Abstract
DNA affinity chromatography with the promoter region of the Corynebacterium glutamicum pck gene, encoding phosphoenolpyruvate carboxykinase, led to the isolation of four transcriptional regulators, i.e., RamA, GntR1, GntR2, and IolR. Determination of the phosphoenolpyruvate carboxykinase activity of the ΔramA, ΔgntR1 ΔgntR2, and ΔiolR deletion mutants indicated that RamA represses pck during growth on glucose about 2-fold, whereas GntR1, GntR2, and IolR activate pck expression about 2-fold irrespective of whether glucose or acetate served as the carbon source. The DNA binding sites of the four regulators in the pck promoter region were identified and their positions correlated with the predicted functions as repressor or activators. The iolR gene is located upstream and in a divergent orientation with respect to a iol gene cluster, encoding proteins involved in myo-inositol uptake and degradation. Comparative DNA microarray analysis of the ΔiolR mutant and the parental wild-type strain revealed strongly (>100-fold) elevated mRNA levels of the iol genes in the mutant, indicating that the primary function of IolR is the repression of the iol genes. IolR binding sites were identified in the promoter regions of iolC, iolT1, and iolR. IolR therefore is presumably subject to negative autoregulation. A consensus DNA binding motif (5′-KGWCHTRACA-3′) which corresponds well to those of other GntR-type regulators of the HutC family was identified. Taken together, our results disclose a complex regulation of the pck gene in C. glutamicum and identify IolR as an efficient repressor of genes involved in myo-inositol catabolism of this organism.
INTRODUCTION
Corynebacterium glutamicum is a facultative anaerobic Gram-positive soil bacterium of the order Corynebacteriales (1) that grows on a variety of carbon sources and has been used for more than 50 years for large-scale production of l-amino acids (2, 3). Recent metabolic engineering studies have shown that C. glutamicum is also capable of producing a variety of other commercially interesting compounds, e.g., other l-amino acids (4), d-amino acids (5), organic acids such as succinate (6–9), diamines such as cadaverine (10, 11) or putrescine (12), biofuels such as ethanol or isobutanol (13–15), and proteins (16–18).
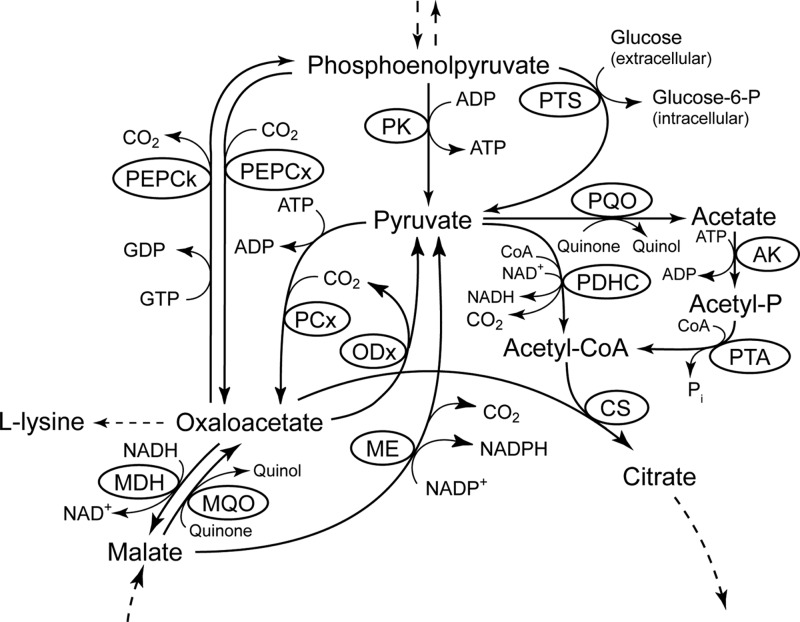
For growth on organic acids such as acetate, gluconeogenic reactions are necessary in order to provide the cells with hexose and pentose sugars (19). Depending on the subset of enzymes present at the phosphoenolpyruvate (PEP)-pyruvate-oxaloacetate node of a given organism, PEP carboxykinase or malic enzyme and/or oxaloacetate decarboxylase in combination with PEP synthetase catalyzed the conversion of C4 intermediates from the tricarboxylic acid (TCA) cycle into PEP (20), the direct precursor for gluconeogenesis. C. glutamicum possesses a rather complex PEP-pyruvate-oxaloacetate node (Fig. 1) compared to other model organisms such as Escherichia coli and Bacillus subtilis and is equipped with PEP carboxykinase as well as with malic enzyme and oxaloacetetate decarboxylase (20). Although the presence of a PEP synthetase was previously proposed for some strains (21, 22), the inability of a defined PEP carboxykinase-negative mutant of C. glutamicum to grow on acetate or lactate (23) argues against a functional PEP synthetase in C. glutamicum and a gluconeogenic function of malic enzyme or oxaloacetate decarboxylase. The validity of this finding is additionally supported by the fact that a pyruvate carboxylase-negative strain was unable to grow on lactate (24). In a PEP synthetase-possessing strain, this enzyme in combination with PEP carboxylase should have overcome the anaplerotic deficiency caused by the absence of pyruvate carboxylase (20).
Fig 1.
The phosphoenolpyruvate (PEP)-pyruvate-oxaloacetate node in C. glutamicum. Abbreviations: AK, acetate kinase; CS, citrate synthase; MDH, malate dehydrogenase; ME, malic enzyme; MQO, malate:quinone oxidoreductase; ODx, oxaloacetate decarboxylase; PCx, pyruvate carboxylase; PDHC, pyruvate dehydrogenase complex; PEPCk, PEP carboxykinase; PEPCx, PEP carboxylase; PK, pyruvate kinase; PQO, pyruvate:quinone oxidoreductase; PTA, phosphotransacetylase; PTS, phosphotransferase system.
PEP carboxykinase catalyzes the reversible conversion between oxaloacetate and PEP (25): oxaloacetate + ATP/GTP ⇄ PEP + CO2 + ADP/GDP. While microbial enzymes often use ATP as a phosphate donor, the C. glutamicum PEP carboxykinase has been shown to be highly specific for GTP (26–29) and thus represents a notable exception. The kinetic analysis of the purified enzyme revealed ATP to inhibit PEP carboxykinase activity in the oxaloacetate-forming reaction (28), indicating that the enzyme mainly functions in gluconeogenesis and not in anaplerosis under physiological conditions. Since PEP carboxykinase of C. glutamicum also shows significant activity in cells grown on glucose (23, 28) and due to the fact that optimization of the cellular oxaloacetate concentration is crucial, especially for improved l-lysine production (30), deletion of the pck gene in a l-lysine-producing strain resulted in an increase in l-lysine productivity by 20% (23). Therefore, besides the activities of pyruvate carboxylase and PEP carboxylase, PEP carboxykinase activity is an important target to modulate the net carbon flux toward oxaloacetate and thus increase precursor supply for l-lysine production.
Expression of the PEP carboxykinase gene pck (sometimes also named pckA) is controlled in different ways in different microorganisms. In most bacteria studied so far, pck is expressed depending on the carbon source in the growth medium, with expression being low during growth with glycolytic substrates and higher during growth with gluconeogenic substrates. In E. coli, the pckA gene as well as the genes encoding enzymes of the alternative C4-decarboxylating route, i.e., maeB (malic enzyme), sfc (malic enzyme), and ppsA (PEP synthase), are subject to glucose repression (31–35). For pckA, glucose repression is relieved via (i) cyclic AMP (cAMP) receptor protein (CRP)-dependent activation in the presence of elevated cytoplasmic cAMP levels in response to glucose starvation (33) and (ii) activation by the catabolite repressor/activator Cra (formerly known as FruR) (36). In B. subtilis, pckA transcription is also repressed in the presence of glucose but is independent of the presence of CcpA, the major transcriptional catabolite repressor of this organism. Instead, the transcriptional regulator CcpN, which is a repressor of pckA expression and directs carbon flow between glycolysis and gluconeogenesis (37), could be identified. In C. glutamicum, carbon source-dependent expression of the pck gene was suggested by the two- and 3-fold-higher specific PEP carboxykinase activities in acetate- and lactate-grown cells, respectively, compared to glucose-grown cells (23). DNA microarray and quantitative reverse transcription-PCR experiments confirmed an acetate-dependent transcriptional regulation of the pck gene (38). In C. glutamicum ATCC 13032, a growth phase-dependent regulation of pck gene expression was not observed by either enzyme activity (23) or quantitative protein measurements (39), whereas in C. glutamicum R, growth-phase-dependent differences in the pck transcript level on glucose were reported, with maxima in the exponential and early stationary phases (40). The presence of a binding site for the CRP homologue GlxR in the pck promoter region and the fact that GlxR binds to the pck promoter in a cAMP-dependent manner in vitro (40, 41) indicate an involvement of GlxR in regulation of pck transcription. In silico analysis of the pck promoter also revealed binding sites for the regulator of acetate metabolism RamA (42). DNA microarray analysis indicated that RamA represses pck transcription during growth on glucose, as the pck mRNA level was increased in a RamA-negative strain, and a direct interaction between RamA and the pck promoter region could be shown (43).
In the present work, DNA affinity chromatography with the pck promoter region was used to search for additional regulators of pck expression. In addition to RamA, we identified the two functionally redundant transcriptional regulators, GntR1 and GntR2 (44), and another GntR-type regulator, Cg0196. The relevance of these regulators for pck expression under glycolytic and gluconeogenic conditions was investigated. Additionally, we analyzed the regulon of Cg0196 by transcriptome analysis of a Δcg0196 mutant and by DNA binding studies with purified Cg0196. Our results clearly showed that the primary function of Cg0196 is the transcriptional control of a variety of iol genes involved in myo-inositol metabolism (45). Therefore, we designated Cg0196 IolR and the corresponding gene iolR.
MATERIALS AND METHODS
Bacteria, plasmids, and culture conditions.
Bacterial strains and plasmids as well as their relevant characteristics and sources are listed in Table 1. Plasmid pK19mobsacB-ΔiolR1 was obtained as follows: the up- and downstream regions of iolR were amplified by PCR using the oligonucleotide pairs iolR_DF1_for/iolR_DF1_rev and iolR_DF2_for/iolR_DF2_rev (see Table S1 in the supplemental material) and chromosomal DNA of the C. glutamicum wild-type (WT) strain as the template. The resulting PCR fragments were then used as the templates for overlap extension PCR with the oligonucleotides iolR_DF1_for and iolR_DF2_rev. The resulting 1.51-kb PCR product was cut with HindIII and BamHI and cloned into pK19mobsacB cut with the same enzymes. DNA sequencing revealed point mutations within a distinct area in the upstream region of iolR. Therefore, a 5′-shortened 1,197-bp fragment was cut out of pK19mobsacB-ΔiolR1 via the use of SalI and BamHI and cloned again into pK19mobsacB to obtain pK19mobsacB-ΔiolR2 without any unintended mutations. For construction of plasmid pET16b-iolR, the iolR coding region was amplified from chromosomal DNA of C. glutamicum WT using the oligonucleotides iolR_pET16b_for and iolR_pET16b_rev. The resulting PCR fragment was digested with NdeI and BamHI and cloned into pET16b cut with the same enzymes. The IolR protein encoded by pET16b-iolR contains an N-terminal extension of 21 amino acids, including a decahistidine tag.
Table 1.
Strains and plasmids used in this studya
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)-U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| BL21(DE3) | F− dcm ompT gal hsdS (rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Novagen |
| C. glutamicum strains | ||
| WT | ATCC 13032, wild-type strain | ATCC |
| ΔramA | ramA (cg2831) deletion mutant of WT | 42 |
| ΔgntR1 | gntR1 (cg2783) deletion mutant of WT | 44 |
| ΔgntR2 | gntR2 (cg1935) deletion mutant of WT | 44 |
| ΔgntR1 ΔgntR2 | gntR1 and gntR2 deletion mutant of WT | 44 |
| ΔiolR | iolR (cg0196) deletion mutant of WT | This study |
| WT (pEK0) | WT carrying plasmid pEK0 | 83 |
| WT (pEK0-pckB) | WT carrying plasmid pEK0-pckB | 23 |
| ΔiolR (pEK0) | iolR deletion mutant of WT carrying plasmid pEK0 | This study |
| ΔiolR (pEK0-pckB) | iolR deletion mutant of WT carrying plasmid pEK0-pckB | This study |
| Plasmids | ||
| pET28a-ramA | Kmr; pET28a carrying ramA gene | 42 |
| pET16b | Apr; vector for overexpression of genes in E. coli, adding a C-terminal decahistidine affinity tag to the synthesized protein, oriV, PT7, lacIq | Novagen |
| pET16b-gntR1 | Apr; pET16b carrying gntR1 gene | 44 |
| pET16b-gntR2 | Apr; pET16b carrying gntR2 gene | 44 |
| pET16b-iolR | Apr; pET16b carrying iolR gene | This study |
| pEK0 | Kmr; E. coli/C. glutamicum shuttle vector | 83 |
| pEK0-pckB | Kmr; pEK0 carrying the pck gene | 23 |
| pK19mobsacB | Kmr; vector for allelic exchange in C. glutamicum, oriV oriT sacB lacZα | 84 |
| pK19mobsacB-ΔiolR2 | Kmr; pK19mobsacB carrying the iolR gene with an internal 593-bp deletion | This study |
Apr, ampicillin; Kmr, kanamycin; ATCC, American Type Culture Collection.
Tryptone-yeast extract medium (46) (2× TY) was used for cultivation of C. glutamicum and E. coli DH5α and Terrific Broth (TB) medium (47) for growth of E. coli BL21(DE3). For growth of C. glutamicum on glucose, myo-inositol, and potassium acetate (at the concentrations indicated in Results), CGXII minimal medium (48) was used. When appropriate, the medium contained kanamycin (50 μg ml−1 for strains carrying plasmids; 25 μg ml−1 for the pK19mobsacB-ΔiolR2 integration mutant) or ampicillin (100 μg ml−1). C. glutamicum was grown aerobically at 30°C and E. coli at 37°C as 60-ml cultures in 500-ml baffled Erlenmeyer flasks on a rotary shaker at 120 rpm. Growth of the bacteria was followed by measuring the optical density at 600 nm (OD600). For screening applications, C. glutamicum was grown as 800 μl-cultures in 48-well baffled microtiter plates (Flowerplates) at 80% humidity and 1,200 rpm using a BioLector system (m2p-labs, Baesweiler, Germany). Growth was monitored online via backscatter [OD600 = (backscatter − 103.5)/50.5].
Recombinant DNA work.
Restriction enzymes, T4 DNA ligase, calf intestinal phosphatase, RNase A, and proteinase K were obtained from Fermentas (St. Leon-Rot, Germany). Isolation of chromosomal and plasmid DNA from C. glutamicum was performed as described previously (49). Preparation of plasmids from E. coli was carried out either according to the method of Birnboim (50) or, for DNA sequence validation, by Eurofins MWG Operon (Ebersberg, Germany), with an Omega Bio-Tek E.Z.N.A. Plasmid minikit (VWR International, Darmstadt, Germany). DNA transfer into C. glutamicum by electroporation was performed essentially as described previously (51) at 2.5 kV, 200 Ω, and 25 μF. Transformation of E. coli was carried out according to the method of Dower et al. (52). PCR experiments were performed using a Biometra Personal Cycler (Biometra, Göttingen, Germany) with Taq DNA polymerase (Fermentas). Oligonucleotides were obtained from Eurofins MWG Operon or from biomers.net (Ulm, Germany). PCR products were separated on agarose gels and purified using a NucleoSpin II extract kit (Macherey-Nagel, Düren, Germany).
Enzyme assays.
In order to determine PEP carboxykinase activity in cell extracts, C. glutamicum cells were grown in minimal medium containing the respective carbon source and harvested in the exponential-growth phase. Cells from a 50-ml culture were washed once with 20 ml buffer A (100 mM HEPES [pH 6.8], 10 mM MgCl2, 0.5 mM MnCl2, 2 mM glutathione) and resuspended in an appropriate volume of the same buffer (about 2 ml/g cell [wet weight]). The cell suspension was transferred into 2-ml screw-cap vials together with 250 mg glass beads (0.1 mm diameter) and subjected to mechanical disruption with a RiboLyser (Hybaid, Heidelberg, Germany) three times for 34 s each time at speed 6.5 and 4°C with intermittent cooling on ice for 5 min. After disruption, the glass beads and cellular debris were removed by centrifugation for 20 min at 20,000 × g and 4°C. The supernatant was used to spectrophotometrically assay PEP carboxykinase activity in a continuous system, essentially as described by Aich (26). The assay mixture contained 0.4 mM NADH, 30 U l-malate dehydrogenase, 4 mM PEP, 50 mM NaHCO3, 10 to 20 μl crude cell extract (corresponding to 150 to 300 μg protein), and 1 mM GDP. Interfering PEP carboxylase activity was blocked by the addition of 50 mM sodium aspartate (53). The protein concentration was determined by using a bicinchoninic acid (BCA) protein assay reagent kit (Pierce, Rockford, IL) with bovine serum albumin (BSA) as the standard.
DNA affinity purification.
The pck promoter region was amplified from chromosomal DNA of the C. glutamicum WT strain using oligonucleotides P_pck_1_for and pck_btac_rev. The resulting 510-bp fragment was used as the template for a second PCR with oligonucleotides P_pck_1_for and bt_anchor. Oligonucleotide bt_anchor was 5′ tagged with biotin via a TEG linker (Eurofins MWG Operon). The resulting 510-bp DNA fragment was coupled to Streptavidin-coated magnetic beads (Dynabeads M-280 Streptavidin; Invitrogen, Darmstadt, Germany) and used for enrichment of proteins binding to the pck promoter region as described previously (42). The eluted proteins were separated by SDS-PAGE, stained with Coomassie, and identified by tryptic peptide mass fingerprinting (54).
Overproduction and purification of GntR1, GntR2, RamA, and IolR.
The C. glutamicum proteins GntR1 and GntR2 were overproduced in E. coli BL21(DE3)/pLysS and purified by nickel chelate affinity chromatography as described previously (44). RamA and IolR were overproduced in E. coli BL21(DE3) using the expression plasmids pET28a-ramA and pET16b-iolR, respectively. Expression was induced at an OD600 of 2.0 by addition of 1 mM isopropyl β-d-thiogalactoside (IPTG). Two hours after induction, cells were harvested by centrifugation, washed once with the appropriate disruption buffer, and stored at −20°C. For purification of RamA, thawed cells were resuspended in 30 ml buffer NNG (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 5% [vol/vol] glycerol) containing 20 mM imidazol (NNIG-20) and disrupted mechanically with a French pressure cell (SLM Aminco, Urbana, IL) at 12.4 MPa five times with intermittent cooling on ice. Intact cells and cell debris were removed by centrifugation for 40 min at 20,000 × g and 4°C. The membranes present in the resulting supernatant were separated by ultracentrifugation for 1.5 h at 150,000 × g and 4°C. The soluble protein fraction was then applied to a HisTrap FF 1-ml column (GE Healthcare Germany, Munich, Germany) previously equilibrated with NNIG-20 buffer for nickel chelate affinity chromatography. Adsorbed proteins were eluted with a stepwise gradient consisting of 75, 308, and 500 mM imidazol in buffer NNG. Fractions containing RamA-His6 collected at 308 mM imidazol were pooled, and the buffer was exchanged with storage buffer (50 mM Tris-HCl [pH 7.4], 100 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol [DTT], 10% [vol/vol] glycerol) by using a 5-ml HiTrap desalting column (GE Healthcare) as described by the manufacturer. IolR protein was purified as described above for RamA except that buffer TNG (50 mM Tris-HCl [pH 7.4], 500 mM NaCl, 5% [vol/vol] glycerol) containing 20 mM imidazol was used for binding and a 20-ml linear gradient of buffer TNG with 56 to 500 mM imidazol was used for elution. Fractions containing IolR-His10, collected at 439 mM imidazol, were pooled and concentrated in a Microcon YM-10 unit (Merck Millipore, Darmstadt, Germany), and the buffer was exchanged with TG buffer (50 mM Tris-HCl [pH 7.4], 5% [vol/vol] glycerol). Protein concentrations were determined as described above, and the purified proteins were stored at −20°C until use.
EMSAs.
For testing the binding of RamA, GntR1, GntR2, and IolR to promoter DNA fragments of putative target genes, various concentrations of purified His-tagged versions of the corresponding proteins were mixed with 40 to 100 ng DNA fragments using either the gel shift buffers described previously for RamA (55) and GntR1/GntR2 (44) or 50 mM Tris-HCl (pH 7.4) with 50 mM KCl, 5 mM MgCl2, 1 mM EDTA, and 10% (vol/vol) glycerol for IolR. After 20 min incubation at room temperature, the samples were loaded onto a native polyacrylamide gel (5% to 15%). Electrophoresis was performed at 150 V for 50 to 90 min using 1× TBE buffer (89 mM Tris-borate, 1 mM EDTA, pH 8.3) for IolR and GntR1/GntR2 and 0.5× TBE buffer for RamA. The gels were stained with ethidium bromide or Sybr green I and visualized with a UV transilluminator (Vilber Lourmat Germany, Eberhardzell, Germany). The DNA fragments used in electrophoretic mobility shift assays (EMSAs) were generated either by PCR using the corresponding oligonucleotides listed in Table S1 in the supplemental material or by annealing two complementary oligonucleotides by incubating them in an equimolar ratio in annealing buffer (10 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA) for 5 min at 95°C, followed by cooling at room temperature. PCR products were purified with a PCR purification kit (Qiagen, Hilden, Germany), and their concentration was measured with a Nanodrop ND-1000 spectrophotometer (Peqlab, Erlangen, Germany).
Global gene expression analysis.
DNA microarray analysis was used to compare genome-wide mRNA concentrations of the mutant strain ΔiolR with those of the WT strain. The strains were grown in CGXII minimal medium with 4% (wt/vol) glucose as the sole carbon source, and RNA was isolated from cells harvested in the early exponential-growth phase (OD600 of about 5). Preparation of RNA and synthesis of fluorescently labeled cDNA were performed as described previously (56). Custom-made DNA microarrays spotted with 70-mer oligonucleotides derived from the genome sequence entry NC_006958.1 (57) were obtained from Operon Biotechnologies (Cologne, Germany). The experimental details for hybridization and washing of these microarrays as well as subsequent data acquisition and analysis were described previously (44). The transcriptome comparison was performed in triplicate starting from independent cultures.
Identification of transcription start sites (TSSs) and promoter regions by RNAseq.
A 5′-end enriched RNAseq library was constructed according to the following procedures. Depletion of stable rRNA and enrichment of mRNA molecules were performed using a Ribo-Zero rRNA removal kit for Gram-positive bacteria (Epicentre Biotechnologies, Madison, WI). The enriched mRNA was fragmented by MgKOAc hydrolysis. Four volumes of RNA solution was mixed with one volume of MgKOAc solution (100 mM KOAc and 30 mM MgOAc in 200 mM Tris-HCl, pH 8.1), and the mixture was incubated for 2.5 min at 94°C. The reaction was stopped by adding an equal volume of 1× TE (10 mM Tris, 1 mM EDTA, pH 8.0) and chilling on ice for 5 min. The fragmented RNA was precipitated by addition of three volumes of 0.3 M NaAc in ethanol with 2 μl glycogen and incubation overnight at −20°C. The precipitated RNA fragments were dissolved in water, and the 5′-end RNA fragments were enriched by using Terminator 5′-phosphate-dependent exonuclease (Epicentre Biotechnologies). After RNA precipitation (as described above), the triphosphates were removed using RNA 5′ polyphosphatase (Epicentre Biotechnologies). After RNA precipitation (as described above), the 5′-enriched, monophosphorylated RNA fragments were used to construct a cDNA library by using a Small RNA Sample Prep kit (Illumina, San Diego, CA). The fragmentation of RNA molecules (fragment sizes were 200 to 500 bp) and RNA concentration were monitored using the RNA 6000 Pico assay on an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Sequencing of the cDNA library was carried out on the Genome Analyzer IIx platform (Illumina). Resulting reads were aligned to the C. glutamicum genomic sequence using the mapping software SARUMAN (58). TSSs and promoter regions were deduced by combining published information about promoter regions in C. glutamicum (59) with 5′-end enriched RNAseq data.
Microarray data accession number.
The microarray data were deposited in the GEO database with the accession number GSE44812.
RESULTS
Isolation of transcriptional regulatory proteins binding to the pck promoter region.
Previous studies have shown carbon source-dependent transcriptional regulation of the pck gene encoding PEP carboxykinase of C. glutamicum with up to 10-fold-higher transcript levels during growth on gluconeogenic carbon sources compared to growth on glucose (38, 40). Two transcriptional regulators, GlxR and RamA, were reported to be involved in regulation of pck transcription (40, 41, 43). However, in the course of characterizing the transcriptional regulator RamA, we speculated about additional regulatory proteins involved in the control of pck expression (43). To identify such proteins, we performed DNA affinity chromatography using the immobilized pck promoter region. For this purpose, 488-bp biotinylated pck promoter fragments (extending from 413 bp upstream to 75 bp downstream of the pck start codon) were bound to streptavidin-coated magnetic beads and incubated with crude extracts obtained from C. glutamicum WT cells grown either in minimal medium with 4% (wt/vol) glucose or in minimal medium with 2% (wt/vol) potassium acetate. Cells were harvested in the exponential-growth phase and, for glucose-grown cells, additionally in the stationary phase. Nonspecifically bound proteins were removed by several low-salt washing steps (0.1 M NaCl) with subsequent magnetic separation. Specifically bound proteins were eluted with buffers containing 0.3 M, 0.5 M, and 1 M NaCl and subjected to SDS-PAGE and Coomassie staining. No striking differences could be observed between either the samples from the different growth phases or the samples from the different carbon sources (data not shown).
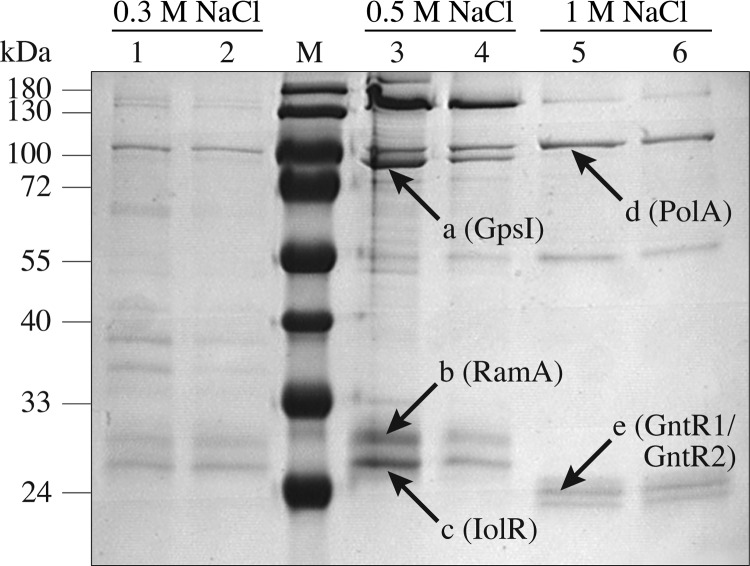
In Fig. 2, an SDS-polyacrylamide gel with the proteins enriched from cell extracts of a C. glutamicum culture growing exponentially on glucose is shown. The five protein bands labeled a to e were isolated and subjected to peptide mass fingerprinting after tryptic digestion. Comparison of the mass lists with the nonredundant NCBI database resulted in the identification of six C. glutamicum proteins with a significant MOWSE score, including four transcriptional regulators. High-molecular-mass proteins a and d were identified as polyribonuclotide phosphorylase GpsI (Cg2166; 12 identified peptides, sequence coverage 21%, score 64) and DNA polymerase I (Cg1525; 8 identified peptides, sequence coverage 15%, score 53), respectively. The dominant proteins of lower molecular mass labeled b (about 30 kDa) and c (about 28 kDa) were identified as RamA (Cg2831; 15 identified peptides, sequence coverage 50%, score 112) and Cg0196, which is annotated in this work as IolR (9 identified peptides, sequence coverage 60%, score 92), respectively. Protein band e (about 24 kDa) was identified to represent the paralogous and functionally redundant transcriptional regulators GntR1 (Cg2783; 13 identified peptides, sequence coverage 58%, score 118) and GntR2 (Cg1935; 8 identified peptides, sequence coverage 37%, score 60).
Fig 2.
Coomassie-stained SDS-polyacrylamide gel of proteins purified by DNA affinity chromatography with the pck promoter region. Extracts of C. glutamicum cells growing exponentially in glucose minimal medium were incubated with a 488-bp biotin-labeled pck promoter fragment immobilized on Streptavidin-coated magnetic beads. Bound proteins were eluted with a buffer containing 0.3 M NaCl (lanes 1 and 2), 0.5 M NaCl (lanes 3 and 4), or 1 M NaCl (lanes 5 and 6). The protein bands labeled a to e were identified by peptide mass as polyribonuclotide phosphorylase GpsI (Cg2166), RamA (Cg2831), IolR (Cg0196), DNA polymerase I PolA (Cg1525), and GntR1 (Cg2783) and GntR2 (Cg1935). Lane M, molecular mass standard.
The iolR gene (cg0196) is located upstream of and divergently orientated with respect to a set of genes encoding enzymes involved in myo-inositol transport and degradation in C. glutamicum (45). The iolR gene encodes a protein of 253 amino acids with a predicted molecular mass of 27.9 kDa, which corresponds well to the mass of the protein isolated by DNA affinity chromatography. The IolR protein is composed of an N-terminal DNA binding domain (amino acid residues 23 to 70) resembling the highly conserved winged helix-turn-helix (wHTH) DNA binding domain of the GntR family of transcriptional regulators and a C-terminal regulatory ligand binding domain for effector-binding/oligomerization (amino acid residues 108 to 244), which is homologous to the UbiC transcription regulator-associated (UTRA) domain. GntR-type regulators are classified into four subfamilies based on their C-terminal domains (60). According to the secondary structure assignment by Define Secondary Structure of Proteins (DSSP) (61) and the crystal structure of residues 105 to 253 (Molecular Modeling Database [MMDB] accession no. 46481; Protein Data Bank [PDB] accession no. 2P19), the C terminus of IolR contains α-helical and β-sheet structures. Due to this structure and a length of the C-terminal domain of about 150 amino acids, IolR can be classified into the HutC subfamily of GntR-type regulators.
In vitro binding of RamA, GntR1, GntR2, and IolR to the pck promoter.
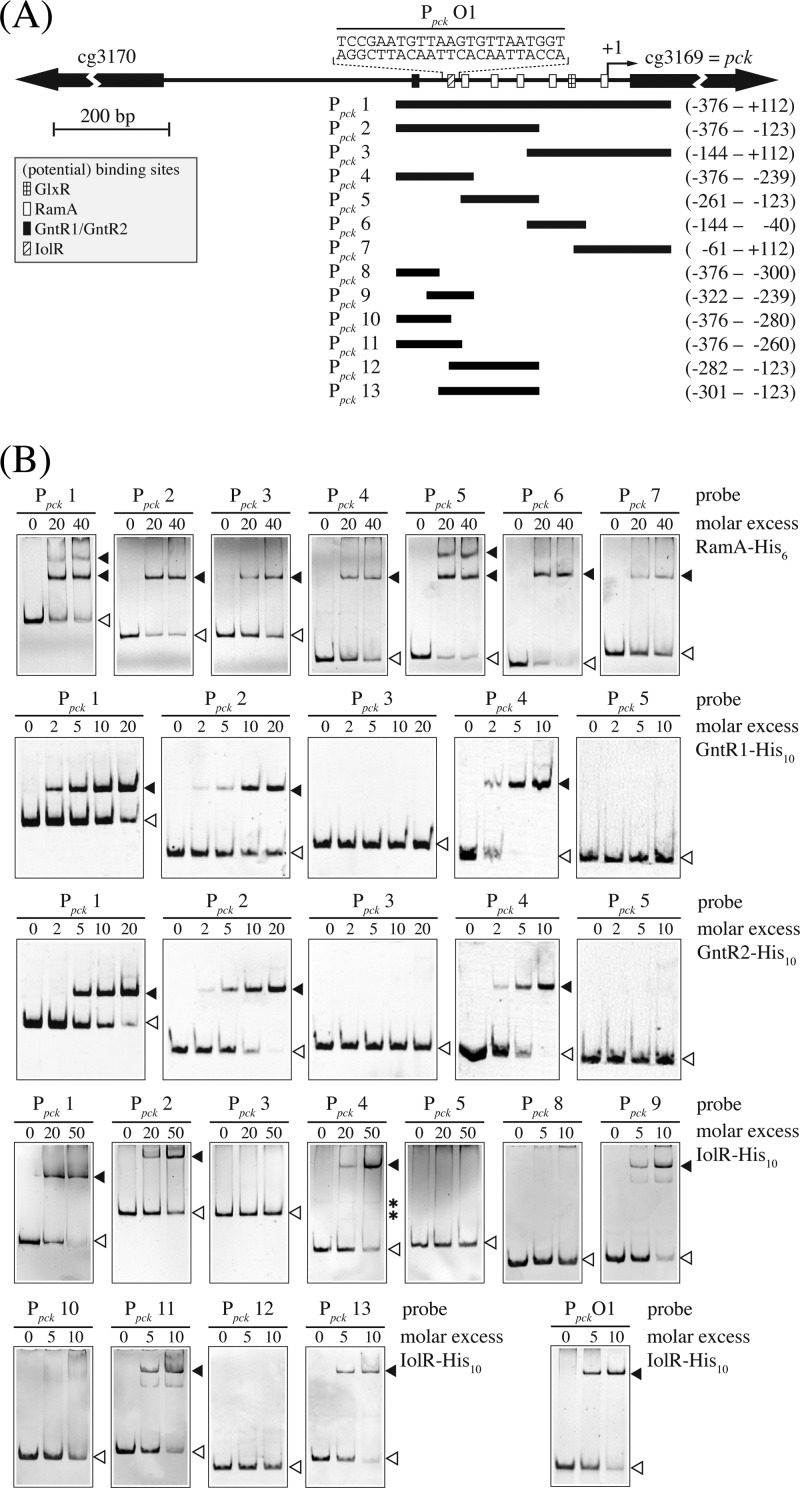
In silico analysis of the C. glutamicum pck promoter used for DNA affinity chromatography indicated the presence of one GntR1/GntR2 and five RamA binding sites. The latter are located at positions −1 to −7, −84 to −90, −150 to −156, −197 to −202, and −247 to −252 relative to the TSS of pck (40), respectively, and totally match the described RamA consensus motifs A/C-G4–6-C/T and A-C4–5-A/G/T (42, 43). The potential GntR1/GntR2 binding site is located at position −326 to −340 relative to the TSS of pck and matches the described consensus motif WWtgaTMNTACYWNt (W = A or T, M = A or C, Y = C or T, N = any base; lowercase letters represent nucleotides that are conserved in at least 75% of the binding sites used for the generation of the motif sequence) (44) except for one mismatch (T versus G at position −335).
To analyze the interaction of RamA, GntR1, GntR2, and IolR with the promoter region of pck in more detail, we overexpressed the corresponding genes in E. coli and purified them as His-tagged RamA-His6, GntR1-His10, GntR2-His10, and IolR-His10. The isolated proteins were used for EMSAs with various pck promoter fragments (Fig. 3). In a first series of experiments, different amounts of the proteins were incubated with the promoter fragment Ppck 1 covering the region from −376 bp to +112 bp with respect to the pck TSS. Each of the four proteins shifted this DNA fragment (Fig. 3). To confirm binding of RamA and GntR1/GntR2 to the predicted binding sites and to identify the IolR binding site within the pck promoter fragment, a second series of EMSAs was performed with a set of six different subfragments of Ppck 1 (Ppck 2 to Ppck 7). With RamA-His6, distinct retardations were observed with all fragments containing at least one predicted RamA binding site, indicating that all of these sites are able to bind RamA. In the case of GntR1 and GntR2, only those fragments containing the predicted binding site (Ppck 2 and Ppck 4) were retarded, whereas the other fragments showed no interaction, supporting the notion of the functionality of the predicted motif (Fig. 3).
Fig 3.
Binding of RamA, GntR1, GntR2, and IolR to the pck promoter region. (A) DNA fragments (Ppck 1 to Ppck 13) used to determine the location of the RamA, GntR1, GntR2, and IolR binding site(s) in the pck promoter are shown. The transcription start site (TSS) of the pck gene is indicated by an arrow labeled +1. Black bars indicate the DNA fragments used for electrophoretic mobility shift assays (EMSAs). The numbers given to the right show the positions of the fragment ends relative to the TSS. The GlxR, RamA, GntR1/GntR2, and IolR binding sites are indicated with checkered, white, black, and striped boxes, respectively. (B) Results of EMSAs using different fragments of the pck promoter region and His-tagged RamA, GntR1, GntR2, and IolR. The DNA fragments used (Ppck 1 to Ppck 13) are indicated at the top of each gel. Free DNA and DNA-protein complexes are indicated with white and black arrowheads, respectively. Nonspecific bands are indicated with asterisks.
In the case of IolR, no preliminary information was available on the position of the DNA binding site. The EMSAs with the subfragments revealed binding to fragments Ppck 2 and Ppck 4, but not to Ppck 3 and Ppck 5, indicating the IolR binding site to be located within fragment Ppck 4, which covers bases −376 to −239 relative to the TSS of the pck gene (Fig. 3). To exactly localize the binding site of IolR, another set of subfragments (Ppck 8 to Ppck 13) was analyzed and shifts were observed with fragments Ppck 9, Ppck 11, and Ppck 13 (Fig. 3). Therefore, the IolR binding site was assumed to be located at position −294 to −272 within the pck promoter region. This finding was substantiated by showing that IolR-His10 binds to a 23-bp double-stranded (ds)-oligonucleotide (Ppck O1; 5′-TCCGAATGTTAAGTGTTAATGGT-3′) representing the overlapping region between fragments Ppck 11 and Ppck 13 (Fig. 3).
Impact of ramA, gntR1 and/or gntR2, and iolR gene deletion on PEP carboxykinase activity.
To test the role of the transcriptional regulators RamA, GntR1, GntR2, and IolR in pck gene expression, we compared the specific PEP carboxykinase activity of cell extracts of C. glutamicum WT, ΔramA, ΔgntR1, ΔgntR2, ΔgntR1 ΔgntR2, and ΔiolR strains (Fig. 4). The cells were grown either with 2% (wt/vol) glucose or with 1% (wt/vol) potassium acetate as the sole carbon source and harvested in the exponential-growth phase. In accordance with previous results (23), the WT strain showed about-2-fold-higher PEP carboxykinase activity during growth with acetate (0.35 ± 0.02 U/mg protein) compared to growth with glucose (0.15 ± 0.01 U/mg protein). As expected, C. glutamicum ΔramA was not able to grow on acetate as the sole carbon source (42). With glucose as the carbon source, the PEP carboxykinase activity of the ΔramA mutant (0.34 ± 0.03 U/mg protein) was comparable to that of the WT during growth with acetate, confirming RamA to be a repressor of pck transcription during growth with glucose. As expected from the functional redundancy of GntR1 and GntR2 (44), the PEP carboxykinase activities of the C. glutamicum ΔgntR1 and ΔgntR2 strains (0.18 ± 0.02 and 0.17 ± 0.02 U/mg protein on glucose; 0.39 ± 0.02 and 0.37 ± 0.02 U/mg protein on acetate) did not significantly differ from the activities of the WT under these conditions. However, deletion of both gntR1 and gntR2 resulted in less than half of the PEP carboxykinase activity seen with the WT, both on glucose (0.07 ± 0.01 U/mg protein) and on acetate (0.17 ± 0.01 U/mg protein). Therefore, we propose identification of GntR1 and GntR2 as activators of pck transcription. Similar to the results observed for the ΔgntR1 ΔgntR2 double mutant, the PEP carboxykinase activity of the iolR-negative strain was also reduced more than 2-fold compared to that of the WT, both on glucose (0.06 ± 0.01 U/mg protein) and on acetate (0.17 ± 0.01 U/mg protein). Consequently, also IolR presumably functions as an activator of pck transcription.
Fig 4.
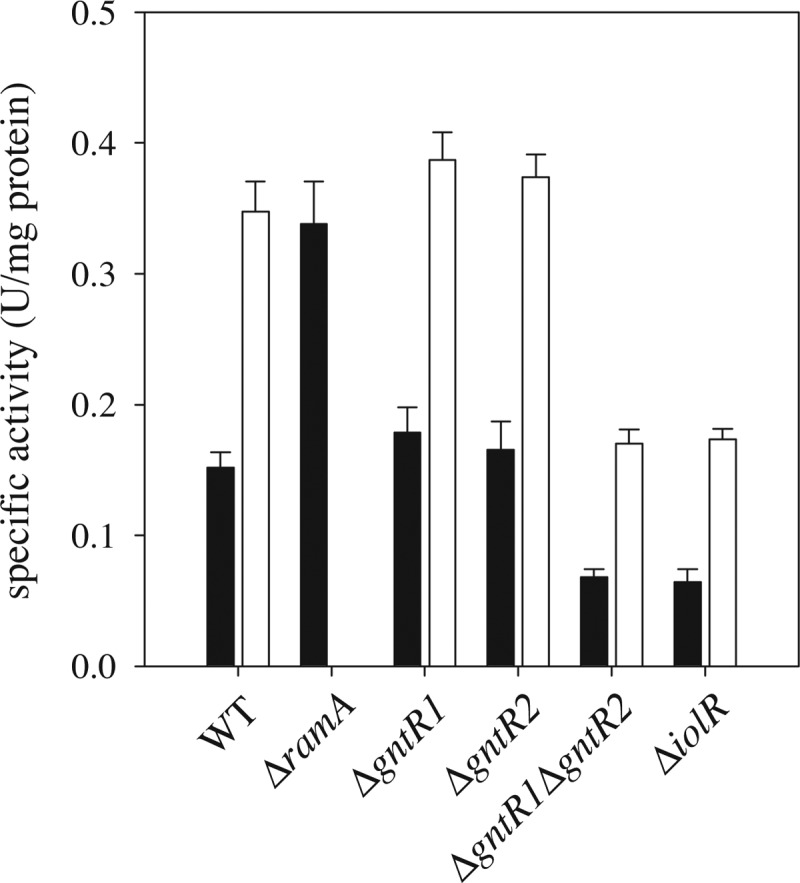
PEP carboxykinase activity of cell extracts obtained from the indicated C. glutamicum strains grown in CGXII minimal medium supplemented with either 2% (wt/vol) glucose (black bars) or 1% (wt/vol) potassium acetate (white bars). The ΔramA mutant is not able to grow on acetate as the sole carbon and energy source. Mean values and standard deviations of the results of three independent cultures are shown.
Transcriptome analysis of the ΔiolR mutant strain.
To identify additional genes that are potentially regulated by IolR, the transcriptome profile of the iolR deletion strain of C. glutamicum was compared to that of the WT strain using DNA microarray analysis. A relatively small number (22) of genes exhibited an at least 3-fold change in their transcript levels (Table 2). Among those genes with a lower mRNA level in the ΔiolR mutant were gapX, encoding the NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, aceA, encoding isocitrate lyase, and, consistent with the lowered PEP carboxykinase activity of the ΔiolR mutant and also with the in vitro binding of IolR to the pck promoter region (see above), the pck gene. With the threshold reduced to 2 (data not shown), aceB, encoding malate synthase, mez, encoding malic enzyme, sucC, encoding the β-subunit of succinyl-coenzyme A (CoA) synthetase, and ptsS, encoding the sucrose-specific IIABC component of the phosphotransferase system, also exhibited significantly lower transcript levels in the ΔiolR mutant. Interestingly, with gapX, pck, mez, aceA, and aceB, expression of a prominent set of genes encoding enzymes linked to gluconeogenesis was negatively affected by the deletion of iolR.
Table 2.
Transcriptome comparison of the C. glutamicum ΔiolR mutant and the wild-type strain using DNA microarrays
| cg locus tag | NCgl locus tag | Known or predicted function of the gene product | Gene | mRNA ratioa (ΔiolR strain/WT strain) |
|---|---|---|---|---|
| cg0197 | NCgl0155 | myo-Inositol catabolism, carbohydrate kinase | iolC | 141.08 |
| cg0198 | NCgl0156 | Hypothetical protein | 156.91 | |
| cg0199 | NCgl0157 | myo-Inositol catabolism, aldehyde dehydrogenase | iolA | 152.50 |
| cg0201 | NCgl0158 | myo-Inositol catabolism | iolB | 103.57 |
| cg0202 | NCgl0159 | myo-Inositol catabolism, thiamine pyrophosphate-requiring enzyme | iolD | 110.34 |
| cg0203 | NCgl0160 | 2-Keto-myo-inositol dehydratase | iolE | 145.21 |
| cg0204 | NCgl0161 | myo-Inositol 2-dehydrogenase, oxidoreductase | iolG | 129.71 |
| cg0205 | NCgl0162 | myo-Inositol catabolism, isomerase/epimerase | iolH | 112.40 |
| cg0206 | NCgl0163 | Efflux carrier, major facilitator superfamily | iolP | 52.56 |
| cg0207 | NCgl0164 | myo-Inositol dehydrogenase, oxidoreductase | oxiA | 445.00 |
| cg0210 | NCgl0167 | LacI-family transcriptional regulator | 10.00 | |
| cg0223 | NCgl0178 | myo-Inositol transporter | iolT1 | 77.05 |
| cg1069 | NCgl0900 | Glyceraldehyde-3-phosphate dehydrogenase | gapX | 0.37 |
| cg1595 | NCgl1353 | Universal stress protein UspA or related nucleotide-binding protein | uspA2 | 6.61 |
| cg1935 | NCgl1650 | Gluconate-responsive repressor of genes involved in gluconate catabolism and the pentose phosphate pathway | gntR2 | 3.16 |
| cg2560 | NCgl2248 | Isocitrate lyase | aceA | 0.23 |
| cg2732 | NCgl2399 | Gluconate kinase | gntK | 109.66 |
| cg3096 | NCgl2698 | Aldehyde dehydrogenase | ald | 0.21 |
| cg3107 | NCgl2709 | Zn-dependent alcohol dehydrogenase | adhA | 0.20 |
| cg3169 | NCgl2765 | Phosphoenolpyruvate carboxykinase (GTP) | pck | 0.32 |
| cg3195 | NCgl2787 | Flavin-containing monooxygenase | 0.34 | |
| cg3216 | NCgl2808 | Gluconate permease | gntP | 4.90 |
The mRNA ratios shown represent mean values of the results of three independent DNA microarray experiments starting from independent cultures grown in CGXII minimal medium with 4% (wt/vol) glucose. All genes listed showed at least a 3-fold change in mRNA levels (increased or decreased) in at least two of the three replicates with P < 0.05 and signal-to-noise (S/N) ratio > 10.
On the other hand, an up to 445-fold increase in mRNA level could be observed for 16 genes in the mutant (Table 2). Twelve of them, mostly exhibiting a more than 100-fold increase in their mRNA level, are located in a large gene cluster encoding proteins for myo-inositol uptake and degradation (see Fig. S1 in the supplemental material) (45). A residual subset of three genes, gntP, gntK, and gntR2, code for gluconate permease, gluconate kinase, and one of the functionally redundant repressors of gluconate metabolism, respectively (44). To analyze whether iolC, iolT1, gntP, gntK, gntR2, aceA, aceB, mez, and gapX are directly regulated by IolR, binding of purified IolR-His10 to the corresponding promoter regions was tested in EMSAs. Obvious retardation could be observed only with the intergenic region between iolR and iolC and the promoter region of iolT1 (see below and data not shown).
Identification of IolR binding sites within the promoter regions of iolC, iolR, and iolT1 and prediction of a conserved IolR binding motif.
The iolC and iolT1 genes are part of a so-called C. glutamicum iol cluster (45), whose genes were named according to the iol cluster of B. subtilis (62). The iolC gene (cg0197) was annotated as 2-deoxy-5-keto-gluconokinase and is the first of 10 genes that are most likely cotranscribed as an operon. The iolT1 gene (cg0223) encoding myo-inositol transporter 1 is located further downstream in a divergent orientation. The iolR gene is located immediately upstream of iolC in the opposite orientation. The TSSs of the respective genes (see Table S2 in the supplemental material) were derived from a RNASeq library generated in the course of another project at the Institute for Genome Research and Systems Biology, University of Bielefeld (unpublished data). For iolR, two neighboring A residues located 136 and 137 bp upstream of the start codon were identified as TSSs. In the case of iolT1, an A residue located 85 bp upstream of the start codon and another A residue located 113 bp upstream of the start codon were found to be the TSSs. For both iolR and iolT, the TSSs closer to the start codon gave rise to a larger amount of transcripts and are labeled +1 in Fig. 5. For iolC, the TSS was located 5 bp downstream of the proposed start codon. Therefore, a reannotation of the iolC coding sequence was required in which the translational start site was shifted from position 168976 to position 169039 according to the NCBI RefSeq NC_006958.1. This reannotation is based on the occurrence of a putative Shine-Dalgarno sequence upstream of the ATG start codon and is consistent with the annotation in the sequence entry NCBI RefSeq NC_003450.3 as well as with the sequence used for crystallization of IolC (MMDB accession no. 86522; PDB accession no. 3PL2). The TSS was located 58 bp upstream of the start codon at position 160039.
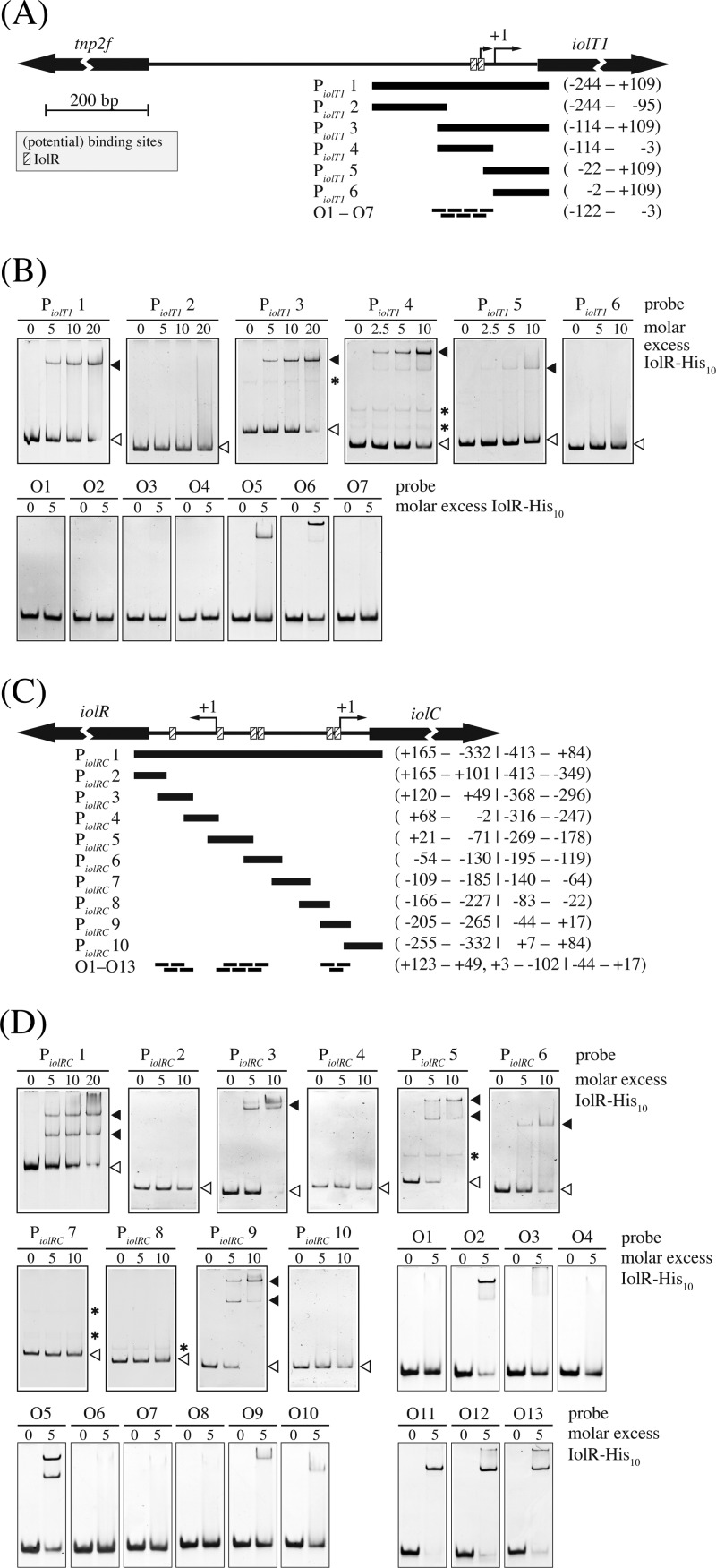
Fig 5.
Binding of IolR to the iolT1 promoter region (A and B) and the intergenic region of iolR and iolC (C and D). In panels A and C, the DNA fragments (PiolT1 1 to PiolT1 6 and PiolRC 1 to PiolRC 10) and ds-oligonucleotides used to determine the location of the IolR binding site(s) are shown as black bars below a map of the promoter regions. The numbers given to the right show the positions of the fragment ends or the region covered by consecutive oligonucleotides relative to the transcription start site (TSS). The dominant TSS of the respective genes is indicated by an arrow labeled +1. The identified IolR binding sites are indicated with striped boxes. Both figures are in the same scale. In panels B and D, the results of electrophoretic mobility shift assays using the different fragments and oligonucleotides and His-tagged IolR are shown. Free DNA and DNA-protein complexes are indicated with white and black arrowheads, respectively. Nonspecific bands are indicated with asterisks.
Binding of IolR-His10 to the promoter region of iolT1 and to the intergenic region between iolR and iolC was tested in gel shift experiments by using a 353-bp fragment of the iolT1 promoter region and a 497-bp fragment covering the region between iolR and iolC. In the case of iolT1, a single protein-DNA complex was formed, whereas in the case of the iolR-iolC region, two distinct protein-DNA complexes were observed (Fig. 5). These results suggested the presence of at least one IolR binding site within the iolT1 promoter and of at least two IolR binding sites in the iolR-iolC intergenic region. To exactly localize the binding sites, subfragments of various sizes were used in a second series of EMSAs (Fig. 5). Within the iolT1 promoter region, a specific interaction was detected with fragments PiolT1 3 and PiolT1 4 (Fig. 5A and B). The slight interaction with fragment PiolT1 5 was abolished when 20 bp was removed from its 5′ end (PiolT1 6), indicating that the IolR binding site is located between positions −114 and −3 with respect to the TSS of iolT1. A set of seven 30-bp ds-oligonucleotides (PiolT1 O1 to PiolT1 O7) was used to identify the IolR binding site(s) within this region. At a 5-fold molar excess of IolR-His10, only oligonucleotides PiolT1 O5 and PiolT1 O6 shifted, suggesting the presence of two IolR binding sites between position −62 and position −18 relative to the TSS of iolT1 that is closer to the start codon. At least three IolR binding sites were predicted within the intergenic region between iolR and iolC due to IolR-His10-mediated retardation of the fragments PiolRC 3, PiolRC 5, PiolRC 6, and PiolRC 9 (Fig. 5C and D). To localize the binding sites within these fragments, a set of 13 30-bp ds-oligonucleotides (PiolRC O1 to PiolRC O13) was used (Fig. 5D). At a 5-fold molar excess of IolR-His10, interaction was detected with oligonucleotides PiolRC O2, PiolRC O5, PiolRC O9, and PiolRC O10, spanning positions +108 to +79, +3 to −27, and −58 to −102 relative to the TSS of iolR and with oligonucleotides PiolRC O11, PiolRC O12, and PiolRC O13, spanning positions −44 to +17 relative to the TSS of iolC. These results suggest the presence of at least five IolR binding sites within the intergenic region between iolR and iolC.
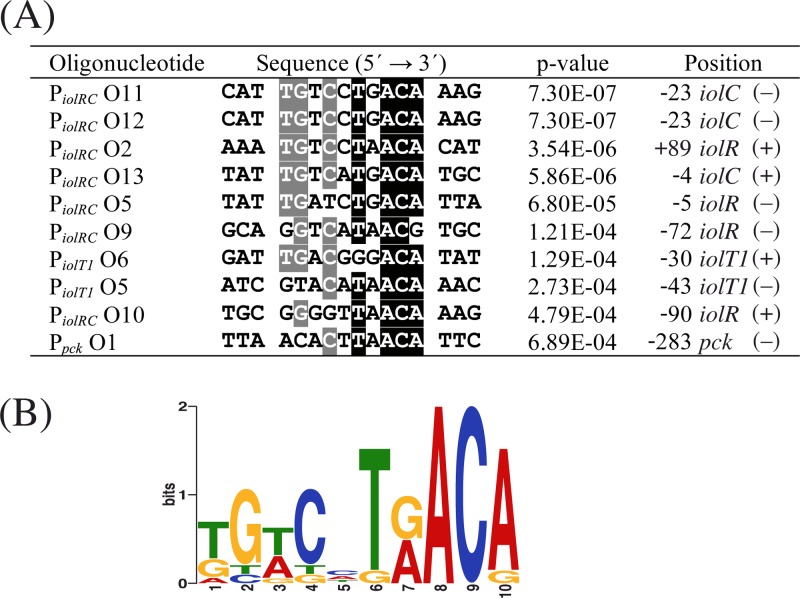
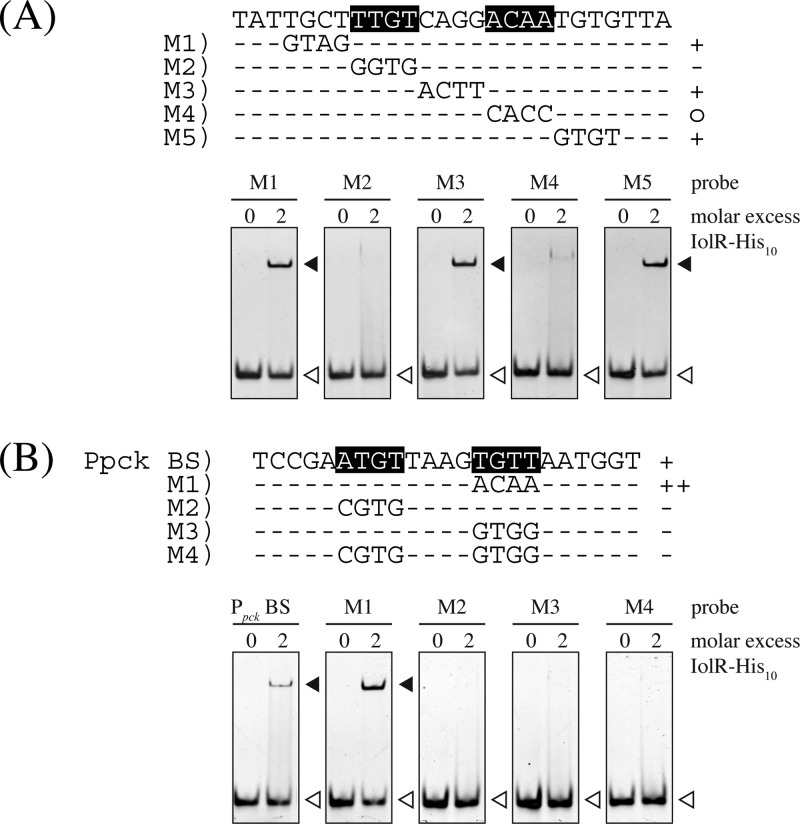
The DNA regions narrowed down as IolR binding sites were analyzed for similarities by the online tool MEME, which resulted in the identification of the consensus motif KGWCHTRACA (K = G or T, W = A or T, H = A or C or T, R = A or G) (Fig. 6). The double shift observed for probe O5 in panel D of Fig. 5 might have been due to a weakly conserved second binding site whose occupation by IolR is enhanced by the well-conserved binding site. The relevance of the identified consensus motif in the binding region of PiolRC O11 was tested by performing EMSAs with DNA fragments in which four nucleotides of the proposed motif were sequentially exchanged. As shown in Fig. 7A, by mutating the half-sites of the highly conserved regions within the palindromic sequence, i.e., TTGT and ACAA, binding of IolR was strongly impaired, whereas no negative impact on binding could be observed by mutation of the center and the neighboring zones. According to the MEME result, the IolR binding site within the pck promoter offers the least conserved region of all fragments (Fig. 6A). To elucidate the role of the sequences ATGT and TGTT, which are positioned at the highly conserved regions of the proposed IolR binding motif in the pck promoter, we performed EMSAs with mutated fragments. As shown in Fig. 7B, exchange of TGTT to ACAA, which matches the proposed consensus motif, enhanced binding of IolR-His10, whereas mutations of one or both sites to sequences inconsistent with the consensus motif led to a complete inhibition of IolR-His10 binding.
Fig 6.
(A) Binding sites predicted by the MEME software within ds-oligonucleotides that were shifted by IolR. Nucleotides indicated by white characters shaded in black and gray are conserved in at least 90% and 60% of all binding sites, respectively. The position numbers indicate the distances from the center of the binding site (fifth bp of the motif) to the transcription start site of iolR, iolC, iolT1, and pck. (+) or (−) indicates whether the sequence shown is derived from the coding strand or on the template strand of the downstream gene, respectively. (B) Consensus motif of IolR derived from the binding sites using the WebLogo tool (http://weblogo.berkeley.edu).
Fig 7.
Mutational analysis of the putative IolR binding motifs within the promoter regions of iolC (A) and pck (B). The importance of the predicted DNA sequence motif (Fig. 6B) for IoR binding was tested in electrophoretic mobility shift assays with DNA fragments in which 4 nucleotides in or next to the proposed motif were exchanged. According to the results of the shifts, the fragments were divided into three categories as follows: ++, mutated fragment shifted with higher affinity than the wild-type (WT) fragment; +, mutated fragment shifted like the WT fragment; o, mutated fragment hardly shifted; −, mutated fragment not shifted at all. Bases indicated with white characters shaded in black are important for IolR binding.
Phenotypic consequences of iolR deficiency in combination with pck overexpression.
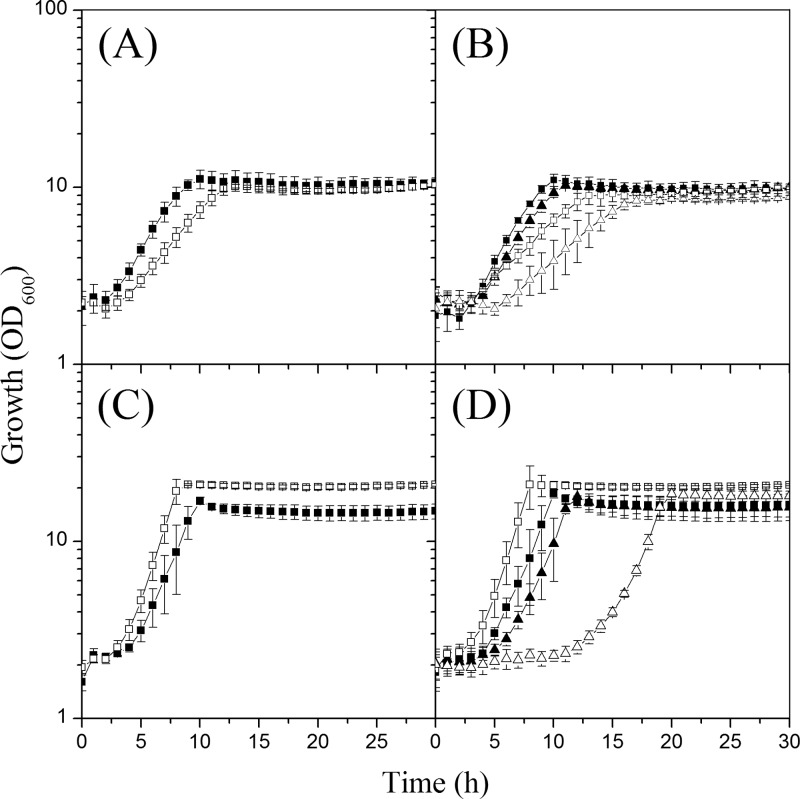
During cultivation of the iolR deletion mutant for the measurement of PEP carboxykinase activities, we noticed a growth rate that was slightly lower than that of the WT in acetate minimal medium. To further elucidate the phenotypic consequences of IolR deficiency, the growth behavior of the ΔiolR mutant was compared to that of the WT in CGXII minimal medium with different carbon and energy sources. No difference was observed with 2% (wt/vol) glucose as the sole carbon and energy source (0.32 ± 0.03 h−1 and 0.33 ± 0.04 h−1). A decreased growth rate was found in media supplemented with 1% (wt/vol) potassium acetate (0.18 ± 0.02 h−1 versus 0.25 ± 0.02 h−1) (Fig. 8A) or 1% (wt/vol) sodium l-lactate (0.11 ± 0.01 h−1 versus 0.14 ± 0.01 h−1). An increased growth rate was observed in medium with 2% (wt/vol) myo-inositol (0.40 ± 0.01 h−1 versus 0.36 ± 0.04 h−1) (Fig. 8C). In this medium, the ΔiolR mutant reached a slightly higher cell density than the WT, whereas in all other media the final OD values were comparable.
Fig 8.
(A) Growth of C. glutamicum wild-type (WT) (black squares) and ΔiolR (white squares) strains in minimal medium with 1% (wt/vol) acetate. (B) Growth of WT (pEK0) (black squares), WT (pEK0_pckB) (black triangles), ΔiolR (pEK0) (white squares), and ΔiolR (pEK0_pckB) (white triangles) strains in minimal medium with 1% acetate. (C) Growth of WT (black squares) and ΔiolR (white squares) strains in minimal medium with 2% (wt/vol) myo-inositol. (D) Growth of WT (pEK0) (black squares), WT (pEK0_pckB) (black triangles), ΔiolR (pEK0) (white squares), and ΔiolR (pEK0_pckB) (white triangles) strains in minimal medium with 2% myo-inositol. The strains were cultivated in microtiter plates using the Biolector system. Mean values and standard deviations of the results of three independent experiments performed with two replicates each are shown; the backscatter was converted to OD600 values as described in Materials and Methods.
To test if the reduced growth rate of the ΔiolR mutant in acetate minimal medium was due to the lowered expression of the pck gene, we overexpressed pck in the WT strain and the ΔiolR mutant by using plasmid pEK0-pckB (23) and compared their growth characteristics with those of the corresponding strains carrying the pEK0 vector. Functional overexpression of pck was confirmed by six-to-10-fold-higher PEP carboxykinase activities in the WT strain and the iolR deletion mutant carrying plasmid pEK0-pckB (1.20 ± 0.30 and 0.66 ± 0.01 U/mg protein, respectively, during growth with glucose; 2.42 ± 0.15 and 1.64 ± 0.28 U/mg protein, respectively, during growth with acetate). The reference WT (pEK0) and ΔiolR (pEK0) strains had PEP carboxykinase activities of 0.15 ± 0.02 and 0.06 ± 0.00 U/mg protein during growth on glucose and of 0.38 ± 0.02 and 0.19 ± 0.06 U/mg protein during growth with acetate. Overexpression of pck did not improve growth on acetate of the ΔiolR (pEK0-pckB) strain compared to the reference ΔiolR (pEK0) strain, as the strains showed comparable growth rates of 0.16 ± 0.04 h−1 and 0.16 ± 0.02 h−1, respectively. Both ΔiolR strains grew slower than the WT (pEK0) and WT (pEK0-pckB) strains, which had growth rates on acetate of 0.25 ± 0.01 h−1 and 0.24 ± 0.02 h−1, respectively (Fig. 8B). Interestingly, during growth with myo-inositol, the ΔiolR (pEK0_pckB) strain revealed a prolonged lag phase of about 10 h and a reduced growth rate of 0.34 ± 0.02 h−1 compared to strain ΔiolR (pEK0) (0.42 ± 0.01 h−1) (Fig. 8D). Similar effects were not observed when these strains were cultivated on glucose (data not shown), indicating that high PEP carboxykinase activity negatively impacts myo-inositol utilization.
DISCUSSION
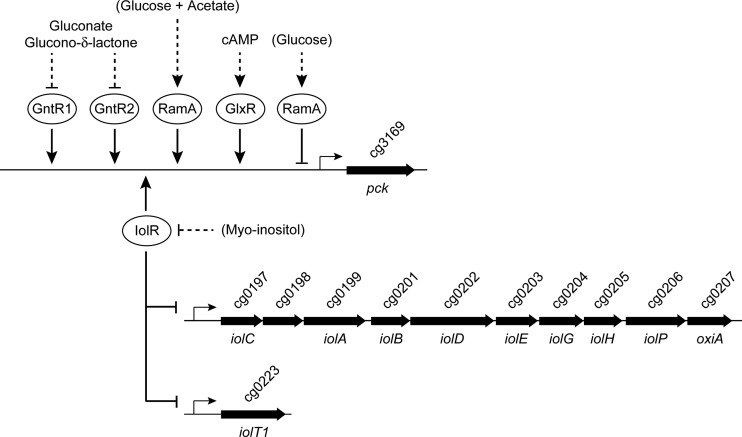
The anaplerotic and gluconeogenic reactions at the PEP-pyruvate-oxaloacetate node are of importance for the production of TCA cycle-derived amino acids and organic acids. In addition to the C3-carboxylating enzymes PEP carboxylase and pyruvate carboxylase, C. glutamicum possesses the C4-decarboxylating enzymes PEP carboxykinase, oxaloacetate decarboxylase, and malic enzyme, and all of them are present with significant specific activities in extracts of cells grown on glucose (20, 63). 13C-based metabolic flux analysis repeatedly revealed that a high C4-decarboxylating flux exists in vivo in C. glutamicum which varies significantly under different growth and/or production conditions and is supposed to be regulated by mechanisms at the enzyme activity and gene expression levels (20). Quantification of the individual in vivo fluxes at the PEP-pyruvate-oxaloacetate node (64) indicated PEP carboxykinase to be the responsible enzyme by which two-thirds of anaplerotically synthesized oxaloacetate are recycled to PEP, resulting in a futile cycle under glycolytic conditions. The advantage of this futile cycling and the physiological function of PEP carboxykinase under glycolytic conditions are far from being understood and require a better understanding of the regulation of the genes and enzymes at the PEP-pyruvate-oxaloacetate node under different metabolic conditions. Here, we focused on transcriptional regulation of the pck gene under glycolytic and gluconeogenic conditions. We report that in addition to GlxR and RamA, the GntR-type regulators GntR1, GntR2, and IolR bind to the pck promoter and are capable of modulating expression of the pck gene. Additionally, we uncovered the role of IolR in regulation of genes involved in myo-inositol catabolism (Fig. 9).
Fig 9.
Overview of the complex regulation of the pck gene by the transcriptional regulators RamA, GlxR, GntR1, GntR2, and IolR and the role of IolR as activator of the pck gene and repressor of genes involved in myo-inositol catabolism. Positive or negative influence on the target promoters is symbolized by arrowheads or bars, respectively. Dual transcriptional regulation of pck by RamA under different growth conditions was described previously (43). Gluconate and glucono-δ-lactone were shown to inhibit DNA binding by GntR1 and GntR2 (44), whereas cAMP was shown to be required for binding of GlxR to the pck promoter (41). In contrast to these regulators, possible effectors of RamA and IolR have not yet been identified. In the case of IolR, it is likely to be myo-inositol itself or a degradation product.
Although direct interaction of GlxR and the pck promoter has been shown (40, 41), the GlxR protein was not enriched with the pck promoter probe in our DNA affinity chromatography experiments. The GlxR protein was initially characterized as a transcriptional regulator that represses the aceB gene of the glyoxylate bypass (65). Additional studies showed that GlxR is a global regulator with dual functionality that is predicted to directly regulate about 14% of the annotated C. glutamicum genes (66–69). Binding of GlxR to several of its target promoters was shown to be strictly dependent on the presence of cAMP in vitro (40, 41, 65, 68, 70–73). The intracellular concentration of cAMP in C. glutamicum seems to vary with the carbon source and the growth phase of the culture, as it is, for instance, high when cells are grown on glucose and low when cells are grown on acetate medium (65, 74). These differences in cAMP levels during growth on glucose and acetate together with the presence of a GlxR binding site in the pck promoter suggest that GlxR is involved in carbon source-dependent regulation of pck. The GlxR binding site is located from position −62 to position −47 relative to the TSS of pck (40, 41), and, according to this position, activation of pck by GlxR has been predicted (41).
Besides GlxR, RamA also is a master regulator in C. glutamicum involved in adjusting the expression levels of genes related to carbohydrate metabolism, specifically, those related to ethanol and acetate metabolism, sugar uptake, glycolysis, TCA cycle, anaplerosis, and gluconeogenesis (43, 75). The ramA gene is upregulated 2- to 3-fold in the presence of acetate in the growth medium, which results in 2-fold-larger amounts of RamA in C. glutamicum cells grown on acetate instead of glucose (76). Independent of the acetate-dependent upregulation, the ramA gene is also subject to negative autoregulation (76). In accordance with the refined RamA consensus DNA binding motifs A/T/C-GGGG-N and A/T/C-CCCC-N (43), we proposed five potential RamA binding sites within the pck promoter fragment used in this study for DNA affinity chromatography, which are located at positions −1 to −7, −84 to −90, −150 to −156, −197 to −202, and −247 to −252 relative to the TSS of pck. The previously observed binding of RamA-His6 to the pck promoter region (43) was confirmed by EMSAs, which showed interaction of RamA with all of the predicted sites. The 1.9-fold-higher mRNA level of the pck gene (43) and the 2.2-fold-higher PEP carboxykinase activity measured in the RamA-deficient strain in comparison to the WT during growth with glucose clearly indicate that RamA functions as a repressor of pck expression under these conditions. However, in a previous study, we also observed RamA to activate pck in cells grown in minimal medium with glucose plus acetate or in complex medium (43). This dual function of RamA as activator and repressor was also observed for the gap, cg3226, sucC, pta, ackA, and ald genes (43) and might be due to interaction or competition with a further (regulatory) protein or an effector(s) specific for the one or the other condition.
C. glutamicum is able to use gluconate as the sole carbon and energy source (68). The two functionally redundant transcriptional regulators GntR1 and GntR2 are involved in coordinating the expression of genes involved in gluconate catabolism and glucose uptake (44). Gluconate and glucono-δ-lactone were shown to interfere with binding of GntR1 and GntR2 to their target promoters, leading to derepression of genes involved in gluconate catabolism (gntP, gntK) and reduced expression of ptsG and ptsS, encoding the glucose- and sucrose-specific enzyme II components of the phosphotransferase system (44). In this study, GntR1 and GntR2 were isolated by DNA affinity chromatography with the pck promoter and binding was confirmed by EMSAs with purified GntR1 and GntR2. As indicated by (i) the position of the binding site far upstream of the pck TSS, (ii) the 2-fold-lower pck transcript levels in a ΔgntR1 ΔgntR2 double mutant grown on glucose (ΔgntR1 ΔgntR2 strain/WT strain mRNA ratio = 0.52; P < 0.05 [44]), and (iii) the about-2-fold-reduced PEP carboxykinase activity of the ΔgntR1 ΔgntR2 double mutant, both GntR1 and GntR2 function as activators of pck transcription in the absence of gluconate or glucono-δ-lactone. A reduced expression of pck during growth on gluconate compared to glucose might be related to the differences in the uptake and catabolism of this sugar acid. In contrast to glucose, gluconate is imported by the permease GntP and phosphorylated by ATP rather than by the PEP-dependent phosphotransferase system. Catabolism of 6-phosphogluconate occurs via the pentose phosphate pathway rather than via glycolysis. These differences might lead to a higher PEP availability and a reduced demand for PEP carboxykinase activity.
Like RamA, GntR1, and GntR2, the GntR-type transcriptional regulator IolR was enriched by DNA affinity chromatography performed with the pck promoter region. The binding site of IolR within the pck promoter is located far upstream of the TSS, and the about-3-fold-lower mRNA level and the about-2-fold-lower specific PEP carboxykinase activity in the ΔiolR mutant than in the WT indicate that IolR acts as an activator. These results are entirely different from those of a recent study by Hyeon et al. (77). Those authors described Cg0196 (IolR) as a novel regulator which negatively regulated pck expression when the cells were grown on glucose as the carbon source, and thus they designated the Cg0196 protein PckR. Repression of pck was reported to be mediated by binding of PckR to the 18-bp motif CTAAAGTTTTAACTAGTT, which overlaps the −35 region of the pck promoter. By DNA microarray analysis, Hyeon et al. solely identified glgA (cg1268), encoding a glycosyl transferase (78), as an additional potential target of PckR besides the pck gene. We also tested the interaction of purified IolR with a DNA probe containing the 18-bp motif; however, in EMSAs we could not observe any binding (Fig. 3, fragment Ppck 3). The genomic location of cg0196 along with the results of our DNA microarray and DNA binding studies clearly show that the primary function of Cg0196 is the repression of genes involved in myo-inositol metabolism. This view is also supported by the fact that a cg0196 deletion mutant showed growth on myo-inositol superior to that of the wild-type strain (Fig. 8C); therefore, we think that the designation of this protein as IolR is appropriate.
The iolR gene is located in the opposite direction immediately upstream of a large cluster of genes (see Fig. S1 in the supplemental material) encoding enzymes essential for myo-inositol metabolism (45). Clustering of iol genes is also common in Clostridium perfringens and B. subtilis, and transcription is negatively regulated by the repressor IolR encoded upstream and in the opposite orientation (79–81). Upon deletion of iolR in C. glutamicum, the mRNA level of the iol genes was increased 50- to 150-fold. We identified several IolR binding sites within the promoters of iolR, iolC, and iolT1 which led to the identification of the consensus motif KGWCHTRACA, which perfectly matches the palindromic binding site of HutC of Pseudomonas putida, the first studied member of all HutC-like GntR family regulators (5′-TTGT.ta.ACAA-3′ [periods represent variable distances between the half-sites of the highly conserved bases of the HutC consensus sequence, and lowercase letters represent nucleotides that are conserved in 80% to 90% of the binding sites used for the generation of the motif sequence]) (82). The location of the IolR binding sites in the direct vicinity of the TSSs of iolR, iolC, and iolT1 is in accordance with the observed function of IolR as repressor of the iol gene cluster and the iolT1 gene and suggests that the iolR gene is subject to negative autoregulation.
Comparing the genes showing altered expression during growth of the WT on myo-inositol versus glucose (45) with the genes showing different mRNA levels in the ΔiolR mutant during growth on glucose (this study), a striking subset of common genes can be found. Among those with reduced mRNA levels during growth of the WT on myo-inositol and during growth of the ΔiolR mutant on glucose are mez, encoding malic enzyme, sucC, encoding the β-subunit of succinyl-CoA synthetase, and ptsS, encoding the sucrose-specific IIABC component of the phosphotransferase system, indicating significant carbon source-dependent regulation of genes of the central metabolism. Additionally, upon iolR deletion we observed reduced mRNA levels of genes generally linked to gluconeogenesis, i.e., pck, aceA, aceB, and gapX, which did not exhibit significantly different transcript levels in the previous study performed with myo-inositol or glucose as the substrate (45). However, with the exception of the pck promoter, no direct interaction of IolR with the promoters of mez, aceA, aceB, and gapX could be observed. Thus, it is very unlikely that IolR is directly responsible for the reduced expression of these genes and the reduced growth rate observed for the iolR-deficient strain during growth on gluconeogenic carbon sources. Although gntR2 was found to be significantly upregulated in the ΔiolR mutant and would have been a likely candidate for cross-regulation, no specific interaction of IolR and the gntR2 promoter region was observed.
The small set of genes directly regulated by IolR together with the strongly elevated levels of the iol genes in the ΔiolR mutant indicate that IolR primarily plays a role in derepressing genes necessary for myo-inositol degradation and additionally slightly modifies pck expression for fine-tuning the metabolic flux at the PEP-pyruvate-oxaloacetate node during myo-inositol degradation. This view is also supported by the fact that high PEP carboxykinase activity seems to have a negative impact on myo-inositol utilization (Fig. 8). Assuming that myo-inositol is funneled into the central metabolism of C. glutamicum via the intermediates dihydroxyacetone phosphate and acetyl-CoA, as described for B. subtilis (62), a high level PEP carboxykinase activity could lead to a decreased concentration of oxaloacetate, which is required as an acceptor for acetyl-CoA in the citrate synthase reaction.
In conclusion, we have elucidated the role of IolR as a transcriptional repressor of genes involved in myo-inositol catabolism and showed that PEP carboxykinase synthesis in C. glutamicum is controlled by at least four different transcriptional regulators, RamA, GntR1, GntR2, and IolR. This complex regulation might be the consequence of the high flexibility at the PEP-pyruvate-oxaloacetate node in this organism and the need to tightly adjust the gluconeogenic and anaplerotic reactions to a given condition.
Supplementary Material
Footnotes
Published ahead of print 19 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00265-13.
REFERENCES
- 1.Gao B, Gupta RS. 2012. Phylogenetic framework and molecular signatures for the main clades of the phylum actinobacteria. Microbiol. Mol. Biol. Rev. 76:66–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hermann T. 2003. Industrial production of amino acids by coryneform bacteria. J. Biotechnol. 104:155–172 [DOI] [PubMed] [Google Scholar]
- 3.Leuchtenberger W, Huthmacher K, Drauz K. 2005. Biotechnological production of amino acids and derivatives: current status and prospects. Appl. Microbiol. Biotechnol. 69:1–8 [DOI] [PubMed] [Google Scholar]
- 4.Wendisch VF, Bott M, Eikmanns BJ. 2006. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr. Opin. Microbiol. 9:268–274 [DOI] [PubMed] [Google Scholar]
- 5.Stäbler N, Oikawa T, Bott M, Eggeling L. 2011. Corynebacterium glutamicum as a host for synthesis and export of D-amino acids. J. Bacteriol. 193:1702–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litsanov B, Brocker M, Bott M. 2012. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl. Environ. Microbiol. 78:3325–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Litsanov B, Brocker M, Bott M. 2013. Glycerol as a substrate for aerobic succinate production in minimal medium with Corynebacterium glutamicum. Microb. Biotechnol. 6:189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litsanov B, Kabus A, Brocker M, Bott M. 2012. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb. Biotechnol. 5:116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H. 2008. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl. Microbiol. Biotechnol. 81:459–464 [DOI] [PubMed] [Google Scholar]
- 10.Kind S, Kreye S, Wittmann C. 2011. Metabolic engineering of cellular transport for overproduction of the platform chemical 1,5-diaminopentane in Corynebacterium glutamicum. Metab. Eng. 13:617–627 [DOI] [PubMed] [Google Scholar]
- 11.Mimitsuka T, Sawai H, Hatsu M, Yamada K. 2007. Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci. Biotechnol. Biochem. 71:2130–2135 [DOI] [PubMed] [Google Scholar]
- 12.Schneider J, Wendisch VF. 2010. Putrescine production by engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 88:859–868 [DOI] [PubMed] [Google Scholar]
- 13.Blombach B, Riester T, Wieschalka S, Ziert C, Youn J-W, Wendisch VF, Eikmanns BJ. 2011. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 77:3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H. 2004. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 8:243–254 [DOI] [PubMed] [Google Scholar]
- 15.Smith KM, Cho K-M, Liao JC. 2010. Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 87:1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi Y, Date M, Yokoyama K, Umezawa Y, Matsui H. 2003. Secretion of active-form Streptoverticillium mobaraense transglutaminase by Corynebacterium glutamicum: processing of the pro-transglutaminase by a cosecreted subtilisin-like protease from Streptomyces albogriseolus. Appl. Environ. Microbiol. 69:358–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meissner D, Vollstedt A, Dijl JM, Freudl R. 2007. Comparative analysis of twin-arginine (Tat)-dependent protein secretion of a heterologous model protein (GFP) in three different Gram-positive bacteria. Appl. Microbiol. Biotechnol. 76:633–642 [DOI] [PubMed] [Google Scholar]
- 18.Scheele S, Oertel D, Bongaerts J, Evers S, Hellmuth H, Maurer KH, Bott M, Freudl R. 2013. Secretory production of an FAD cofactor-containing cytosolic enzyme (sorbitol-xylitol oxidase from Streptomyces coelicolor) using the twin-arginine translocation (Tat) pathway of Corynebacterium glutamicum. Microb. Biotechnol. 6:202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kornberg HL. 1966. Anaplerotic sequences and their role in metabolism, p 1–31 In Campell PN, Greville GD. (ed), Essays in biochemistry. Academic Press, New York, NY [Google Scholar]
- 20.Sauer U, Eikmanns BJ. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29:765–794 [DOI] [PubMed] [Google Scholar]
- 21.Jetten MSM, Pitoc GA, Follettie MT, Sinskey AJ. 1994. Regulation of phospho(enol)-pyruvate- and oxaloacetate-converting enzymes in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 41:47–52 [Google Scholar]
- 22.Vallino JJ, Stephanopoulos G. 2000. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Reprinted from Biotechnol. Bioeng., vol. 41:pp 633-646 (1993). Biotechnol. Bioeng. 67:872–885 [PubMed] [Google Scholar]
- 23.Riedel C, Rittmann D, Dangel P, Möckel B, Petersen S, Sahm H, Eikmanns BJ. 2001. Characterization of the phosphoenolpyruvate carboxykinase gene from Corynebacterium glutamicum and significance of the enzyme for growth and amino acid production. J. Mol. Microbiol. Biotechnol. 3:573–583 [PubMed] [Google Scholar]
- 24.Peters-Wendisch PG, Kreutzer C, Kalinowski J, Pátek M, Sahm H, Eikmanns BJ. 1998. Pyruvate carboxylase from Corynebacterium glutamicum: characterization, expression and inactivation of the pyc gene. Microbiology 144:915–927 [DOI] [PubMed] [Google Scholar]
- 25.Utter MF, Kolenbrander HM. 1972. Formation of oxaloacetate by CO2 fixation on phosphoenolpyruvate, p 117–170 In Boyer PD. (ed), The enzymes. Academic Press, New York, NY [Google Scholar]
- 26.Aich S, Imabayashi F, Delbaere LTJ. 2003. Expression, purification, and characterization of a bacterial GTP-dependent PEP carboxykinase. Protein Expr. Purif. 31:298–304 [DOI] [PubMed] [Google Scholar]
- 27.Aich S, Prasad L, Delbaere LTJ. 2008. Structure of a GTP-dependent bacterial PEP-carboxykinase from Corynebacterium glutamicum. Int. J. Biochem. Cell Biol. 40:1597–1603 [DOI] [PubMed] [Google Scholar]
- 28.Jetten MSM, Sinskey AJ. 1993. Characterization of phosphoenolpyruvate carboxykinase from Corynebacterium glutamicum. FEMS Microbiol. Lett. 111:183–188 [Google Scholar]
- 29.Peters-Wendisch PG, Eikmanns BJ, Thierbach G, Bachmann B, Sahm H. 1993. Phosphoenolpyruvate carboxylase in Corynebacterium glutamicum is dispensable for growth and lysine production. FEMS Microbiol. Lett. 112:269–274 [Google Scholar]
- 30.Blombach B, Seibold GM. 2010. Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of L-lysine production strains. Appl. Microbiol. Biotechnol. 86:1313–1322 [DOI] [PubMed] [Google Scholar]
- 31.Geerse RH, Van der Pluijm J, Postma PW. 1989. The repressor of the PEP:fructose phosphotransferase system is required for the transcription of the pps gene of Escherichia coli. Mol. Gen. Genet. 218:348–352 [DOI] [PubMed] [Google Scholar]
- 32.Goldie H. 1984. Regulation of transcription of the Escherichia coli phosphoenolpyruvate carboxykinase locus: studies with pck-lacZ operon fusions. J. Bacteriol. 159:832–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gosset G, Zhang Z, Nayyar S, Cuevas WA, Saier MH., Jr 2004. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J. Bacteriol. 186:3516–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh M-K, Liao JC. 2000. Gene expression profiling by DNA microarrays and metabolic fluxes in Escherichia coli. Biotechnol. Prog. 16:278–286 [DOI] [PubMed] [Google Scholar]
- 35.Oh M-K, Rohlin L, Kao KC, Liao JC. 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277:13175–13183 [DOI] [PubMed] [Google Scholar]
- 36.Saier MH, Jr, Ramseier TM. 1996. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 178:3411–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Servant P, Le Coq D, Aymerich S. 2005. CcpN (YqzB), a novel regulator for CcpA-independent catabolite repression of Bacillus subtilis gluconeogenic genes. Mol. Microbiol. 55:1435–1451 [DOI] [PubMed] [Google Scholar]
- 38.Gerstmeir R, Wendisch VF, Schnicke S, Ruan H, Farwick M, Reinscheid D, Eikmanns BJ. 2003. Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 104:99–122 [DOI] [PubMed] [Google Scholar]
- 39.Voges R, Noack S. 2012. Quantification of proteome dynamics in Corynebacterium glutamicum by 15N-labeling and selected reaction monitoring. J. Proteomics 75:2660–2669 [DOI] [PubMed] [Google Scholar]
- 40.Han SO, Inui M, Yukawa H. 2007. Expression of Corynebacterium glutamicum glycolytic genes varies with carbon source and growth phase. Microbiology 153:2190–2202 [DOI] [PubMed] [Google Scholar]
- 41.Kohl TA, Baumbach J, Jungwirth B, Pühler A, Tauch A. 2008. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: in silico and in vitro detection of DNA binding sites of a global transcription regulator. J. Biotechnol. 135:340–350 [DOI] [PubMed] [Google Scholar]
- 42.Cramer A, Gerstmeir R, Schaffer S, Bott M, Eikmanns BJ. 2006. Identification of RamA, a novel LuxR-type transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 188:2554–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Auchter M, Cramer A, Hüser A, Rückert C, Emer D, Schwarz P, Arndt A, Lange C, Kalinowski J, Wendisch VF, Eikmanns BJ. 2011. RamA and RamB are global transcriptional regulators in Corynebacterium glutamicum and control genes for enzymes of the central metabolism. J. Biotechnol. 154:126–139 [DOI] [PubMed] [Google Scholar]
- 44.Frunzke J, Engels V, Hasenbein S, Gätgens C, Bott M. 2008. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol. Microbiol. 67:305–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krings E, Krumbach K, Bathe B, Kelle R, Wendisch VF, Sahm H, Eggeling L. 2006. Characterization of myo-inositol utilization by Corynebacterium glutamicum: the stimulon, identification of transporters, and influence on L-lysine formation. J. Bacteriol. 188:8054–8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 47.Tartof KD, Hobbs CA. 1987. Improved media for growing plasmid and cosmid clones. Bethesda Res. Lab. Focus 9:12–14 [Google Scholar]
- 48.Eikmanns BJ, Metzger M, Reinscheid D, Kircher M, Sahm H. 1991. Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl. Microbiol. Biotechnol. 34:617–622 [DOI] [PubMed] [Google Scholar]
- 49.Eikmanns BJ, Thum-Schmitz N, Eggeling L, Lüdtke KU, Sahm H. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817–1828 [DOI] [PubMed] [Google Scholar]
- 50.Birnboim HC. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243–255 [DOI] [PubMed] [Google Scholar]
- 51.van der Rest ME, Lange C, Molenaar D. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541–545 [DOI] [PubMed] [Google Scholar]
- 52.Dower WJ, Miller JF, Ragsdale CW. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mori M, Shiio I. 1985. Synergistic inhibition of phosphoenolpyruvate carboxylase by aspartate and 2-oxoglutarate in Brevibacterium flavum. J. Biochem. 98:1621–1630 [DOI] [PubMed] [Google Scholar]
- 54.Schaffer S, Weil B, Nguyen VD, Dongmann G, Günther K, Nickolaus M, Hermann T, Bott M. 2001. A high-resolution reference map for cytoplasmic and membrane-associated proteins of Corynebacterium glutamicum. Electrophoresis 22:4404–4422 [DOI] [PubMed] [Google Scholar]
- 55.Toyoda K, Teramoto H, Inui M, Yukawa H. 2009. Involvement of the LuxR-type transcriptional regulator RamA in regulation of expression of the gapA gene, encoding glyceraldehyde-3-phosphate dehydrogenase of Corynebacterium glutamicum. J. Bacteriol. 191:968–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Möker N, Brocker M, Schaffer S, Krämer R, Morbach S, Bott M. 2004. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54:420–438 [DOI] [PubMed] [Google Scholar]
- 57.Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Möckel B, Pfefferle W, Pühler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegräbe I, Tauch A. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5–25 [DOI] [PubMed] [Google Scholar]
- 58.Blom J, Jakobi T, Doppmeier D, Jaenicke S, Kalinowski J, Stoye J, Goesmann A. 2011. Exact and complete short-read alignment to microbial genomes using graphics processing unit programming. Bioinformatics 27:1351–1358 [DOI] [PubMed] [Google Scholar]
- 59.Pátek M, Nešvera J. 2011. Sigma factors and promoters in Corynebacterium glutamicum. J. Biotechnol. 154:101–113 [DOI] [PubMed] [Google Scholar]
- 60.Rigali S, Derouaux A, Giannotta F, Dusart J. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507–12515 [DOI] [PubMed] [Google Scholar]
- 61.Kabsch W, Sander C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637 [DOI] [PubMed] [Google Scholar]
- 62.Yoshida K, Yamaguchi M, Morinaga T, Kinehara M, Ikeuchi M, Ashida H, Fujita Y. 2008. myo-Inositol catabolism in Bacillus subtilis. J. Biol. Chem. 283:10415–10424 [DOI] [PubMed] [Google Scholar]
- 63.Cocaign-Bousquet M, Guyonvarch A, Lindley ND. 1996. Growth rate-dependent modulation of carbon flux through central metabolism and the kinetic consequences for glucose-limited chemostat cultures of Corynebacterium glutamicum. Appl. Environ. Microbiol. 62:429–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petersen S, De Graaf AA, Eggeling L, Möllney M, Wiechert W, Sahm H. 2000. In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J. Biol. Chem. 275:35932–35941 [DOI] [PubMed] [Google Scholar]
- 65.Kim H-J, Kim T-H, Kim Y, Lee H-S. 2004. Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum. J. Bacteriol. 186:3453–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jungwirth B, Sala C, Kohl TA, Uplekar S, Baumbach J, Cole ST, Pühler A, Tauch A. 2013. High-resolution detection of DNA binding sites of the global transcriptional regulator GlxR in Corynebacterium glutamicum. Microbiology 159:12–22 [DOI] [PubMed] [Google Scholar]
- 67.Kohl TA, Tauch A. 2009. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: detection of the corynebacterial core regulon and integration into the transcriptional regulatory network model. J. Biotechnol. 143:239–246 [DOI] [PubMed] [Google Scholar]
- 68.Letek M, Valbuena N, Ramos A, Ordóñez E, Gil JA, Mateos LM. 2006. Characterization and use of catabolite-repressed promoters from gluconate genes in Corynebacterium glutamicum. J. Bacteriol. 188:409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toyoda K, Teramoto H, Inui M, Yukawa H. 2011. Genome-wide identification of in vivo binding sites of GlxR, a cyclic AMP receptor protein-type regulator in Corynebacterium glutamicum. J. Bacteriol. 193:4123–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bussmann M, Emer D, Hasenbein S, Degraf S, Eikmanns BJ, Bott M. 2009. Transcriptional control of the succinate dehydrogenase operon sdhCAB of Corynebacterium glutamicum by the cAMP-dependent regulator GlxR and the LuxR-type regulator RamA. J. Biotechnol. 143:173–182 [DOI] [PubMed] [Google Scholar]
- 71.Han SO, Inui M, Yukawa H. 2008. Effect of carbon source availability and growth phase on expression of Corynebacterium glutamicum genes involved in the tricarboxylic acid cycle and glyoxylate bypass. Microbiology 154:3073–3083 [DOI] [PubMed] [Google Scholar]
- 72.Jungwirth B, Emer D, Brune I, Hansmeier N, Pühler A, Eikmanns BJ, Tauch A. 2008. Triple transcriptional control of the resuscitation promoting factor 2 (rpf2) gene of Corynebacterium glutamicum by the regulators of acetate metabolism RamA and RamB and the cAMP-dependent regulator GlxR. FEMS Microbiol. Lett. 281:190–197 [DOI] [PubMed] [Google Scholar]
- 73.Panhorst M, Sorger-Herrmann U, Wendisch VF. 2011. The pstSCAB operon for phosphate uptake is regulated by the global regulator GlxR in Corynebacterium glutamicum. J. Biotechnol. 154:149–155 [DOI] [PubMed] [Google Scholar]
- 74.Polen T, Schluesener D, Poetsch A, Bott M, Wendisch VF. 2007. Characterization of citrate utilization in Corynebacterium glutamicum by transcriptome and proteome analysis. FEMS Microbiol. Lett. 273:109–119 [DOI] [PubMed] [Google Scholar]
- 75.Arndt A, Eikmanns BJ. 2008. Regulation of carbon metabolism in Corynebacterium glutamicum, p 155–182 In Burkovski A. (ed), Corynebacteria: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 76.Cramer A, Eikmanns BJ. 2007. RamA, the transcriptional regulator of acetate metabolism in Corynebacterium glutamicum, is subject to negative autoregulation. J. Mol. Microbiol. Biotechnol. 12:51–59 [DOI] [PubMed] [Google Scholar]
- 77.Hyeon JE, Kang DH, Kim YI, You SK, Han SO. 2012. GntR-type transcriptional regulator PckR negatively regulates the expression of phosphoenolpyruvate carboxykinase in Corynebacterium glutamicum. J. Bacteriol. 194:2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tzvetkov M, Klopprogge C, Zelder O, Liebl W. 2003. Genetic dissection of trehalose biosynthesis in Corynebacterium glutamicum: inactivation of trehalose production leads to impaired growth and an altered cell wall lipid composition. Microbiology 149(Pt 7):1659–1673 [DOI] [PubMed] [Google Scholar]
- 79.Kawsar HI, Ohtani K, Okumura K, Hayashi H, Shimizu T. 2004. Organization and transcriptional regulation of myo-inositol operon in Clostridium perfringens. FEMS Microbiol. Lett. 235:289–295 [DOI] [PubMed] [Google Scholar]
- 80.Yoshida K, Aoyama D, Ishio I, Shibayama T, Fujita Y. 1997. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J. Bacteriol. 179:4591–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshida K, Shibayama T, Aoyama D, Fujita Y. 1999. Interaction of a repressor and its binding sites for regulation of the Bacillus subtilis iol divergon. J. Mol. Biol. 285:917–929 [DOI] [PubMed] [Google Scholar]
- 82.Hu L, Allison SL, Phillips AT. 1989. Identification of multiple repressor recognition sites in the hut system of Pseudomonas putida. J. Bacteriol. 171:4189–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eikmanns BJ, Kleinertz E, Liebl W, Sahm H. 1991. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene 102:93–98 [DOI] [PubMed] [Google Scholar]
- 84.Schäfer A, Tauch A, Jäger Kalinowski WJ, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.