulfide, thiosulfate, or elemental sulfur, but not in cells exposed to sulfite (Table 3).
The relatively large changes for mRNA levels in cells grown on sulfide of dsrA (97-fold) or of dsrE (90-fold) compared to dsrC (14-fold), dsrR (1.7-fold), or dsrS (1.3-fold) corresponded to the values detected by Grimm et al., who applied the qRT-PCR technique (43). Those authors determined absolute mRNA values and found all studied dsr genes to be constitutively transcribed on a low level, even under photoorganoheterotrophic growth conditions. Furthermore, dsrC and dsrS were established to be constitutively transcribed from additional, separate promoter sites, and the respective transcript amounts under photoheterotrophic conditions were found to be much higher than those for the other studied dsr genes. In accordance with these findings, the changes in relative mRNA levels observed in our work appeared less pronounced for dsrC and dsrS than for dsrA and dsrE, although the absolute mRNA amounts during growth on sulfide were quite similar, especially for dsrE, dsrC, and dsrS (43).
Compared to sgpB and sgpC, relative changes for sgpA mRNA levels were small on all sulfur sources (Table 3). This finding generally matched previous quantitative RT-PCR results (44), which had not indicated a significant regulation of sgpA transcription, whereas expression levels of sgpB and sgpC have been found to be clearly enhanced in the presence of sulfide (44).
The relative mRNA levels of the genes encoding the proteins of the indirect cytoplasmic sulfite oxidation pathway, namely, APS reductase (Alvin_1119 to _1121) and ATP sulfurylase (Alvin_1118), were found to be enhanced up to 10-fold on sulfide, while the maximum increase on sulfite was only 2.4-fold for the APS reductase membrane anchor gene aprM (Table 3). We showed previously that the APS reductase pathway is not essential for sulfite oxidation in A. vinosum (2), although its presence is advantageous for cells under certain growth conditions (17).
With regard to thiosulfate oxidation, the genes for the Sox proteins SoxXAK and SoxL showed a general tendency for an increase of relative mRNA levels in the presence of elemental sulfur or sulfite, with a maximum increase of 5.8-fold observed for soxX on elemental sulfur. Changes upon exposure to thiosulfate, the only established substrate of the proteins in A. vinosum, stayed below the threshold, with the exception of soxX relative mRNA levels, which increased 2-fold in the presence of this sulfur substrate (Table 3). Relative mRNA level of the tsdA gene (Alvin_0091), which encodes the tetrathionate-forming thiosulfate dehydrogenase of A. vinosum (4), appeared more or less unaffected by the presence of reduced sulfur compounds (Table 3).
The same holds true for the proteins thought to be important for the oxidation of sulfide, namely, flavocytochrome c sulfide dehydrogenase FccAB (Alvin_1092 and _1093) and the two different sulfide:quinone oxidoreductases SqrD (Alvin_2145) and SqrF (Alvin_1195). The obvious constitutive expression of these genes is in accordance with the well-established capacity of A. vinosum cultures to oxidize sulfide, with maximum rates observed immediately after addition of this substrate.
Transcriptional responses of genes encoding proteins putatively involved in oxidative sulfur metabolism.
Elevated relative mRNA levels were observed on at least one or two sulfur sources for a number of genes that have previously been speculated to be involved in dissimilatory sulfur metabolism. Especially pronounced changes were detected on elemental sulfur for Alvin_1324 (15-fold), which encodes a redoxin domain protein (Prx/Grx peroxiredoxin), and the adjacent gene, Alvin_1323 (3-fold), which encodes glutathione amide disulfide reductase (Table 2) (45, 46). The latter has been suggested to be involved in the metabolism of an organic perthiol, possibly glutathione amide perthiol, which functions in transport of sulfane sulfur across the cytoplasmic membrane (7). In this context, it is noteworthy that significant changes in mRNA levels were not observed for Alvin_0481 to _0482 or Alvin_1430 to _1433, which encode probable glutathione transporters (47, 48).
A cluster of three genes located in the immediate vicinity of the glutathione amide reductase gene, namely, Alvin_1317 to _1319, also showed significantly elevated relative mRNA levels on sulfide (Table 2). The genes encode the three subunits of a putative sulfur or polysulfide reductase, with highest similarities to SreA, SreB, and SreC of the thermoacidiphilic archaeon Acidianus ambivalens. SreABC has been shown to reduce not only elemental sulfur but also polysulfides in vitro (49). For A. vinosum, the respective gene products could in principle be involved in the oxidation of the polysulfides that occur as intermediates during the oxidation of sulfide to sulfur that is stored in sulfur globules, just as has been speculated for the related proteins from Chlorobaculum tepidum (50). In both cases, the iron-sulfur cluster-containing β-subunit as well as the active site molybdopterin-containing α-subunit is predicted to be oriented toward the bacterial periplasm.
Similar changes as those noted for the putative sulfur reductase genes were observed for Alvin_2489 to _2491 (3-fold increase in mRNA on sulfide). The latter genes also encode a complex iron-sulfur molybdoprotein that could be involved in oxidative sulfur metabolism. Due to the predicted orientation of the active site toward the bacterial cytoplasm not only in A. vinosum but also in green sulfur bacteria, it has been speculated that the protein functions in the oxidation of sulfite to sulfate (7).
We also paid special attention to genes encoding proteins with probable functions in sulfur relay systems. In addition to dsrC, A. vinosum contains four more genes for TusE/DsrC/DsvC family sulfur relay proteins (3). Among these, Alvin_0345 (25-fold increase in mRNA on elemental sulfur) and Alvin_1508 (6-fold increase on sulfide) showed highly elevated relative mRNA levels in the presence of sulfur compounds (Table 2). The genes Alvin_0028 and Alvin_0732 revealed less pronounced but still significantly increased relative mRNA levels on elemental sulfur and sulfite. A. vinosum contains a number of genes that encode proteins with probable sulfurtransferase functions, i.e., rhodanese (thiosulfate sulfurtransferase) domain-containing proteins (Table 4), that could also play a role in dissimilatory sulfur metabolism (51). Indeed, Alvin_0866 and Alvin_1587 revealed changes of relative mRNA levels of considerably more than 2-fold during growth on elemental sulfur and sulfide, respectively (Table 4). Recently, we established a sulfur transfer function also for the heterohexameric DsrEFH protein encoded in the A. vinosum dsr locus (51). The related protein TusBCD from E. coli interacts efficiently with the protein TusA in a sulfur relay system during thiouridine biosynthesis (52). In A. vinosum, Alvin_2600, which encodes a TusA-like protein, and Alvin_2601, which encodes a membrane-bound DsrE domain-containing protein, are situated immediately upstream of a gene for a rhodanese domain-containing protein. While an effect on the relative mRNA level of Alvin_2599 was not observed on any of the sulfur compounds tested, levels of the tusA and dsrE-like genes were enhanced up to 7-fold during growth on sulfide and up to 4-fold during growth on thiosulfate (Table 2), pointing to a possible function in oxidative sulfur metabolism.
Table 4.
Relative transcription of genes encoding rhodanese domain containing proteins
| Locus tag | Annotation | Strand | Avg fold change after growth on: |
|||
|---|---|---|---|---|---|---|
| Sulfide | Thiosulfate | Sulfur | Sulfite | |||
| Alvin_0258 | Rhodanese domain protein | + | 0.50 | 0.68 | 1.95 | 0.94 |
| Alvin_0866 | Rhodanese domain protein | − | 0.23 | 1.14 | 3.00 | 1.86 |
| Alvin_0868 | Rhodanese domain protein | − | 2.13 | 1.51 | 1.35 | 1.46 |
| Alvin_1587 | Rhodanese domain protein | − | 3.80 | 0.88 | 0.86 | 1.28 |
| Alvin_2599 | Rhodanese domain protein | + | 1.79 | 1.64 | 0.68 | 0.83 |
| Alvin_3028 | Rhodanese domain protein | + | 0.94 | 0.75 | 0.47 | 0.88 |
Strongly affected genes with unknown functions.
The observed transcriptional responses of A. vinosum to different sulfur compounds support wide portions of the current model of sulfur oxidation in purple sulfur bacteria (Fig. 1). The microarray approach therefore appears promising for the identification of new sulfur metabolism-related genes. A number of genes with unknown functions revealed at least a 2-fold-higher relative mRNA level in the presence of all four tested reduced sulfur compounds, implicating either a general role for the encoded proteins in sulfur oxidation or of the metabolic pathways switched upon the change from organoheterotrophic to lithoautotrophic growth (Table 2). Alvin_0284 encodes a protein with the conserved nucleic acid-binding motif RNP1 (53) and could be involved in sulfur-dependent regulation. Alvin_1305 and Alvin_1306 are part of a larger gene cluster comprising Alvin_1301 to _1309. Relative mRNA levels for all of these genes were significantly elevated on sulfide, sulfur, and thiosulfate, pointing to the possibility that the products are involved in processes upstream of sulfite oxidation. Besides probable protein-encoding genes, four RNA genes were also found to be upregulated on all sulfur compounds. The respective tRNAs bind the acidic, negatively charged amino acids aspartic acid and glutamic acid and the corresponding amidated forms, asparagine and glutamine, respectively (Table 2).
Relative transcript levels of some hypothetical genes were increased on three of the four sulfur compounds, e.g., Alvin_1743 on sulfide, sulfur, and sulfite, or Alvin_2010 on sulfide, thiosulfate, and sulfite. The latter is part of the cluster Alvin_2010 to _2013. Alvin_2011 and _2012 also showed positive response to the presence of sulfide. Hypothetical genes that showed elevated relative mRNA levels on thiosulfate, sulfur, and sulfite included Alvin_2705, annotated as a hemerythrin-like metal-binding protein (gene cluster Alvin_2703 to _2705) and Alvin_2093, which encodes a transmembrane protein (Table 2). Functions can at this point not be predicted for any of these genes/proteins.
A role in the metabolism of a specific sulfur compound may be indicated by specifically elevated relative mRNA levels. We therefore analyzed genes with unknown functions that revealed increased relative transcript levels of more than 5-fold during growth on at least one of the applied sulfur sources (Table 2). According to their genomic location next to the dsr operon and the sox gene cluster, respectively, increased relative mRNA levels of Alvin_1250 on elemental sulfur (10-fold) and Alvin_2107 on sulfide (11-fold) and thiosulfate (6-fold) were especially noteworthy.
On elemental sulfur, the most prominent responses of genes for hypothetical proteins were observed for Alvin_1196, Alvin_1197, and Alvin_1468 (Table 2). The genes Alvin_1196 and Alvin_1197 are located immediately downstream of and transcribed in the same direction as Alvin_1195, which encodes SqrF, a sulfide:quinone oxidoreductase of type F that is responsible for the oxidation of sulfide when present in high concentrations in C. tepidum (11). The gene Alvin_1468 encodes a periplasmic protein that belongs to the YceI-like family of proteins (54).
The most pronounced effects on relative mRNA levels of hypothetical genes in the presence of sulfide were recorded for Alvin_3072 and _3073 (76-fold and 14-fold), Alvin_2136 and _2137 (50-fold and 3-fold), Alvin_2515 (28-fold), Alvin_1394 and _1395 (9-fold and 27-fold), and Alvin_0476 (21-fold) (Table 2). It is noteworthy that Alvin_1394 and Alvin_1395 are transcribed in the same direction and encode a b-type cytochrome and an octaheme cytochrome c, respectively. BLAST searches revealed high similarities of the latter to tetrathionate reductases (55). In addition, a complete gene cluster (Alvin_1737 to _1741) exhibited enhanced relative mRNA levels exclusively on sulfide, with values up to 18-fold higher (Table 2). BLAST searches for Alvin_1737 and Alvin_1740 revealed similarities to CT1276 of Chlorobaculum tepidum TLS. For CT1276, a similar response on sulfide was reported recently (50). The adjacent gene, CT1277, exhibited a strong increase in the relative transcription level on sulfide as well, and for this gene's product, a role in sulfide-dependent gene expression changes was suggested, based on its DNA-binding domain (50). The corresponding gene in A. vinosum is encoded by Alvin_1742, which is located adjacent but transcribed in the opposite direction to the gene cluster of A. vinosum. Just like CT1277, Alvin_1742 revealed an elevated relative mRNA level on sulfide (5-fold); however, this value was based on only one evaluable experiment (see Table S3 in the supplemental material).
On thiosulfate, the most pronounced effects on relative mRNA levels for hypothetical genes that have not been mentioned or discussed so far were recorded for Alvin_3032, which encodes a 74-amino-acid protein. This gene is also found in several other sulfur-oxidizing organisms, such as Thiocystis violascens DSM 198, Thiorhodococcus drewsii AZ1, Thiorhodovibrio sp. 970, and Thioalkalivibrio sulfidophilus HL-EbGr7. In the latter, the corresponding gene is located immediately adjacent to a dsrC-like gene situated close to a dsr gene cluster. The relative mRNA abundance of Alvin_3032 was also drastically elevated on sulfide (Table 2). On sulfite, additional hypothetical genes with transcriptional changes of more than 5-fold were not detected (Table 2).
Validation of differential gene expression.
The ultimate goal of our microarray approach was the identification of so-far-unknown genes related to oxidative sulfur metabolism. It was therefore strictly necessary to validate the data acquired from the DNA microarray technique via qRT-PCR. As mentioned above, effects on relative mRNA levels observed in this work for several dsr and sgp genes matched previously published RT-PCR data (43, 44) and provided a first validation for our global transcriptional profiling approach. In addition, we have now reexamined relative mRNA levels of several conspicuous genes (Alvin_1196 and _1197, Alvin_0258, and Alvin_3028, which encode rhodaneses, and Alvin_2600 and Alvin_2601, which encode TusA and a DsrE-like protein, respectively) by qRT-PCR. The gene Alvin_0486, which encodes a uroporphyrinogen decarboxylase, was chosen as an endogenous reference because its relative transcription rate was always found to be at a value of 1 throughout all DNA microarray experiments (see Table S3 in the supplemental material).
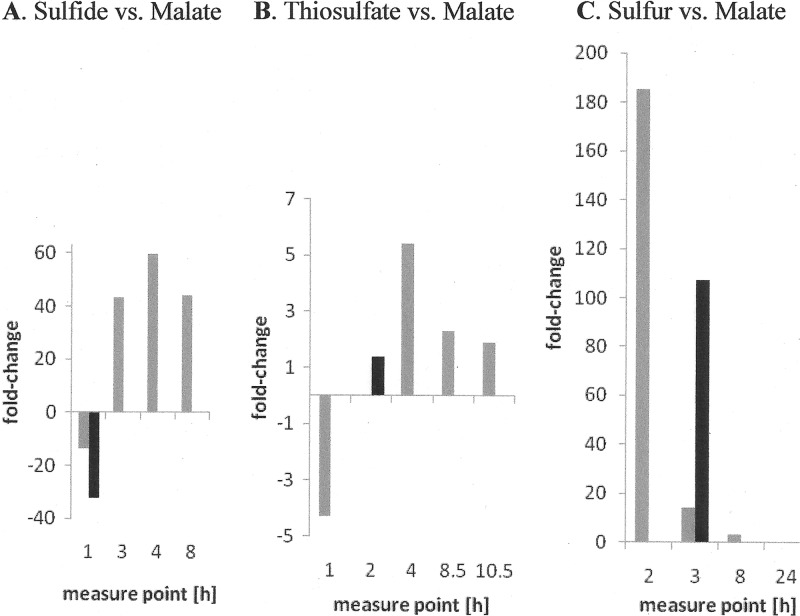
qRT-PCR for Alvin_1196 and _1197 was performed with RNA extracted at several time points during a 24-h exposure of A. vinosum cells to sulfide, thiosulfate, elemental sulfur, or malate. In this way, potential dynamic changes of the relative amount of the respective mRNA over time were covered. For the other four genes, relative mRNA levels were determined at time points corresponding exactly to those of the microarray experiment. A comparison of the data compiled in Fig. 2 and Table 5 shows that the qRT-PCR experiments verified the DNA microarray results. The only slight discrepancy was that for the relative mRNA level of Alvin_1196 and _1197 during growth on elemental sulfur compared to growth on malate (Fig. 2C). Here, the change measured 3 h after sulfur addition was only 15-fold via qRT-PCR compared to 116-fold via the microarray approach. However, a change in the latter range was determined via qRT-PCR 2 h after substrate addition. The observed difference is most likely due to the slightly different growth conditions applied, namely, culturing the cells in culture bottles and thermostatted glass fermentors, respectively.
Fig 2.
Relative mRNA levels of Alvin_1196 and _1197 during growth on malate compared to growth on sulfide (A), thiosulfate (B), or elemental sulfur (C), as assessed by qRT-PCR (gray columns). Results were adjusted using Alvin_0486, which encodes a uroporphyrinogen decarboxylase, as an endogenous reference. Results generated by the DNA microarray experiments are shown as black columns.
Table 5.
Relative mRNA levels of selected A. vinosum genes encoding potential sulfur relay proteins, as determined by the DNA microarray technique and qRT-PCR
| Locus tag | Annotation | Strand | Avg fold change after growth on: |
|||
|---|---|---|---|---|---|---|
| Sulfide |
Sulfur |
|||||
| Array | qRT-PCR | Array | qRT-PCR | |||
| Alvin_0258 | Rhodanese domain protein | + | NDa | ND | 1.95 | 1.23 |
| Alvin_3028 | Rhodanese domain protein | + | 0.93 | 0.55 | 0.47 | 0.59 |
| Alvin_2600 | SirA family protein | + | 6.84 | 6.81 | ND | ND |
| Alvin_2601 | Hypothetical protein Alvin_2601 | + | 5.74 | 3.28 | 1.34 | 2.03 |
ND, not determined.
It should be stressed that time-response qRT-PCR experiments indeed revealed dynamic changes in relative mRNA levels for some genes. For example, the DNA microarray experiment performed on sulfide (1 h after substrate addition) did not identify genes Alvin_1196 and Alvin_1197 as being affected by the presence of sulfide. However, when incubation of the cells on sulfide was extended to 3, 4, and 8 h, qRT-PCR analyses clearly indicated a correlation of the transcription of both genes with the oxidation of sulfide (Fig. 2). In contrast to growth on thiosulfate and elemental sulfur, relative mRNA levels of both genes continuously increased on sulfide, with changes of up to 60-fold over the examined period of 8 h (Fig. 2A).
Transcriptome-based identification of new sulfur-regulated genes by insertional inactivation.
Four sets of genes with relative mRNA levels strongly affected by elemental sulfur and sulfide, namely, Alvin_1196 and Alvin_1197, Alvin_1468, Alvin_2093, and Alvin_3072, were independently replaced by a kanamycin Ω cassette (56). All mutant strains were able to grow photoorganoheterotrophically on malate, either in RCV medium or in 0 medium (Table 6; see also Table S4 in the supplemental material). We can thus exclude general growth defects and effects on sulfate assimilation for these mutants. As a next step, metabolism of different reduced sulfur compounds was studied. We did not find any differences between the mutants and the wild type when elemental sulfur (50 mM) or sulfite (5 mM) was supplied as the electron donor (see Table S4). We can thus confidently state that the respective proteins play neither a direct nor an indirect role in metabolism of externally supplied elemental sulfur or the oxidation of sulfite to sulfate. However, all mutants exhibited a pronounced phenotype on sulfide and to some extent on thiosulfate, especially at the step of sulfur globule formation (Table 6; see also Table S4).
Table 6.
Features of Alvin_1196 and Alvin_1197, Alvin_1468, Alvin_2093, and Alvin_3072 and the corresponding proteins
| Feature | Alvin_1196/97 | Alvin_1468 | Alvin_2093 | Alvin_3072 |
|---|---|---|---|---|
| Phenotype of deletion mutant in presence of 8 mM sulfide | ∼60% reduced sulfur formation rate | Sulfur formation not possible | 60% reduced sulfur formation rate | >90% reduced sulfur formation rate |
| Phenotype of deletion mutant in presence of 4 mM thiosulfate | ∼70% reduced sulfur formation rate | ∼55% reduced sulfur formation rate | ∼65% reduced sulfur formation rate | ∼35% reduced sulfur formation rate |
| Photoorganoheterotrophic growth on malate | + | + | + | + |
| Sulfate assimilation | + | + | + | + |
| Genomic localization | Next to sqrF | Inconspicuous | Adjacent to gene encoding sulfotransfer domain-containing protein | Adjacent to gene encoding potential sulfite efflux pump |
| Relative mRNA level | Up on S2−, S0 and SO32− | Up on S0 and SO32− | Up on S0 and SO32− | Up on S2− |
| Subcellular localization | Membrane | Periplasm | Membrane | Membrane |
| No. of transmembrane helices | 1 | None | 5 | 2 |
| Length (amino acids) | 32 | 198 (174 without signal peptide) | 262 | 61 |
| Cysteine residues | 1, conserved | No | 1, not conserved | 1, conserved |
| Annotation/functional group | None | YceI family protein | Hypothetical protein, DUF2189 family | Hypothetical protein |
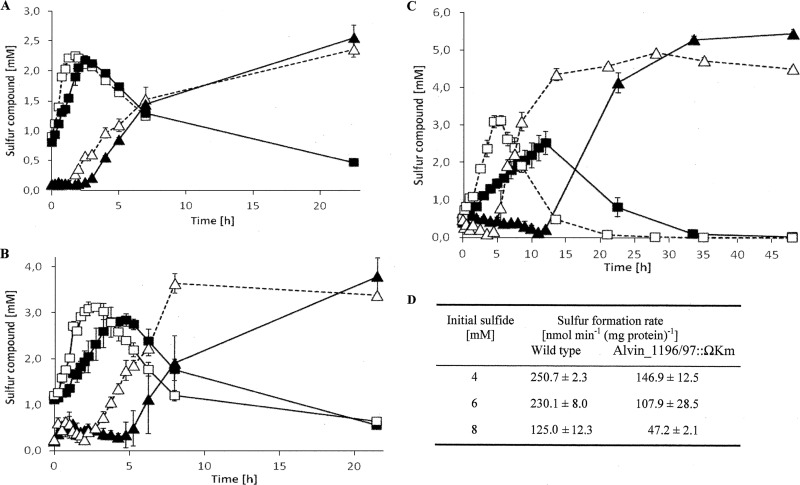
For mutant A. vinosum Alvin_1196/97::ΩKm, the rate of intracellular sulfur production from sulfide was significantly lower than that of the wild type, and this effect became more pronounced with increased sulfide concentrations (Fig. 3). This phenotype indicates that both Alvin_1196 and Alvin_1197 are involved in the oxidation of sulfide, at least when it is provided in high concentrations. Hints to the possible function of Alvin_1196 and _1197 come from analyses of neighboring genes and the encoded amino acid sequences (Table 6; see also Fig. S3 in the supplemental material). The products of Alvin_1196 and Alvin_1197 are completely identical and predicted to each consist of nothing more than a single transmembrane α-helix (Fig. S3b). BLAST searches for these proteins revealed only two hits, namely, for Thiorhodococcus drewsii AZ1 (two copies) and Thiorhodovibrio sp. 970 (one copy), two more purple sulfur bacteria of the family Chromatiaceae (see Fig. S3c). In A. vinosum, the adjacent gene, Alvin_1195, encodes a sulfide:quinone oxidoreductase of type F (see Fig. S3a). Transfer of the electrons resulting from SqrF-catalyzed sulfide oxidation into the quinone pool requires a connection of this enzyme to the membrane. However, transmembrane helix predictions for A. vinosum SqrF yield unclear results, just as elements involved in membrane binding are not apparent for the related protein SqrE from the archaeon Acidianus ambivalens (57). SQR from Aquifex aeolicus has been described as a periplasmic monotopic membrane protein (58). Two regions (Lys376-Asn395 and Pro400-Lys412) are responsible for membrane attachment in the Aquifex protein. While the first is present, the latter has no equivalent in the A. vinosum SqrF sequence. Based on the lack of regions suited for membrane attachment or insertion in A. vinosum SqrF, it is tempting to speculate that Alvin_1196 and Alvin_1197 are involved in this function. Our suggestion is corroborated by first analyses of a SqrF-deficient mutant that has a phenotype almost indiscernible from that of the Alvin_1196/97::ΩKm strain (R. Zigann and C. Dahl, unpublished data).
Fig 3.
Phenotypic characterization of A. vinosum Alvin_1196/97::ΩKm. (A to C) Intracellular sulfur (wild type [□] and mutant [■]) and extracellular sulfate concentrations (wild type [△] and mutant [▲]) of A. vinosum strain Rif50 (dotted line) and mutant A. vinosum Alvin_1196/97::ΩKm (solid line) during growth on 4 mM (A), 6 mM (B), or 8 mM sulfide (C). Note the different time scale for panel C. (D) Sulfur formation rates for the wild type and mutant are compared. Initial protein concentrations: 71.5 ± 0.5 μg ml−1 for the wild type and 73 ± 3 μg ml−1 for the mutant. Protein concentrations after 48 h on 8 mM sulfide: 92 μg ml−1 and 95.5 ± 0.5 μg ml−1 for the wild type and mutant, respectively.
Mutant A. vinosum Alvin_1468::ΩKm revealed an exceptionally strong response with regard to the formation of sulfur globules. This strain was completely unable to form sulfur globules when exposed to 8 mM sulfide and exhibited a sulfur formation rate reduced by 55% on 4 mM thiosulfate (see Table S4 in the supplemental material). The localization of the gene is inconspicuous; genes related to sulfur metabolism are not apparent in its immediate vicinity (see Fig. S4 in the supplemental material). The Alvin_1468 protein is synthesized with a signal peptide for periplasmic localization and thus resides in the same cellular compartment as the sulfur globules that can no longer efficiently be formed in its absence. Alvin_1468 falls into the YceI-like family of proteins; however, there is no sequence homology to E. coli YceI, a periplasmic protein induced by high pH, and the only structurally characterized member of the family, TT1927b from Thermus thermophilus. The latter binds polyprenyl pyrophosphate and structurally resembles the lipocalin fold (59). Lipocalins bind a range of small hydrophobic molecules and have been reported to participate in transport of insoluble substrates in Eubacteria (60). Alvin_1468 may therefore be able to bind hydrophobic sulfur compounds, such as polysulfanes, organylsulfanes, or even elemental sulfur, that occur in purple sulfur bacterial sulfur globules or during their formation (44, 61, 62).
The A. vinosum mutant lacking Alvin_2093 was also specifically and severely affected with regard to formation of sulfur globules in the presence of sulfide (8 mM) and thiosulfate (4 mM). The respective rates reached not more than 40% of those of the wild type (see Table S4 in the supplemental material). The adjacent, adversely transcribed gene, Alvin_2092, encodes a cytoplasmic protein containing a sulfotransfer domain. Notably, Alvin_2095 conforms to COG0715, which comprises periplasmic components of ABC-type nitrate/sulfonate/bicarbonate transport systems. The protein derived from Alvin_2093 is predicted to be membrane bound and contains a nonconserved cysteine residue facing the cytoplasm (see Fig. S5b and c in the supplemental material). Speculations about its in vivo function are currently premature.
Exposure of mutant A. vinosum Alvin_3072::ΩKm to 8 mM sulfide resulted in a two-stage response very different from that of the wild type. The sulfur formation rate of the Alvin_3072 deficient mutant reached 57% of the wild-type rate (71.3 ± 22 nmol min−1 mg−1 [mean ± standard deviation]) for the first 30 min of the experiment and then dropped dramatically to only about 8% (9.8 ± 3.4 nmol min−1 mg−1) of the wild-type rate (Fig. 4; see also Table S4). The maximum sulfur content of the cells reached for the Alvin_3072 deficient mutant was only 17.8 ± 0.8 μmol mg−1, i.e., 58% of that found for the wild type (see Table S4). The gene Alvin_3072 resides in a cluster comprising Alvin_3069 to Alvin_3073 (see Fig. S6a). The encoded proteins are annotated as a thioredoxin domain-containing protein, a hypothetical protein, a Crp/Fnr family transcriptional regulator, a hypothetical protein, and a C4-dicarboxylate transporter/malic acid transport protein, respectively. Only Alvin_3072 and the adjacent transporter gene revealed increased relative mRNA levels on sulfide implying cotranscription of these two but not of the other genes with them (Table 2). Both genes are separated by only 48 nucleotides. The kanamycin Ω cassette replacing Alvin_3072 therefore most likely also prevents formation of the Alvin_3073-encoded transporter, and more experiments are needed to exactly dissect the effect exerted by the interposon. The protein Alvin_3073 belongs to a family of transporters that includes a malate uptake system in the yeast Schizosaccharomyces pombe (63), a tellurite resistance protein in Escherichia coli (64), and notably, a sulfite efflux pump in Saccharomyces cerevisiae and other fungi, including Aspergillus fumigatus (65). Alvin_3072 is conserved in several sulfide-oxidizing bacteria (see Fig. S6b). It is found in both purple sulfur bacteria, e.g., Halorhodospira halophia, and green sulfur bacteria, e.g., C. tepidum. The corresponding proteins share a conserved cysteine residue (see Fig. S6b) and are predicted to consist of two transmembrane domains with the cysteine residue located near the end of the second transmembrane helix oriented toward the cytoplasm (see Fig. S6c).
Fig 4.
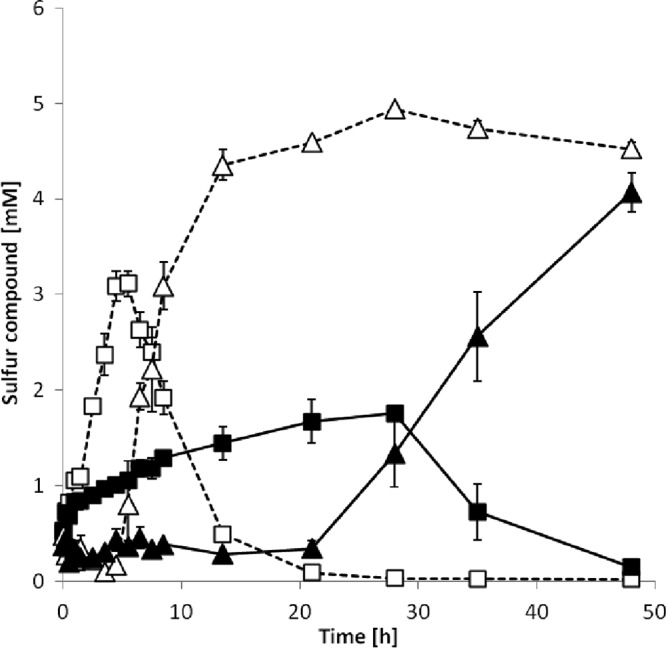
Intracellular sulfur and extracellular sulfate concentrations of A. vinosum wild type (dotted lines) and mutant A. vinosum Alvin_3072::ΩKm (solid lines) during growth on 8 mM sulfide. □, intracellular sulfur in the wild type; ■, intracellular sulfur in the mutant; △, extracellular sulfate in the wild type; ▲, extracellular sulfate in the mutant. Initial protein concentrations: 71.5 ± 0.5 μg ml−1 for the wild type and 72.5 ± 0.5 μg ml−1 for the mutant.
Conclusions.
Transcription profiles obtained for the purple sulfur bacterium A. vinosum upon exposure to sulfide, thiosulfate, elemental sulfur, or sulfite compared to photoorganoheterotrophic growth on malate provided global insights into changes in relative mRNA levels triggered by reduced sulfur compounds. The data generated during this study confirmed the important roles of established pathways, e.g., the Dsr pathway. Furthermore, Alvin_1196 and _1197, Alvin_1468, Alvin_2093, and Alvin_3072 are obvious examples that the DNA microarray technique constitutes an excellent method for the identification of new sulfur metabolism-related genes, not only in A. vinosum but surely also in other sulfur-oxidizing bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (grant 351/6-1).
We thank Renate Zigann for excellent technical assistance.
Footnotes
Published ahead of print 19 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00154-13.
REFERENCES
- 1.Pattaragulwanit K, Dahl C. 1995. Development of a genetic system for a purple sulfur bacterium: conjugative plasmid transfer in Chromatium vinosum. Arch. Microbiol. 164:217–222 [Google Scholar]
- 2.Dahl C. 1996. Insertional gene inactivation in a phototrophic sulphur bacterium: APS-reductase-deficient mutants of Chromatium vinosum. Microbiology 142:3363–3372 [DOI] [PubMed] [Google Scholar]
- 3.Weissgerber T, Zigann R, Bruce D, Chang Y-J, Detter JC, Han C, Hauser L, Jeffries CD, Land M, Munk AC, Tapia R, Dahl C. 2011. Complete genome sequence of Allochromatium vinosum DSM 180T. Stand. Genomic Sci. 5:311–330. 10.4056/sigs.2335270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denkmann K, Grein F, Zigann R, Siemen A, Bergmann J, van Helmont S, Nicolai A, Pereira IAC, Dahl C. 2012. Thiosulfate dehydrogenase: a wide-spread unusual acidophilic c-type cytochrome. Environ. Microbiol. 14:2673–2688 [DOI] [PubMed] [Google Scholar]
- 5.Grein F, Venceslau SS, Schneider L, Hildebrandt P, Todorovic S, Pereira IAC, Dahl C. 2010. DsrJ, an essential part of the DsrMKJOP complex in the purple sulfur bacterium Allochromatium vinosum, is an unusual triheme cytochrome c. Biochemistry 49:8290–8299 [DOI] [PubMed] [Google Scholar]
- 6.Hensen D, Sperling D, Trüper HG, Brune DC, Dahl C. 2006. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol. Microbiol. 62:794–810 [DOI] [PubMed] [Google Scholar]
- 7.Frigaard N-U, Dahl C. 2009. Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54:103–200 [DOI] [PubMed] [Google Scholar]
- 8.Imhoff JF. 2005. Family I. Chromatiaceae Bavendamm 1924, 125AL emend. Imhoff 1984b, 339, p 3–40 In Brenner DJ, Krieg NR, Staley JT, Garrity GM. (ed), Bergey's manual of systematic bacteriology. Springer, New York, NY [Google Scholar]
- 9.Pattaragulwanit K, Brune DC, Trüper HG, Dahl C. 1998. Molecular genetic evidence for extracytoplasmic localization of sulfur globules in Chromatium vinosum. Arch. Microbiol. 169:434–444 [DOI] [PubMed] [Google Scholar]
- 10.Reinartz M, Tschäpe J, Brüser T, Trüper HG, Dahl C. 1998. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch. Microbiol. 170:59–68 [DOI] [PubMed] [Google Scholar]
- 11.Gregersen LH, Bryant DA, Frigaard N-U. 2011. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front. Microbiol. 2:116. 10.3389/fmicb.2011.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welte C, Hafner S, Krätzer C, Quentmeier AT, Friedrich CG, Dahl C. 2009. Interaction between Sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Lett. 583:1281–1286 [DOI] [PubMed] [Google Scholar]
- 13.Pott AS, Dahl C. 1998. Sirohaem-sulfite reductase and other proteins encoded in the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144:1881–1894 [DOI] [PubMed] [Google Scholar]
- 14.Dahl C, Engels S, Pott-Sperling AS, Schulte A, Sander J, Lübbe Y, Deuster O, Brune DC. 2005. Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol. 187:1392–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sander J, Engels-Schwarzlose S, Dahl C. 2006. Importance of the DsrMKJOP complex for sulfur oxidation in Allochromatium vinosum and phylogenetic analysis of related complexes in other prokaryotes. Arch. Microbiol. 186:357–366 [DOI] [PubMed] [Google Scholar]
- 16.Lübbe YJ, Youn H-S, Timkovich R, Dahl C. 2006. Siro(haem)amide in Allochromatium vinosum and relevance of DsrL and DsrN, a homolog of cobyrinic acid a,c diamide synthase for sulphur oxidation. FEMS Microbiol. Lett. 261:194–202 [DOI] [PubMed] [Google Scholar]
- 17.Sanchez O, Ferrera I, Dahl C, Mas J. 2001. In vivo role of APS reductase in the purple sulfur bacterium Allochromatium vinosum. Arch. Microbiol. 176:301–305 [DOI] [PubMed] [Google Scholar]
- 18.Franz B, Lichtenberg H, Hormes J, Modrow H, Dahl C, Prange A. 2007. Utilization of solid “elemental” sulfur by the phototrophic purple sulfur bacterium Allochromatium vinosum: a sulfur K-edge XANES spectroscopy study. Microbiology 153:1268–1274 [DOI] [PubMed] [Google Scholar]
- 19.Weaver PF, Wall JD, Gest H. 1975. Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 105:207–216 [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21.Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1997. Current protocols in molecular biology. John Wiley & Sons, New York, NY [Google Scholar]
- 22.Bazaral M, Helinski DR. 1968. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J. Mol. Biol. 36:185–194 [DOI] [PubMed] [Google Scholar]
- 23.Dahl C, Speich N, Trüper HG. 1994. Enzymology and molecular biology of sulfate reduction in the extremely thermophilic archaeon Archaeoglobus fulgidus. Methods Enzymol. 243:331–349 [DOI] [PubMed] [Google Scholar]
- 24.Seibl R, Höltke H-J, Rüger R, Meindl A, Zauchau HG, Raßhofer R, Wolf H, Arnold N, Wienberg J, Kessler C. 1990. Nonradioactive labeling and detection of nucleic acids. Biol. Chem. Hoppe-Seyler 371:939–951 [DOI] [PubMed] [Google Scholar]
- 25.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784–791 [Google Scholar]
- 26.Bartlett JK, Skoog DA. 1954. Colorimetric determination of elemental sulfur in hydrocarbons. Anal. Chem. 26:1008–1011 [Google Scholar]
- 27.Sörbo B. 1987. Sulfate: turbidometric and nephelometric methods. Methods Enzymol. 143:3–6 [DOI] [PubMed] [Google Scholar]
- 28.Brazma A. 2009. Minimum information about a microarray experiment (MIAME): successes, failures, challenges. Sci. World J. 9:420–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polen T, Wendisch VF. 2004. Genomewide expression analysis in amino acid-producing bacteria using DNA microarrays. Appl. Biochem. Biotechnol. 118:215–232 [DOI] [PubMed] [Google Scholar]
- 30.van Gemerden H. 1968. Growth measurements of Chromatium cultures. Arch. Mikrobiol. 64:103–110 [DOI] [PubMed] [Google Scholar]
- 31.Viale AM, Kobayashi H, Akazawa T. 1990. Distinct properties of Escherichia coli products of plant-type ribulose-1,5-bisphosphate carboxylase/oxygenase directed by two sets of genes from the photosynthetic bacterium Chromatium vinosum. J. Biol. Chem. 265:18386–18392 [PubMed] [Google Scholar]
- 32.Hanson TE, Tabita FR. 2001. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 98:4397–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuller RC, Kornberg HL, Sisler EC, Smillie RM. 1961. Carbon metabolism in Chromatium. J. Biol. Chem. 236:2140–2149 [PubMed] [Google Scholar]
- 34.Dahl C, Rákhely G, Pott-Sperling AS, Fodor B, Takács M, Tóth A, Kraeling M, Győrfi K, Kovács A, Tusz J, Kovács K. 1999. Genes involved in hydrogen and sulfur metabolism in phototrophic sulfur bacteria. FEMS Microbiol. Lett. 180:317–324 [DOI] [PubMed] [Google Scholar]
- 35.Grein F, Pereira IAC, Dahl C. 2010. The Allochromatium vinsosum DsrMKJOP transmembrane complex: biochemical characterization of individual components aids understanding of complex function in vivo. J. Bacteriol. 192:6369–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogata H, Kellers P, Lubitz W. 2010. The crystal structure of the [NiFe] hydrogenase from the photosynthetic bacterium Allochromatium vinosum: characterization of the oxidized enzyme (Ni-A state). J. Mol. Biol. 402:428–444 [DOI] [PubMed] [Google Scholar]
- 37.Herter SM, Kortlüke CM, Drews G. 1998. Complex I of Rhodobacter capsulatus and its role in reverted electron transport. Arch. Microbiol. 169:98–105 [DOI] [PubMed] [Google Scholar]
- 38.Giuliani M-C, Jourlin-Castelli C, Leroy G, Hachani A, Giudici-Orticoni MT. 2010. Characterization of a new periplasmic single domain rhodanese encoded by a sulfur-regulated gene in a hyperthermophilic bacterium Aquifex aeolicus. Biochimie 92:388–397 [DOI] [PubMed] [Google Scholar]
- 39.Pibernat IV, Abella CA. 1996. Sulfide pulsing as the controlling factor of spinae production in Chlorobium limicola strain UdG 6038. Arch. Microbiol. 165:272–278 [DOI] [PubMed] [Google Scholar]
- 40.Brooke JS, Thompson JB, Beveridge TJ, Koval SF. 1992. Frequency and structure of spinae on Chlorobium spp. Arch. Microbiol. 157:319–322 [Google Scholar]
- 41.Neumann S, Wynen A, Trüper HG, Dahl C. 2000. Characterization of the cys gene locus from Allochromatium vinosum indicates an unusual sulfate assimilation pathway. Mol. Biol. Rep. 27:27–33 [DOI] [PubMed] [Google Scholar]
- 42.Kredich NM. 1992. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol. Microbiol. 6:2747–2753 [DOI] [PubMed] [Google Scholar]
- 43.Grimm F, Dobler N, Dahl C. 2010. Regulation of dsr genes encoding proteins responsible for the oxidation of stored sulfur in Allochromatium vinosum. Microbiology 156:764–773 [DOI] [PubMed] [Google Scholar]
- 44.Prange A, Engelhardt H, Trüper HG, Dahl C. 2004. The role of the sulfur globule proteins of Allochromatium vinosum: mutagenesis of the sulfur globule protein genes and expression studies by real-time RT PCR. Arch. Microbiol. 182:165–174 [DOI] [PubMed] [Google Scholar]
- 45.Van Petegem F, De Vos D, Savvides S, Vergauwen B, van Beeumen J. 2007. Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase. J. Mol. Biol. 374:883–889 [DOI] [PubMed] [Google Scholar]
- 46.Vergauwen B, Pauwels F, Jacquemotte F, Meyer TE, Cusanovich MA, Bartsch RG, van Beeumen JJ. 2001. Characterization of glutathione amide reductase from Chromatium gracile. Identification of a novel thiol peroxidase (Prx/Grx) fueled by glutathione amide redox cycling. J. Biol. Chem. 276:20890–20897 [DOI] [PubMed] [Google Scholar]
- 47.Pittman MS, Robinson HC, Poole RK. 2005. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J. Biol. Chem. 280:32254–32261 [DOI] [PubMed] [Google Scholar]
- 48.Suzuki H, Koyanagi T, Izuka S, Onishi A, Kumagai H. 2005. The yliA, -B, -C, and -D genes of Escherichia coli K-12 encode a novel glutathione importer with an ATP-binding cassette. J. Bacteriol. 187:5861–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laska S, Lottspeich F, Kletzin A. 2003. Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology 149:2357–2371 [DOI] [PubMed] [Google Scholar]
- 50.Eddie BJ, Hanson TE. 2012. Chlorobaculum tepidum TLS displays a complex transcriptional respsonse to sulfide addition. J. Bacteriol. 195:399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stockdreher Y, Venceslau SS, Josten M, Sahl HG, Pereira IAC, Dahl C. 2012. Cytoplasmic sulfurtransferases in the purple sulfur bacterium Allochromatium vinosum: evidence for sulfur transfer from DsrEFH to DsrC. PLoS One 7(7):e40785. 10.1371/journal.pone.0040785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. 2006. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell 21:97–108 [DOI] [PubMed] [Google Scholar]
- 53.Clery A, Blatter M, Allain FHT. 2008. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 18:290–298 [DOI] [PubMed] [Google Scholar]
- 54.Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mowat CG, Rothery E, Miles CS, McIver L, Doherty MK, Drewette K, Taylor P, Walkinshaw MD, Chapman SK, Reid GA. 2004. Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation. Nat. Struct. Mol. Biol. 11:1023–1024 [DOI] [PubMed] [Google Scholar]
- 56.Fellay R, Frey J, Krisch HM. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vivo insertional mutagenesis of Gram-negative bacteria. Gene 52:147–154 [DOI] [PubMed] [Google Scholar]
- 57.Brito JA, Sousa FL, Stelter M, Bandeiras TM, Vonrhein C, Teixeira M, Pereira MM, Archer M. 2009. Structural and functional insights into sulfide:quinone oxidoreductase. Biochemistry 48:5613–5622 [DOI] [PubMed] [Google Scholar]
- 58.Marcia M, Ermler U, Peng GH, Michel H. 2009. The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc. Natl. Acad. Sci. U. S. A. 106:9625–9630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Handa N, Terada T, Doi-Katayama Y, Hirota H, Tame JR, Park SY, Kuramitsu S, Shirouzu M, Yokoyama S. 2005. Crystal structure of a novel polyisoprenoid-binding protein from Thermus thermophilus HB8. Protein Sci. 14:1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bishop RE. 2000. The bacterial lipocalins. Biochim. Biophys. Acta 1482:73–83 [DOI] [PubMed] [Google Scholar]
- 61.Prange A, Arzberger I, Engemann C, Modrow H, Schumann O, Trüper HG, Steudel R, Dahl C, Hormes J. 1999. In situ analysis of sulfur in the sulfur globules of phototrophic sulfur bacteria by X-ray absorption near edge spectroscopy. Biochim. Biophys. Acta 1428:446–454 [DOI] [PubMed] [Google Scholar]
- 62.Franz B, Gehrke T, Lichtenberg H, Hormes J, Dahl C, Prange A. 2009. Unexpected extracellular and intracellular sulfur species during growth of Allochromatium vinosum with reduced sulfur compounds. Microbiology 155:2766–2774 [DOI] [PubMed] [Google Scholar]
- 63.Osawa H, Matsumoto H. 2006. Cytotoxic thio-malate is transported by both an aluminum-responsive malate efflux pathway in wheat and the MAE1 malate permease in Schizosaccharomyces pombe. Planta 224:462–471 [DOI] [PubMed] [Google Scholar]
- 64.Liu MF, Turner RJ, Winstone TL, Saetre A, Dyllick-Brenzinger M, Jickling G, Tari LW, Weiner JH, Taylor DE. 2000. Escherichia coli TehB requires S-adenosylmethionine as a cofactor to mediate tellurite resistance. J. Bacteriol. 182:6509–6513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nardi T, Corich V, Giacomini A, Blondin B. 2010. A sulphite-inducible form of the sulphite efflux gene SSU1 in a Saccharomyces cerevisiae wine yeast. Microbiology 156:1686–1696 [DOI] [PubMed] [Google Scholar]
- 66.Hipp WM, Pott AS, Thum-Schmitz N, Faath I, Dahl C, Trüper HG. 1997. Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology 143:2891–2902 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.