Abstract
Toxigenic Clostridium sordellii causes uncommon but highly lethal infections in humans and animals. Recently, an increased incidence of C. sordellii infections has been reported in women undergoing obstetric interventions. Pathogenic strains of C. sordellii produce numerous virulence factors, including sordellilysin, phospholipase, neuraminidase, and two large clostridial glucosylating toxins, TcsL and TcsH. Recent studies have demonstrated that TcsL toxin is an essential virulence factor for the pathogenicity of C. sordellii. In this study, we identified and characterized TcsR as the toxin gene (tcsL) regulator in C. sordellii. High-throughput sequencing of two C. sordellii strains revealed that tcsR lies within a genomic region that encodes TcsL, TcsH, and TcsE, a putative holin. By using ClosTron technology, we inactivated the tcsR gene in strain ATCC 9714. Toxin production and tcsL transcription were decreased in the tcsR mutant strain. However, the complemented tcsR mutant produced large amounts of toxins, similar to the parental strain. Expression of the Clostridium difficile toxin gene regulator tcdR also restored toxin production to the C. sordellii tcsR mutant, showing that these sigma factors are functionally interchangeable.
INTRODUCTION
Clostridium sordellii, an anaerobic, Gram-positive, spore-forming bacterium, is a common inhabitant of soil and the animal gastrointestinal tract. Virulent strains of C. sordellii are recognized as the causative agents of a broad spectrum of human diseases, including myonecrosis, uterine infections, and sepsis. C. sordellii is also known to cause lethal infections in several animal species, including sheep, foals, and lambs (1–4). Recently, fatal cases of C. sordellii endometritis following medical abortions caused by mifepristone-misoprostol combinations have been reported (5). It has been suggested that mifepristone-misoprostol may facilitate colonization of C. sordellii in uterine tissue, trigger toxin expression, and induce hypotension and systemic shock by deregulating the host's immune response (6).
Pathogenic C. sordellii strains produce up to seven identified exotoxins (7). Of these, the two major toxins, lethal toxin (TcsL) and the hemorrhagic toxin (TcsH), are regarded as major virulence factors (8, 9). The lethal toxin produced by C. sordellii was shown to evoke enteritis in animals and proved essential for the virulence of C. sordellii (9, 10). TcsH and TcsL are members of the large clostridial cytotoxin (LCC) family, with predicted molecular masses of 300 kDa and 250 kDa, respectively (8, 9). The C. sordellii toxins were reported to be similar to Clostridium diffcile toxins A and B, both in terms of biological activity and antigenicity (11). To date, only the TcsL-encoding gene has been sequenced; it was found to be 76% identical to the C. difficile toxin B gene (tcdB).
In this study, we sequenced two C. sordellii strains, ATCC 9714 and VPI 9048, by high-throughput techniques, and we identified many open reading frames (ORFs) surrounding the tcsL gene. Consistent with previous reports, the VPI 9048 strain carries both the TcsL- and TcsH-encoding genes, whereas strain ATCC 9714 encodes only the lethal toxin TcsL. In the region of the toxin genes, we identified a small ORF with similarities to RNA polymerase sigma factors, including TcdR, the sigma factor that transcribes C. difficile toxin genes. We named the apparent sigma factor ORF tcsR and report here its role in toxin gene regulation. A C. sordellii tcsR mutant was found to be defective in toxin production, due to reduced transcription of tcsL. Further, we complemented the mutant with a functional tcsR gene and found that toxin production was restored. The C. sordellii tcsR mutant could also be complemented by the C. difficile toxin gene regulator, tcdR, showing that these sigma factors are closely related to each other. This is the first known report on the toxin locus region in the C. sordellii genome and its regulator.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. sordellii strains VPI 9048 (TechLab, VA) (12), ATCC 9714 (13) (Table 1), and the tcsR mutant strain were grown anaerobically (10% H2, 10% CO2, and 80% N2) in tryptose-yeast extract (TY) broth or TY agar. E. coli strain SL-17, used for conjugation, was cultured aerobically in LB medium. When necessary, E. coli cultures were supplemented with chloramphenicol or ampicillin at 30 μg ml−1 and 100 μg ml−1, respectively. All routine plasmid constructions were carried out using standard procedures.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) and/or origin | Source or reference |
|---|---|---|
| C. sordelii strains | ||
| ATCC 9714 | TcsL+ TcsH− | American Type Culture Collection (13) |
| VPI 9048 | TcsL+ TcsH+ | Tec Lab (VA) (12) |
| E. coli strains | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | New England BioLabs, MA |
| S17-1 | Favors conjugation | 43 |
| Plasmids | ||
| pMTL007C-E5 | ClosTron plasmid | 14 |
| pTUM007::Cdi-tcsR-342 | pMTL007C-E5 carrying tcsR-specific intron | This study |
| pRPF185 | pMTL960, Cmr Tmr, gusA+, inducible tetracycline (Tet) promoter | 17 |
| pRGL153A | pRPF185 with a promoterless gusA gene | This study |
| pRGL100 | Tet promoter in pRPF185 replaced with tcsL promoter | This study |
| pRGL161 | Tet promoter in pRPF185 replaced with tcsR promoter | This study |
| pRGL162 | Tet promoter in pRPF185 replaced with tcsH promoter | This study |
| pRGL163 | Tet promoter in pRPF185 replaced with tcsE promoter | This study |
| pRGL154 | pRPF185 without a gusA gene | This study |
| pRGL145-1 | pRGL154 with wild-type tcsR under inducible Tet promoter | This study |
| pRGL144-1 | pRGL154 carrying wild-type tcdR under inducible Tet promoter | This study |
High-throughput genome sequencing, assembly, and annotation.
The genomes of C. sordellii ATCC 9714 and VPI 9048 were sequenced using the FLX genome sequencer (Roche 454 Life Science, Branford, CT) and the Illumina (San Diego, CA) genome analyzer following the manufacturers' instructions. For Roche 454 sequencing, the shotgun library was prepared with 5 μg of genomic DNA using the standard DNA library preparation kit (04852265001; Roche). Nebulized, purified, and adaptor-ligated single-stranded DNA fragments were clonally amplified using the emulsion PCR kit I (04852290001; Roche). Sequencing on the GS FLX was performed using the standard LR70 sequencing kit (04932315001; Roche). The images were processed using the genome sequencer FLX data processing pipeline 1.1.02.15, and sequences generated were assembled using the Newbler assembler (Roche). For Illumina sequencing, the DNA template library was prepared using the Illumina genomic DNA sample prep kit. Briefly, 5 μg of genomic DNA was broken into fragments of approximately 100 bp by nebulization. After performing end repairing and adaptor ligation, the samples were gel purified to recover fragments of 150 to 250 bp that were then PCR amplified for 15 cycles. The DNA template library was then used for flow cell preparation using the standard cluster generation kit (Illumina). Sequencing on the Illumina genome analyzer was performed using genomic DNA sequencing primer V2 for 36 cycles. At the end of the run, images were processed using the Solexa data analysis pipeline 0.2.2.6. Reads from Roche 454 and Illumina systems were mapped to contigs using the SOAP package and default parameters. Assembled contigs were submitted to for annotation service to The Institute for Genomic Sciences at the University of Maryland, where it was run through the prokaryotic annotation pipeline. Along with gene finding, results of Glimmer, Blast-extend-repraze (BER) searches, HMM searches, TMHMM searches, and SignalP predictions, and automatic annotations from Auto-Annotate were included in the annotation pipeline.
Construction of the tcsR mutant.
A tcsR mutant was generated in C. sordellii ATCC 9714 by the insertion of a bacterial group II intron using the ClosTron gene knockout system as described by Heap et al. (14). The insertion site in the antisense orientation between nucleotides 234 and 235 of the tcsR ORF was selected to design the retargeting intron. The intron was designed using the Perutka algorithm, a Web-based design tool available at the Clostron site. It was then synthesized and cloned in plasmid pMTL007-E5C. The resulting plasmid, pTUM007: Cdi-tcsR-234a, was transferred to C. sordellii strain ATCC 9714 by conjugation as described previously (14). Thiamphenicol-resistant transconjugants were resuspended in 200 μl of TY broth and plated on TY agar plates containing erythromycin (5 μg ml−1) to select potential Ll.ltrB insertions. The putative tcsR mutants were then screened by PCR using tcsR-specific primers (ORG94 and ORG95) in combination with the EBS-U universal primer (see Table S1 in the supplemental material).
Southern blot analysis.
Southern blot analysis was performed as described previously to verify a specific single integration of the group II intron into the genome (15). Ten micrograms of genomic DNA was digested with EcoRV enzyme and separated on a 0.8% agarose gel by electrophoresis. DNA was transferred onto an Immobilon-NY+ nylon membrane (Millipore, Bedford, MA) by the capillary transfer method. Prehybridization of the filter was conducted for 2 h at 60°C in 5× SCC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt solution, and 100 mg/ml of salmon sperm DNA. Probes specific for the group II intron ermB gene and tcsR genes were radiolabeled ([32P]dATP) using a High Prime kit (Roche) and hybridized overnight in 10 ml fresh prehybridization buffer at 60°C. The hybridized membrane was washed twice for 30 min in 2× SCC, 0.5% SDS, for 30 min and in 1× SSC, 0.5% SDS, and then analyzed using a phosphorimage screen and a Typhoon 9410 scanner (GE Healthcare).
Growth measurement with BioScreenC plate reader.
The growth patterns of parent and tcsR mutant strains were studied in TY medium by using a Bioscreen C plate reader that was kept inside the anaerobic chamber. Ten-fold-diluted overnight-grown bacterial cultures (15 μl) were used to inoculate 150 μl TY medium into each well. Ten wells were inoculated with the parent strain and another 10 with the mutant strain to monitor their growth over 24 h. The plate temperature was maintained at 37°C throughout the growth period, and the optical density at 600 nm (OD600) was measured every 30 min after 10 s of shaking of the plate.
Toxin assay.
For the toxin assay, C. sordellii ATCC 9714 and its tcsR mutant cultures were grown in TY for 10 h, and the bacterial cells were collected after centrifugation. Cell pellets were resuspended in 10 mM Tris buffer (pH 8.0) containing a protease inhibitor cocktail (Roche, Mannheim, Germany). The cytosolic contents were obtained by sonicating the cells, followed by a brief centrifugation to remove the unbroken cells and cell debris. The total protein concentration was determined using the Bio-Rad protein assay reagent. Equal amounts of cytosolic proteins (50 μg) were assayed for their relative toxin contents by using the C. difficile Premier Toxin A&B enzyme-linked immunosorbent assay (ELISA) kit from Meridian Diagnostics Inc. (Cincinnati, OH). This ELISA kit is known to recognize C. sordellii toxins as well (16).
RNA extraction and QRT-PCR.
Total RNA was extracted from C. sordellii cultures grown for 10 h in TY medium, following a protocol described previously (10, 14). After treating the total RNA with DNase (Turbo; Ambion), reverse transcription (RT) was performed using avian myeloblastosis virus reverse transcriptase (Promega) and random hexamer oligonucleotide primers with1 μg of template RNA. The cDNA samples were then stored at −20°C until needed. Primers specific for tcsL, tcsE, and tcsR (see Table S1 in the supplemental material) were designed using Primer 3 software (Geneious Software). Quantitative RT-PCR (QRT-PCR) was performed using the iQPCR real-time PCR instrument (Bio-Rad). Reactions were carried out using SYBR green master mix (Bio-Rad) with 20 ng of cDNA as the template. Samples were normalized using C. sordellii 16S rRNA.
Construction of reporter plasmids and β-glucuronidase assay.
Approximately 600 bp of the upstream DNA regions of tcsL, tcsH, tcsR, or tcsE genes, along with their potential ribosomal-binding sites (RBS), were PCR amplified using specific primers with KpnI and SacI recognition sequences (see Table S1) using ATCC 9714 chromosomal DNA as a template. Plasmid pRPF185 carries a gusA gene for β-glucuronidase under the tetracycline-inducible (tet) promoter (17). Using KpnI and SacI digestion, we removed the tet promoter and replaced it with either tcsL, tcsR, tcsH, or tcsE upstream regions to create plasmids pRGL100, pRGL161, pRGL162 and pRGL163, respectively (Table 1). To create plasmid pRGL153A with promoterless gusA, we removed the tet promoter from plasmid pRPF185 with KpnI, SacI digestions and then the construct was self-ligated after creating blunt ends. Plasmids pRGL100, pRGL161, pRGL162, pRGL163, and pRGL153A (control) were introduced into ATCC 9714 and its tcsR mutant through conjugation as described above. The transconjugants were then grown in TY medium in the presence of thioamphenicol (15 μg/ml) overnight. Overnight cultures were used as inocula at a 1:100 dilution to start a new culture. Bacterial cultures were harvested at 10 h of growth, and the amount of β-glucuronidase activity was assessed as described elsewhere (18) with minor modifications. Briefly, the cells were washed, suspended in 0.8 ml of Z buffer (60 mM Na2HPO4 · 7H2O [pH 7.0], 40 mM NaH2PO4 · H2O, 10 mM KCl, 1 mM MgSO4 · 7H2O, 50 mM 2-mercaptoethanol), and sonicated. The enzyme reaction was started by the addition of 0.16 ml of 6 mM p-nitrophenyl β-d-glucuronide (Sigma) to the broken cells and stopped by the addition of 0.4 ml of 1.0 M NaCO3. β-Glucuronidase activity was calculated as described earlier (18, 19).
Complementation of the C. sordellii tcsR mutant.
The tcsR ORF along with its RBS were PCR amplified from ATCC 9714 chromosomal DNA by using primers ORG203 and ORG204 (see Table S1 in the supplemental material). Similarly, the tcdR ORF with its RBS were amplified from JIR8094 chromosomal DNA by using primers ORG208 and ORG209. The resulting PCR products digested with SacI and BamHI were eventually cloned into vector pRFP185 (17) under a tetracycline-inducible promoter to create plasmids pRGL145-1(with tcsR) and pRGL144-1 (with tcdR), which were then introduced into the tcsR mutant strain. Transconjugants carrying either pRGL145-1, pRGL144-1, or the vector pRGL154 alone (pRPF185 without gusA) were grown overnight in TY medium supplemented with thiamphenicol. Fresh 10-ml cultures were initiated using 0.1 ml of overnight culture and were grown for 4 h in TY medium up to an OD600 of 0.5 with thiamphenicol before the induction with ATc (anhydrotetracycline) at a concentration of 50 ng/ml. Cultures were harvested 4 h after induction, and cytosolic proteins were extracted for the detection of toxins via ELISA.
Nucleotide sequence accession numbers.
The nucleotide sequences and the corresponding automated annotations for the first versions of the genomes of C. sordellii strains ATCC 9714 and VPI 9048 were submitted to GenBank and assigned accession numbers APWR00000000 and AQGJ00000000, respectively.
RESULTS
Sequencing and de novo assembly of C. sordellii genomes.
Genome sequences for two C. sordellii strains were generated via the 454 and Illumina sequencing technologies. Strain ATCC 9714 is known to produce only TcsL, whereas strain VPI 9048 produces both TcsL and TcsH toxins. The Roche 454 GS FLX system was used to generate sequences of strain ATCC 9714, which were assembled using the Newbler assembler (Roche). A total of 637,164 reads with an average length of 387 bases was obtained and was assembled into 164 contigs with an average contig size of 21,629 bp. Strain ATCC 9714 was resequenced using Illumina technology along with strain VPI 9048. Totals of 3.563 and 3.616 million reads of 35 bases in length were obtained for ATCC 9714 and VPI 9048, respectively. Sequences from both 454 and Illumina were assembled using the SOAP package. This resulted in 104 and 166 contigs for strains ATCC 9714 and VPI 9048, respectively. Gaps in genome coverage were not filled in with manual sequencing due to resource constraints. The overall characteristics of the draft C. sordellii genomes are summarized in Table 2.
Table 2.
Salient features of C. sordellii draft genomes and other Clostridium spp. strain genomes
| Feature | C. sordellii ATCC 9714a | C. sordellii VPI 9048a | C. difficile 630b | C. difficile R20291b | C. difficile 196b | C. perfringens strain 13b | C. botulinum ATCC 3502b |
|---|---|---|---|---|---|---|---|
| Size (Mbp) | ∼3.03 | ∼3.32 | 4.29 | 4.19 | 4.11 | 3. 03 | 3.88 |
| G+C % | 27.4 | 27.3 | 29.06 | 28.8 | 28.6 | 28.57 | 28.24 |
| Protein-coding genes | 3,271c | 3,985c,d | 3,798 | 3,757 | 3,454 | 2,723 | 3,590 |
| RNA | |||||||
| tRNA genes | 66c | 36c | 87 | 82 | 82 | 96 | 80 |
| rRNA 23S | 12c | 6c | 11 | 10 | 10 | 10 | 9 |
| rRNA 16S | 12c | 6c | 11 | 9 | 10 | 10 | 9 |
| rRNA 5S | 12c | 6c | 10 | 8 | 9 | 10 | 9 |
Draft genome.
Genome information available as of May 2013.
Predicted value; the number may change in the future.
Only ORFs with more than 50 amino acid residues are included in this value.
Features of C. sordellii toxin gene locus and its similarity to the C. difficile PaLoc.
Bacterial virulence-associated genes are often found in mobile genetic elements. In clostridia, the tetanus toxin gene of Clostridium tetani is carried on a plasmid, while the genes for botulinum toxins in Clostridium botulinum strains are within bacteriophage genomes (20, 21). The gene for Clostridium novyi alpha-toxin, which shows high homology to TcsL of C. sordellii and TcdA of C. difficile, is also carried by a phage (22). However, the pathogenicity locus (PaLoc) in C. difficile is distinct and is not associated with an actively mobile genetic element (23). The C. difficile PaLoc includes five genes, tcdR, tcdB, tcdE, tcdA, and tcdC and is found in the same locus in all C. difficile toxigenic strains (Fig. 1C). Regions adjacent to the C. difficile PaLoc are not similar to any known transposon, plasmid, or phage-like element (23). However, the base composition of the PaLoc differs from that of the genome as a whole, suggesting that it was acquired by horizontal transfer (23).
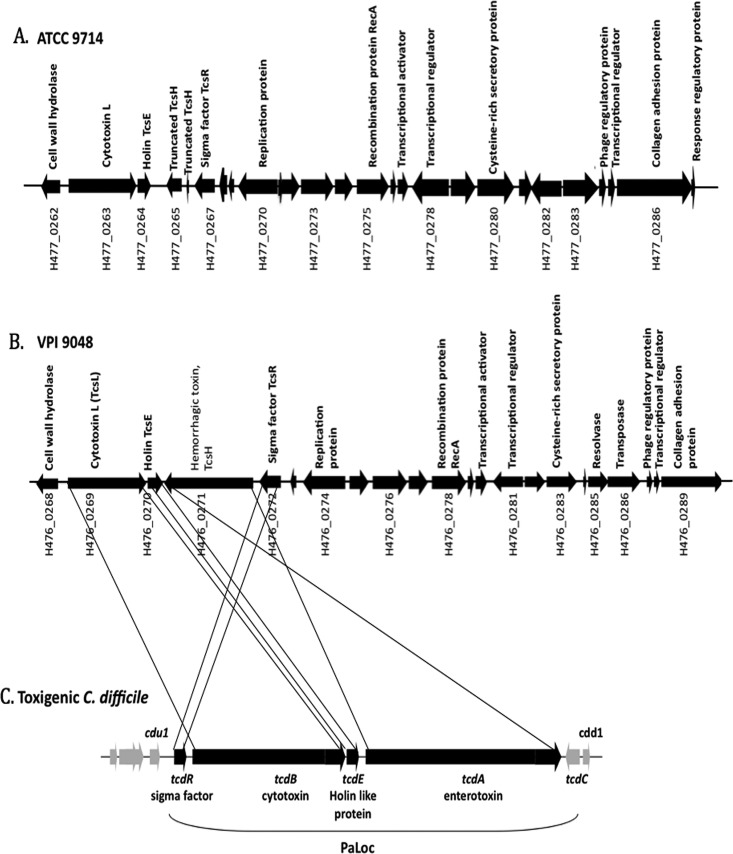
Fig 1.
(A and B) Genetic organization of genes near tcsL and tcsH in C. sordellii strains ATCC 9714 (A) and VPI 9048 (B). (C) Genetic organization of genes in the C. difficile PaLoc. The ORFs are indicated by arrowheads pointing in the direction of transcription. Black lines drawn between the genes of C. sordellii (VPI 9048) and the C. difficile PaLoc represent regions with sequence similarity between the two bacteria.
In C. sordellii, the lethal toxin-encoding tcsL gene is 76% similar to tcdB, and the hemorrhagic toxin-encoding gene tcsH is 78% similar to tcdA. We identified the tcsL gene within the 57,746-bp contig 88 in strain ATCC 9714 and within the 77,359-bp contig 152 of the VPI 9048 strain. Most of the genes surrounding the toxin-encoding genes are conserved in strains VPI 9048 and ATCC 9714 (Fig. 1A and B). Immediately downstream of the tcsL gene is tcsE, a gene that encodes a holin-like protein that is homologous to the tcdE gene of C. difficile. TcdE is essential for the efficient secretion of toxins by C. difficile (24). In strain VPI 9048, tcsH, the hemorrhagic toxin-encoding gene, lies downstream of tcsE, but in strain ATCC 9714 only a truncated tcsH gene is present. Immediately upstream of tcsH is tcsR, a gene that is homologous to several sigma factor-encoding genes, including tcdR of C. difficile. Organization of the toxin genes in C. sordellii differs from that of the C. difficile pathogenicity locus. In C. difficile, the toxin genes tcdA and tcdB are transcribed in the same direction, but in C. sordellii, tcsL and tcsH are transcribed in opposite directions. In C. difficile, tcdR is upstream of the tcdB gene and is transcribed in the same direction as the toxin genes. In C. sordellii the tcsR gene is upstream of the tcsH gene and is transcribed in the same direction as the tcsH gene. In C. difficile, tcdC, a gene downstream of tcdA, codes for an anti-sigma factor that affects toxin gene transcription by regulating TcdR activity. We were unable to identify any tcdC homologue near the toxin locus of C. sordellii or at any other location in the incompletely sequenced genomes. This suggests that the toxin genes in C. sordellii may be regulated differently from C. difficile.
Unlike the case for the C. difficile PaLoc, the genes adjacent to the toxin genes in C. sordellii show several hallmarks of a mobile genetic element. Specifically, the toxin locus of C. sordellii shows signatures of integrative and conjugative elements (ICEs) (Fig. 1A and B; see also Tables S2 and S3 in the supplemental material). ICEs are self-transmissible, mobile, genetic elements that encode the machinery for conjugation as well as for the regulatory systems to control their excision from the chromosome and their conjugative transfer. Unlike conjugative plasmids, ICEs do not replicate autonomously; instead, they integrate into the host chromosome. Predicted coding sequences in the toxin locus show homology to conjugal transfer proteins, plasmid replication proteins, transposases, recombinases, and resolvases (see Tables S2 and S3). The proteins encoded by genes VPI 9048 H476_0274 and ATCC 9714 H477_0270, which lie upstream of tcsR, show homology to various plasmid replication proteins (see Tables S2 and S3). The genes (VPI 9048 H476_0297, VPI 9048 H476_0302, ATCC 9714 H477_0297, and ATCC 9714 H477_0298) appear to encode a type IV secretory system conjugative DNA transfer family protein that is also part of the toxin locus. In addition, a type IV secretion system-coupling DNA-binding domain protein was identified near the toxin-encoding genes. Moreover, a TraB homologue (VPI 9048 H476_0319) that plays a role in DNA transfer in other bacteria is also present in the toxin loci, indicating the possibility of conjugal transfer of the genetic element. The presence of transposase-encoding genes (VPI 9048 H476_0321 and VPI 9048 H476_0286) along with conjugative elements suggests that the toxin loci in C. sordellii may be part of an integrative conjugative element. All ICEs encode an integrase, which enables their integration into the host chromosome by site-specific recombination. In the C. sordellii toxin loci, we couldn't identify any genes likely to encode an integrase, but we did find a recA-type gene that might encode a protein involved in homologous recombination (see Tables S2 and S3 in the supplemental material). This is an unusual signature for an ICE and requires further functional characterization.
Besides the main pathogenicity factors TcsH and TcsL, the toxin locus in C. sordellii codes for several proteins that may be involved in virulence-associated processes during infection. The genes H476_0289 in VPI 9048 and H477_0286 in ATCC 9714 code for a possible collagen-binding protein with predicted CNA peptide repeats in the C-terminal region. In Staphylococcus aureus, the CNA repeat protein mediates bacterial adherence to collagenous tissues, such as cartilage, a process that is important in the pathogenesis of septic arthritis caused by staphylococci (25). The strain VPI 9048 toxin locus also includes a gene that codes for a probable GNAT family acetyltransferase (H476_0290). Such proteins include aminoglycoside acetyl transferases that confer resistance to the antibiotics kanamycin and gentamicin (26, 27). The gene VPI 9048 H476_0305 appears to encode a cell wall protein with an endopeptidase domain and may be involved in the invasion of host cells.
Mutation in tcsR affects cytotoxin production in C. sordellii.
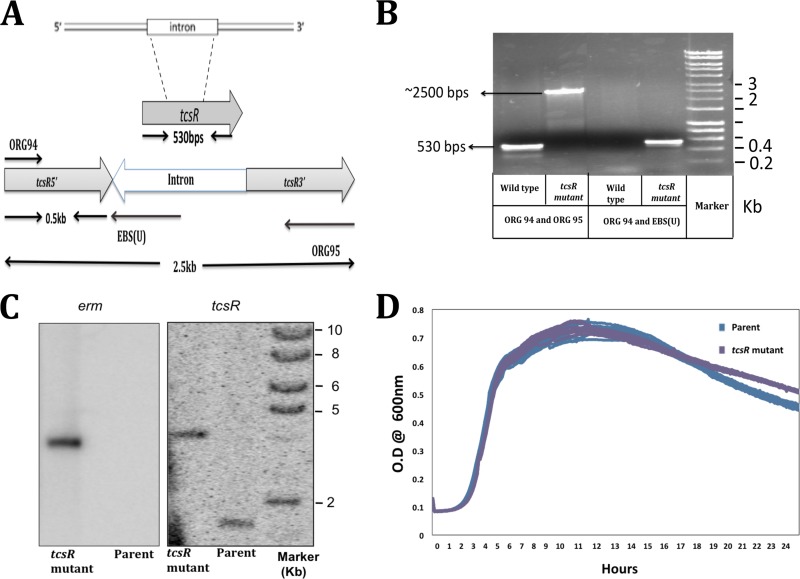
We initiated functional characterization of the toxin loci genes by characterizing the tcsR gene, which is present downstream of tcsL. TcsR appears to be a 174-residue protein that shows 34% of homology to C. difficile TcdR, an alternative sigma factor that drives transcription of the toxin genes tcdA and tcdB (19, 28). To determine whether TcsR is necessary for high-level expression of C. sordellii tcsL, a tcsR mutant was created in the ATCC 9714 strain by using the ClosTron technique (14). Intron insertion sites in the tcsR gene were identified using the Perutka algorithm available from the ClosTron site (http://clostron.com/ ). The group II intron in plasmid pMTL007C-E5 was retargeted to integrate within the tcsR coding sequence at position bp 342 on the DNA sense strand (Fig. 2A). The tcsR-retargeted plasmid pTUM007::Cdi-tcsR-342 was introduced into C. sordellii ATCC 9714 by conjugation with E. coli, and thioamphenicol-resistant transconjugants were selected. We could not introduce plasmids into VPI 9048, even with repeated attempts. Hence, our tcsR characterization study was limited to the ATCC 9714 strain. To confirm the successful inactivation of the tcsR gene, PCR was performed using gene-specific primers (ORG94 and ORG95) and an intron-specific primer (EBS universal primer). When tcsR-specific primers were used, amplified bands of 2.5 kb and 0.5 kb were obtained from the tcsR mutant and parent strains, respectively (Fig. 2B). The presence of the 2.5-kb amplification product indicates the presence of a 2.0-kb intron within the tcsR gene. PCR was also performed using the intron-specific primer EBS universal and the tcsR-specific primer ORG94. A PCR product of 0.5-kb was observed only in the tcsR mutant (Fig. 2B). Furthermore, Southern blot hybridizations were performed to confirm a single integration site of the group II intron within the tcsR gene in the mutant strain chromosome. Chromosomal DNA from strain ATCC 9714 and its tcsR mutant strain was digested with EcoRV and subjected to Southern blot hybridization using 32P-labeled tcsR and ermB probes. As expected, the tcsR probe hybridized with both the mutant and the parent strains. In the parent strain, a band at 1.9-kb was observed, and in the mutant the probe hybridized with a band of 4 kb, consistent with the insertion of the intron into the tcsR gene. The intron-specific ermB probe hybridized only with the tcsR mutant strain in the same 4-kb band, further confirming the presence of the intron within the tcsR gene (Fig. 2C). Growth curves of the parent and tcsR mutant strains in TY medium over a 24-hour period were essentially identical (Fig. 2D). To see if TcsR plays a role in toxin production, toxin enzyme-linked immunosorbent assays (ELISAs) were performed with equal amounts of cytosolic proteins (50 μg/well) from the 10-h-old parent and tcsR mutant strains. Absorbance recorded at 405 nm represented the toxin titer. The absorbance for mutant strain samples was approximately 4-fold lower than for the parent strain. This result suggests that TcsR is required for maximal toxin production in C. sordellii (Fig. 3A).
Fig 2.
Construction and characterization of the tcsR mutant in C. sordellii ATCC 9714. (A) Schematic representation of ClosTron (group II intron)-mediated disruption of the tcsR gene in C. sordellii. (B) PCR verification of the intron insertion, conducted with gene-specific primers ORG94 and ORG95 or the intron-specific primer EBS universal [EBS(U)] with ORG94. (C) Southern blot analysis of genomic DNA from C. sordellii ATCC 9714 and tcsR mutant strains with erm (intron-specific) and tcsR probes. Chromosomal DNA was digested with EcoRV. (D) Growth curves of parent ATCC 9714 and the tcsR mutant in TY medium.
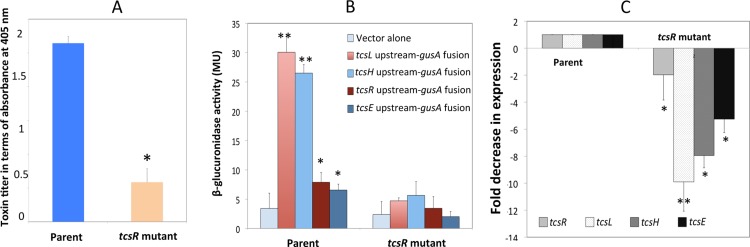
Fig 3.
TcsR mediates the transcription of toxin genes. (A) Quantification of toxins in parent ATCC 9714 and tcsR mutant strains. Toxin titers in cytoplasmic proteins harvested from parent and tcsR mutant were determined by ELISA, and the signal from the test was recorded as the absorbance at 405 nm. The data shown are means ± standard errors of three replicate samples. Student's t test was used for statistical analysis. *, P < 0.05. (B) Expression of β-glucuronidase in parent ATCC 9714 and tcsR mutant strains carrying plasmids with gusA as the reporter gene fused to the promoters of tcsL, tcsH, tcsR, and tcsE. Strains carrying a promoterless gusA plasmid (pRGL153A) were used as control. Data represent the means ± standard errors of the means (SEM) (n = 3). (C) Comparison of transcript levels of tcsL, tcsH, tcsR, and tcsE in parent and tcsR mutant strains based on QRT-PCR. Data represent the mean fold change in expression ± SEM (n = 3) compared to the parent ATCC 9714 strain. Ten-hour-old bacterial cultures were used in all the experiments presented.
TcsR affects tcsL transcription in C. sordellii.
To verify that TcsR regulates toxin gene transcription in C. sordellii, a tcsL promoter-gusA fusion was constructed. A 600-bp region upstream of the tcsL gene was PCR amplified and cloned in the vector pRGL153A to create plasmid pRGL100, which was then introduced into the parent strain ATCC 9714 and its tcsR mutant strain by transconjugation. Similarly, constructs with tcsH, tcsR, and tcsE promoter-gusA fusion constructs were also made and introduced into the parent and tcsR mutant strains. Strains carrying promoter-gusA fusions (pRGL100, pRGL161, pRGL162, or pRGL163) or vector alone (pRGL153A) were grown in TY medium with thioamphenicol, and a β-glucuronidase assay was performed using samples collected after 10 h of growth (late exponential phase). A 6-fold-higher level of β-glucuronidase activity was recorded for the parent strain than for the tcsR mutant strain (Fig. 3B). Similarly, a tcsH-gusA fusion was expressed at a 5-fold-higher level in the parent strain than in the tcsR mutant. Approximately 2-fold-higher expression levels of the tcsR-gusA and tcsE-gusA fusions were recorded in the parent strain than in the tcsR mutant. These results provided evidence that TcsR positively influences the transcription of tcsL, tcsH, tcsR, and tcsE genes in C. sordellii.
QRT-PCR was also performed with the RNA extracted from 10-h cultures of the parent and tcsR mutant strains. With QRT-PCR, the transcript levels for tcsL and tcsH were 10-fold and 8-fold lower, respectively, in the tcsR mutant than in the parent strain (Fig. 3C). We also compared the transcript levels of tcsR and tcsE in the tcsR mutant versus the parent strain. C. difficile TcdR positively regulates its own production (18, 28). Six-fold and 2-fold decreases in transcript levels of tcsE and tcsR, respectively, were recorded in the tcsR mutant versus the parent strain (Fig. 3C). These results suggest that TcsR may activate its own transcription and of the tcsE gene in C. sordellii.
Complementation of the tcsR mutant.
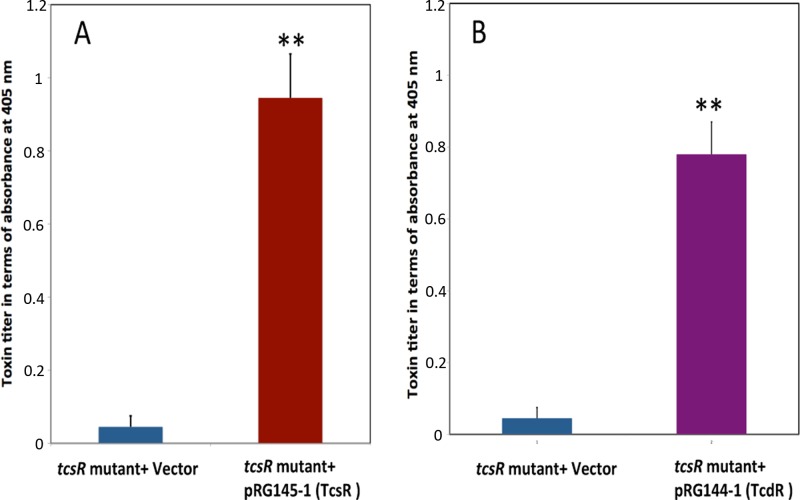
To further confirm that disruption of the tcsR gene causes underexpression of the toxin genes, the tcsR mutant was complemented with the wild-type tcsR gene from ATCC 9714. The tcsR gene was cloned under the control of a tetracycline-inducible promoter in the vector pRGL154, and the resulting construct, pRGL145-1, was then introduced into the tcsR mutant by conjugation. The tcsR mutant with pRGL154 alone served as a control in these experiments. Bacterial strains were grown in TY medium with thioamphenicol to an OD600 of 0.5, and ATc was added to 50 ng/ml to induce expression of TcsR. Three hours postinduction, bacterial cultures were harvested and equal amounts of cytosolic proteins (50 μg/well) from each strain were used for toxin ELISAs. The complemented tcsR mutant strain had a toxin titer nearly 20-fold higher than that of the tcsR mutant with the vector alone (Fig. 4A).
Fig 4.
Complementation of the tcsR mutant with C. sordellii tcsR (A) or C. difficile tcdR (B). The tcsR or the tcdR genes were cloned under a tetracycline-inducible promoter. The resulting plasmid constructs and the vector alone were introduced into the tcsR mutant for complementation. Bacterial cultures at an OD600 of 0.5 were induced for 4 h, and the toxins in the cytoplasm were quantified by ELISA. The signal from the test was recorded as the absorbance at 405 nm. The data shown are means ± standard errors of the means of three replicate samples. Student's t test was used for statistical analysis. **, P ≤ 0.01.
C. difficile tcdR can complement the C. sordellii tcsR mutant.
The clostridial sigma factors TcdR, TetR, BotR, and UviA are similar to the ECF sigma factor family (group 4 of the σ70 family), but they differ enough in structure and function that they have been assigned to their own group (group 5) (29). Since they belong to a similar group, these sigma factors are interchangeable in terms of activation of transcription by RNA polymerase core enzyme in vitro and are partially interchangeable in vivo (29, 30). To determine whether TcdR is interchangeable with TcsR, we complemented the C. sordellii tcsR mutant with C. difficile tcdR. The tcdR gene was cloned under the control of the tetracycline-inducible promoter in pRGL154, and the resulting plasmid, pRGL144-1, was introduced into the tcsR mutant by conjugation. Cytosolic proteins (50 μg) collected from cultures that had been induced for 3 h with ATc were tested for their toxin content in an ELISA. The C. sordellii tcsR mutant complemented with tcdR produced nearly 16-fold more toxin than the control (tcsR mutant with vector alone) (Fig. 4B). This result shows that TcdR can function in C. sordellii to drive transcription of the tcsL toxin gene, implying that TcsR is also a sigma factor.
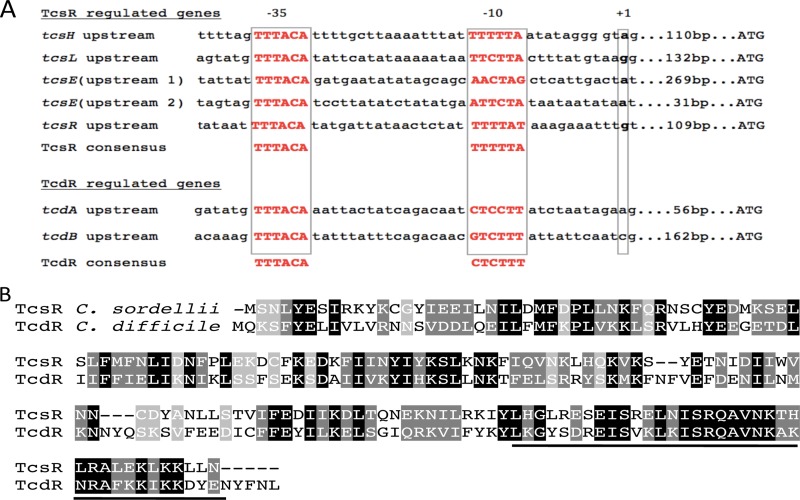
Promoters recognized by TcdR and its most closely related sigma factor, UviA, are thought to have a conserved TTTACA hexanucleotide motif in the −35 region and the sequence CTC/TTTT in the −10 region (29). The amino acid sequences of TcsR and TcdR showed high conservation in the putative region 4.2, which interacts with the −35 sequence (Fig. 5B). Moreover, the regions upstream of the tcsL, tcsH, tcsE, and tcsR genes contain the highly conserved TTTACA sequence and less-well-conserved potential −10 sequences (Fig. 5A). Our complementation studies, along with the sequence analysis, suggest strongly that TcsR is a new member of the group 5 sigma factors, all of which to date have been discovered in Clostridium spp.
Fig 5.
(A) Sequence alignment of the predicted promoter regions of the cytotoxin and hemorrhagic toxic genes (tcsL and tcsH), the regulator tcsR, and two predicted promoters of the tcsE gene. TcdR-regulated tcdA, tcdB, and tcdR promoters are also presented. Predicted −35 and −10 regions and the +1 site are boxed. The distance of the ATG codon from the +1 site is indicated. The promoter consensus sequences recognized by TcsR and TcdR sigma factor are shown at the bottom. (B) Alignment of TcsR from C. sordellii with TcdR from C. difficile. Shaded in black are identical residues, shaded in gray are similar residues, and the dashes represent gaps in the alignment. Underlined are the region 4.2 sequences of the group 5 sigma factors that are predicted to interact with the highly conserved −35 region.
DISCUSSION
Clostridium sordellii is known to cause lethal infections in animals and humans, including in women undergoing medically induced abortions with mifepristone-misoprostol (5, 31). Incidences of C. sordellii infections in intravenous heroin users have also been reported (32). Although C. sordellii infections are relatively rare in humans, the high mortality rate (approximately 70%) associated with these infections makes C. sordellii one of the important pathogens requiring more attention (33).
In this study, we have presented the draft genome sequences of C. sordellii strains ATCC 9714 and VPI 9048. High-throughput sequencing and subsequent assembly generated 104 and 166 contigs of the ATCC 9714 and VPI 9048 genomes, respectively. Gaps in genome coverage were not filled in with manual sequencing due to resource constraints. This approach is consistent with de novo sequencing and the publication of other pathogen genomes, given that the lengths of the draft genomes were consistent with other sequenced clostridial genomes (Table 2) and that the two strains whose genome sequences reported here are vastly similar. Gaps are typically caused by large (greater than the library insert size) fragments, which tend to be rRNA operons, large mobile elements, or duplicated regions, and likely do not materially detract from the quality of the data analysis presented here. The nearly complete genome sequences of the two C. sordellii strains that we have reported here will help in the further characterization of C. sordellii pathogenesis. While functional analysis is ultimately required to elucidate the roles of individual genes in pathogenesis, sequence information can assist greatly in this effort.
Similar to those of many related clostridial pathogens, the C. sordellii genome is highly A+T-rich (G+C content of 27%). Manatee annotation analysis of the C. sordellii genomes showed that only 1% of the genome represents mobile genetic elements. This feature of the C. sordellii genome is in drastic contrast to its close relative, C. difficile, in which nearly 11% of the genome consists of mobile genetic elements (34). The mobile genetic elements in the ATCC 9714 and VPI 9048 genomes are primarily cryptic phages. In many C. difficile strains and in Bacillus subtilis, a skin element (sigK intervening sequence) is inserted within the gene sigK that codes for a sporulation-specific sigma factor (35, 36). skin is required for efficient sporulation, and its excision occurs at the onset of sporulation in C. difficile (36). skin is absent in C. sordellii; the SigK-encoding ORF (VPI 9048 H476_1977; ATCC 9714 H477_0922) is intact without any insertion elements. C. difficile, in the presence of glucose and other metabolizable sugars, downregulates toxin gene transcription (18, 19). The carbon catabolite repressor CcpA is responsible for this carbon catabolite repression (CCR) of toxin genes in C. difficile (37). C. sordellii carries a CcpA-encoding gene (VPI 9048 H476_1939; ATCC 9714 H477_3163) that is 80% identical to the C. difficile CcpA. Preliminary data from our lab show that toxin production in C. sordellii is repressed in the presence of glucose (data not shown) and suggest a possible role for CcpA-mediated transcriptional repression of toxin genes. CodY is another global regulator that controls gene expression in response to nutrient availability (38). CodY is widely present in many Gram-positive bacterial pathogens, including C. difficile (39), Staphyloccocus aureus (40), and Listeria monocytogenes (41). A C. difficile codY mutant expresses high levels of toxins during growth in rich medium, and CodY was found to repress the expression of tcdR, the alternate sigma factor that is specific for the toxin genes (39). C. sordellii also encodes a CodY homologue (VPI 9048 H476_0550; ATCC 9714 H477_0862), which is 78% identical to C. difficile CodY and may play a role in virulence gene expression.
For many years, the lack of molecular and genetic tools to manipulate C. sordellii has made it difficult to study the importance of potential virulence factors. Recently, the cytotoxin TcsL-encoding gene in C. sordellii was inactivated using Targetron technology (42), which demonstrates the feasibility of genetic manipulations in C. sordellii. Using similar technology, in this work we have inactivated the putative sigma factor-encoding gene tcsR and have shown that TcsR is needed for the transcription of the toxin-encoding genes tcsL and tcsH. Our promoter-reporter fusion studies and the quantitative real-time PCR analysis provided evidence that TcsR is required for the transcription of genes tcsL, tcsH, tcsR, and tcsE. Genetic experiments in this study showed that TcsR and TcdR are interchangeable in regulating toxin gene transcription on C. sordellii and suggest TcsR to be a new member in group 5 of clostridal sigma factors. Furthermore, an in vitro transcription experiment that tested the ability of purified TcsR and RNA polymerase core enzyme to initiate transcription from the tcsL or tcsH promoters is needed to conclusively prove TcsR is a sigma factor. Such experiments are currently under progress in our lab.
In conclusion, we have sequenced and presented genomes of two C. sordellii strains. Sequencing information revealed that the toxin genes tcsL and tcsH are part of a region that shows signatures of an integrative conjugative element. Mutational and computational analyses revealed TcsR to belong to the group 5 sigma factors. The availability of the genome sequence from these two different C. sordellii strains now will facilitate the identification of more virulence-associated factors in this pathogen. More functional studies on these putative virulence factors in C. sordellii may well help us to determine their contribution to bacterial pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michelle Giglio and Suvarna Nadendla, Institute for Genome Sciences, University of Maryland, for their assistance in genome annotations and gene databank submissions. We thank Robert Carman, Tech Lab, for providing the VPI 9048 strain; Linc Sonenshein, Tufts University, for his advice on the manuscript; Nigel Minton, University of Nottingham, for the plasmid pMTL007C E5; Robert Fagan for plasmid pRPF185; and Rebekah Nicols and Sterling Braun for their assistance throughout the study.
This work was supported by funds from start-up grants to R.G. from KINBRE, supported by the National Center for Research Resources (P20RR016475) and the National Institute of General Medical Sciences (P20GM103418), and from COBRE grant P30 GM103326 to Joe Lutkenhaus, University of Kansas.
Footnotes
Published ahead of print 19 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00711-13.
REFERENCES
- 1.Clark S. 2003. Sudden death in periparturient sheep associated with Clostridium sordellii. Vet. Rec. 153:340. [PubMed] [Google Scholar]
- 2.Lewis CJ, Naylor R. 1996. Sudden death in lambs associated with Clostridium sordellii infection. Vet. Rec. 138:262. [PubMed] [Google Scholar]
- 3.Lewis CJ, Naylor RD. 1998. Sudden death in sheep associated with Clostridium sordellii. Vet. Rec. 142:417–421 [DOI] [PubMed] [Google Scholar]
- 4.Ortega J, Daft B, Assis RA, Kinde H, Anthenill L, Odani J, Uzal FA. 2007. Infection of internal umbilical remnant in foals by Clostridium sordellii. Vet. Pathol. 44:269–275 [DOI] [PubMed] [Google Scholar]
- 5.Fischer M, Bhatnagar J, Guarner J, Reagan S, Hacker JK, Van Meter SH, Poukens V, Whiteman DB, Iton A, Cheung M, Dassey DE, Shieh WJ, Zaki SR. 2005. Fatal toxic shock syndrome associated with Clostridium sordellii after medical abortion. N. Engl. J. Med. 353:2352–2360 [DOI] [PubMed] [Google Scholar]
- 6.Aronoff DM, Hao Y, Chung J, Coleman N, Lewis C, Peres CM, Serezani CH, Chen GH, Flamand N, Brock TG, Peters-Golden M. 2008. Misoprostol impairs female reproductive tract innate immunity against Clostridium sordellii. J. Immunol. 180:8222–8230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith LD. 1975. The pathogenic anaerobic bacteria, 2nd ed, p 291–298 Thomas Publishing, Springfield, IL [Google Scholar]
- 8.Martinez RD, Wilkins TD. 1988. Purification and characterization of Clostridium sordellii hemorrhagic toxin and cross-reactivity with Clostridium difficile toxin A (enterotoxin). Infect. Immun. 56:1215–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popoff MR. 1987. Purification and characterization of Clostridium sordellii lethal toxin and cross-reactivity with Clostridium difficile cytotoxin. Infect. Immun. 55:35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter GP, Awad MM, Hao Y, Thelen T, Bergin IL, Howarth PM, Seemann T, Rood JI, Aronoff DM, Lyras D. 2011. TcsL is an essential virulence factor in Clostridium sordellii ATCC 9714. Infect. Immun. 79:1025–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez RD, Wilkins TD. 1992. Comparison of Clostridium sordellii toxins HT and LT with toxins A and B of C. difficile. J. Med. Microbiol. 36:30–36 [DOI] [PubMed] [Google Scholar]
- 12.Green GA, Schue V, Girardot R, Monteil H. 1996. Characterisation of an enterotoxin-negative, cytotoxin-positive strain of Clostridium sordellii. J. Med. Microbiol. 44:60–64 [DOI] [PubMed] [Google Scholar]
- 13.Hall IC, Scott JP. 1927. Bacillus sordellii, a cause of malignant edema in man. J. Infect. Dis. 41:329–335 [Google Scholar]
- 14.Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452–464 [DOI] [PubMed] [Google Scholar]
- 15.Govind R, Fralick JA, Rolfe RD. 2006. Genomic organization and molecular characterization of Clostridium difficile bacteriophage ϕCD119. J. Bacteriol. 188:2568–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meridian Bioscience 2013. EIA detection of C. difficile toxins A & B. Meridian Bioscience, Cincinnati, OH: http://www.meridianbioscience.com/diagnostic-products/c-difficile/premier/premier-toxins-a-and-b.aspx [Google Scholar]
- 17.Fagan RP, Fairweather NF. 2011. Clostridium difficile has two parallel and essential Sec secretion systems. J. Biol. Chem. 286:27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mani N, Lyras D, Barroso L, Howarth P, Wilkins T, Rood JI, Sonenshein AL, Dupuy B. 2002. Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J. Bacteriol. 184:5971–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupuy B, Sonenshein AL. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol. Microbiol. 27:107–120 [DOI] [PubMed] [Google Scholar]
- 20.Eklund MW, Poysky FT, Reed SM, Smith CA. 1971. Bacteriophage and the toxigenicity of Clostridium botulinum type C. Science 172:480–482 [DOI] [PubMed] [Google Scholar]
- 21.Bruggemann H. 2005. Genomics of clostridial pathogens: implication of extrachromosomal elements in pathogenicity. Curr. Opin. Microbiol. 8:601–605 [DOI] [PubMed] [Google Scholar]
- 22.Eklund MW, Poysky FT, Peterson ME, Meyers JA. 1976. Relationship of bacteriophages to alpha toxin production in Clostridium novyi types A and B. Infect. Immun. 14:793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel-Streiber C. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29–38 [DOI] [PubMed] [Google Scholar]
- 24.Govind R, Dupuy B. 2012. Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog. 8(6):e1002727. 10.1371/journal.ppat.1002727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rich RL, Demeler B, Ashby K, Deivanayagam CC, Petrich JW, Patti JM, Narayana SV, Hook M. 1998. Domain structure of the Staphylococcus aureus collagen adhesin. Biochemistry 37:15423–15433 [DOI] [PubMed] [Google Scholar]
- 26.Draker KA, Northrop DB, Wright GD. 2003. Kinetic mechanism of the GCN5-related chromosomal aminoglycoside acetyltransferase AAC(6′)-Ii from Enterococcus faecium: evidence of dimer subunit cooperativity. Biochemistry 42:6565–6574 [DOI] [PubMed] [Google Scholar]
- 27.Burk DL, Ghuman N, Wybenga-Groot LE, Berghuis AM. 2003. X-ray structure of the AAC(6′)-Ii antibiotic resistance enzyme at 1.8 Å resolution; examination of oligomeric arrangements in GNAT superfamily members. Protein Sci. 12:426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mani N, Dupuy B. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc. Natl. Acad. Sci. U. S. A. 98:5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dupuy B, Matamouros S. 2006. Regulation of toxin and bacteriocin synthesis in Clostridium species by a new subgroup of RNA polymerase sigma-factors. Res. Microbiol. 157 201–205 [DOI] [PubMed] [Google Scholar]
- 30.Dupuy B, Mani N, Katayama S, Sonenshein AL. 2005. Transcription activation of a UV-inducible Clostridium perfringens bacteriocin gene by a novel sigma factor. Mol. Microbiol. 55:1196–1206 [DOI] [PubMed] [Google Scholar]
- 31.Miech RP. 2005. Pathophysiology of mifepristone-induced septic shock due to Clostridium sordellii. Ann. Pharmacother. 39:1483–1488 [DOI] [PubMed] [Google Scholar]
- 32.Kimura AC, Higa JI, Levin RM, Simpson G, Vargas Y, Vugia DJ. 2004. Outbreak of necrotizing fasciitis due to Clostridium sordellii among black-tar heroin users. Clin. Infect. Dis. 38:e87–e91 [DOI] [PubMed] [Google Scholar]
- 33.Aldape MJ, Bryant AE, Stevens DL. 2006. Clostridium sordellii infection: epidemiology, clinical findings, and current perspectives on diagnosis and treatment. Clin. Infect. Dis. 43:1436–1446 [DOI] [PubMed] [Google Scholar]
- 34.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 35.Stragier P, Kunkel B, Kroos L, Losick R. 1989. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243:507–512 [DOI] [PubMed] [Google Scholar]
- 36.Haraldsen JD, Sonenshein AL. 2003. Efficient sporulation in Clostridium difficile requires disruption of the σK gene. Mol. Microbiol. 48:811–821 [DOI] [PubMed] [Google Scholar]
- 37.Antunes A, Martin-Verstraete I, Dupuy B. 2011. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 79:882–899 [DOI] [PubMed] [Google Scholar]
- 38.Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203–207 [DOI] [PubMed] [Google Scholar]
- 39.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66:206–219 [DOI] [PubMed] [Google Scholar]
- 40.Montgomery CP, Boyle-Vavra S, Roux A, Ebine K, Sonenshein AL, Daum RS. 2012. CodY deletion enhances in vivo virulence of community-associated methicillin-resistant Staphylococcus aureus clone USA300. Infect. Immun. 80:2382–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobel L, Sigal N, Borovok I, Ruppin E, Herskovits AA. 2012. Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence. PLoS Genet. 8(9):e1002887. 10.1371/journal.pgen.1002887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao Y, Senn T, Opp JS, Young VB, Thiele T, Srinivas G, Huang SK, Aronoff DM. 2010. Lethal toxin is a critical determinant of rapid mortality in rodent models of Clostridium sordellii endometritis. Anaerobe 16:155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teng F, Murray BE, Weinstock GM. 1998. Conjugal transfer of plasmid DNA from Escherichia coli to enterococci: a method to make insertion mutations. Plasmid 39:182–186 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.