Abstract
Campylobacter jejuni NCTC11168 does not produce any endogenous siderophores of its own yet requires the CfrA enterobactin transporter for in vivo colonization. In addition, the genome of C. jejuni NCTC11168 contains three distinct TonB energy transduction systems, named TonB1, TonB2, and TonB3, that have not been tested for their role in siderophore uptake or their functional redundancy. We demonstrate that C. jejuni NCTC11168 transports ferric-enterobactin in an energy dependant manner that requires TonB3 for full activity with TonB1 showing partial functional redundancy. Moreover C. jejuni NCTC11168 can utilize a wide variety of structurally different catechol siderophores as sole iron sources during growth. This growth is solely dependent on the CfrA enterobactin transporter and highlights the wide range of substrates that this transporter can recognize. TonB3 is also required for growth on most catechol siderophores. Furthermore, either TonB1 or TonB3 is sufficient for growth on hemin or hemoglobin as a sole iron source demonstrating functional redundancy between TonB1 and TonB3. In vivo colonization assays with isogenic deletion mutants revealed that both TonB1 and TonB3 are required for chick colonization with TonB2 dispensable in this model. These results further highlight the importance of iron transport for efficient C. jejuni colonization.
Introduction
Iron is an essential micronutrient for almost all bacteria [1]. Although iron is one of the most abundant elements on earth, its availability within the host is extremely limited. Iron limitation arises due to its poor solubility under aerobic conditions at physiological pH and due to the presence of the host iron-binding glycoproteins lactoferrin and transferrin [1]. Bacteria thus employ a variety of strategies to acquire the iron sources needed for their growth, including the import of ferrous ions though the high affinity FeoB system [1]. In addition, many bacteria produce iron chelators known as siderophores to circumvent the host’s attempts to sequester free iron. The prototypical siderophore is the catecholate enterobactin produced by E. coli [2]. Enterobactin and other catecholate siderophores such as salmochelin and bacillibactin are powerful iron chelating molecules and are often virulence factors for pathogenic bacteria [3]. Siderophores are secreted into the extracellular medium, where they complex iron and are then imported by highly specific outer membrane proteins (OMPs) in Gram negative bacteria [2–5].
Bacteria that produce multiple siderophores often express separate OMPs dedicated to the transport of each specific siderophore. This is seen in E. coli and Salmonella sp. that express FepA and IroN for enterobactin and salmochelin transport respectively[6]. However some transporters can recognize and transport multiple siderophores [7]: Salmonella IroN can also transport enterobactin and bacillibactin, while B. subtilis contains an import pathway that recognizes itoic acid, bacillibactin and enterobactin [8, 9]. The energy required to transport these ligands across the outer membrane in Gram negative bacteria is provided by the TonB-ExbB-ExbD energy transduction system [4, 10, 11]. Through currently undefined mechanisms, the TonB protein transduces the energy from the proton motif force of the inner membrane to the outer membrane receptors to allow for ligand import [4, 12]. While some bacteria such as E. coli contain a single TonB complex; many bacteria possess multiple TonB systems [11]. The presence of multiple TonB systems within a single organism implies that each has its own specialized function within the cell. However, given the structural homology of each TonB complex within a single organism, these TonB systems are often able to compensate for each other [10, 13–16].
The food-borne pathogen Campylobacter jejuni is a major cause of bacterial enteritis in humans world-wide [17]. Analysis of the C. jejuni NCTC11168 genome reveals the presence of multiple OMP iron transporters and the presence of three different TonB-ExbB-ExbD systems [18–21]. C. jejuni has been shown to acquire iron through enterobactin, hemin, transferrin/lactoferrin and through ferrous ion import [21–26]. Iron acquisition has also been demonstrated to be an important colonization and virulence determinant in C. jejuni [21, 22, 27]. Although C. jejuni is unable to synthesize siderophores such as enterobactin, it is able to utilize exogenous enterobactin as an iron source through the action of the CfrA or CfrB transporters [21, 25–27]. Structural characterization of CfrA coupled with homology modelling has suggested that CfrA may interact with enterobactin differently from the E. coli FepA receptor and that CfrA may exhibit a broad siderophore recognition range [28].
It is likely that the transport of ferric enterobactin, heme and other iron compounds is TonB dependent in C. jejuni, although the role of each specific TonB in iron transport has not yet been experimentally demonstrated. The aim of this study was to investigate the role of TonB1, TonB2, and TonB3 in iron uptake within C. jejuni. We report the construction of isogenic deletion mutants into these genes and their functional characterization using siderophore-mediated growth promotion assays. We also assessed the role of each TonB in chick gut colonization. Our work demonstrates that C. jejuni is capable of using a wide variety of structurally different catecholate siderophores as sole iron sources. Moreover the majority of these siderophores are transported through the outer membrane protein CfrA and are dependent on TonB3 for energy transduction. Unlike E. coli, C. jejuni is able to use both enterobactin and enantioenterobactin as sole iron sources [29]. In addition, while both TonB1 and TonB3 are required for commensal chick colonization, TonB2 is dispensable in this colonization model.
Methods
Bacterial Growth
C. jejuni NCTC11168 was routinely cultured at 37°C in a MACS-VA500 microaerophilic workstation (Don Whitley, West Yorkshire, England) under 83% N2, 4% H2, 8% O2 and 5% CO2 on Mueller-Hinton (MH) agar plates. The C. jejuni strains were tested for motility on 0.4% MH agar plates before performing in vivo animal experiments. For the in vitro growth assays, the C. jejuni wild type and mutant strains were grown in MH broth, MEMα or MEMα supplemented with 40 μM FeSO4 at 37°C under microaerophilic conditions. Luria-Bertani (LB) broth and agar plates were used to culture E. coli DH5α at 37°C under aerobic conditions. The strains used in this study are listed in Supplementary Table 1. The construction of the ΔcfrA mutant [21] has been previously described and the ΔchuA mutant was constructed as described in Ridley et al. [23].
Single tonB mutant construction
Single deletion mutants of tonB1, tonB2 and tonB3 were constructed as follows (see Supplemental Figure 1 for the genomic location of the various tonB genes). The tonB1 gene was PCR amplified from C. jejuni NCTC11168 chromosomal DNA using primers TonB11 and TonB12 (Supplementary Table 2). The resulting PCR product was purified, digested with BglII and ligated to a BamHI restricted pUC19 to yield pAS16 (Supplementary Table 1). Inverse PCR was performed with the TonB13 and TonB14 primers (containing BamHI restriction sites) to delete a 675 bp region within the tonB1 gene. The PCR product was purified, digested with BamHI, self-ligated and transformed into E. coli DH5α to yield plasmid pAS20. The kanamycin resistant cassette (KmR) from the pILL600 plasmid was amplified, digested with BamHI and cloned into BamHI digested pAS20 to yield pAS23. The orientation of the KmR cassette was confirmed by DNA sequencing and then transformed into C. jejuni NCTC11168 by natural transformation as previously described [30]. Briefly, C. jejuni wild-type cells were grown to mid-log-phase in biphasic flasks and 50 μL aliquots were spotted onto MH plates and allowed to grow at 37°C under microaerophilic conditions for 12 hours. The final construct was added on to the C. jejuni lawns and incubated for an additional 12 hours at 37°C under microaerophilic conditions. Thereafter, the lawns were resuspended in 200 μL MH broth and spread onto MH plates containing kanamycin (30 μg/ml). Positive colonies were confirmed by PCR amplification of C. jejuni mutant chromosomal DNA using primer combinations amplifying the KmR cassette and the respective tonB genes. The ΔtonB2 (AS240) and ΔtonB3 (AS239) mutants were constructed using the same protocol, except using the chloramphenicol resistant cassette (CmR) from pRY111 as the selective marker for the ΔtonB2 mutant. The transformants were selected on MH agar plates containing kanamycin (10 μg/mL) or chloramphenicol (20μg/mL). The use of an alternate antibiotic cassette was to facilitate the construction of double deletion mutants. We also constructed an independent deletion mutant of tonB3 using the chloramphenicol resistant cassette (CmR) to further confirm the tonB3 mutant phenotype (AS1135).
Complementation of tonB3
Complementation of the C. jejuni NCTC11168 ΔtonB3::cm mutant strain was constructed as described previously [30, 31]. The tonB3 gene was amplified from extracted C. jejuni genomic DNA using Phusion high fidelity polymerase (Finnzymes) and the tonB3_SE_comp and tonB3_AS_comp primers (Supplementary Table 2). The amplified gene product was purified using the PureLink PCR Purification Kit (Invitrogen) and subsequently directionally cloned into XhoI (Invitrogen) digested pRRK using the In-fusion Dry-down cloning kit (Clontech) [30]. The pRRK+tonB3 plasmid was sequenced to confirm the absence of mutations. The C. jejuni ΔtonB3 mutant strain was then naturally transformed with the final construct and successful transformants were selected for on MH agar plates supplemented with chloramphenicol (20 μg/mL) and kanamycin (30 μg/mL). Positive colonies were confirmed by PCR using the kanamycin cassette specific primer AR56 and ribosomal region specific primers ak233-ak235 (Supplementary Table 2).
Double tonB mutant construction
The C. jejuni NCTC11168 ΔtonB1ΔtonB2 (AS241) mutant was constructed by naturally transforming the C. jejuni ΔtonB2 mutant (described above) with the final plasmid construct of the ΔtonB1 mutant. The double tonB mutants of C. jejuni NCTC11168 ΔtonB2ΔtonB3 (AS242) and ΔtonB1ΔtonB3 (AS355) were constructed by naturally transforming the C. jejuni ΔtonB1, and ΔtonB2 single mutant strains (described above) with the final plasmid construct for the ΔtonB3 mutant (Supplementary Table 1). Positive transformants were selected for on kanamycin (30 μg/ml) and chloramphenicol (20 μg/mL) MH agar plates grown under microaerophilic conditions at 37 °C and confirmed by PCR.
Siderophore synthesis and purification
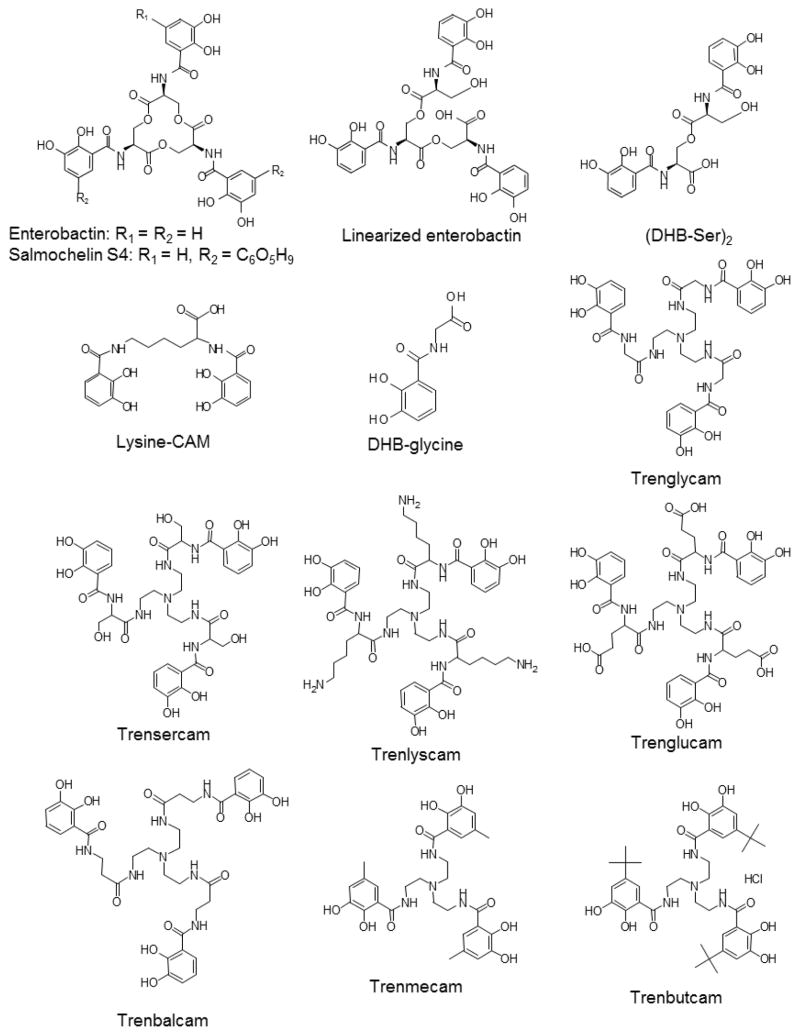
Enterobactin was purified from E. coli supernatants as previously described [32]. Salmochelin, linearized enterobactin (Trimer) and (DHB-Ser)2 (Dimer) were obtained from Prof. Günter Winkelmann (University of Tübingen). Lysine-catecholate was synthesized as previously described [7]. The synthesis of DHB-glycine has been described previously [33]. Synthesis of the siderophore analogs TRENGlyCAM (TGC), TRENSerCAM (TSG), TRENLysCAM (TKC), TRENGluCAM (TEC), TRENbalCAM (TAC), TRENmeCAM(TMC) and TRENbutCAM(TBC) was performed as described elsewhere [34]. The structures of these siderophores and synthetic siderophore analogs are depicted Figure 1.
Figure 1. Structures of natural siderophores and synthetic siderophore analogs used in this study.
Chemical models of siderophore analogs used in this study. DHB = 2,3-dihydroxybenzoic acid. The TREN (tris(2-aminoethyl)amine) based analogs consist of a TREN backbone, a linker region (gly=glycine, ser = serine, lys = lysine, glu=glutamate, bal= β-alanine) and a CAM (catecholamide) group. TRENmeCAM and TRENbutCAM have additional methyl (me) or butyl (but) groups respectively on the catecholamide ring.
55Fe3+-enterobactin uptake assay
Mid-log-phase C. jejuni cells grown in iron-limited MEMα medium were pelleted and washed in 10 mM Tris buffer (pH 7.4). The cells were resuspended in uptake buffer to an OD600= 0.6 (~109 bacteria per ml) and kept on ice. The uptake buffer was composed of 5 g/L Na2HPO4, 5 g/L KH2PO4, 1.18 g/L NH4Cl, 0.089 g/L Na2SO4, 0.042 g/L MgCl2 hexahydrate and 10 g/L casamino acids. The resuspended cells were allowed to equilibrate at 37°C for 10 minutes before performing the uptake assay. The 55Fe3+-enterobactin complex used as the iron source was prepared as follows: 10 μL of 10 mM enterobactin was added to 5 μL of 3.6 mM 55FeCl3, followed by the addition of 182.18 μL water and 2.88 μL of 25 mM FeCl3. The 55Fe3+-enterobactin complex was incubated at room temperature for 30 minutes followed by the addition of 100 μL of 500 mM sodium phosphate buffer and 700 μL of ddH2O for a final volume of 1000 μL. This solution was centrifuged at 13000 rpm for 10 minutes and the supernatant was collected. The 55Fe3+-enterobactin solution was diluted by taking 100 μL of the supernatant and mixing with 9.9 mL uptake buffer. This diluted 55Fe3+-enterobactin complex was then filtered twice using a 0.45 μm nitrocellulose filter and used for the uptake assay. The uptake assay was performed using a Millipore filtering assembly (Millipore Corporation, Ledford, MS). One mL of the filtered 55Fe3+-enterobactin complex was added to 10 mL of C. jejuni cells (18 nM final 55Fe3+-enterobactin complex concentration). Aliquots of 1 mL were taken at 0, 3, 6, 9 and 12 minutes after the addition of the ferric-enterobactin complex to the cells. The aliquots were passed through a 0.45 μm nitrocellulose filter by vacuum filtration and the filters were washed with 5 mL of 0.1 M citric acid. The filters were then allowed to dry and were subsequently immersed in 5 mL of scintillation cocktail, (Scintiverse, USA), vortexed and counted in a Beckmann LS5000 TD scintillation counter. Final CPM counts were corrected for background. Additional uptake experiments were performed for the wild-type strain as described above except that the protonophore carbonyl-cyanide-m-chlorphenylhydrazone (CCCP) was added to the cells (final concentration 33 μM) three minutes after uptake was initiated. The uptake experiments were repeated at least three times for each condition. Iron uptakes were statistically compared at specific time points using a Student’s t test and P values below 0.05 were considered significant.
C. jejuni growth with various sole sources of iron
The ability of the C. jejuni wild-type and mutant strains to utilize different iron sources was assessed on iron-limited assay plates as described previously [21]. Briefly, the bacterial strains to be tested were grown to mid-log-phase in MH medium, harvested by centrifugation, and resuspended in MH medium to an OD of 1.0 at 600 nm. One mL of this bacterial suspension was added to 25 mL of cooled molten MH agar containing 20 μM desferri-ferrioxamine, poured into petri plates and allowed to solidify. Sterile disc papers containing 10 μL of 10 mM ferric-enterobactin, hemin, hemoglobin and other iron siderophore sources were laid on the surface of the plate and the diameter of the zone of growth promotion was measured after 24 hours of incubation at 37°C under microaerophilic conditions. In addition, 10 mM FeSO4 was used as a positive control for C. jejuni growth. The zone of growth for each mutant was normalized to the corresponding wild type control preformed on the same date and reported as a percentage of growth promotion relative to the wild-type strains. The growth promotion assay was repeated at least three times for each iron source and each bacterial strain and the data was statistically analyzed using single-factor analysis of variance (ANOVA). A P-value < 0.05 was considered significant. To note, the chloramphenicol deletion construct of tonB3 (AS11132) displayed the same phenotype with regards to enterobactin usage as the kanamycin deletion construct (AS239).
Chick colonization assays
One day old specific pathogen free chicks (Tyson farms, Arkansas) were checked upon arrival using cloacal swabs to ensure that they were Campylobacter free. The chicks were housed at 25°C in a brooder that maintained a temperature in the range of 33–35°C. Commercial chicken starter diet and water were given ad libitum to the chicks. For challenge, C. jejuni wild-type and tonB mutant strains were first grown on motility MH-agar plates under microaerophilic conditions and then transferred to MH biphasic medium. Mid-log-phase bacteria were harvested and resuspended in sterile PBS buffer. An inoculum of 0.25 mL of a bacterial suspension (containing approximately ~104–105 viable bacteria) was fed orally to each bird. Five birds per group were inoculated separately for the wild-type strain or each mutant. The colonization potential was determined four days post challenge as described previously [21]. Briefly, the ceca were collected following euthanasia, weighed and the contents were homogenized and serially diluted onto selective Karmali agar plates. After incubation at 37°C for 72 hours in a microaerophilic chamber, the bacterial load was determined and expressed as CFU/per gram of ceca. Statistical analysis was done using the non-parametric Mann-Whitney Rank sum test with a p<0.05 considered significant.
Competitive in vivo chick colonization assays
The competitive colonization studies were done using five chicks per group. Each bird was inoculated with 0.5 mL of a bacterial suspension containing a 1:1 mixture of wild-type to tonB mutant strain. Approximately 105 viable bacteria of both wild-type and mutant were present in the mixed inoculums as confirmed by plating serial dilutions of the inoculum on MH agar with and without kanamycin and/or chloramphenicol. The ceca were collected four days post inoculation and processed as described above. The homogenized cecal contents were serially diluted in PBS buffer, and plated on Karmali agar plates and Karmali agar plates supplemented with kanamycin and/or chloramphenicol as required. Colonies were counted after the plates were incubated at 37°C for 72 hours in a microaerophilic chamber. The mutant titer was obtained from the CFU recovered on Karmali agar based plates containing kanamycin and/or chloramphenicol, and the wild type bacterial load was calculated by subtracting the number of mutants from the total number of bacteria recovered on Karmali agar plates without antibiotics. Finally, the in vivo competitive index, which corresponds to the ratio of output mutant to wild type bacteria recovered divided by the ratio of input mutant to wild type bacteria inoculated was calculated for each bird. Statistical analysis of the data was performed using a single sample Student t test with a p<0.05 considered significant.
Results
Construction of tonB single and double mutants
C. jejuni NCTC11168 contains three different tonB genes, tonB1, tonB2 and tonB3. We therefore set out to determine if each TonB system was required for siderophore transport and chick colonization. Since the TonB systems could overlap in function, we generated specific mutants in each system and double mutants that would render only one TonB system functional. Each tonB gene was deleted by allelic exchange with antibiotic resistance cassettes. These mutants were also used to construct the double deletion tonB constructs (ΔtonB1/B2, ΔtonB1/B3, ΔtonB2/B3). Attempts to construct a triple tonB mutant lacking all the three known TonB systems and to complement the double tonB mutants were not successful due to the lack of the appropriate genetic tools in C. jejuni.
Transport of ferric-siderophores is tonB specific
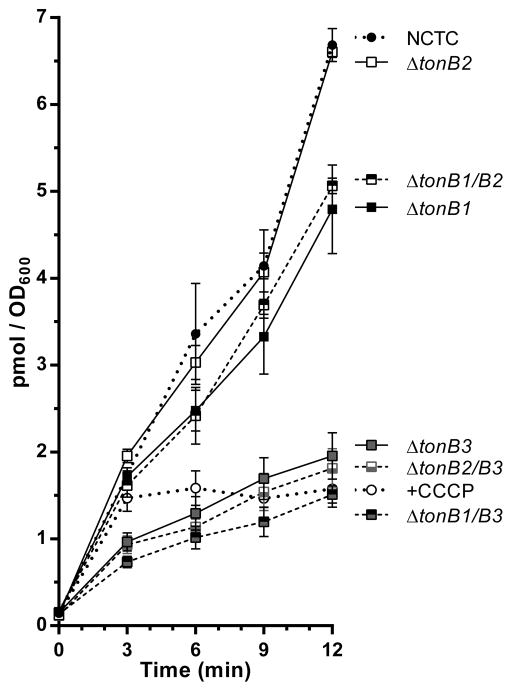
We then proceeded to test if the tonB mutants were defective in ferric-enterobactin uptake. 55Fe3+-enterobactin uptake assays were performed using the wild-type strain and the various tonB mutants. We used cells treated with the protophore cyanide-m-chlorophenylhydrazone (CCCP) to control for radioactivity associated with the cell surface and not actively transported into the cell. As shown in Figure 2, there was a statistically significant difference (p<0.05) in the ferric-enterobactin uptake for all of the single and double tonB mutants; with the exception of the tonB2 mutant. Strains that contained a deletion of tonB1 in the absence of an additional deletion in tonB3 (ie ΔtonB1, ΔtonB1/B2) were slightly (but still significantly) affected in enterobactin uptake. However, all the strains that contained a deletion of tonB3 (ie ΔtonB3, ΔtonB1/B3, ΔtonB2/B3) were completely abrogated in enterobactin uptake and mimicked the +CCCP phenotype.
Figure 2. Transport of ferric-enterobactin is TonB specific.
Uptake of 55Fe3+-enterobactin by C. jejuni NCTC11168 and tonB mutant constructs. In the +CCCP sample, NCTC11168 cells were exposed to CCCP 3 min after 55Fe3+-enterobactin addition. Experimental conditions were as described in Materials and Methods.
C. jejuni can scavenge iron from wide variety of siderophores
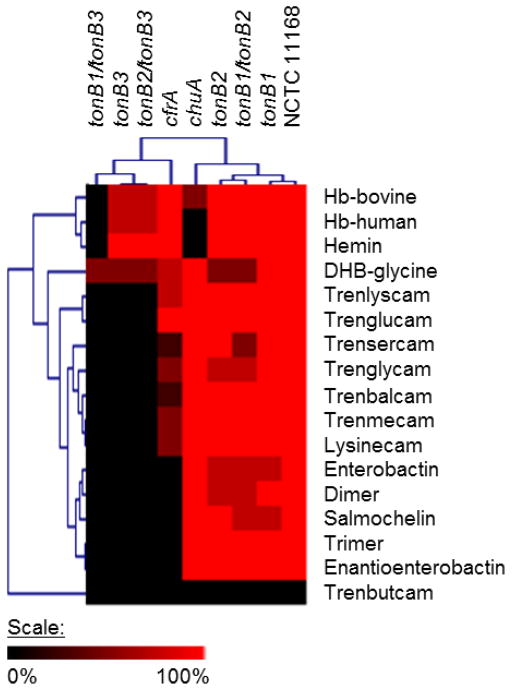
We performed growth promotion assays on a wide variety of different catecholate siderophores (Figure 3), as well as hemin and haemoglobin, to determine C. jejuni’s ability to use these chelators as sole iron sources. The tonB mutants were also assayed for growth using the same iron sources. Previous work has established that CfrA is required for enterobactin growth and ChuA is required for hemin growth in C. jejuni [21, 23]. Isogenic deletion mutants of these two transporters were included as negative controls. Siderophore mediated growth promotions of the mutants were normalized to mean value of those from the wild-type strain for each specific iron source. The results were subsequently clustered according to their siderophore utilization profile (Figure 3). The mutants segregated into two main clusters based on their iron source usage, with ΔcfrA, ΔtonB3 and tonB3 double knockouts in one cluster and ΔchuA, ΔtonB1, ΔtonB2, and ΔtonB1/B2 in the other. The first cluster is characterized by the inability of the mutants to effectively use enterobactin (and related derivatives), the TRENCAM derivatives and Lys-CAM as iron sources. The ΔtonB3 mutants (single or double) show no growth on any of these compounds and ΔcfrA showed limited growth on Lys-CAM and the TRENCAM derivatives. This indicates that CfrA is the sole transporter for enterobactin and related siderophores, although another transporter(s) is able to partially compensate for cfrA deletion with regards to TRENCAM/Lys-CAM siderophore usage. TonB3 is absolutely required for both enterobactin and TRENCAM/Lys-CAM related siderophore utilization. Interestingly, TonB3 also contributes to heme/hemin transport. All tonB3 mutants were affected for heme/hemin utilization within C. jejuni with the ΔtonB1/B3 double mutant completely inhibited in its ability to use heme/hemin as an iron source. The second cluster is composed of ΔchuA and ΔtonB1, ΔtonB2, and ΔtonB1/B2. The ChuA transporter was only required for hemoglobin/hemin utilization and ΔtonB2 displayed minor defects in the utilization of DHB-Gly, TRENgluCAM (TGC), enterobactin and dimer (DHB-Ser)2. The ΔtonB1 mutant showed a minor defect in utilization of enterobactin and salmochelin. Since the ΔtonB3 mutant displayed the most apparent phenotype, we complemented the tonB3 mutation by inserting a functional copy of tonB3 into the deletion mutant’s genome. The complemented ΔtonB3+tonB3 exhibited fully restored ability to use enterobactin as a sole iron source. This result demonstrates tonB3’s specific role in enterobactin transport.
Figure 3. C. jejuni can scavenge iron from wide variety of sources.
Selected mutant strains were immobilised in MH agar treated with DFO and provided with various iron sources to determine their ability to use each source. The mean growth promotions of each mutant were normalized to those measured for the wild-type strain. The intensity of the red color output for the figure is a function of the relative growth promotion. Black denotes the absence of growth promotion. The results from these experiments were subjected to hierarchical clustering analysis using Pearson’s correlation coefficient.
The tonB mutants are significantly affected in their ability to colonize chicks
The ability of the tonB mutants to colonize the gastrointestinal tract was tested in the commensal (chick) animal model for C. jejuni. Firstly, the colonization ability of the tonB mutants and the wild-type strain were assessed by orally challenging five birds per group separately with approximately 104 viable bacteria. Four days post inoculation, the birds were sacrificed, the ceca were recovered and the cecal contents checked for the bacterial load. All the tonB mutants were found to be significantly attenuated in colonization as compared to the wild-type strain, with the exception of tonB2 (Figure 4A). These results indicate that tonB1 and tonB3 are essential for cecal colonization of C. jejuni. The level of colonization indicated that the double mutants were more severely affected than the single mutants, with the ΔtonB1tonB3 double mutant below the detection limit of 100 CFU/gram ceca. Secondly, the ability of the tonB mutants to colonize the chicken ceca was further tested in competition with the wild-type C. jejuni NCTC11168 strain. The birds were inoculated with a 1:1 mixture of the wild-type strain and the individual tonB mutants. Five birds per group were tested for the colonization potential of the mutant and the wild-type strain. Four days after inoculation, the ceca were recovered and the contents were then plated on selective media to calculate the competitive index for each bird. As shown in Figure 4B, with the exception of the single tonB2 mutant, all the other single and double tonB mutants were outcompeted by the wild-type strain in the gastrointestinal tract of birds (p<0.05). These results corroborated the findings of the chick colonization assays with individual strains. The double mutants were the most severely attenuated followed by the tonB3 and tonB1 single mutants. The competitive indices of the mutants were also determined in vitro, and found to be less affected than in vivo (Figure 4B). Therefore, the inability of the mutants to colonize in vivo is indicative of an in vivo specific defect.
Figure 4. TonB mutants are impaired in their ability to colonize the chick gut.
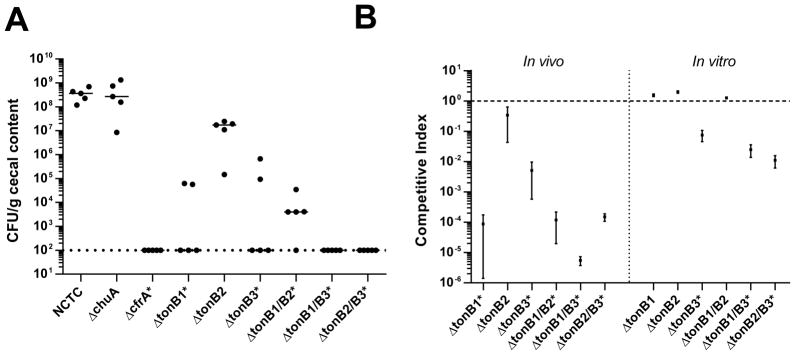
Panel A: Chick colonization studies with NCTC11168 and selected mutants. Five chicks per group were infected with C. jejuni. The data points indicate the level of colonization in each chick with the black bar representing the median and the dashed line the detection limit. Strains that showed significant differences as compared to the wildtype (p<0.05) are indicated with an asterisk. Panel B: Competitive indices of each mutant as compared to the wild type control in competitive association studies with chicks and in vitro. Strains that showed significant differences are indicated with an asterisk (p<0.05).
Discussion
In this study we report the construction and characterization of the TonB energy transduction systems in C. jejuni NCTC11168 (Supplemental Table 3). We have previously demonstrated that the enterobactin transporter CfrA is the sole transporter of ferric-enterobactin in C. jejuni NCTC11168 and is essential for in vivo colonization [21]. Mutants in tonB1 and tonB3 showed statistically significant reductions in their ability to transport enterobactin while the tonB2 mutant was unaffected. While the ΔtonB1 showed a slight reduction in enterobactin transport, the ΔtonB3 phenotype was severely affected and mimicked the +CCCP phenotype. This indicates that TonB3 is the primary energy transducer of enterobactin mediated iron transport through CfrA with TonB1 playing a relatively minor role.
C. jejuni cannot produce enterobactin endogenously and presumably scavenges enterobactin produced by the host’s intestinal microbiota [21]. Given this opportunistic use of enterobactin by C. jejuni, we speculated that CfrA may have evolved a broad substrate spectrum to better take advantage of multiple endogenously produced siderophores. To investigate this hypothesis we tested the C. jejuni wild type, ΔtonB mutants, and ΔcfrA mutant for their ability to utilize a variety of enterobactin-like iron sources. These included salmochelin (enterobactin glycosylated at rings 1 and 2), enantioenterobactin and enterobactin breakdown products (linearized enterobactin, (DHB-Ser)2 and DHB-Gly). We also probed the specificity of this transport by utilizing a series of TREN based analogs that differ in charge, arm-length and bulkiness. In addition, we tested the strains for their ability to utilize lysine bis-catecholate (Lys-CAM). Lys-CAM is structurally related to enterobactin and is a tetradentate chelator as compared to enterobactin’s hexadentate chelation. The intracellular iron sources, hemin and hemoglobin were also screened for utilization by the tonB mutants as previous work in V. cholerae found that multiple TonB systems could allow proper heme uptake and bacterial growth [15, 16].
C. jejuni NCTC11168 was able to utilize all of the iron sources tested, with the exception of TBC (TRENbutCAM). It is likely that the bulky butyl groups on the catecholate rings prevented its uptake through outer membrane transporters. Deletion of cfrA resulted in complete abrogation of enterobactin utilization and greatly reduced use of TRENCAM derivatives (with the exception of TRENgluCAM). This indicates that there may be another transporter for TRENCAM derivatives. However both CfrA and this putative transporter are both completely dependent on TonB3 for energy transduction as deletion of tonB3 resulted in no growth with enterobactin/TRENCAM derivatives and Lys-CAM. CfrA is able to accommodate a number of different substitutions in the TRENCAM derivatives that would change bulkiness, denticity, and/or overall charge of the ferric-siderophore complex. Substitutions affecting the length and charge of the TREN arms did not impede siderophore usage, but the addition of butyl groups to the catecholate groups on TBC prevented the use of the siderophore. These results suggest that CfrA is a promiscuous transporter that can transport a wide variety of structurally different catecholate siderophores and this transport is dependent on TonB3 activity. TonB1 was found to be partially redundant with TonB3 for hemoglobin and hemin use as deletion of both tonBs was required to abolish heme utilization. Deletion of tonB1 also resulted in a slight growth defect when enterobactin or salmochelin were the sole iron sources which may be related to the slight decrease in enterobactin uptake seen in the ΔtonB1 mutant (Figure 2). The tonB2 deletion mutant showed minor defects in the use of TGC, enterobactin and (DHB-Ser)2.
Interestingly, C. jejuni was able to grow by using enantioenterobactin as an iron source. Previous studies have demonstrated that while E. coli FepA can transport enantioenterobactin, E. coli cannot use this siderophore analog for growth [29]. This lack of growth is presumably due to the fact that the enterobactin esterase Fes is stereospecific and cannot cleave the tri-D-serine triactone ring present in enantioenterobactin [29]. The bacillibactin esterase BesA displays a similar phenotype in that it can hydrolyze the tri-L-serine triactone rings in enterobactin and bacillibactin, but not the tri-D-serine triactone ring present in enantioenterobactin [29]. Our results indicate that C. jejuni does not have these limitations in its siderophore usage and may therefore possess novel pathways for iron release from enterobactin that are not stereospecific.
Recent work by Zeng et al. have provided the first experimental evidence for the presence of novel pathways in C. jejuni enterobactin usage [27]. This study has proposed that there are two pathways for enterobactin assimilation in C. jejuni species. The authors propose that in C. jejuni NCTC11168, ferric-enterobactin is taken up through the CfrA OMP and primarily transported to the cytoplasm via the CeuBCDE ABC-transporter system. Once ferric-enterobactin is within the cytosol Zeng et al. propose that the ferric ion is reduced to the ferrous form and thus the iron is released from enterobactin without enterobactin cleavage as recently demonstrated in E. coli [35]. Given the relative novelty of this enterobactin assimilation pathway, it would be interesting to investigate if the postulated ferric reductase could reduce both enterobactin and enantioenterbactin. In addition, this study also identified a C. jejuni enterobactin esterase (Cee) that displayed similar enterobactin cleavage kinetics as E. coli Fes and is responsible for C. jejuni NCTC11168’s ability to use enterobactin as a sole iron source in the absence of a functional CeuBCDE ABC-transporter system [21, 27]. It would also be interesting to test if Cee shows the same stereospecificity as Fes or whether Cee can also cleave enantioenterobactin. Either of these two enterobactin assimilation pathways could be responsible for C. jejuni NCTC11168’s ability to grow on enantioenterobactin.
The presence of multiple tonB genes has been reported for several microbes including Vibrio cholerae, Vibrio anguillarum, Vibrio vulnificus, and Pseudomonas aeruginosa [5, 10, 11, 14–16]. Studies in these diverse species have shown that the different TonB systems have both overlapping and unique functions. This can result in distinctive in vivo phenotypes when the different TonB systems are deleted. For example, V. anguillarum requires only one of its two TonB systems for virulence [14]. However in V. vulnificus each of its three TonB systems can compensate for each other and single deletion mutants are unaffected in virulence [36]. V. vulnificus requires double or triple tonB knockouts before virulence defects are observed [36]. In contrast, the deletion of either tonB1 or tonB2 in V. cholerae results in virulence defects [16].
Our results show that C. jejuni deletion mutants in either TonB1 or TonB3 are significantly impaired for chick gut colonization (Figure 4). TonB3’s importance in chick colonization is likely related to its role in siderophore transport, in agreement with the similar in vivo phenotype of the ΔtonB3 and ΔcfrA mutants. In addition, the inability of either TonB1 or TonB2 to compensate for TonB3 in vivo or in vitro highlights the important function that TonB3 plays in C. jejuni physiology. The defect in the ΔtonB1 mutant’s chick colonization could be due to the slight decrease in enterobactin uptake seen in vitro, however it should be noted that the ΔtonB1 mutant growth defect was comparatively minor in our growth promotion assays. Thus, the ΔtonB1 phenotype could also be due to a defect in the uptake of an unidentified nutrient. Classically, TonB systems have been shown to be involved in the transport of iron complexes (e.g. siderophores) or cobalamin (Vitamin B12) [37]. However, recent work has revealed new TonB-dependant substrates such as copper, nickel and various sugars and thus the potential role for TonB1 in chick colonization may extend beyond iron metabolism [37–39]. The ΔtonB2 mutant displayed no significant defects when colonizing the chick gut indicating that TonB2 is dispensable in this animal model. In addition, the ΔchuA mutant was not affected for colonization and this indicates that C. jejuni does not require hemin/hemoglobin during commensal chick colonization.
Significantly, while most C. jejuni strains contain three TonB genes, certain strains only have one TonB system. In particular, C. jejuni 81–176 does not have TonB1 or TonB3 and solely encodes for TonB2 [40]. However C. jejuni 81–176 is able to use hemin/hemoglobin as a sole iron source [41]. Given that C. jejuni NCTC11168 TonB2 cannot support growth on hemin (Figure 3) this suggests that the functionality of different TonB systems may not be necessarily conserved across C. jejuni strains and that different C. jejuni strains have developed unique functions for their respective TonB systems based on their own specific environmental requirements. Presumably, the sole TonB in C. jejuni 81–176 is constrained in its specificity as it is the only energy transducing system available in this strain. In contrast, the presence of multiple TonB systems in C. jejuni NCTC11168 has allowed each TonB system to become more specialized in the transporters that they energize. This specialization has progressed such that the TonB2 system in C. jejuni NCTC11168 no longer recognizes the same targets as the TonB2 system in C. jejuni 81–176.
Conclusions
This work demonstrates the ability of CfrA to transport a wide variety of structurally dissimilar siderophores and siderophore analogs. These findings highlight the high degree of adaptability present in C. jejuni strains and this adaptability may contribute to C. jejuni’s ability to successfully colonize a wide variety of hosts and ecological niches. This work has also shown that specific TonB systems are absolutely required for the transport of certain ligands (e.g. TonB3 and enterobactin), while other ligand transport can be energized by multiple TonB systems (e.g. TonB1/TonB3 and hemin). Finally, C. jejuni NCTC11168 TonB2’s functional divergence from C. jejuni 81–176 TonB2 highlights the inherent difficulty in translating results from one C. jejuni strain to another, as the role of specific proteins may differ based on the unique context of each genotype.
Supplementary Material
Acknowledgments
This work was supported by NIH (RO1-AI055612) and CIHR (MOP#84224) to A.S., NIH (RO1 AI11744) to K.N.R and a CIHR-Banting graduate scholarship to J.B.
References
- 1.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27(2–3):215–37. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A. 2003;100(7):3584–8. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischbach MA, et al. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol. 2006;2(3):132–8. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 4.Krewulak KD, Vogel HJ. TonB or not TonB: is that the question? Biochem Cell Biol. 2011;89(2):87–97. doi: 10.1139/o10-141. [DOI] [PubMed] [Google Scholar]
- 5.Cornelis P, Matthijs S, Van Oeffelen L. Iron uptake regulation in Pseudomonas aeruginosa. Biometals. 2009;22(1):15–22. doi: 10.1007/s10534-008-9193-0. [DOI] [PubMed] [Google Scholar]
- 6.Muller SI, Valdebenito M, Hantke K. Salmochelin, the long-overlooked catecholate siderophore of Salmonella. Biometals. 2009;22(4):691–5. doi: 10.1007/s10534-009-9217-4. [DOI] [PubMed] [Google Scholar]
- 7.Stintzi A, et al. Microbial iron transport via a siderophore shuttle: a membrane ion transport paradigm. Proc Natl Acad Sci U S A. 2000;97(20):10691–6. doi: 10.1073/pnas.200318797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dertz EA, et al. Bacillibactin-mediated iron transport in Bacillus subtilis. J Am Chem Soc. 2006;128(1):22–3. doi: 10.1021/ja055898c. [DOI] [PubMed] [Google Scholar]
- 9.Rabsch W, et al. Salmonella typhimurium IroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J Bacteriol. 1999;181(11):3610–2. doi: 10.1128/jb.181.11.3610-3612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuehl CJ, Crosa JH. The TonB energy transduction systems in Vibrio species. Future Microbiol. 2010;5(9):1403–12. doi: 10.2217/fmb.10.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu BC, Peacock RS, Vogel HJ. Bioinformatic analysis of the TonB protein family. Biometals. 2007;20(3–4):467–83. doi: 10.1007/s10534-006-9049-4. [DOI] [PubMed] [Google Scholar]
- 12.Gresock MG, et al. Death of the TonB Shuttle Hypothesis. Front Microbiol. 2011;2:206. doi: 10.3389/fmicb.2011.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kustusch RJ, Kuehl CJ, Crosa JH. The ttpC gene is contained in two of three TonB systems in the human pathogen Vibrio vulnificus, but only one is active in iron transport and virulence. J Bacteriol. 2012;194(12):3250–9. doi: 10.1128/JB.00155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stork M, et al. Two tonB systems function in iron transport in Vibrio anguillarum, but only one is essential for virulence. Infect Immun. 2004;72(12):7326–9. doi: 10.1128/IAI.72.12.7326-7329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mey AR, Payne SM. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol Microbiol. 2001;42(3):835–49. doi: 10.1046/j.1365-2958.2001.02683.x. [DOI] [PubMed] [Google Scholar]
- 16.Seliger SS, et al. The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol Microbiol. 2001;39(3):801–12. doi: 10.1046/j.1365-2958.2001.02273.x. [DOI] [PubMed] [Google Scholar]
- 17.Zilbauer M, et al. Campylobacter jejuni-mediated disease pathogenesis: an update. Trans R Soc Trop Med Hyg. 2008;102(2):123–9. doi: 10.1016/j.trstmh.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 18.Gundogdu O, et al. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics. 2007;8:162. doi: 10.1186/1471-2164-8-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parkhill J, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403(6770):665–8. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 20.Holmes K, et al. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology. 2005;151(Pt 1):243–57. doi: 10.1099/mic.0.27412-0. [DOI] [PubMed] [Google Scholar]
- 21.Palyada K, Threadgill D, Stintzi A. Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol. 2004;186(14):4714–29. doi: 10.1128/JB.186.14.4714-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naikare H, et al. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect Immun. 2006;74(10):5433–44. doi: 10.1128/IAI.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridley KA, et al. Heme utilization in Campylobacter jejuni. J Bacteriol. 2006;188(22):7862–75. doi: 10.1128/JB.00994-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller CE, et al. Utilization of lactoferrin-bound and transferrin-bound iron by Campylobacter jejuni. J Bacteriol. 2008;190(6):1900–11. doi: 10.1128/JB.01761-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu F, et al. Identification and characterization of a new ferric enterobactin receptor, CfrB, in Campylobacter. J Bacteriol. 2010;192(17):4425–35. doi: 10.1128/JB.00478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng X, Xu F, Lin J. Molecular, antigenic, and functional characteristics of ferric enterobactin receptor CfrA in Campylobacter jejuni. Infect Immun. 2009;77(12):5437–48. doi: 10.1128/IAI.00666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng X, et al. Identification and characterization of a periplasmic trilactone esterase, Cee, revealed unique features of ferric enterobactin acquisition in Campylobacter. Mol Microbiol. 2013;87(3):594–608. doi: 10.1111/mmi.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carswell CL, Rigden MD, Baenziger JE. Expression, purification, and structural characterization of CfrA, a putative iron transporter from Campylobacter jejuni. J Bacteriol. 2008;190(16):5650–62. doi: 10.1128/JB.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abergel RJ, et al. Enzymatic hydrolysis of trilactone siderophores: where chiral recognition occurs in enterobactin and bacillibactin iron transport. J Am Chem Soc. 2009;131(35):12682–92. doi: 10.1021/ja903051q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid AN, et al. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl Environ Microbiol. 2008;74(5):1583–97. doi: 10.1128/AEM.01507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlyshev AV, Wren BW. Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni. Appl Environ Microbiol. 2005;71(7):4004–13. doi: 10.1128/AEM.71.7.4004-4013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young IG, Gibson F. Isolation of enterochelin from Escherichia coli. Methods Enzymol. 1979;56:394–8. doi: 10.1016/0076-6879(79)56037-6. [DOI] [PubMed] [Google Scholar]
- 33.Rodgers SJ, et al. Ferric Ion Sequestering Agents. 15. Synthesis, Solution Chemistry, and Electrochemistry of a New Cationic Analog of Enterobactin. Inorg Chem. 1987;26(10):1622–1625. [Google Scholar]
- 34.Dertz EA, Xu J, Raymond KN. Tren-based analogues of bacillibactin: structure and stability. Inorg Chem. 2006;45(14):5465–78. doi: 10.1021/ic060321x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miethke M, Hou J, Marahiel MA. The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli. Biochemistry. 2011;50(50):10951–64. doi: 10.1021/bi201517h. [DOI] [PubMed] [Google Scholar]
- 36.Alice AF, Naka H, Crosa JH. Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus. Infect Immun. 2008;76(9):4019–37. doi: 10.1128/IAI.00208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schauer K, Rodionov DA, de Reuse H. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem Sci. 2008;33(7):330–8. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 38.Schauer K, et al. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol Microbiol. 2007;63(4):1054–68. doi: 10.1111/j.1365-2958.2006.05578.x. [DOI] [PubMed] [Google Scholar]
- 39.Blanvillain S, et al. Plant carbohydrate scavenging through tonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One. 2007;2(2):e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofreuter D, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun. 2006;74(8):4694–707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pickett CL, et al. Iron acquisition and hemolysin production by Campylobacter jejuni. Infect Immun. 1992;60(9):3872–7. doi: 10.1128/iai.60.9.3872-3877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.