Abstract
Salicylidene acylhydrazides (SAHs) inhibit the type III secretion system (T3S) of Yersinia and other Gram-negative bacteria. In addition, SAHs restrict the growth and development of Chlamydia species. However, since the inhibition of Chlamydia growth by SAH is suppressed by the addition of excess iron and since SAHs have an iron-chelating capacity, their role as specific T3S inhibitors is unclear. We investigated here whether SAHs exhibit a function on C. trachomatis that goes beyond iron chelation. We found that the iron-saturated SAH INP0341 (IS-INP0341) specifically affects C. trachomatis infectivity with reduced generation of infectious elementary body (EB) progeny. Selection and isolation of spontaneous SAH-resistant mutant strains revealed that mutations in hemG suppressed the reduced infectivity caused by IS-INP0341 treatment. Structural modeling of C. trachomatis HemG predicts that the acquired mutations are located in the active site of the enzyme, suggesting that IS-INP0341 inhibits this domain of HemG and that protoporphyrinogen oxidase (HemG) and heme metabolism are important for C. trachomatis infectivity.
INTRODUCTION
Chlamydiae are Gram-negative bacteria that cause common respiratory diseases, sexually transmitted diseases, and the eye disease trachoma (1). Antibiotics are effective for treating most chlamydial infections; however, new antibiotic drugs or treatment approaches may be needed to combat future infections to limit the emergence of secondary infections (2). Persistent chlamydial infections might exist in clinical practice but have not yet been successfully validated. However, “persistent” Chlamydia forms can be generated in the laboratory during antibiotic-induced stress, viral coinfection, and amino acid and iron limitation (3).
Chlamydiae are obligate intracellular pathogens with a biphasic developmental cycle. The pathogen exists in two distinct forms: the environmentally stable and infectious elementary body (EB) and the replicative reticulate body (RB). The EB form attaches to epithelial cells and enters by endocytosis. Once inside the host cell, the EB transitions to the metabolically active RB, which replicates within the confines of the pathogen-containing vacuole (or inclusion). Midway through the developmental cycle, the RB forms begin to transition back to EB forms in an asynchronous manner (4). Finally, EBs are released either by cell lysis or by an inclusion extrusion mechanism into the extracellular milieu (5, 6). At all stages of infection, chlamydiae manipulate the host cell by secreting effector proteins that help establish a replicative niche and suppress innate immune responses (7). Many of these effectors are likely substrates of type III secretion (T3S) and are synthesized at early, middle, and late stages in the developmental cycle (8, 9). T3S systems are well-characterized delivery systems for virulence factors in Gram-negative bacteria and are important for bacterial avoidance of professional phagocytes, suppression of innate immunity, and promotion of uptake into epithelial cells (10). Structurally, T3S systems resemble “injection needles” consisting of a basal apparatus that spans the inner and outer membranes (11).
A role for T3S in Chlamydia development has been proposed (12, 13), but formal proof has been hampered by the lack of practical genetic tools. The use of small inhibitory molecules provides an alternative approach to study the role played by T3S in Chlamydia development (14). Small-molecule screens performed by Kauppi et al. identified salicylidene acylhydrazides (SAHs) as inhibitors of the Yersinia pseudotuberculosis T3S (15, 16). Subsequent studies indicate that SAHs block Chlamydia growth but not entry into cells (17–21), which supports the prevalent notion that the T3S is essential during the middle and late stages of the Chlamydia developmental cycle (22). Subsequent studies indicate that secretion or localization of predicted T3S effectors is altered by SAHs (19, 23–26). Because the growth inhibition of Chlamydia by SAHs is reversed by exogenous iron, it has been postulated that iron chelation by SAHs may be responsible for their antichlamydial properties (27, 28). However, the SAH INP0406, which does not inhibit T3S, retains iron-chelating properties, yet it cannot inhibit Chlamydia growth (28).
In this study, we have investigated how SAHs affect Chlamydia trachomatis development and secretion, in both the presence and absence of exogenously added iron. We found that iron-saturated INP0341 (IS-INP0341) inhibits the generation of infectious C. trachomatis EBs and that mutations in hemG mediate resistance to INP0341.
MATERIALS AND METHODS
Chemicals and INP compounds.
The chemical compounds INP0010 (also known as ME0052) (16, 29) and INP0341 (30) were synthesized and purified from commercially available hydrazides and salicylaldehydes, as described previously (31). The SAHs were dissolved in dimethyl sulfoxide (DMSO; Sigma) to a final concentration of 20 mM and stored at room temperature without exposure to light for a maximum of 3 weeks. FeSO4 (Merck), FeCl3 (Sigma), and deferoxamine methanesulfonate (DFO; Sigma) were diluted in distilled water and filter sterilized (0.22-μm-pore size polyethersulfone [PES] membrane; Corning). For iron saturation experiments, the SAHs were diluted in prewarmed RPMI medium prior to supplementation of iron sulfate. Compound INP0341 was iron saturated by mixing equal molar equivalents of compound and FeCl3. The mixture was incubated for 1 h at room temperature prior to supplementation to prewarmed RPMI medium.
Cells lines and Chlamydia strains.
HeLa cell lines (DSMZ) were grown in RPMI 1640 medium (Sigma) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma) and 20 mM HEPES (pH 8.0). Vero cells (ATCC CCL-81) were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco/Invitrogen) supplemented with 10% FBS at 37°C (5% CO2). C. trachomatis serovar LGV-L2 434/Bu (ATCC VR902B) was propagated in HeLa cells and purified as described by Caldwell et al. (32). Cells and bacteria were negative for mycoplasma infection, as determined by a mycoplasma detection kit (Stratagene).
Chlamydia infections and determination of infectivity.
HeLa cells were infected by C. trachomatis in Hank's balanced salt solution (HBSS; Gibco/Invitrogen) at a multiplicity of infection (MOI) of 0.1 to 1 for 1 h at 37°C in 5% CO2. HBSS was removed, and RPMI medium containing DMSO, INP0341, INP0010, or DFO was added to the infected cells (lacking cycloheximide). Infectivity was determined by harvesting infectious EB progeny at 44 to 48 h postinfection (p.i.). Harvested bacteria were diluted in HBSS, fresh HeLa cells were infected, and ∼40 h after reinfection C. trachomatis inclusions were counted. Data are represented as the relative amount of EBs in treated infections compared to the amount of EBs in DMSO-treated infections. When multiple strains were analyzed, infectivity was determined by normalization to input inclusion-forming units (IFUs) as previously described (33).
Immunofluorescence analysis.
In samples fixed with methanol, C. trachomatis was detected with a rabbit anti-major outer membrane protein (MOMP) antibody and a lissamine rhodamine sulfonyl chloride (LRSC)-conjugated anti-rabbit antibody (Jackson ImmunoResearch Laboratories). Chlamydial protease-like activity factor (CPAF) and IncA were detected in samples that were fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.1% Triton-X (34). Thereafter, an anti-CPAF antibody (34) was detected with an anti-rabbit LRSC-conjugated antibody and a mouse anti-IncA (gift from Daniel D. Rockey, Oregon State University) antibody with an anti-mouse fluorescein isothiocyanate (FITC)-conjugated antibody (both secondary antibodies were from Jackson ImmunoResearch laboratories). Host and bacterial DNA were stained with 200 nM 4′,6-diamidino-2-phenylindole (DAPI). Images were acquired by laser scanning confocal microscopy (Nikon Eclipse C1 plus). The signal from uninfected cells was used to set the background levels. Digital images were processed using Adobe Photoshop C6 software (Adobe Systems, Inc.).
TEM.
At 44 h p.i., infected cells were processed for transmission electron microscopy (TEM) as previously described (33). Briefly, cells were fixed with 2.5% glutaraldehyde–0.05% malachite green (EMS) in 0.1 M sodium cacodylate buffer (pH 6.8) and then postfixed with 0.5% osmium tetroxide–0.8% potassium ferricyanide in 0.1 M sodium cacodylate, 1% tannic acid, and 1% uranyl acetate. Samples were dehydrated with graded amounts of ethanol, embedded in Spurr's resin, and subsequently imaged on a Tecnai G2 Twin microscope (FEI).
Determination of inclusion size.
Infected cells were fixed at 44 h p.i. and stained with an anti-MOMP antibody as described above. Inclusion membranes were visualized with a 440-nm diode laser and a specific detector to obtain transmission photographs (see Fig. S1 in the supplemental material). Subsequently, each inclusion area was measured using EZ-C1 software (Nikon). When C. trachomatis strains were grown in the absence of the SAHs, it was possible to determine inclusion sizes by using a Cellomics ArrayScan Vti HCS automated fluorescent imaging system (ThermoFisher) as previously described (33).
Selection of INP0341-resistant strains.
HeLa cells grown in large flasks were infected with C. trachomatis LGV-L2 at an MOI of 5 (∼108 IFUs). Infected cells were treated with compound INP0341 at a concentration ranging from 20 to 35 μM and excess iron sulfate or with iron-saturated INP0341 at a concentration ranging from 25 to 40 μM for serial passages. Infectious progeny were collected at 44 to 48 h p.i. and subsequently used for reinfection (see Fig. 5). During the first four to six passages, the cells were infected at an MOI of 2 or higher in the presence of SAHs; thereafter, the selection continued at an MOI of 1 or lower. When mutant populations were passaged in the absence of INP0341, the MOI was kept between 0.5 and 2 to allow genetic recombination. Clonal strains were collected by a plaquing assay on cell monolayers as previously described (33). Briefly, confluent Vero cells were grown in six-well plates and infected with 100 to 10 IFUs from the mutant populations for 2 h, and infected cells were overlaid with agarose-DMEM. Plaques in the monolayers were picked 10 to 20 days after infection and propagated in HeLa cells.
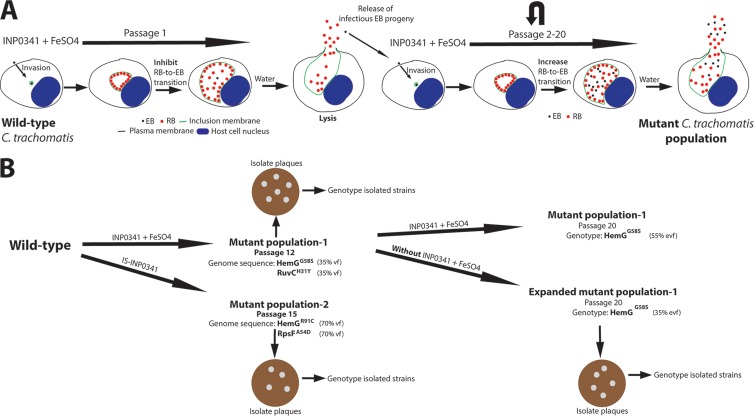
Fig 5.
Strategy to select and isolate C. trachomatis mutant strains. (A) HeLa cells infected with wild-type C. trachomatis LGV-L2 were treated with INP0341 in the presence of exogenously added iron sulfate (250 μM). At 44 to 48 h p.i., infected cells were lysed with ice-cold water to release infectious EB progeny which were used to infect fresh HeLa cells for a new round of selection. This procedure was repeated, and selection was continued for up to 20 passages. Notably, the selected C. trachomatis mutant population had increased RB-to-EB transition in the presence of INP0341 and iron sulfate. (B) Wild-type C. trachomatis LGV-L2 was serially passaged as described above. Note that the first mutant population (passage 12) was further grown in both the presence and absence of selective pressure for serial passages. At the indicated passages of the mutants, bacterial DNA was purified and subjected to whole-genome sequencing (WGS) or genotyped by capillary sequencing. Identified mutations via WGS were verified with capillary sequencing. Infectious EB progeny from respective mutant populations were used to infect Vero cells for the formations of plaques. Formed plaques were amplified in HeLa cells, and the clonal C. trachomatis strains were subsequently genotyped and characterized. vf, variant frequency; evf, estimated variant frequency.
WGS and genotyping.
C. trachomatis genomic DNA prepared for whole-genome sequencing (WGS) was purified from density gradient-purified bacteria with a DNeasy Blood and Tissue Kit (Qiagen) (purification of total DNA from Gram-negative bacteria) according to the manufacturer's instructions. DNA was concentrated by sodium acetate-ethanol precipitation, and the pellets were dissolved in AE buffer (elution buffer for genomic DNA; Qiagen) to avoid aggregation. For WGS, 1 μg of enriched chlamydial DNA was fragmented with an Adaptive Focused Acoustics S220 instrument (Covaris, Inc., Woburn, MA), and DNA sequencing libraries were prepared with a library construction kit (TruSeq DNA Sample Preparation Kit, version 2; Illumina, Inc., San Diego, CA) according to the manufacturer's instructions. Libraries were sequenced on a MiSeq DNA Sequencing Platform (Illumina, Inc., San Diego, CA) at Duke University's Institute for Genome Sciences and Policy (IGSP) DNA core sequencing facility. Genome assembly and single-nucleotide variant (SNV) identification were performed with Geneious, version 6, from Biomatters (San Francisco, CA). The C. trachomatis L2 434/Bu genome (GenBank number NC_010287) was used as the reference sequence. A mutation is herein defined as a nucleotide variant present at a frequency above 15% and a strand bias below 80%, detectable by both WGS and capillary sequencing in a bacterial population. In the second derived mutant population, a mutation in phosphatidylcholine-hydrolyzing phospholipase D (CTL0413) was detected by WGS at a variant frequency of 33% and strand bias of 75%. Attempts to read through this GC-rich region were not possible using conventional capillary sequencing (data not shown). Therefore, it is possible that this mutant population contains a mutation in CTL0413. To identify mutations by capillary sequencing, templates were PCR amplified with the primers listed in Table S1 in the supplemental material. PCR products were sequenced by capillary sequencing (BigDye; Applied Biosystems). The intensities of the fluorescence peaks detected by capillary DNA sequencing were used as estimates of the relative abundance of any single nucleotide variant in the bacterial population. Estimated frequencies were consistent with the obtained values from WGS (data not shown).
Structural prediction.
Template candidates, structurally homologous to C. trachomatis HemG (CtHemG), were identified through hidden Markov model (HMM)-HMM comparisons using the HHpred (35) tool kit with default settings; MODELLER (36) and the MPI tool kit were used with default settings to create the three-dimensional (3D) models. The STRAP (37) tool kit was used to make structural alignments and structure superposition. Structural visualization was made using PyMOL (38). A montage of images was prepared in Adobe Photoshop C6 (Adobe Systems, Inc.).
RESULTS
Salicylidene acylhydrazides inhibit C. trachomatis infectivity but not growth following addition of excess iron.
We recently reported that salicylidene acylhydrazides (SAHs) act as inhibitors of the Yersinia pseudotuberculosis T3S (15, 16). We further showed that the SAH variants INP0341 and INP0010 also inhibit Chlamydia growth and development, observations that have been corroborated by other groups (17, 18, 20, 28). These findings suggest that T3S activity is essential for Chlamydia (22). It has previously been shown that excess iron in the cell culture medium can suppress the negative impact of SAHs on C. trachomatis growth, suggesting that the iron chelation properties of SAH may be partially responsible for the observed growth inhibition (28). However, INP0406, an SAH that does not inhibit the T3S yet retains the capacity to chelate iron, similar to INP0341, does not restrict C. trachomatis growth (28). We compared the antimicrobial activities of INP0341 and INP0010 on C. trachomatis and contrasted them with the effects induced by the iron-specific chelator DFO (39). HeLa cells were infected with C. trachomatis for 1 h, and the medium was exchanged with medium containing the indicated inhibitors (see Fig. 1). At 44 h p.i., the infected cells were fixed, stained, and analyzed by laser scanning confocal microscopy. Addition of INP0010 (10 and 30 μM) and INP0341 (10 and 30 μM) to infected cells resulted in the formation of enlarged, aberrant C. trachomatis forms, as had been previously observed with iron chelators (39) (Fig. 1). Indeed, DFO treatment also induced the formation of these aberrant forms although significantly higher concentrations were required (400 μM) (Fig. 1). Addition of 250 μM iron sulfate suppressed the effects of INP0341, INP0010, and DFO, resulting in no visible aberrant C. trachomatis, which indicates that the iron limitation induced by these compounds is reduced. However, in the presence of excess iron and INP0341, the bacteria appeared to aggregate at the center of the inclusion (Fig. 1; see also Fig. S1 in the supplemental material). Addition of INP0010 alone or DFO in the presence or absence of excess iron did not cause this redistribution (Fig. 1). We next measured inclusions sizes, which can be used to indirectly assess bacterial growth (40). Addition of excess iron to INP0010-, INP0341-, and DFO-treated infected cells resulted in similar inclusion sizes; however, these inclusions were significantly larger than those under iron-free conditions (Fig. 2). These results indicate that excess iron at least partly suppresses the growth-inhibitory effect of SAHs and DFO. Interestingly, in the presence of excess iron, INP0341 treatment resulted in detachment of bacteria from the inclusion membrane. Together, these data indicate that INP0341 exhibits an additional effect(s) that is distinguishable from its iron chelating effects.
Fig 1.
The salicylidene acylhydrazides and DFO induce an enlargement of C. trachomatis cells that can be suppressed by addition of excess iron. HeLa cells were infected with C. trachomatis and treated throughout the infection (0 to 44 h p.i.) as indicated. At 44 h p.i., infected cells were fixed with methanol, and C. trachomatis was detected with an anti-MOMP antibody. DAPI was used to detect host and bacterial DNA. Images were acquired with a Nikon Eclipse C1 confocal laser scanning microscope. Images are representative of at least two independent biological experiments prepared in duplicates. DFO, deferoxamine methanesulfonate.
Fig 2.
Iron suppresses the growth-inhibitory effect of the SAHs and DFO. HeLa cells were infected with C. trachomatis and treated as indicated on the figure. At 44 h p.i., infected cells were fixed with methanol, and C. trachomatis was detected with an anti-MOMP antibody. Photographs were produced using Nikon Eclipse C1 confocal laser scanning microscopy. Each inclusion area was measured by using the EZ-C1 software (Nikon). Horizontal lines indicate mean values of 50 inclusions, and values for DMSO-treated inclusions were arbitrarily set to 1. Differences in inclusion sizes were determined to be statistically significant by using a Mann-Whitney U test (***, P < 0.001, two-tailed, between indicated treatment in the absence or presence of 250 μM FeSO4). SAHs, salicylidene acylhydrazides; DFO, deferoxamine methanesulfonate; ns, not significant; AU, arbitrary units.
Next, we quantified the yield of infectious EBs after treating infected cells with INP0010, INP0341, and DFO in the presence or absence of excess iron. Compounds were added immediately after infection, EBs were harvested at 44 h p.i., and titers were determined by infecting a monolayer of HeLa cells. Addition of excess iron almost completely suppressed the effect of DFO (45% of DMSO control) while the effects of INP0010 and INP0341 were partly suppressed (6% and 0.15% of DMSO control, respectively) for the generation of infectious EBs (Fig. 3). These results show that the effect of DFO on Chlamydia infectivity is suppressed by excess iron, while the effect of treatment with SAHs is only partly suppressed. This further suggests that the SAHs, particularly INP0341, have an effect on C. trachomatis development that is not linked to iron chelation. The fact that compound INP0341 possesses a stronger effect on C. trachomatis than INP0010 might be explained by more efficient uptake into host cells or binding to a molecular target(s).
Fig 3.

Excess iron partially suppresses the effect by SAHs on C. trachomatis infectivity. HeLa cells were infected with C. trachomatis and treated with DMSO, 30 μM INP0341, 30 μM INP0010, and 400 μM DFO in the absence or presence of FeSO4 using concentrations ranging from 100 to 800 μM. At 44 h p.i., infected cells were lysed with ice-cold distilled water and diluted in HBSS. Diluted bacteria were used for reinfection of new HeLa cells to determine the amount of infectious progeny generated; ∼40 h after reinfection cells were fixed and stained, and subsequently inclusions were counted. Values for DMSO-treated infections were normalized to 100%. The value from each treatment represents the mean of at least two independent experiments. *, P < 0.05; **, P < 0.01, between SAHs and DFO-treated infections in the presence of 250 μM FeSO4 (unpaired Student's t test, two-tailed). SAHs, salicylidene acylhydrazides; DFO, deferoxamine methanesulfonate.
Secretion of CPAF or IncA is not inhibited by INP0341.
The above results suggest that SAHs restrict the generation of infectious C. trachomatis EBs. The simplest interpretation is that SAHs target the T3S in C. trachomatis because earlier studies identified INP0341 as a potent inhibitor of T3S in Yersinia and other Gram-negative bacteria (16, 41). We therefore investigated the localization of the secreted proteins IncA and CPAF after treatment with INP0341 or DFO following excess addition of iron. IncA is a T3S effector known to be secreted to the chlamydial inclusion membrane during the midportion of development (42); CPAF is a mid-late effector secreted via the general secretion pathway to the host cell cytoplasm (34). We found that CPAF was secreted to the host cell cytoplasm in the majority of infected cells after INP0341 or DFO treatment, analogous to the case of IncA, which was localized to the inclusion membrane independent of INP0341 treatment (Fig. 4A and B). These data suggest that INP0341 does not inhibit secretion of IncA or CPAF, which is consistent with the growth alleviation that follows addition of excess iron to INP0341-treated C. trachomatis. Thus, these data suggest that the effect of INP0341 is not related to secretion.
Fig 4.
Secretion of IncA and CPAF is not affected by INP0341 following addition of iron. (A) HeLa cells were infected with C. trachomatis and treated with DMSO, INP0341, or DFO in the presence of FeSO4. At 42 h p.i., infected cells were fixed with 4% PFA and subsequently incubated with anti-CPAF and anti-IncA; DAPI was used to detect host and bacterial DNA. The images were acquired with Nikon Eclipse C1 confocal laser scanning microscopy. (B) IncA and CPAF secretion was manually quantified in 50 infected cells from two independent experiments in samples prepared as described above. DFO, deferoxamine methanesulfonate.
Isolation of SAH-resistant C. trachomatis mutant strains.
To define the mechanism of action of INP0341, we isolated spontaneous INP0341-resistant populations of C. trachomatis LGV-L2 by serial passage of bacteria through several rounds of infection in HeLa cells in the presence of INP0341 and excess iron (Fig. 5). This strategy has been previously used to understand the mode of action for new antibacterial compounds (43, 44). INP0341-resistant C. trachomatis began to emerge after six passages, and stable resistance was achieved by passage 12. Genomic DNA was isolated from this mutant population and subjected to whole-genome sequencing (WGS). Two mutations were identified (35% variant frequency per mutation) after INP0341 selection. One mutation was in ruvC, a gene that encodes the Holliday junction resolvase known to facilitate the last step in DNA recombination (45). The mutation in ruvC leads to a histidine-to-tyrosine substitution at residue 31. The second mutation was in hemG, a gene encoding the protoporphyrinogen oxidase (PPO), an enzyme that catalyzes the aromatization of protoporphyrinogen IX to protoporphyrin IX, a central step in heme biosynthesis (46). The mutation in hemG changes a glycine to serine at residue 58. The genome of C. trachomatis encodes all the components for heme biosynthesis, indicating an important role for heme in oxidative catalysis and respiration during C. trachomatis development (47). We next plaque purified clonal strains from this mutant population (Fig. 5) and genotyped them for the hemG and ruvC mutant alleles. Almost all strains contained both the HemGG58S and the RuvCH31Y substitutions (Table 1), indicating that both mutations are present within individual SAH-resistant strains in this population.
Table 1.
Isolated mutant strains
| C. trachomatis straina | Amino acid substitution(s) | No. of mutants with the substitution(s)/total no. of strains | Origin (passage no.) |
|---|---|---|---|
| Mutant 1 | HemGG58S, RuvCH31Y | 5/6 | Mutant population 1 (12) |
| Mutant 2 | HemGG58S | 2/5 | Expanded mutant population 1 (20) |
| Mutant 3 | HemGR91C, RpsFA54D | 2/4 | Mutant population 2 (15) |
Plaque-purified strains isolated from the indicated mutant populations, genotyped by capillary sequencing.
Because mutants in a population can be positively or negatively selected depending on the fitness cost of the individual mutation, we decided to further passage our INP0341-resistant mutant population for an additional eight passages in the presence or absence of INP0341. Interestingly, in both populations, the RuvCH31Y substitution was negatively selected while the HemGG58S substitution was retained (Fig. 5), suggesting that the HemGG58S substitution is primarily responsible for INP0341 resistance and implying that the RuvCH31Y substitution had a high fitness cost. Plaque-purified strains were collected from the expanded mutant population that had been grown in the absence of INP0341 (named expanded mutant population P20, where P20 indicates passage 20), and clonal isolates were genotyped for the hemG and ruvC mutant alleles. We obtained strains bearing only a mutation in hemG (Table 1). Next, we characterized these mutant strains by assessing growth and production of infectious EBs in the absence of the compound. In comparison to the wild-type strain, the strain with the RuvCH31Y substitution delayed growth, with significantly smaller inclusions at 24 and 36 h p.i. The mutant strain carrying only the HemGG58S substitution formed slightly smaller inclusions at 24 h p.i., while at later time points the sizes were comparable to those of wild-type strains (see Fig. S2A in the supplemental material). The yields of infectious EBs were also quantified, and the double mutant produced significantly fewer infectious EBs at 36 and 48 h p.i., than both the wild type and the strain carrying only the HemGG58S substitution (see Fig. S2B). These data are consistent with the observation that the RuvCH31Y substitution has a high fitness cost while the HemGG58S substitution has a relatively low cost.
Because SAHs chelate iron (28) yet some of their antichlamydial functions appear to be independent from iron sequestration (Fig. 3), we decided to use a genetic approach to determine if we could identify mutants resistant to iron-loaded SAHs. We first tested if INP0341 could be iron saturated prior to addition to the cell culture medium. We found that a ratio of 1:1 (iron chloride to INP0341) was needed to avoid precipitation of INP0341 and that 30 μM iron-saturated INP0341 (IS-INP0341) affected the intrainclusion distribution of C. trachomatis to a similar degree as the addition of excess iron to medium containing 30 μM INP0341 (data not shown). This approach was therefore employed to further investigate if INP0341 has an effect beyond iron chelation and as well to avoid potential secondary effects by the addition of excess iron to the cell culture medium. We started a new selection for C. trachomatis LGV-L2 variants that would be resistant to IS-INP0341 and were able to enrich for a resistant population by passage 15. WGS of this population identified two mutations present at 70% frequency (Fig. 5), including a new mutation in hemG leading to an arginine-to-cysteine change at residue 91. The second mutation was in rpsF, a gene that encodes the 30S ribosomal protein S6 (Fig. 5), leading to an alanine-to-aspartic acid substitution at residue 54. Altogether, these data indicate that mutation in hemG is strongly associated with resistance to IS-INP0341.
Mutations in hemG mediate IS-INP0341 resistance with increased infectivity.
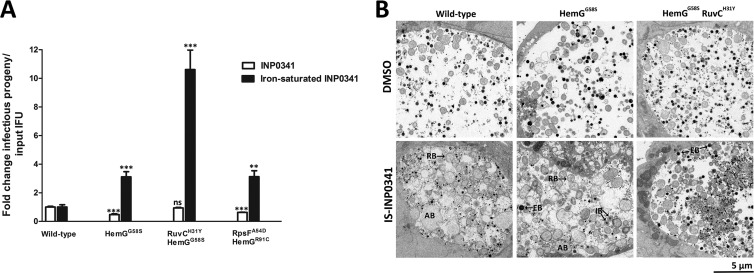
The above data suggest that mutations in hemG mediate resistance to the effect of INP0341 separate from the iron chelation feature, resulting in increased generation of infectious EBs. To verify this resistance, we first tested the collected mutant strains (Table 1) for the generation of infectious EBs in the presence of IS-INP0341. The strain with the HemGG58S substitution displayed a 3-fold increased yield of infectious EBs. A similar increase was observed for the strain with the HemGR91C and the RpsFA54D substitutions. The strain with both the HemGG58S and the RuvCH31Y mutations resulted in a further increase (∼10-fold) of infectious EBs (Fig. 6A). In the case of iron-saturated INP0010, all the strains generated similar yields of infectious EBs (data not shown). Next, we characterized the effect of IS-INP0341 on the mutant strains by transmission electron microscopy and determined that IS-INP0341 treatment of the mutant strains did not induce the formation of aberrant RBs to the same extent as in wild-type C. trachomatis (Fig. 6B). Together, these data suggest that IS-INP0341 inhibits some aspect of the RB-to-EB transition and that the hemG mutant strains at least partly overcome this inhibitory effect, with more intermediate bodies (IBs) and EBs observed (Fig. 6B). Finally, we tested the strains for sensitivity to iron limitation induced by INP0341 by assessing the infectivity at 48 h p.i. In comparison with the wild-type strain, all the strains displayed similar or slightly more sensitivity to iron limitation induced by INP0341 (Fig. 6A). These data show that the mutant strains are not resistant to iron limitation induced by INP0341, and therefore we conclude that INP0341 has an effect beyond iron chelation.
Fig 6.
Mutant strains are resistant to IS-INP0341 but not to iron limitation induced by INP0341. (A) HeLa cells were infected with the indicated C. trachomatis strains at an MOI of 0.1 and subsequently treated with 30 μM IS-INP0341 or 10 μM nonsaturated INP0341, which inhibits C. trachomatis development to a similar degrees as 30 μM IS-INP0341 and therefore was used to analyze the mutant strains in a comparative susceptibility assay. In parallel each strain was titrated to normalize the infectivity to the exact number of input IFUs. Infectious progeny was collected at 48 h p.i. and subsequently used for reinfection. Data presented were acquired from an experiment performed in triplicate which is representative of at least two independent experiments. The treated wild-type value was set to 1 for each treatment. **, P < 0.01; ***, P < 0.001 between treated wild-type and mutant strains for each treatment (unpaired Student's t test, two-tailed). (B) Morphological analysis after treatment with DMSO only or 30 μM IS-INP0341 using transmission electron microscopy. Infected cells (MOI of 1) were fixed at 44 h p.i. and processed as described in Materials and Methods. RB, reticulate bodies; EB, elementary bodies; IB, intermediate bodies; AB, aberrant bodies. IS-INP0341, iron-saturated INP0341.
Structural prediction of C. trachomatis HemG.
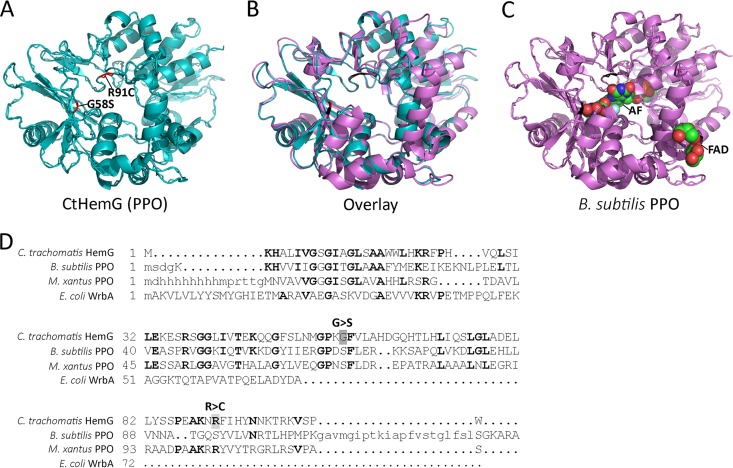
The crystal structure of protoporphyrinogen oxidase (PPO) from Myxococcus xanthus (MxPPO) (48) was predicted to have the highest structural homology to C. trachomatis HemG (CtHemG), and a structural model was built from that template (Fig. 7A). Based on structural prediction, PPO from Bacillus subtilis (BsPPO) (49) is also predicted to have a high structural homology to CtHemG (Fig. 7B). BsPPO has recently been cocrystalized with flavine-adenine dinucleotide (FAD) and the inhibitor acifluorfen (AF); the latter is proposed to mimic half of the PPO substrate protoporphyrinogen XI and therefore binds to the active site of BsPPO (48, 49). This structural information allowed us to map the mutations that confer IS-INP0341 resistance onto a structural model of the Chlamydia HemG. In this manner, we determined that the mutation that confers IS-INP0341 resistance occurs in close vicinity to the putative catalytic site (Fig. 7C).
Fig 7.
Predicted structure of C. trachomatis HemG. (A) Protoporphyrinogen oxidase (PPO) from Myxococcus xanthus was used as a template to model C. trachomatis HemG (CtHemG). (B) Protoporphyrinogen oxidase (PPO) from Bacillus subtilis (BsPPO) and the predicted structure of CtHemG were overlaid. (C) BsPPO cocrystalized with FAD and the inhibitor AF, the latter of which binds to the active site of BsPPO. Red residues in CtHemG indicate where IS-INP0341-resistant substitutions are localized, and the corresponding residues in BsPPO are indicated in black. (D) Structural alignment of CtHemG, BsPPO, MxPPO, and WrbA, regarded as a potential target of SAHs (54). Notably all four proteins contain a FAD-binding domain (the first ∼55 amino acids). G>S and R>C indicate resistance substitutions in CtHemG.
DISCUSSION
New methods that enable the isolation and rapid mapping of mutations in Chlamydia genes, combined with recent reports of successful transformation of C. trachomatis with plasmid DNA (33, 50, 51, 52), indicate that genetic approaches will soon yield new insight into the biology of this obligate intracellular pathogen. However, these genetic approaches still remain somewhat limited. Thus, the use of small organic molecules with inhibitory properties presents an alternative approach to study the molecular function of Chlamydia gene products. By selecting C. trachomatis mutant strains resistant to novel inhibitory compounds, the target or mode of action of the compound may be revealed. In addition, this method can also lead to the isolation of isogenic mutant strains with phenotypes separate from drug resistance. Thus, an extension of this procedure can complement genetic approaches such as chemical mutagenesis.
Selected SAH compounds have been used to study T3S of Chlamydia and other Gram-negative bacteria (41, 53–55). These compounds inhibit chlamydial growth, development, and secretion, suggesting a role for the T3S throughout the intracellular developmental cycle (17, 18, 19–21, 23, 24, 25, 26, 56). However, Slepenkin et al. reported that addition of excess iron restored Chlamydia growth during SAH treatment, suggesting that these compounds affect chlamydial growth via iron chelation (28). On the other hand, Prantner and Nagarajan argued that the SAHs might have an effect separate from iron chelation, possibly affecting late T3S activity in Chlamydia (27). Since addition of excess iron to SAHs does not restore secretion of T3S substrates in Yersinia (data not shown), we wanted to determine if these compounds had an effect on Chlamydia development beyond iron chelation.
In this report we confirm previously published data that excess iron suppresses the effect of SAHs on bacterial growth (27, 28) (Fig. 1 and 2). However, despite the presence of iron, compound INP0341 still caused distinct phenotypes, including the accumulation of bacteria within regions of the inclusion lumen (see Fig. S1 in the supplemental material), and reduced the yield of infectious EB progeny (Fig. 3). In addition, we found that INP0341 did not affect the secretion of IncA (T3S) or CPAF (Fig. 4A and B). In aggregate, our results suggest that INP0341 has an effect on C. trachomatis beyond iron chelation that likely includes a partial block in the RB-to-EB transition. Such a block in differentiation is consistent with our observation by electron microscopy whereby the RB-to-EB transition appears to be impaired in IS-INP0341-treated infections (Fig. 6B).
We collected independent INP0341-resistant C. trachomatis populations and showed that mutations in hemG primarily mediate the resistance observed (Fig. 5). This gene encodes protoporphyrinogen oxidase (PPO or HemG), an enzyme that catalyzes the second to last step of heme biosynthesis (46, 57). Chlamydia codes for all components for the biosynthesis of heme (47), an important cofactor of peroxidases, catalases, sensor molecules, and cytochromes (46). Cytochromes are essential for respiration and are the most abundant proteins that contain heme in bacteria (58). Thus, HemG activity is indirectly coupled to respiration. In Escherichia coli it has been suggested that HemG activity also has a direct role in respiration (59, 60). At this stage, it is unclear how the specific hemG mutations identified provide resistance to IS-INP0341. The simplest interpretation is that the mutations in hemG impair a potential interaction between IS-INP0341 and HemG. Alternatively, these are gain-of-function mutations that allow the bacteria to cope with oxidative stress caused by IS-INP0341 or inhibited heme biosynthesis. Together, these arguments strongly suggest that HemG activity plays an important role in the generation of C. trachomatis progeny, at least in vitro and under these experimental conditions.
In order to isolate isogenic strains, we took advantage of the fact that the RuvCH31Y substitution has a high fitness cost. Specifically, the mutant population that contained both the HemGG58S and RuvCH31Y substitutions was grown in the absence of INP0341 for multiple passages. The clonal strains collected from this expanded mutant population were isogenic for the mutation in hemG (Table 1). This approach represents a promising strategy to collect panels of isogenic C. trachomatis mutant strains.
Secretion of the T3S effector IncA is not inhibited by IS-INP0341, suggesting that the chlamydial T3S is not a direct target of this compound; however, a potential scenario is that the chlamydial T3S has a role in control host iron homeostasis such that it feeds RB metabolism and that addition of excess iron to SAHs would partially suppress this inhibitory effect. This scenario is difficult to determine because both SAHs and DFO affect both IncA and CPAF secretion (data not shown); thus, all secretion appears to be impaired during iron chelation. Furthermore, IS-INP0341 affects HemG and late C. trachomatis development, and IS-INP0341 might, via an effect on HemG, impair secretion of T3S effectors that are expressed late during infection.
By using a biochemical approach, we identified WrbA in E. coli as a potential target of the SAHs (54); both WrbA and C. trachomatis HemG contain a FAD-binding domain (Fig. 7D), which could indicate that this domain plays a role in drug-target interaction. The fact that we acquired resistance mutations in HemG from two independent selections and that these mutations were located in close vicinity to the FAD-binding domain further suggests that HemG is the bona fide target of IS-INP0341.
In summary, we successfully isolated C. trachomatis mutant strains that are resistant to IS-INP0341. Our data further show that mutations in hemG primarily mediate a resistant phenotype with increased infectivity. Altogether, our data suggest a novel link between HemG and C. trachomatis infectivity. In addition, HemG and heme metabolism represent an interesting and attractive target for the development of antibacterial therapeutics to treat chlamydial infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Alfaro for critically reading the manuscript and S. Arreljung for technical assistance.
S.B., M.E., and H.W.-W. were funded by the Swedish Research Council, Å.G. was funded by Västerbottens county council, K.A.F. was supported by a Public Health Service grant (AI065530) from the National Institutes of Health, and R.H.V was supported by Public Health Service grants (AI100759 and AI081694).
Footnotes
Published ahead of print 12 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00506-13.
REFERENCES
- 1.Horn M. 2008. Chlamydiae as symbionts in eukaryotes. Annu. Rev. Microbiol. 62:113–131 [DOI] [PubMed] [Google Scholar]
- 2.Valdivia RH. 2012. Thinking outside the box: new strategies for antichlamydial control. Future Microbiol. 7:427–429 [DOI] [PubMed] [Google Scholar]
- 3.Wyrick PB. 2010. Chlamydia trachomatis persistence in vitro: an overview. J. Infect. Dis. 201(Suppl 2):S88–S95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fields KA, Hackstadt T. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 18:221–245 [DOI] [PubMed] [Google Scholar]
- 5.Cocchiaro JL, Valdivia RH. 2009. New insights into Chlamydia intracellular survival mechanisms. Cell Microbiol. 11:1571–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hybiske K, Stephens RS. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U. S. A. 104:11430–11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betts HJ, Wolf K, Fields KA. 2009. Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal. Curr. Opin. Microbiol. 12:81–87 [DOI] [PubMed] [Google Scholar]
- 8.Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37:913–925 [DOI] [PubMed] [Google Scholar]
- 9.Valdivia RH. 2008. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 11:53–59 [DOI] [PubMed] [Google Scholar]
- 10.Cornelis GR. 2000. Type III secretion: a bacterial device for close combat with cells of their eukaryotic host. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galan JE, Wolf-Watz H. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567–573 [DOI] [PubMed] [Google Scholar]
- 12.Hoare A, Timms P, Bavoil PM, Wilson DP. 2008. Spatial constraints within the chlamydial host cell inclusion predict interrupted development and persistence. BMC Microbiol. 8:5. 10.1186/1471-2180-8-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson DP, Timms P, McElwain DL, Bavoil PM. 2006. Type III secretion, contact-dependent model for the intracellular development of chlamydia. Bull. Math. Biol. 68:161–178 [DOI] [PubMed] [Google Scholar]
- 14.Puri AW, Bogyo M. 2009. Using small molecules to dissect mechanisms of microbial pathogenesis. ACS Chem. Biol. 4:603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kauppi AM, Nordfelth R, Uvell H, Wolf-Watz H, Elofsson M. 2003. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10:241–249 [DOI] [PubMed] [Google Scholar]
- 16.Nordfelth R, Kauppi AM, Norberg HA, Wolf-Watz H, Elofsson M. 2005. Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73:3104–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey L, Gylfe Å, Sundin C, Muschiol S, Elofsson M, Nordström P, Henriques-Normark B, Lugert R, Waldenström A, Wolf-Watz H, Bergström S. 2007. Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. FEBS Lett. 581:587–595 [DOI] [PubMed] [Google Scholar]
- 18.Engström P, Bailey L, Önskog T, Bergström S, Johansson J. 2010. A comparative study of RNA and DNA as internal gene expression controls early in the developmental cycle of Chlamydia pneumoniae. FEMS Immunol. Med. Microbiol. 58:244–253 [DOI] [PubMed] [Google Scholar]
- 19.Muschiol S, Bailey L, Gylfe Å, Sundin C, Hultenby K, Bergström S, Elofsson M, Wolf-Watz H, Normark S, Henriques-Normark B. 2006. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 103:14566–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muschiol S, Normark S, Henriques-Normark B, Subtil A. 2009. Small molecule inhibitors of the Yersinia type III secretion system impair the development of Chlamydia after entry into host cells. BMC Microbiol. 9:75. 10.1186/1471-2180-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf K, Betts HJ, Chellas-Gery B, Hower S, Linton CN, Fields KA. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61:1543–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters J, Wilson DP, Myers G, Timms P, Bavoil PM. 2007. Type III secretion a la Chlamydia. Trends Microbiol. 15:241–251 [DOI] [PubMed] [Google Scholar]
- 23.Chellas-Gery B, Wolf K, Tisoncik J, Hackstadt T, Fields KA. 2011. Biochemical and localization analyses of putative type III secretion translocator proteins CopB and CopB2 of Chlamydia trachomatis reveal significant distinctions. Infect. Immun. 79:3036–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong S, Lei L, Chang X, Belland R, Zhong G. 2011. Chlamydia trachomatis secretion of hypothetical protein CT622 into host cell cytoplasm via a secretion pathway that can be inhibited by the type III secretion system inhibitor compound 1. Microbiology 157:1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobolt-Pedersen AS, Christiansen G, Timmerman E, Gevaert K, Birkelund S. 2009. Identification of Chlamydia trachomatis CT621, a protein delivered through the type III secretion system to the host cell cytoplasm and nucleus. FEMS Immunol. Med. Microbiol. 57:46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Lei L, Gong S, Chen D, Flores R, Zhong G. 2011. The chlamydial periplasmic stress response serine protease cHtrA is secreted into host cell cytosol. BMC Microbiol. 11:87. 10.1186/1471-2180-11-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prantner D, Nagarajan UM. 2009. Role for the chlamydial type III secretion apparatus in host cytokine expression. Infect. Immun. 77:76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slepenkin A, Enquist PA, Hägglund U, de la Maza LM, Elofsson M, Peterson EM. 2007. Reversal of the antichlamydial activity of putative type III secretion inhibitors by iron. Infect. Immun. 75:3478–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ur-Rehman T, Nordfelth R, Blomgren A, Zetterström CE, Elofsson M, Gylfe Å. 2012. Preliminary pharmacokinetics of the bacterial virulence inhibitor n′-(3,5-dibromo-2-hydroxy-benzylidenene)-nicotinic acid hydrazide. Adv. Exp. Med. Biol. 954:349–356 [DOI] [PubMed] [Google Scholar]
- 30.Slepenkin A, Chu H, Elofsson M, Keyser P, Peterson EM. 2011. Protection of mice from a Chlamydia trachomatis vaginal infection using a salicylidene acylhydrazide, a potential microbicide. J. Infect. Dis. 204:1313–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahlgren MK, Zetterström CE, Gylfe S, Linusson A, Elofsson M. 2010. Statistical molecular design of a focused salicylidene acylhydrazide library and multivariate QSAR of inhibition of type III secretion in the Gram-negative bacterium Yersinia. Bioorg. Med. Chem. 18:2686–2703 [DOI] [PubMed] [Google Scholar]
- 32.Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen BD, Valdivia RH. 2012. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc. Natl. Acad. Sci. U. S. A. 109:1263–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen I, Bednar MM, Amin V, Davis BK, Ting JP, McCafferty DG, Valdivia RH. 2011. The Chlamydia protease CPAF regulates host and bacterial proteins to maintain pathogen vacuole integrity and promote virulence. Cell Host Microbe 10:21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. 1995. Evaluation of comparative protein modeling by MODELLER. Proteins 23:318–326 [DOI] [PubMed] [Google Scholar]
- 37.Gille C, Frommel C. 2001. STRAP: editor for structural alignments of proteins. Bioinformatics 17:377–378 [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. 2004. The PyMOL user's manual. DeLano Scientific, San Carlos, CA [Google Scholar]
- 39.Raulston JE. 1997. Response of Chlamydia trachomatis serovar E to iron restriction in vitro and evidence for iron-regulated chlamydial proteins. Infect. Immun. 65:4539–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tietzel I, El-Haibi C, Carabeo RA. 2009. Human guanylate binding proteins potentiate the anti-chlamydia effects of interferon-gamma. PLoS One 4:e6499. 10.1371/journal.pone.0006499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keyser P, Elofsson M, Rosell S, Wolf-Watz H. 2008. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 264:17–29 [DOI] [PubMed] [Google Scholar]
- 42.Subtil A, Parsot C, Dautry-Varsat A. 2001. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol. Microbiol. 39:792–800 [DOI] [PubMed] [Google Scholar]
- 43.Balakrishnan A, Patel B, Sieber SA, Chen D, Pachikara N, Zhong G, Cravatt BF, Fan H. 2006. Metalloprotease inhibitors GM6001 and TAPI-0 inhibit the obligate intracellular human pathogen Chlamydia trachomatis by targeting peptide deformylase of the bacterium. J. Biol. Chem. 281:16691–16699 [DOI] [PubMed] [Google Scholar]
- 44.Sandoz KM, Eriksen SG, Jeffrey BM, Suchland RJ, Putman TE, Hruby DE, Jordan R, Rockey DD. 2012. Resistance to a novel antichlamydial compound is mediated through mutations in Chlamydia trachomatis secY. Antimicrob. Agents Chemother. 56:4296–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connolly B, Parsons CA, Benson FE, Dunderdale HJ, Sharples GJ, Lloyd RG, West SC. 1991. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc. Natl. Acad. Sci. U. S. A. 88:6063–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinemann IU, Jahn M, Jahn D. 2008. The biochemistry of heme biosynthesis. Arch. Biochem. Biophys. 474:238–251 [DOI] [PubMed] [Google Scholar]
- 47.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759 [DOI] [PubMed] [Google Scholar]
- 48.Corradi HR, Corrigall AV, Boix E, Mohan CG, Sturrock ED, Meissner PN, Acharya KR. 2006. Crystal structure of protoporphyrinogen oxidase from Myxococcus xanthus and its complex with the inhibitor acifluorfen. J. Biol. Chem. 281:38625–38633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin X, Sun L, Wen X, Yang X, Tan Y, Jin H, Cao Q, Zhou W, Xi Z, Shen Y. 2010. Structural insight into unique properties of protoporphyrinogen oxidase from Bacillus subtilis. J. Struct. Biol. 170:76–82 [DOI] [PubMed] [Google Scholar]
- 50.Binet R, Maurelli A. 2009. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc. Natl. Acad. Sci. U. S. A. 106:292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, Nelson DE, Caldwell HD. 2011. Generation of targeted Chlamydia trachomatis null mutants. Proc. Natl. Acad. Sci. U. S. A. 108:7189–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Watkins H, Zhou B, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD. 2013. The Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence associated genes. Infect. Immun. 81:636–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Layton AN, Hudson DL, Thompson A, Hinton JC, Stevens JM, Galyov EE, Stevens MP. 2010. Salicylidene acylhydrazide-mediated inhibition of type III secretion system-1 in Salmonella enterica serovar Typhimurium is associated with iron restriction and can be reversed by free iron. FEMS Microbiol. Lett. 302:114–122 [DOI] [PubMed] [Google Scholar]
- 54.Wang D, Zetterström CE, Gabrielsen M, Beckham KS, Tree JJ, Macdonald SE, Byron O, Mitchell TJ, Gally DL, Herzyk P, Mahajan A, Uvell H, Burchmore R, Smith BO, Elofsson M, Roe AJ. 2011. Identification of bacterial target proteins for the salicylidene acylhydrazide class of virulence-blocking compounds. J. Biol. Chem. 286:29922–29931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veenendaal AK, Sundin C, Blocker AJ. 2009. Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J. Bacteriol. 191:563–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chin E, Kirker K, Zuck M, James G, Hybiske K. 2012. Actin recruitment to the Chlamydia inclusion is spatiotemporally regulated by a mechanism that requires host and bacterial factors. PLoS One 7:e46949. 10.1371/journal.pone.0046949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayfield JA, Dehner CA, DuBois JL. 2011. Recent advances in bacterial heme protein biochemistry. Curr. Opin. Chem. Biol. 15:260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panek H, O'Brian MR. 2002. A whole genome view of prokaryotic haem biosynthesis. Microbiology 148:2273–2282 [DOI] [PubMed] [Google Scholar]
- 59.Boynton T, Daugherty L, Dailey T, Dailey H. 2009. Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity. Biochemistry 48:6705–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mobius K, Arias-Cartin R, Breckau D, Hannig AL, Riedmann K, Biedendieck R, Schroder S, Becher D, Magalon A, Moser J, Jahn M, Jahn D. 2010. Heme biosynthesis is coupled to electron transport chains for energy generation. Proc. Natl. Acad. Sci. U. S. A. 107:10436–10441 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.