Abstract
Tolerant self-antigen–specific CD8 T cells fail to proliferate in response to antigen, thereby preventing autoimmune disease. By using an in vivo mouse model, we show that tolerant T cells proliferate and become functional under lymphopenic conditions, even in a tolerogenic environment. However, T cell rescue is only transient, with tolerance reimposed upon lymphorepletion even in the absence of tolerogen (self-antigen), challenging the prevailing paradigm that continuous antigen exposure is critical to maintain tolerance. Genome-wide messenger RNA and microRNA profiling revealed that tolerant T cells have a tolerance-specific gene profile that can be temporarily overridden under lymphopenic conditions but is inevitably reimposed, which suggests epigenetic regulation. These insights into the regulatory mechanisms that maintain or break self-tolerance may lead to new strategies for the treatment of cancer and autoimmunity.
T cell tolerance to self-antigens is required to prevent autoimmunity and arises through both central and peripheral mechanisms. Central tolerance occurs in the thymus through the process of negative selection, where developing thymocytes are deleted if they react too strongly to self-antigens presented by major histocompatability complex (MHC). Central tolerance is incomplete, however, because not all peripheral self-antigens are adequately presented in the thymus. Thus, self-reactive T cells that escape negative selection must be inactivated in the periphery by deletion, suppression by regulatory T cells, and/or cell-intrinsic programs to create a state of functional unresponsiveness (1). Peripheral tolerant CD8 T cells are phenotypically similar to antigen-experienced CD8 T cells but cannot proliferate in response to antigen stimulation (2). The rules governing this proliferative defect are not fully understood, but continuous exposure to self-antigen (tolerogen) is thought to be required (3–5). Many cancer antigens are self-antigens, and tolerance to these proteins can impede antitumor T cell responses. A critical challenge in tumor immunology is to develop strategies that break T cell tolerance to tumor/self-antigens without causing unacceptable autoimmune injury. Thus, it is necessary to define the molecular program underlying functional impairment of tolerant T cells and to understand the regulatory mechanism(s) that maintain or break self-tolerance.
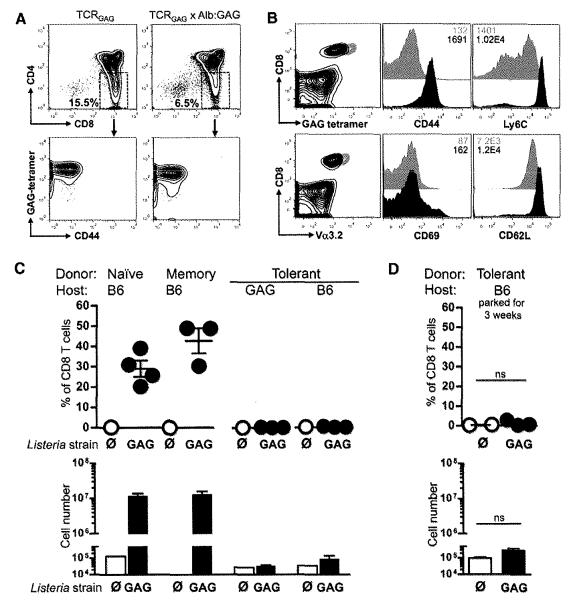
We developed a double transgenic mouse model of CD8 T cell tolerance to a self-antigen that is also a potential target tumor antigen (6, 7). T cell receptor transgenic mice (TCRGAG) were engineered with CD8 T cells specific for the Friend murine leukemia virus (FMuLV) GAG epitope, an immunodominant antigen from the FMuLV-transformed mouse leukemia cell line, FBL (fig. S1A). These mice were crossed to a second transgenic mouse strain, Alb:GAG, which selectively expresses the gag transgene in hepatocytes under control of the albumin promoter (fig. S1B). In double transgenic TCRGAG×Alb:GAG mice, TCRGAG CD8 T cells are only partially deleted in the thymus (Fig. 1A), yielding peripheral CD8 T cells expressing high levels of the GAG-specific TCR that appear antigen-experienced as evidenced by their Ly6Chi and CD44hi immunophenotype (Fig. 1B). Despite the presence of high avidity, self-antigen–specific TCRGAG CD8 T cells in the periphery, TCRGAG×Alb:GAG mice show no signs of autoimmune liver injury. The peripheral T cells from TCRGAG×Alb:GAG mice, in contrast to functional naïve or CD44hi/CD62Lhi memory TCRGAG CD8 T cells, are tolerant and unable to proliferate in response to immunization with a highly immunogenic recombinant Listeria monocytogenes strain expressing GAG (LM-GAG) (Fig. 1C).
Fig. 1.
Peripheral tolerant TCRGAG CD8 T cells are phenotypically different and functionally impaired compared with naïve and memory TCRGAG CD8 T cells. (A) Flow cytometric analysis of thymocytes from TCRGAG (naïve) and TCRGAG×Alb:GAG (tolerant) mice. Thymocytes were stained for CD4 and CD8, and single-positive CD8 thymocytes were analyzed for expression of CD44 and TCR by H2-Db/GAG-tetramer binding. (B) Flow cytometric analysis of splenocytes from naïve (gray) and tolerant (black) mice. All CD8+ cells are GAG-tetramer+ and Vα3.2+, and histograms are gated on CD8+ cells. Inset numbers indicate mean fluorescence intensity (MFI). (C) Totals of 106 naïve, 105 memory, or 106 tolerant T cells were transferred intravenously into B6 and/or Alb:GAG hosts and infected 1 day later with 3 × 107 colony-forming units (cfu) of LM-GAG (solid circle) or LM-∅ (open circle) as control. Percent (top) and cell numbers (bottom) of donor T cells in spleens were determined 7 days postinfection. (D) A total of 5 × 105 tolerant T cells (Thy1.2) were transferred intravenously into B6 hosts (Thy1.1), and 3 weeks later mice were infected with 3 × 107 cfu of LM-GAG (solid circle) or control LM-∅ (open circle). At 7 days postinfection, splenocytes were analyzed for cell expansion. All results are representative of at least two or three independent experiments with two to four mice per group. Error bars show SEM. ns, not significant.
Maintenance of T cell tolerance has been reported to require continual exposure of T cells to self-antigen (3–5). Therefore, we transferred tolerant T cells into wild-type B6 hosts. Three weeks after transfer, recipient mice were infected with LM-GAG. Transferred tolerant T cells did not expand, demonstrating that the function of self-reactive CD8 T cells is not necessarily restored by removal from the tolerizing environment (Fig. 1D).
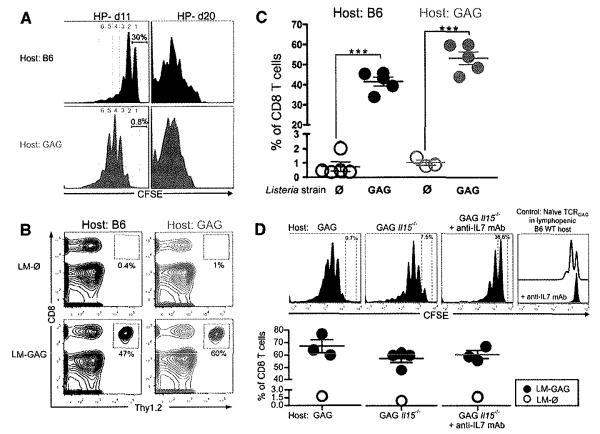
We previously showed that rescue could be achieved by inducing proliferation through alternative signaling pathways, for example, exogenous interleukin-15 (IL-15) in vitro (8, 9). Because “forcing” tolerant T cells to proliferate appeared to abrogate tolerance, we investigated whether tolerant T cells could be rescued in vivo by lymphopenia-mediated homeostasis-driven proliferation (HP). During lymphopenia-driven HP, T cells proliferate independently of cognate antigen encounter, driven by interactions with MHCs expressing nondeleting self-peptides and/or by homeostatic cytokines (10). Tolerant T cells were transferred into sublethally irradiated congenic B6 or Alb:GAG recipients (fig. S2). Tolerant T cells underwent HP in both nontolerogenic and tolerogenic hosts (Fig. 2A). Proliferation was more extensive in GAG self-antigen–expressing hosts, suggesting that TCR signaling, even by a tolerogen, augmented lymphopenia-driven HP. To determine whether the proliferating tolerant T cells were rescued, lymphopenic Alb:GAG or B6 recipients were immunized in vivo 21 days posttransfer with LM-GAG (fig. S2B). Tolerant T cells expanded dramatic6ally to LM-GAG not only in B6 but also in tolerogenic Alb:GAG recipients (Figs. 2, B and C, and 3B, left). To evaluate whether HP and rescue were dependent on the homeostatic cytokines IL-15 and/or IL-7, tolerant T cells were transferred into lymphopenic (i) Alb:GAG mice, (ii) Alb:GAG Ill5−/− mice, or (iii) Alb:GAG Ill5−/− mice treated with neutralizing IL-7 monoclonal antibody (mAb). Although HP was less efficient in Ill5−/− and anti–IL-7–treated Ill5−/− hosts (Fig. 2D, top), tolerant T cells still expanded in response to LM-GAG (Fig. 2D, bottom), suggesting that neither IL-15 nor IL-7 was absolutely required for HP-mediated rescue of tolerant T cells.
Fig. 2.
Tolerant T cells undergo HP and become functional under lymphopenic conditions, even in a tolerogenic environment. (A) About 2 × 105 to 3 × 105 carboxy-fluorescein-succinimidyl ester (CFSE)–labeled tolerant T cells were transferred (intravenously) into lymphopenic Alb:GAG or B6 WT mice (Thy1.1) 1 day after total body irradiation (TBI; 5Gy). At 11 and 20 days later, CFSE dilution of transferred tolerant T cells was analyzed by flow cytometry. Histograms are gated on CD8+ Thy1.2+ cells. Inset numbers show % of cells that did not proliferate, and numbers of cell divisions are indicated. (B and C) At 22 days after adoptive transfer of 0.5 × 105 to 1 × 105 tolerant T cells into lymphopenic Alb:GAG or B6 hosts, mice were immunized with 3 × 107 cfu of LM-GAG or LM-∅. At 7 days postinfection, peripheral blood was analyzed for expansion of transferred tolerant T cells. Results are representative of at least five independent experiments. ***P < 0.0001. (D) (Top) CFSE-labeled tolerant T cells were transferred into lymphopenic (Thy1.1) Alb:GAG mice, Alb:GAG Ill5−/− mice, or Alb:GAG Ill5−/− mice treated with neutralizing mAb against IL-7 (anti-IL7) 1 day after TBI, and spleen cells were analyzed 12 days later (top). To confirm the efficacy of anti-IL7 treatment, we transferred CFSE-labeled, naïve T cells into lymphopenic B6 wild-type (WI) mice (open histogram) or B6 mice treated with neutralizing anti-IL7 (solid histogram); 12 days later spleen cells were analyzed. Histograms are gated on CD8+ Thy1.2+ cells. (Bottom) At day 22 after transfer, mice were immunized with LM-GAG or LM-∅, and 7 days later spleens were assessed for expansion of donor T cells. Results are pooled from two independent experiments with a total of five to seven mice per group.
Fig. 3.
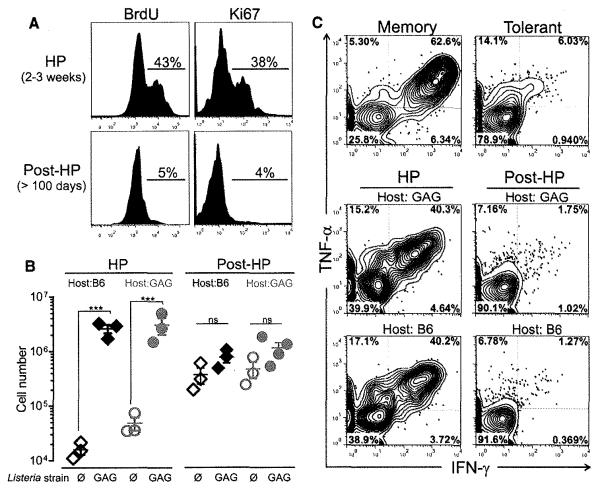
Rescued T cells reacquire tolerance in lymphoreplete hosts even in the absence of the tolerogen. (A) Flow cytometric analysis of proliferation of tolerant T cells determined by BrdU incorporation (left) and Ki67 staining (right) 2 to 3 weeks after transfer into lymphopenic mice (top) and at >3 to 4 months after transfer, when recipients had become lymphoreplete (bottom). Histograms are gated on CD8+ Thy1.2+ splenocytes. Percents of BrdU-positive and Ki67-positive transferred T cells are shown. (B) At 3 weeks (HP) or >3 to 4 months (post-HP) after transfer into lymphopenic Alb:GAG or B6 (Thy1.1) mice, mice were immunized with 3 × 107 cfu of LM-GAG or LM-∅. At 7 days postinfection, splenocytes were analyzed for expansion of transferred T cells. Data show mean ± SEM. For B6 hosts, ***P < 0.0001; for GAG hosts, ***P = 0.0007. (C) Intracellular TNF-α and IFN-γ production by tolerant T cells isolated 10 to 14 days (HP) or >3 to 4 months after transfer (post-HP) into irradiated Alb:GAG or B6 hosts. Memory and tolerant T cells are shown as control (top). Results are representative of at least three independent experiments.
Sublethally irradiated lymphopenic mice slowly recover lymphocyte numbers and are lymphoreplete by 3 to 4 months (fig. S2A). At this time, very few transferred tolerant T cells incorporated bromodeoxyuridine (BrdU) or expressed Ki67, suggesting that the cells were no longer actively cycling (Fig. 3A). To determine whether rescued tolerant T cells post-HP remained functional, we immunized lymphoreplete hosts with LM-GAG 3 to 4 months posttransfer. T cells rescued in tolerizing Alb:GAG hosts and functional at day 22 (Fig. 3B, left) were once again tolerant (Fig. 3B, right, and fig. S3). Unexpectedly, tolerant T cells rescued by HP in nontolerogenic B6 mice also reacquired tolerance. “Retolerized” T cells isolated from lymphoreplete B6 or Alb:GAG mice produced only limited amounts of the pro-inflammatory cytokines interferon (IFN)-γ and tumor necrosis factor (TNF)–α upon peptide stimulation, in contrast to rescued T cells (Fig. 3C). Thus, lymphopenia-mediated tolerance rescue was transient, with tolerance reestablished in lymphoreplete hosts even in the absence of the tolerogen.
Eukaryotic cells, including T cells, do not divide indefinitely and ultimately enter a phase of replicative senescence, known as the Hayflick limit (11). To determine whether tolerance reacquisition post-HP resulted from replicative senescence, we transferred retolerized T cells from lymphoreplete mice into a second set of lymphopenic recipients. After a second round of HP, retolerized T cells again responded to LM-GAG immunization, indicating that retolerized T cells were not senescent and could be “re-rescued” (fig. S4).
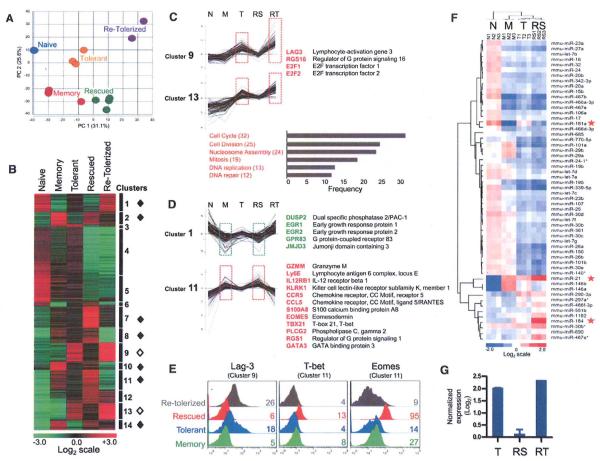
The unexpected finding that tolerant T cells undergoing HP become functional in tolerogenic hosts and reacquire tolerance in nontolerogenic hosts suggested that peripheral tolerance is maintained by TCR-independent signaling pathways. Molecular signatures have been identified for T cells in functionally unresponsive states, including exhausted CD8 T cells during chronic viral jnfection (12, 13), tumor-specific CD8 T cells in metastases (14), and anergic CD4 T cells (15), but the gene expression profile of tolerant self-antigen–specific T cells has not been determined. We performed microarray analysis of naive, memory, tolerant, rescued (lymphopenic tolerogenic hosts), and retolerized (lymphoreplete nontolerogenic hosts) TCRGAG CD8 T cells (fig. S5). Principal component analysis (PCA) (Fig. 4A) and K-means clustering (Fig. 4B and table S1) revealed that tolerant T cells harbor a tolerance-specific gene signature markedly distinct from those of naïve and memory T cells. To identify genes and pathways critical for maintenance of unresponsiveness, we determined which genes were preferentially expressed in tolerant T cells compared with their naïve and memory counterparts: Clusters 9 and 13 (164 genes) were identified as “tolerance-specific” gene sets representing uniquely overexpressed genes, including negative regulators of cell signaling and proliferation, transcription factors, and phosphatases (Fig. 4, B and C, and fig. S6A) (16). Lag3, an inhibitory co-receptor detected on exhausted CD8 T cells during chronic viral infection and tumor-infiltrating T cells, was one of the most highly expressed genes in tolerant T cells (17, 18) (Fig. 4, C and E). Gene ontology classification of tolerance-specific genes revealed that genes modulating cell cycle, cell division, nucleosome assembly, mitosis, and DNA replication were also highly overrepresented and up-regulated. These genes included histone proteins, minichromosome maintenance proteins, kinesins, cell division cycle proteins, and proteins of the spindle assembly and mitotic checkpoint complex (Fig. 4C and fig. S6, A and B). When tolerant T cells underwent HP in lymphopenic tolerogenic hosts, the tolerance gene signature was largely replaced by a signature remarkably similar to that of memory T cells (Fig. 4, A to E). Rescued T cells not only down-regulated tolerance-specific genes but expressed 475 additional genes at levels similar to those in memory T cells. We classified these genes into seven “rescue-associated” gene sets (clusters 1,2, 7, 8, 10, 11, and 14), suggesting that functional impairment of tolerant T cells is due to the expression of tolerance-specific genes and lack of rescue-associated genes. Genes down-regulated in rescued and memory T cells (cluster 1, 84 genes) included the negative regulators and transcription factors Dusp2 and Egr1/Egr2 and the histone demethylase Jmjd3 (19,20) (Fig. 4D and fig. S6C). Genes significantly up-regulated in rescued and memory T cells (clusters 2, 7, 8, 10, 11, 14; 391 genes) included (i) effector molecules Infg, Prf1, Gzmm, and Grn; (ii) master transcription factors controlling effector T cell function Tbx21 (21), Eomes (22), Gata3, and Stat4 (23); and (iii) chemokine and cytokine molecules Cxcr3, Ccr5, Cc15, and Ill2rβ (Fig. 4, D and E, and fig. S6C). Rescued T cells retolerized in nontolerogenic hosts lost the rescue-associated gene signature and reestablished a program similar to that of tolerant T cells (Fig. 4, B to E, and fig. S6, A to C). Thus, T cells “remember” the tolerance program established during the initial encounter(s) with self-antigen in the periphery, raising the question of how such memory is encoded. This likely reflects epigenetics, the regulation of gene expression by mechanisms other than changes in the DNA sequence that is organized at multiple levels involving DNA methylation, histone modifications, nucleosome organization, and noncoding RNAs [e.g., micro-RNA (miRNA)]. Indeed, we found that transcripts of numerous genes regulating chromatin modification were enriched in tolerant and retolerized T cells (e.g., Jmjd3, Dnmt1, Hat1, Hdac2, Hdac3, etc.; fig. S7).
Fig. 4.
Genome-wide mRNA and miRNA expression profiling of naïve, memory, tolerant, rescued, and retolerized TCRGAG CD8 T cells. (A to D) mRNA expression profiling. (A) PCA of the indicated cell populations. A total of 56.9% of the variation in samples was revealed in the first two principal components. For naïve, memory, and tolerant T cells, n = 3; for rescued, n = 4; and for retolerized, n = 2. (B) K-means clustering was used to partition differentially expressed genes into 14 distinct clusters. Transcript levels for all clusters are shown for naïve, memory, tolerant, rescued, and retolerized T cell subsets. The heat map shows log2-transformed expression intensities that were median-centered at the probe level. Higher expression is displayed in red, and lower expression in green. Rescue-associated gene sets indicated by solid diamonds; tolerance-specific gene clusters, open diamonds. (C) (Top) Selected genes from tolerance-specific clusters 9 and 13. (Bottom) Biological processes [Gene Ontology (GO) terms] enriched in tolerant and retolerized T cells. Genes associated within each cluster and GO are listed in fig. S6, A and B. (D) Selected genes from rescue-associated clusters 1 and 11. Additional clustering data are provided in fig. S6C. (E) Flow cytometric validation of Lag-3, T-bet, and Eomes expression in memory, tolerant, rescued, and retolerized T cells. Numbers indicate MFI. Data are representative for three independent experiments. (F) MiRNA microarray. Heat map of two-way hierarchical clustering of miRNAs and T cell samples. Each row represents a miRNA, and each column represents a sample [naïve (N); memory (M); tolerant (T); rescued (RS) T cells]. Higher expression is displayed in red and lower expression in blue; P < 0.001. Red stars indicate the miRNAs that are differently expressed in T compared with RS. (G) Expression of miR-181a in T, RS, and retolerized (RT) T cells determined by quantitative real-time PCR and normalized to miR-34a and miR-let7a. Error bars show SEM.
We focused on one critical aspect of epigenetic regulation of gene expression by asking whether specific miRNA expression changes might contribute to distinct mRNA expression profiles (24). Genome-wide miRNA profiling of naïve (N), memory (M), tolerant (T), and rescued (RS) T cells revealed that N, M, and T had distinct miRNA expression patterns. RS only differed from T in expression of three miRNAs: miR-21, miR-184, and miR-181a (Fig. 4F). miR-21 and miR-184 were overexpressed in RS compared with T, whereas miR-181a was decreased. miR-21 and miR-184 regulate cell proliferation and are overexpressed in many human cancers (25), suggesting that increased levels of miR-21 and miR-184 in RS may be due to ongoing proliferation rather than specifically associated with rescue. We therefore focused on miR-181a and confirmed by quantitative real-time polymerase chain reaction (PCR) that miR-181a was highly expressed in T and RT but low in M and RS (Fig. 4G and fig. S8A). Furthermore, miR-181a expression in M, RS, T, and RT inversely correlated with mRNA levels of known and predicted target genes compiled from published reports pointing to miR-181a as a possible key epigenetic regulator of tolerance (fig. S8, B to D). Previous studies showed that increasing miR-181a expression in CD4 T cells decreases TCR signaling threshold and increases antigen sensitivity (26, 27). Our finding suggests that miR-181a plays a different role in CD8 T cells, negatively regulating cell function.
Many eukaryotic somatic cells, including T cells, exist in a state of cellular quiescence—a reversible nonproliferative state. Cognate antigen stimulation triggers T cells (e.g., naïve and memory) to exit the quiescent state, enter the cell cycle, and undergo clonal expansion. This ability to maintain quiescence while retaining the capacity to rapidly proliferate is tightly orchestrated by transcription factors, cell cycle regulators, and checkpoints, including DNA replication, chromatin organization, and chromosome segregation proteins (28, 29). In contrast to naïve or memory T cells, whose transcriptional machinery is poised to enter the cell cycle upon antigen stimulation, we propose that TCR signaling in tolerant T cells is disengaged from cell cycle reentry control mechanisms. Lymphopenia provides a window of opportunity during which alternative signaling pathways enable tolerant T cells to proliferate independently of cognate antigen encounter, creating a permissive state where epigenetic control is temporarily superseded, allowing tolerant T cells to respond to antigen. However, after the mediator(s) of lymphopenia-mediated rescue disappear, T cells exit the cell cycle and become quiescent, and epigenetic control is once again reimposed, leading to restoration of the tolerant phenotype (fig. S9).
We have delineated the molecular program of tolerant, self-antigen–specific CD8 T cells and identified lymphopenia as a therapeutic opportunity to rescue, expand, and use tolerant T cells for cancer immunotherapy. Because lymphopenia-mediated rescue is only transient, with reestablishment of tolerance dictated by epigenetic memory, permanent rescue of tolerant T cells will require strategies to erase tolerance-specific epigenetic memory. However, an alternative strategy, repeated induction of lymphopenia leading to re-rescue of tolerant T cells, could be the basis for a new approach to cancer immunotherapy. Conversely, lymphopenia has been associated with exacerbation of autoimmunity (30), including graft-versus-host disease after autologous stem cell transplant (auto-GVHD), which is usually self-limited (31). Our findings not only point to a cell-intrinsic molecular mechanism for lymphopenia-associated autoimmune diseases but also explain their transient nature.
Supplementary Material
Acknowledgments
We thank S. Funk, C. Fowler, X. Tan, C. Chou, the other members of the Greenberg lab, and M. Philip for technical support and helpful discussion; the Abkowitz lab for providing the hemavet; M. Black from the UW Flow Cytometry Core Lab; J. Cao from the Immune Monitoring Shared Resource; and A. Dawson from the Genomics Shared Resource at the FHCRC. This work was supported by NIH grants R01 CA033084 (to P.D.G.), K01 CA117985 (to J.N.B.), and P30 CA015704-35 (to J.J.D. and R.S.B.); a grant FND 7008-08 from the Korea Research Institute of Bioscience and Biotechnology (to P.D.G.); and a Pilot and Feasibility Project Award (from P30 DK 56465 to B. Torok-Storb) (to A.S.). A.S. was supported by the Irvington Institute Fellowship Program of the Cancer Research Institute. All of the data reported in this paper are tabulated in the main text and in the supporting online material. The microarray data have been deposited in the Gene Expression Omnibus (GEO accession number GSE32025).
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/science.1214277/DC1 Materials and Methods Figs. S1 to S9 Table S1 References (32–42)
References and Notes
- 1.Mueller DL. Nat. Immunol. 2010;11:21. doi: 10.1038/ni.1817. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz RH. Annu. Rev. Immunol. 2003;21:305. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 3.Choi S, Schwartz RH. Semin. Immunol. 2007;19:140. doi: 10.1016/j.smim.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramsdell F, Fowlkes BJ. Science. 1992;257:1130. doi: 10.1126/science.257.5073.1130. [DOI] [PubMed] [Google Scholar]
- 5.Rocha B, Tanchot C, Von Boehmer H. J. Exp. Med. 1993;177:1517. doi: 10.1084/jem.177.5.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohlén C, et al. J. Exp. Med. 2002;195:1407. doi: 10.1084/jem.20011063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morimoto J, Tan X, Teague RM, Ohlén C, Greenberg PD. J. Immunol. 2007;178:6849. doi: 10.4049/jimmunol.178.11.6849. [DOI] [PubMed] [Google Scholar]
- 8.Teague RM, et al. Nat. Med. 2006;12:335. doi: 10.1038/nm1359. [DOI] [PubMed] [Google Scholar]
- 9.Teague RM, et al. Immunity. 2008;28:662. doi: 10.1016/j.immuni.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams KM, Hakim FT, Gress RE. Semin. Immunol. 2007;19:318. doi: 10.1016/j.smim.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayflick L, Moorhead PS. Exp. Cell Res. 1961;25:585. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 12.Wherry EJ, et al. Immunity. 2007;27:670. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Quigley M, et al. Nat. Med. 2010;16:1147. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baitsch L, et al. J. Clin. Investig. 2011;121:2350. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macián F, et al. Cell. 2002;109:719. doi: 10.1016/s0092-8674(02)00767-5. [DOI] [PubMed] [Google Scholar]
- 16.Murga M, et al. Immunity. 2001;15:959. doi: 10.1016/s1074-7613(01)00254-0. [DOI] [PubMed] [Google Scholar]
- 17.Blackburn SD, et al. Nat. Immunol. 2009;10:29. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosso JF, et al. J. Clin. Investig. 2007;117:3383. doi: 10.1172/JCI31184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Safford M, et al. Nat. Immunol. 2005;6:472. doi: 10.1038/ni1193. [DOI] [PubMed] [Google Scholar]
- 20.Yin Y, Liu YX, Jin YJ, Hall EJ, Barrett JC. Nature. 2003;422:527. doi: 10.1038/nature01519. [DOI] [PubMed] [Google Scholar]
- 21.Intlekofer AM, et al. Nat. Immunol. 2005;6:1236. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 22.Pearce EL, et al. Science. 2003;302:1041. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 23.Thierfelder WE, et al. Nature. 1996;382:171. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 24.Guo H, Ingolia NT, Weissman JS, Bartel DP. Nature. 2010;466:835. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medina PP, Nolde M, Slack FJ. Nature. 2010;467:86. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 26.Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM. Nat. Immunol. 2009;10:1162. doi: 10.1038/ni.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li QJ, et al. Cell. 2007;129:147. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Yusuf I, Fruman DA. Trends Immunol. 2003;24:380. doi: 10.1016/s1471-4906(03)00141-8. [DOI] [PubMed] [Google Scholar]
- 29.Cairns BR. Nature. 2009;461:193. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 30.Datta S, Sarvetnick N. Trends Immunol. 2009;30:430. doi: 10.1016/j.it.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Jones RJ, et al. Lancet. 1989;333:754. doi: 10.1016/s0140-6736(89)92575-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.