Mesenchymal stem cells (MSCs) are adherent multipotent stem cells from bone marrow and potentially numerous other tissues[1[ that serve as an attractive model system for evaluating the influence of extracellular cues on stem cell differentiation. MSCs have been shown to commit to several lineages including: bone, cartilage, fat and smooth muscle as well as transdifferentiation to skeletal muscle and neural fates.[1b, 2] Recently, our group[3[ and others[2a, 2b, 4] have used MSCs to demonstrate the importance of cytoskeletal tension and actomyosin contractility during lineage specification and commitment. For example, MSCs that were cultured either on stiff substrates[2b] or patterned surfaces that promoted cell spreading or cytoskeletal tension[3, 4b] all favored an osteogenic program that depended on increased contractility of the acto-mysoin cytoskeleton. Other reports have demonstrated the use of materials that are modified with cell adhesion ligands to promote MSC osteogenesis;[5[ we reasoned that the molecular characteristics of the adhesion ligands—including the affinity and density—would influence the cytoskeleton of the cell and may therefore serve to direct the differentiation pathways of adherent MSCs. Here we report that the biomolecular interactions between cells and their substrates can be tuned to promote osteogenesis, myogenesis or neurogenesis of cultured MSCs. This work provides an example of the use of molecular engineering to control the influence that materials have in regulating cell function.
We used self-assembled monolayers (SAMs) of alkanethiolates on gold as model substrates because these surfaces allow excellent control over the ligand-receptor interactions that mediate cell adhesion, in part because they are structurally well-defined and in part because the use of monolayers that are terminated in the oligo(ethylene glycol) group are highly effective at preventing non-specific adsorption of protein.[6[ Monolayers to which short peptide adhesion ligands are immobilized have been used to study several aspects of cell adhesion, including that of embryonic and mesenchymal stem cells, and are an established model for these applications.[7[ We prepared substrates by immobilizing either the linear peptide GRGDSC (linRGD) or the cyclic peptide RGDfC (cycRGD, and where f denotes a F residue having the D configuration) to monolayers presenting a maleimide group at a density of 1% (high density) or 0.1% (low density) against a background of tri(ethylene glycol) groups (Figure 1a).[8[ The cyclic peptide has an approximately two orders of magnitude higher affinity for the αvβ3 integrin—an important receptor in the adhesion and osteogenesis of MSCs[9[—than does the linear peptide.[10[ We used self-assembled monolayer desorption ionization (SAMDI) mass spectrometry to confirm immobilization of the peptides to the maleimide group (Figure S1).
Figure 1.
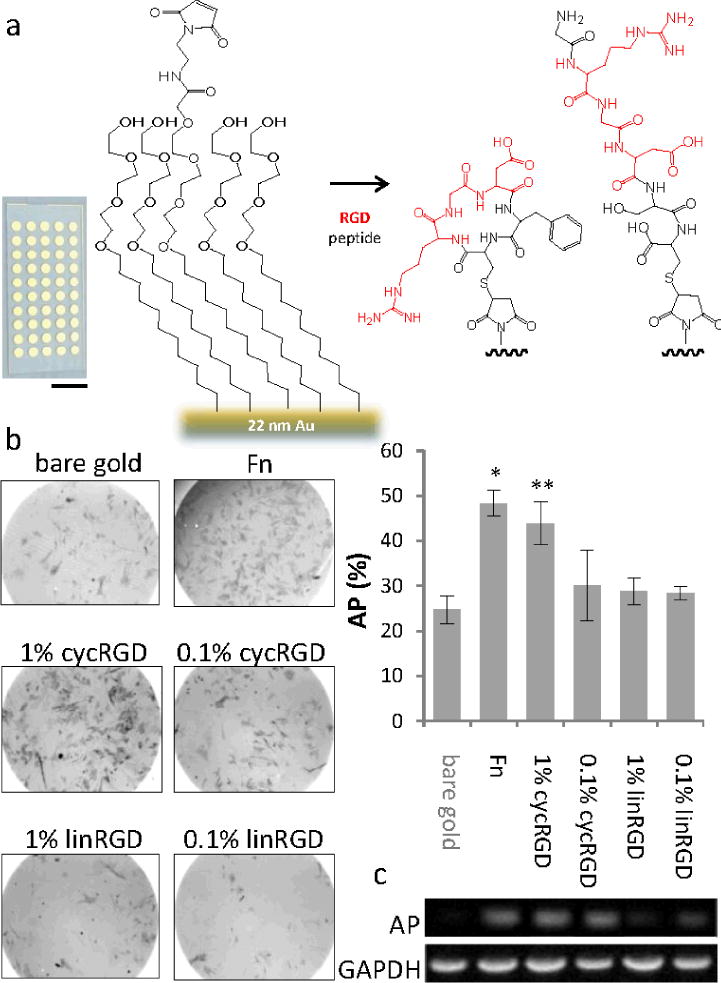
Monolayers presenting a high affinity adhesion ligand promote osteogenesis in adherent mesenchymal stem cells. (a) An array of monolayers were prepared on a glass slide having gold islands (scale bar = 1 cm) and present themaleimidegroup to allow immobilization of peptides. (b) Phase contrast images of mesenchymal stem cells stained for the osteogenesis marker alkaline phosphatase (AP, dark grey) after 10 days of culture on monolayers presenting the linear and cyclic RGD peptides at two surface densities. Right: Quantitation of the fraction of cells expressing AP on the different monolayers. (c) RT-PCR of AP transcript expression compared to GAPDH. Error bars represent standard deviations of at least three experiments. Statistical significance compared to bare gold: *p-value < 0.001, **p-value < 0.02
We cultured MSCs under standard growth conditions (see Supporting Materials) for ten days on substrates having either a bare gold film, a fibronectin-coated gold film or on monolayers presenting either linRGD, cycRGD, or a scrambled form of the linear peptide that has been demonstrated in our laboratory to beinactive (KRDGVC)[11[; for the monolayers, peptides were present at a density of 1% relative to total alkanethiolate (the ‘high’ density) or 0.1% (the ‘low’ density). We then fixed and stained the cells to observe alkaline phosphatase (AP), which is an early marker for osteogenesis. We observed elevated levels of AP expression for cells on the fibronectin (where 48% of the cells stained for AP) and cycRGD substrates (44% on high density, 30% on low density of ligand) compared to a lower level of expression for cells adherent to the bare gold film (25%)(Figure 1b). MSCs cultured on monolayers presenting linRGD at either high or low density expressed levels of AP that are comparable to cells cultured on the unmodified bare gold. Control experiments that used monolayers presenting no peptide or the scrambled RDG peptide showed insignificant levels of cell adhesion and were not included in the analysis. We confirmed these trends by using RT-PCR to quantitate the amount of mRNA coding for AP, and again found higher expression for cells cultured on monolayers presenting cycRGD and the fibronectin-coated substrate relative to those cultured on the linRGD-terminated monolayers (Figure 1c). These results suggest that fibronectin and the monolayers presenting cycRGD promote osteogenesis.
To further investigate the influence of the monolayers on differentiation, we performed immunofluorescence staining of several lineage-specific markers. For example, cells that are differentiating towards osteoblasts express the runt-related transcription factor 2 (Runx2). MSCs cultured on fibronectin and cycRGD surfaces show a higher degree of nuclear Runx2 compared to cells cultured on the linRGD and bare gold surfaces (Figure 2a and b), in agreement with the observations made using staining and RT-PCR of alkaline phosphatase (Figure 1b and c). To determine if the linRGD surface is promoting differentiation towards other lineages, we immunostained cells for a skeletal muscle marker, the early myogenic regulatory transcription factor MyoD. MSCs cultured on monolayers presenting linRGD at high density showed the highest level of MyoD expression. Cells cultured on monolayers presenting cycRGD (at both high and low densities of ligand) or monolayers presenting the linRGD peptide at low density show elevated expression of MyoD compared to cells cultured on bare gold and fibronectin-coated gold but significantly less than cells cultured on monolayers presenting linRGD at high density (Figure 2b). We also immunostained cells for the neurogenic marker β3-tubulin to assess whether certain combinations of peptide affinity and density could promote differentiation to neuron-like cells. As the affinity and density of the peptide ligand were decreased we observed an increase in β3-tubulin expression with the highest percentage of β3-tubulin expressing cells occurring on the monolayers that present linRGD at low density (11%, Figure 2a and b). We performed RT-PCR to verify the trends in immunofluorescence and found a decrease in Runx2 expression as the affinity and density of the immobilized ligands were reduced (Figure 2c). In contrast, expression of the myogenic marker genes (Desmin, MyoD) increased in these samples and showed a maximum expression in cells that were cultured on monolayers presenting linRGD at high density. Expression of the neuronal marker (β3-tubulin, CEND1) was observed primarily in cells cultured on monolayers presenting linRGD at low density although we did observe expression of CEND1 in cells cultured on the bare gold. In addition to neural specific markers, we note that the expression of myogenic markers is elevated on the low density linRGD surface which suggests that a fraction of cells on this peptide surface are specifying markers associated with muscle differentiation. Taken together, these results suggest that the monolayers that present high affinity ligands promote osteogenesis and monolayers that present the low affinity ligand primarily promote myogenesis at high density and neurogenesis at low density of ligand (Figure 2d).
Figure 2.
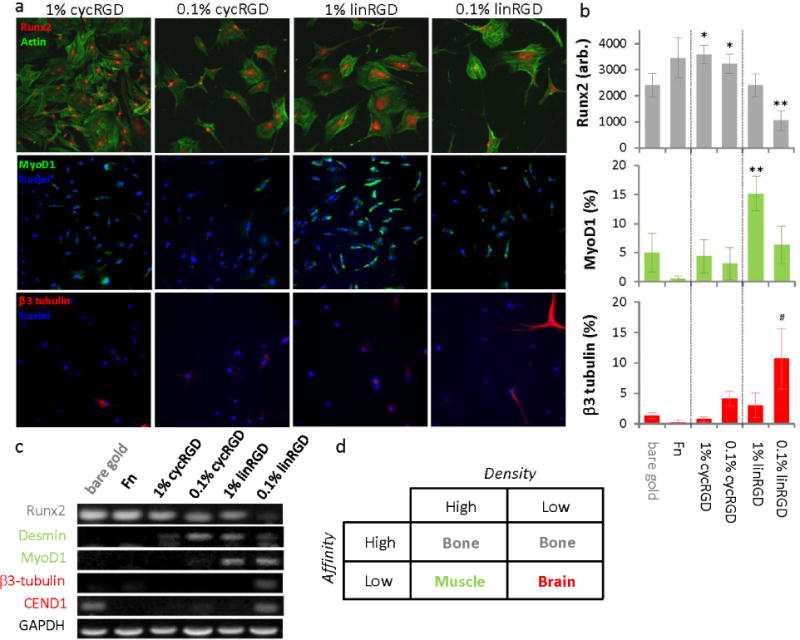
The density and affinity of an adhesion ligand influence the differentiation of mesenchymal stem cells. Cells were cultured on monolayers presenting either the cyclic or linear RGD peptide at a high or low density and differentiation was analyzed using(a, b) immunofluorescence imaging of markers for osteogenesis (Runx2), myogenesis (MyoD1) and neurogenesis (b3-tubulin) and (c) expression analysis by RT-PCR of lineage specific transcripts. (d) Table summarizing the preferred differentiation outcome for celsl cultured on the four monolayer surfaces. Error bars represent standard deviations from three replicates. Statistical significance compared tobare gold: *p-value < 0.02, **p-value < 0.002, #p-value < 0.05 as determined using students t-test. Scale bar is 20 μm.
Since cell morphology has been previously suggested as a qualitative marker of differentiation,[2b] we characterized the changes in shape for MSCs that were cultured on the different substrates. A greater number of cells initially adhered to the bare gold, fibronectin coated gold and cyclic peptide surfaces (Figure S2). Cells showed a higher proliferation rate on surfaces coated with fibronectin when compared to bare gold or peptide modified surfaces. MSCs cultured for one week on monolayers presenting cycRGD remained well spread with a cuboidal phenotype while cells cultured on the linRGD displayed an elongated morphology reminiscent of myoblasts (Figure S2a). For the monolayers that present linRGD at low density, many of the cells extended long processes characteristic of neuronal-like cells. These morphological differences are consistent with previous reports of MSC commitment to the osteogenesis, myogenesis and neurogenesis programs.[2b] We also note a decrease in total cell area and nuclear area after one week in culture for cells adherent to the linRGD surfaces(Figure S2b).Changes in nuclear area have previously been shown to influence gene expression and cell differentiation.[12[
The influence of ligand affinity and density on differentiation are consistent with a model wherein substrates presenting ligands of higher affinity and at higher density lead to more spreading of cells and greater tension in the cytoskeleton, which favors an osteogenic outcome. Previous work has demonstrated the importance of cytoskeletal tension and focal adhesion assembly in directing the differentiation of MSCs.[2b, 3, 4b] We immunostained MSCs for filamentous actin and the focal adhesion protein vinculin. Cells that were adherent to monolayers presenting cycRGD displayed a higher degree of spreading, more stress fibers, and more focal adhesion structures as compared to cells on monolayers presenting the linRGD peptide (Figure 3a–d). We also immunostained MSCs for non-muscle myosin IIa and IIb to evaluate differences in contractility in cells on the different substrates. After normalizing the fluorescence data, a greater fraction of cells expressed high levels of myosin IIb when cultured on monolayers presenting cycRGD (36% on high density, 40% on low density) as compared to cells cultured on monolayers presenting linRGD (13% on high density, 25% on low density, Figure 4a). Since cells cultured on the linRGD surfaces display the highest expression of myogenic markers, we immunostained the cells for the muscle specific myosin heavy chain (MYH). The number of cells expressing high levels of MYH increased significantly as the affinity and density of the cell adhesion peptide decreased, consistent with a model where decreasing cell contractility correlates with myogenesis. For myosin IIa, we observed a comparable level of total fluorescence in cells cultured on the various monolayers (Figure S3). This result agrees with a report that showed expression of this isoform to be relatively insensitive to variations in substrate elasticity.[2b]
Figure 3.
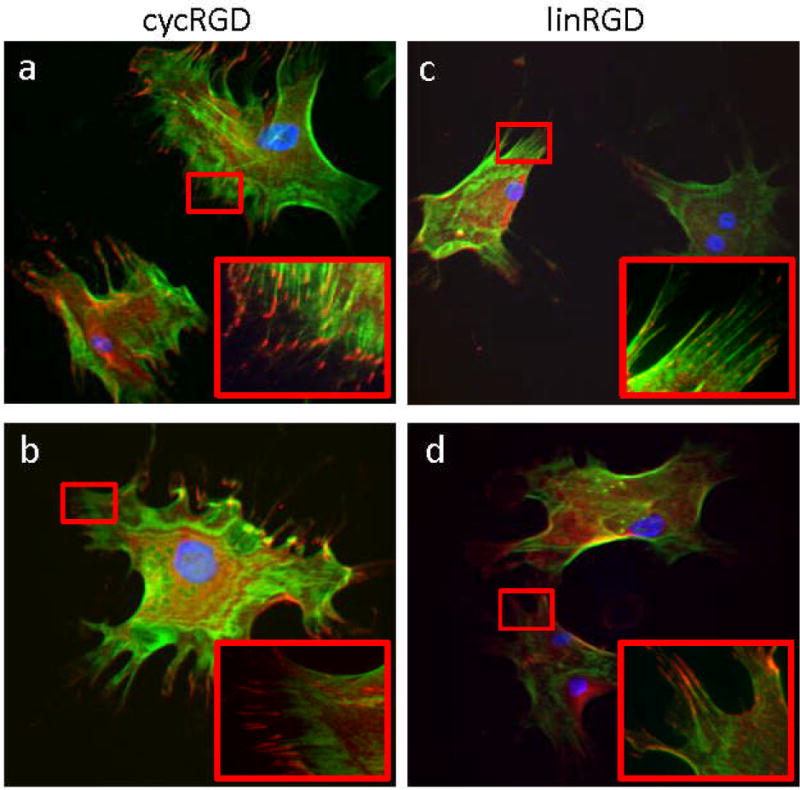
The affinity and density of a cell adhesion peptide influences focal adhesion and stress fiber formation. Immunofluorescence images of mesenchymal stem cells stained for filamentous actin (green), vinculin (red) and nuclei (blue) on (a, b) surfaces presenting 1% and 0.1% cyclic RGD peptide and (c, d) surfaces presenting 1% and 0.1% linear RGD peptide.
Figure 4.
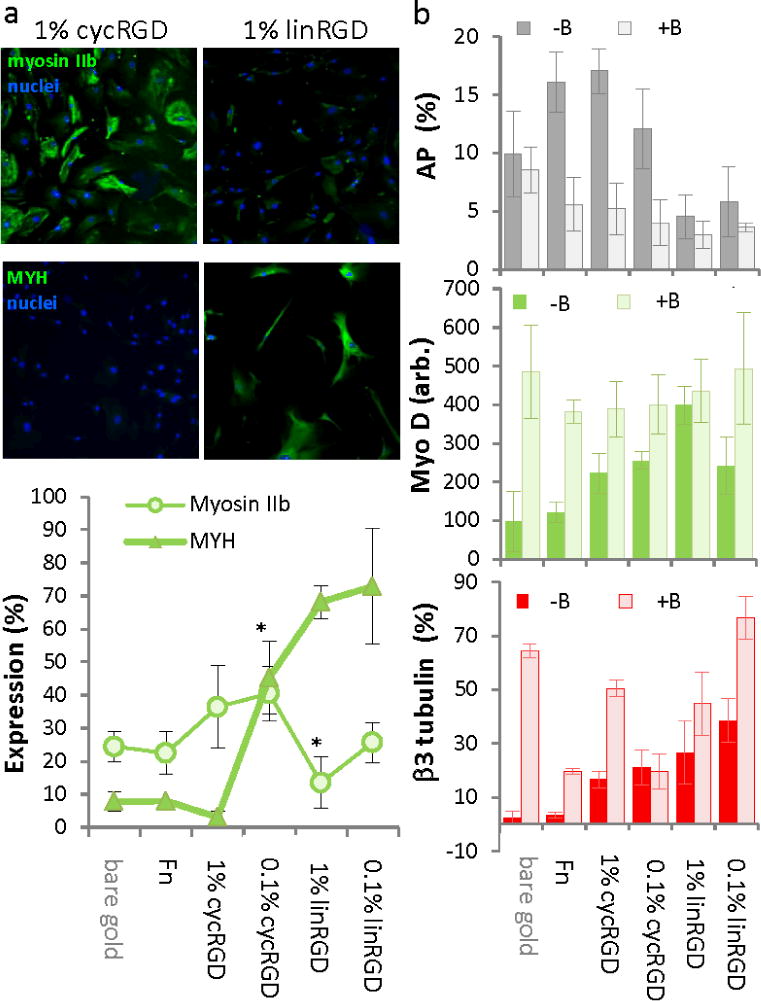
The differentiation of mesenchymal stem cells on peptide surfaces is influenced by non-muscle myosin. (a) Immunofluorescence images and quantitation of cells stained for non-muscle myosin IIb and muscle-specific myosin heavy chain (MYH). *statistical significance p-value < 0.005. (b) Lineage marker expression as determined by immunofluorescence for MSCs exposed to the non-muscle myosin inhibitor blebbistatin (+B) and control cultures that were not exposed (−B): osteogenesis (AP), myogenesis (MyoD1) and neurogenesis (β3 tubulin). Error bars represent standard deviations of at least three replicates. Scale bar is 20 μm.
To confirm the important role that cell contractility plays in differentiation, we cultured MSCs in a medium that was supplemented with blebbistatin, an inhibitor of non-muscle myosins that has little effect on muscle-specific isoforms.[2b] Cells were allowed to fully adhere and then exchanged with medium containing blebbistatin at a concentration that does not significantly perturb cell shape. We found that treated cells showed a decrease in osteogenesis as determined by alkaline phosphatase staining and an increase in expression of MyoD and β3-tubulin (Figure 4b and Figure S4). This result supports our hypothesis that the monolayers having a high affinity ligand promote a more contractile cytoskeleton in adherent cells, which is necessary for the osteogenic preference exhibited by theese cells.
Engler and Discher demonstrated the influence of substrate stiffness on MSC differentiation and found that stiff matrices promoted osteogenesis, those of intermediate stiffness promoted myogenesis and soft matrices promoted neurogenesis.[2b] Our study demonstrates a similar trend in lineage specification where monolayers presenting high affinity peptides promote osteogenesis, those presenting a low affinity peptide at high density promote myogenesis and those that present a low affinity peptide at low density promote neurogenesis. In both examples, the differentiation outcomes depend on the ability of the substrate to oppose the traction forces exerted by the adherent cells. Substrates that present ligands that have a higher affinity for the integrin adhesion receptors are known to increase the spreading of adherent cells, with a corresponding increase in the traction forces applied by the cell.[13[ This idea is further supported by a recent report by Mooney and coworkers that demonstrated the importance of traction-mediated reorganization of the matrix to direct MSC fate within 3D hydrogels.[9a] Osteogenesis was found to occur preferentially at an optimal stiffness where MSCs can maximize the number of adhesive contacts with the surrounding matrix.
This report illustrates a molecular approach to engineering substrates used in stem cell cultures; here, we show that the affinity and density of ligands at the cell-biomaterial interface can be engineered to influence stem cell fate. This demonstration provides another tool that expands on the set of materials science-based approaches that have been used to direct stem cell fate. The relationships identified in this work might also serve as design rules for the modification of other materials used in stem cell cultures. It further suggests that optimal adhesive microenvironments that favor specific differentiation outcomes may exist. Understanding the relationship between adhesion and lineage could assist efforts in defining the MSC niche as well as studies aimed at elucidating the factors that control stem cell lineage guidance and differentiation. We expect that the use of structurally well-defined mimics of the extracellular matrix will similarly be important for controlling cellular activities in a broad range of contexts.
Supplementary Material
Footnotes
This work was supported by the National Cancer Institute of the National Institutes of Health. K.A.K. was supported by a Ruth L. Kirschstein National Research Service Award Number F32GM087048 from the National Institute of General Medical Sciences of the National Institutes of Health.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Kristopher A. Kilian, Email: kakilian@illinois.edu, Current Address: Department of Materials Science and Engineering, University of Illinois at Urbana-Champaign.
Milan Mrksich, Email: milan.mrksich@northwestern.edu, Department of Chemistry, Department of Cell and Molecular Biology, Department of Biomedical Engineering, Northwestern University, Fax: 847-467-3057.
References
- 1.a) Crisan M, Yap S, Casteilla L, Chen C-W, Corselli M, Park Tea S, Andriolo G, Sun B, Zheng B, Zhang L, Norotte C, Teng P-N, Traas J, Schugar R, Deasy Bridget M, Badylak S, Buhring H-J, Giacobino J-P, Lazzari L, Huard J, Peault B. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]; b) Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 2.a) Gao L, McBeath R, Chen CS. Stem Cells. 2010;28:564–572. doi: 10.1002/stem.308. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]; c) Rowlands AS, George PA, Cooper-White JJ. Am J Physiol. 2008;295:C1037–C1044. doi: 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- 3.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Proc Natl Acad Sci U S A. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. Nat Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]; b) McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]; c) Oh S, Brammer KS, Li YSJ, Teng D, Engler AJ, Chien S, Jin S. Proc Natl Acad Sci U S A. 2009;106:2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Park J, Bauer S, Von Mark K, Schmuki P. Nano Lett. 2007;7:1686–1691. doi: 10.1021/nl070678d. [DOI] [PubMed] [Google Scholar]
- 5.a) Hsiong SX, Boontheekul T, Huebsch N, Mooney DJ. Tissue Eng., Part A. 2009;15:263–272. doi: 10.1089/ten.tea.2007.0411. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Martino MM, Mochizuki M, Rothenfluh DA, Rempel SA, Hubbell JA, Barker TH. Biomaterials. 2009;30:1089–1097. doi: 10.1016/j.biomaterials.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Petrie TA, Raynor JE, Reyes CD, Burns KL, Collard DM, Garcia AJ. Biomaterials. 2008;29:2849–2857. doi: 10.1016/j.biomaterials.2008.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Mrksich M. Acta Biomater. 2009;5:832–841. doi: 10.1016/j.actbio.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gurard-Levin ZA, Kilian KA, Kim J, Bahr K, Mrksich M. ACS Chem Biol. 2010;5:863–873. doi: 10.1021/cb100088g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Derda R, Musah S, Orner BP, Klim JR, Li L, Kiessling LL. J Am Chem Soc. 2010;132:1289–1295. doi: 10.1021/ja906089g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Derda R, Li L, Orner BP, Lewis RL, Thomson JA, Kiessling LL. ACS Chem Biol. 2007;2:347–355. doi: 10.1021/cb700032u. [DOI] [PubMed] [Google Scholar]; c) Hudalla GA, Murphy WL. Langmuir. 2010;26:6449–6456. doi: 10.1021/la1008208. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Hudalla GA, Murphy WL. Langmuir. 2009;25:5737–5746. doi: 10.1021/la804077t. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Luo W, Chan EWL, Yousaf MN. J Am Chem Soc. 2010;132:2614–2621. doi: 10.1021/ja907187f. [DOI] [PubMed] [Google Scholar]; f) Koepsel JT, Murphy WL. Langmuir. 2009;25:12825–12834. doi: 10.1021/la901938e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato M, Mrksich M. Biochemistry. 2004;43:2699–2707. doi: 10.1021/bi0352670. [DOI] [PubMed] [Google Scholar]
- 9.a) Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Su JL, Chiou J, Tang CH, Zhao M, Tsai CH, Chen PS, Chang YW, Chien MH, Peng CY, Hsiao M, Kuo ML, Yen ML. J Biol Chem. 285:31325–31336. doi: 10.1074/jbc.M109.087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mas-Moruno C, Rechenmacher F, Kessler H. Anticancer Agents Med Chem. 2011;10:753–768. doi: 10.2174/187152010794728639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Cortes J, Mrksich M. ACS Chemical Biology. 2011;6:1078–1086. doi: 10.1021/cb200186j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas CH, Collier JH, Sfeir CS, Healy KE. Proc Natl Acad Sci U S A. 2002;99:1972–1977. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houseman BT, Mrksich M. Biomaterials. 2001;22:943–955. doi: 10.1016/s0142-9612(00)00259-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


