Summary
Potassium (K+) plays a vital role in bacterial physiology, including regulation of cytoplasmic pH, turgor pressure, and transmembrane electrical potential. Here, we examine the Staphylococcus aureus Ktr system uniquely comprised of two ion-conducting proteins (KtrB and KtrD) and only one regulator (KtrA). Growth of Ktr system mutants was severely inhibited under K+-limitation, yet detectable after an extended lag phase, indicating the presence of a secondary K+ transporter. Disruption of both ktrA and the Kdp-ATPase system, important for K+ uptake in other organisms, eliminated regrowth in 0.1 mM K+, demonstrating a compensatory role for Kdp to the Ktr system. Consistent with K+ transport mutations, S. aureus devoid of the Ktr system became sensitive to hyperosmotic conditions, exhibited a hyperpolarized plasma membrane, and increased susceptibility to aminoglycoside antibiotics and cationic antimicrobials. In contrast to other organisms, the S. aureus Ktr system was shown to be important for low-K+ growth under alkaline conditions, but played only a minor role in neutral and acidic conditions. In a mouse competitive index model of bacteremia, the ktrA mutant was significantly outcompeted by the parental strain. Combined, these results demonstrate a primary mechanism of K+ uptake in S. aureus and a role for this system in pathogenesis.
Keywords: Staphylococcus aureus, potassium homeostasis, alkaline growth, hyperosmotic stress, antibiotic resistance, bacteremia
Introduction
Despite the limited availability of potassium (K+) in the environment, this essential macroelement is the most abundant intracellular cation in all living organisms. Unlike other ions, K+ can accumulate to very high concentrations in bacteria and plays a primary role in regulation of cytoplasmic pH (Bakker & Mangerich, 1981), transmembrane electrical potential (Booth, 1985), and turgor pressure (Epstein, 1986) in non-halophilic bacteria (for review see (Bakker, 1993)). Bacterial cells must therefore possess the ability to effectively import K+ against steep transmembrane concentration gradients to maintain homeostatic K+ concentrations required for growth.
Three major, multicomponent systems have been found to mediate K+ uptake in bacteria: Trk, Ktr, and Kdp (Epstein, 2003). The K+-conducting proteins of these systems (TrkH, KtrB, and KdpA, respectively) are members of the superfamily of K+ transporters (SKT), evolutionarily related to small, two transmembrane K+ channels (Doyle et al., 1998). For SKT protein function, the channel must be activated (opened) via interaction with its regulatory subunit. Peripheral SKT regulatory proteins, such as KtrA within the Ktr system, contain dinucleotide binding sites which, when bound, induce a conformational shift, opening the K+-conducting protein gate (Roosild et al., 2002, Albright et al., 2006, Roosild et al., 2009). As a result, KtrA confers import velocity and ion specificity to KtrB. All well-characterized Ktr systems encode one regulatory protein that specifically activates one channel protein (Nakamura et al., 1998, Zulkifli et al., 2010). For example, Bacillus subtilis encodes two complete Ktr systems (KtrAB and KtrCD), and there appears to be no cross talk between these systems, indicating that each regulator is specific for its channel protein (Holtmann et al., 2003).
Ktr systems appear to be constitutively expressed and responsible for moderate-affinity (mM) K+ uptake requiring sodium ions (Na+) for transport, suggesting Na+/K+ symport activity (Matsuda et al., 2004, Tholema et al., 1999). In contrast, Kdp systems are efficient K+ scavenging pumps, making use of ATP hydrolysis to perform high-affinity K+ uptake. The KdpFABC system is transcriptionally-regulated by the KdpDE two-component system, which senses extracellular K+ (Voelkner et al., 1993, Laermann et al., 2013). Kdp systems are utilized during times of K+ starvation and/or fluctuating ionic conditions when other transporters cannot meet the cellular requirements for K+ (Heermann & Jung, 2010, Walderhaug et al., 1992). Importantly, bacteria typically contain more than one K+ uptake system, likely to ensure that adequate levels of this essential element are present in the cell (Corratge-Faillie et al., 2010).
Mechanisms of K+ transport have been characterized in a variety of bacterial pathogens and implicated in the virulence of these organisms (Alkhuder et al., 2010, Mason et al., 2006, Stingl et al., 2007, Su et al., 2009). However, no studies have clearly elucidated the mechanism of K+ uptake in any staphylococcal species. Staphylococcus aureus is a commensal of the human skin as well as a pathogen known for its ability to cause a wide spectrum of diseases including skin and soft tissue abscesses, bacteremia, and implanted medical device infections (DeLeo et al., 2010). Recent work by Xue et al. (Xue et al., 2011) describing a Kdp system encoded by S. aureus revealed that this system has no significant role in K+ transport. Additionally, a KtrA-like protein was recently found to be required for S. aureus growth under K+-limiting conditions (Corrigan et al., 2013); however, its role in the regulation of Ktr-specific transport proteins was not investigated. In the current study we examined in detail the function of this unique Ktr K+ transport system, which consists of one regulatory protein and two ion-conducting channels required for K+-limited growth of S. aureus in alkaline conditions. We also demonstrated that the Ktr system plays a major role in maintaining cell physiology, including regulating membrane potential and cellular osmotic tolerance. Finally, the Ktr system was shown to play a significant role in antimicrobial resistance and fitness within an S. aureus bacteremia model of infection.
Results
A novel Ktr system in Staphylococcus aureus
To identify potential K+ transport proteins in S. aureus, we aligned the amino acid sequences of the ion-conducting proteins, KtrB and KtrD, from B. subtilis strain 168 with the S. aureus strain FPR3757 proteins in the GenBank database using the Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/). Two hypothetical S. aureus proteins were identified (locus numbers SAUSA300_1979 and SAUSA300_0924) that shared amino acid sequence similarity with both B. subtilis KtrB and KtrD (43% and 50% identity, respectively; see Table 1). Based on the sequence similarities observed, along with the functional studies described below, we renamed the SAUSA300_1979 and SAUSA300_0924 open reading frames, ktrB and ktrD, respectively. Each of the putative S. aureus KtrB and KtrD proteins contain the four glycine residues that form the selectivity filter in Ktr channel proteins (Zhou et al., 2001, Lu & Miller, 1995, Tholema et al., 1999, Tholema et al., 2005) and lack predicted N-terminal transmembrane segments classically found in Trk ion-conducting proteins (Fig. S1). Interestingly, only one protein in S. aureus FRP3757, previously designated KtrA (Corrigan et al., 2013), shared sequence similarity with B. subtilis KtrA and KtrC, with a higher level of identity observed with KtrC (Table 1). Analysis of the KtrA protein revealed that it contains one predicted C-terminal dinucleotide-binding site (KTN/RCK domain), similar to other Ktr regulators (Fig. S2).
Table 1. Identification of staphylococcal Ktr proteins.
| Loc# | KtrB | KtrD | Loc# | KtrA | KtrC | |
|---|---|---|---|---|---|---|
| S. aureus USA300_FPR3757 | 1979 | 43% | 33% | 0988 | 54% | 65% |
| 0924 | 33% | 50% | ||||
| S. epidermidis ATCC 12228 | 0571 | 42% | 33% | 0786 | 53% | 64% |
| 0724 | 33% | 50% | ||||
| S. haemolyticus JCSC1435 | 2567 | 41% | 33% | 1867 | 53% | 64% |
| 1935 | 33% | 52% | ||||
| S. lugdunensis HKU09-01 | 1845 | 33% | 50% | 1788 | 33% | 66% |
| S. saprophyticus ATCC 15305 | 2353 | 41% | 32% | 1703 | 53% | 64% |
| 1762 | 33% | 49% | ||||
| S. carnosus TM300 | 0441 | 42% | 31% | 0709 | 54% | 65% |
| 0626 | 32% | 49% | ||||
| S. pseudintermedius HKU10-03 | 0870 | 43% | 32% | 0795 | 51% | 66% |
| 0719 | 33% | 48% |
Loc# represents locus ID for the given Staphylococcus sp.
Percentage represents the percent amino acid identity to the respective B. subtilis Ktr protein.
All three ktr genes in S. aureus are located separately on the chromosome (Fig. S3) and a number of other S. aureus isolates whose genomes have been sequenced contain these genes (data not shown). The ktrA, ktrB, and ktrD genes also appear to be conserved in several other staphylococcal species, with the one exception being S. lugdunensis, which contains only one putative Ktr transporter system, with the highest level of similarity to the B. subtilis KtrC and KtrD proteins (Table 1). All staphylococcal ktrD genes listed in Table 1 are downstream relative to a gene encoding an HtrA-like serine protease, and all ktrA genes are located downstream of a gene encoding a cytochrome d ubiquinol oxidase subunit-like protein (data not shown). From these in silico analyses, we hypothesize that S. aureus encodes a novel one regulator/two ion-conducting domain Ktr system in which KtrA regulates the K+-transport activity of both KtrB and KtrD.
Requirement for Ktr in alkaline conditions
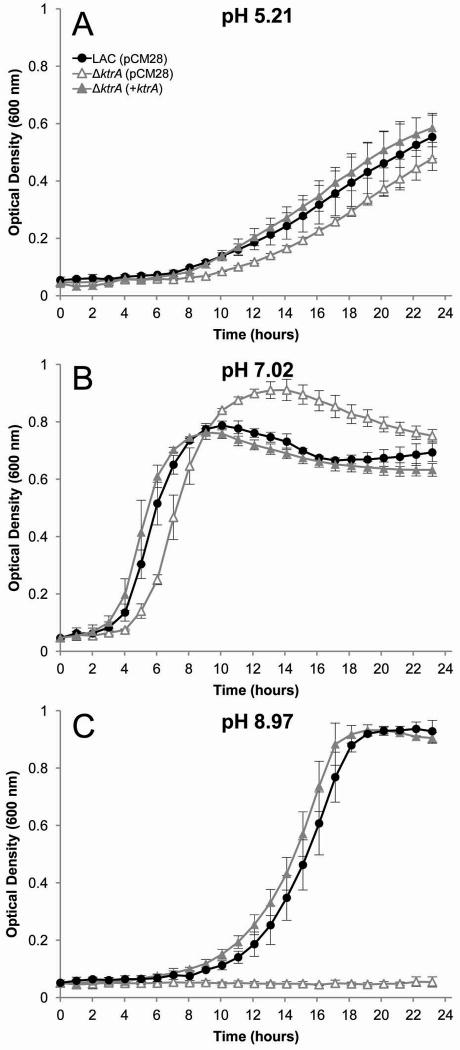
K+ uptake is important for acid stress survival by compensating for H+ extrusion and resulting alkalinaization of the cytoplasm (Kitko et al., 2010). Conversely, K+ efflux (i.e. via K+/H+ antiport) is necessary for alkaline stress survival and acidification of the cytoplasm (Padan et al., 2005, Booth, 1985, Krulwich et al., 2009). Since previous studies in other organisms have demonstrated a role for Ktr-mediated transport at different pH ranges (Zulkifli & Uozumi, 2006, Tholema et al., 2005, Kawano et al., 2000), we initially tested the role of the S. aureus Ktr system by generating a ΔktrA mutant and culturing in K+-limited medium at a range of starting pH conditions. As shown in figure 1, the ΔktrA mutant exhibited a clear defect in growth in alkaline pH, K+-limited medium, a phenotype that was restored by expression of the ktrA gene from a plasmid. Based on these results, subsequent experimentation was performed under alkaline conditions.
Fig. 1.
Growth of S. aureus ΔktrA mutant in 0.5 mM KCl at acidic, neutral, and alkaline pH. Wild-type S. aureus (solid circles) and the ΔktrA mutant (open triangles) were inoculated from overnight cultures to OD600=0.05 in K-CDM supplemented with 0.5 mM KCl.
A and B. Growth in acidic (pH 5.21) or neutral (pH 7.02) conditions had minor effects on growth.
C. At pH 8.97, the ΔktrA strain was unable to grow.
A-C. Expression of ktrA from plasmid pCM28 in ΔktrA (solid triangles) fully complemented the growth defects in all conditions tested.
Data represents the mean of at least two independent experiments, with ± standard deviation.
Low K+ growth requires Ktr
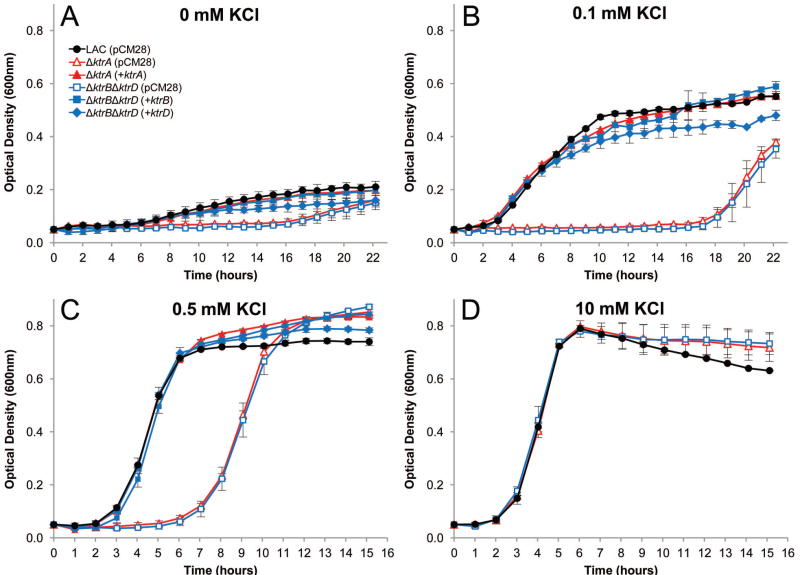
To further examine the role of S. aureus ktr genes in K+ transport, we tested the growth of isogenic ktr mutants in S. aureus strain LAC in the presence of various KCl concentrations. As shown in figure 2A, the parental LAC strain grew slightly in K+-deprived medium (K-CDM), and a 16 hour delay was seen after deletion of the putative Ktr regulator, ktrA (ΔktrA), or both ktrB and ktrD (ΔktrBΔktrD). Growth of the ΔktrA and ΔktrBΔktrD strains in K-CDM supplemented with 0.1 mM KCl remained undetectable for 17 hours after which the cells began to grow (Fig. 2B). When supplemented with 0.5 mM KCl, a 6-hour lag phase was observed before the ΔktrA and ΔktrBΔktrD strain were able to resume a growth rate and yield similar to the wild-type strain (Fig. 2C). When the culture medium was supplemented with 10 mM KCl, all strains grew equally well (Fig. 2D). Notably, deletion of either ktrB or ktrD alone had no impact on growth in K+-limited medium (Fig. S4), demonstrating that only one ion-transporting protein (KtrB or KtrD) and the KtrA regulator are required for growth in low K+ conditions.
Fig. 2.
Growth of S. aureus Ktr mutants in low K+. Wild-type S. aureus (solid black circles) and mutant strains ΔktrA (open red triangles) and ΔktrBΔktrD (open blue squares) were inoculated from overnight cultures to OD600=0.05 in K-CDM supplemented with KCl to the desired concentration.
A and B. Without supplemented KCl (A) and in 0.1 mM supplemented KCl (B), the ΔktrA and ΔktrBΔktrD strains had a severe growth delay compared to wild-type.
C. In 0.5 mM KCl, the ΔktrA and ΔktrBΔktrD strains had an extended lag phase before entering a growth rate and yield similar to wild-type.
D. With KCl supplemented to 10 mM, all stains grew equally.
A-D. Expression of ktrA from plasmid pCM28 (solid red triangles) fully complemented the ΔktrA mutant growth defects, and expression of either ktrB (solid blue diamond) or ktrD (solid blue squares) from pCM28 fully complemented growth of the ΔktrBΔktrD strain in all conditions tested.
Data represents the mean of three independent experiments, with ± standard deviation.
To exclude the possibility that secondary site mutations were responsible for the phenotypes observed, we tested the ability of complementation plasmids to restore growth under K+-limiting conditions. Expression of ktrA from the plasmid pCM28 restored growth of the ktrA mutant to wild-type levels in all K+ concentrations tested. Additionally, expression of either ktrB or ktrD alone fully restored the growth of the ΔktrBΔktrD strain in all K+ concentrations, further demonstrating the requirement for only one channel protein under these growth conditions. Based on these results, we conclude that KtrA regulates the activity of both KtrB and KtrD in S. aureus.
A role for Kdp in low K+ growth
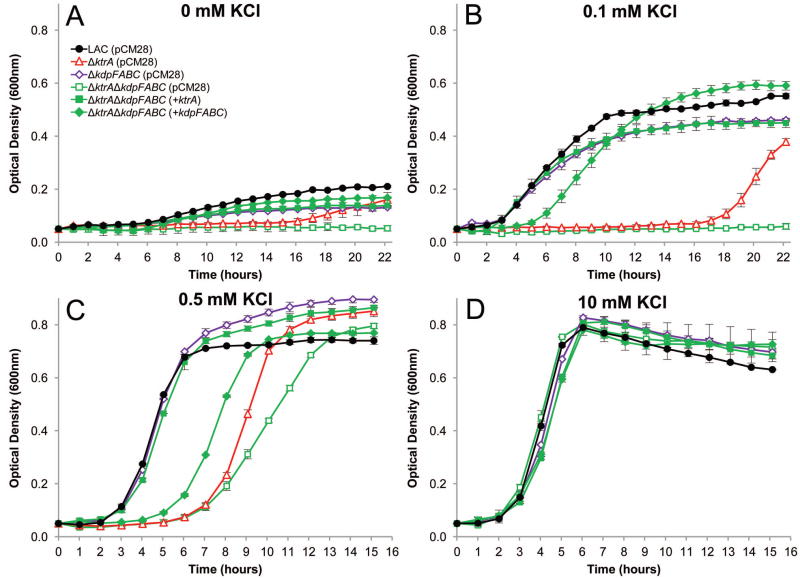
Considering the observation that growth in low K+ resumed in the ΔktrA and ΔktrBΔktrD mutants after extended incubation periods, we hypothesized that expression of the kdpFABC operon is induced under these conditions, thus, providing an alternate mechanism to import K+. To test this hypothesis, we generated kdpFABC mutants, with and without the ktrA mutation, and tested the ability of these strains to grow under low K+ conditions. Consistent with the results of Xue et al. (Xue et al., 2011), disruption of kdpFABC (ΔkdpFABC) had an unremarkable effect on the growth of S. aureus (Fig. 3A-D). However, the presence of both ktrA and kdpFABC mutations (ΔktrAΔkdpFABC) eliminated the growth recovery observed for the ktrA mutant in 0 mM and 0.1 mM KCl (Fig. 3A and B) and reduced the growth rate of the recovery observed in the presence of 0.5 mM KCl (Fig. 3C). In contrast, normal growth was observed in 10 mM KCl (Fig. 3D) suggesting that alternate mechanisms exist to transport K+ under these conditions. As expected, expression of ktrA from a plasmid in the ΔktrAΔkdpFABC strain restored growth in all conditions tested. Interestingly, plasmid-based expression of kdpFABC partially complemented the growth defects of the ΔktrAΔkdpFABC strain in low K+ (Fig. 3A-D), likely due to the multi-copy state of the plasmid and over-expression of kdpFABC. Together, these results reveal a function for Kdp in K+ import, but only in the absence of the Ktr system.
Fig. 3.
Growth of S. aureus Kdp and Ktr mutants in low K+. Wildtype S. aureus (solid black circles) and mutant strains ΔkdpFABC (open purple diamonds) and ΔktrAΔkdpFABC (open green squares) were inoculated from overnight cultures to OD600=0.05 in K-CDM supplemented with KCl to the desired concentration.
A and B. Without supplemented KCl (A) and in 0.1 mM supplemented KCl (B), the ΔktrAΔkdpFABC double mutant was unable to grow, while the ΔkdpFABC single mutant had only a slight growth defect compared to wild-type. The ΔktrA (open red triangles) mutant is shown for reference.
C. In 0.5 mM added KCl, the ΔktrAΔkdpFABC strain had a slower rate of growth than the single ΔktrA mutant.
D. With KCl supplemented to 10 mM, all stains grew similarly.
A-D. Expression of ktrA (solid green squares) from plasmid pCM28 fully complemented the ΔktrAΔkdpFABC double mutant growth defects, and expression of kdpFABC (solid green diamonds) partially complemented growth of the ΔktrAΔkdpFABC strain in all conditions.
Data represents the mean of three independent experiments, with ± standard deviation.
Ktr contributes to osmotic tolerance
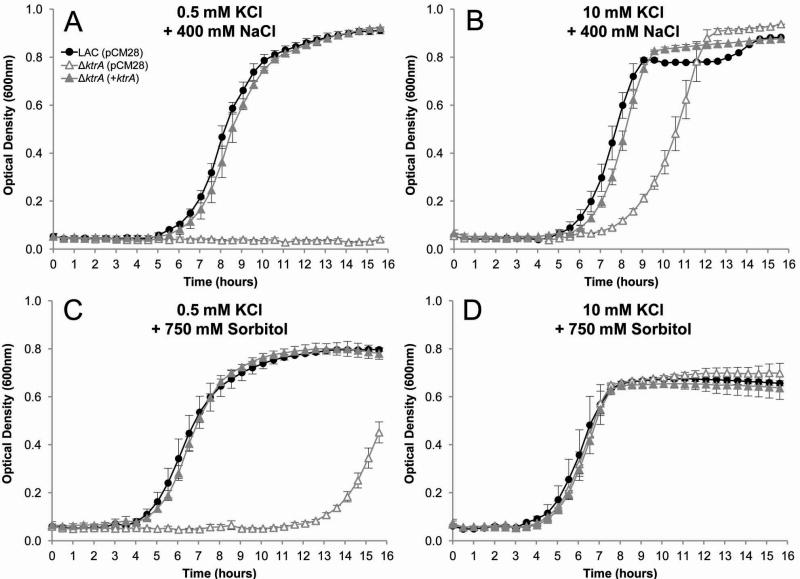
Cellular processes require constant cytoplasmic turgor pressure, necessitating bacteria to cope with changes in external osmolarity for survival. The initial adaptation of bacteria to high osmotic stress is rapid accumulation of K+ (Matsuda et al., 2004, Holtmann et al., 2003, Whatmore et al., 1990), increasing osmotic tolerance. To examine the role of the S. aureus Ktr system in hyperosmotic tolerance, we determined the effects of high concentrations of dissolved solutes on growth of the parental and ΔktrA strains. As shown in figure 4A, growth of the ktrA mutant was completely inhibited when cultured in K-CDM supplemented with 0.5 mM KCl and 400 mM NaCl. Increasing the available K+ by supplementing the growth medium with 10 mM KCl recovered growth of the ktrA mutant (Fig. 4B).
Fig. 4.
Ktr contributes to high osmotic growth. Wild-type (solid circles), ΔktrA (open triangles), and ΔktrA(+ktrA) (solid triangles) strains were inoculated from overnight cultures to OD600=0.05 in K-CDM supplemented with KCl and either NaCl or Sorbitol to the desired concentration.
A and B. Growth in high NaCl. The ΔktrA strain was unable to grow in K-CDM supplemented with 0.5 mM KCl and 400 mM NaCl (A), but recovered growth to near wild-type levels with added 10 mM KCl (B).
C and D. Growth in high sorbitol. As an alternative solute, 750 mM sorbitol also severely delayed the growth of the ΔktrA mutant in 0.5 mM KCl (C), but was fully restored in 10 mM KCl (D).
Expression of ktrA from a pCM28 fully complemented the growth defects of the ΔktrA mutant in all conditions.
Data presented represent the mean of at least two independent experiments, with ± standard deviation.
To ensure that the observed growth defect was due to an osmotic effect of NaCl and not an ionic effect, we examined the growth of our S. aureus strains in high concentrations of sorbitol. Cultured in K-CDM supplemented with 0.5 mM KCl and 750 mM sorbitol, the ΔktrA strain exhibited a severe growth delay compared to the parental and complemented strains (Fig. 4C). As observed with growth in the presence of 400 mM NaCl, supplementing the growth medium with 10 mM KCl restored growth of the ΔktrA strain (Fig. 4D). Similar results were observed using the ΔktrBΔktrD strain and this phenotype was complementable with expression of either ktrB or ktrD from a plasmid (data not shown). Consistent with its role in low K+ growth, disruption of ktrA and kdpFABC extended the lag phase of the ΔktrA strain in 0.5 mM KCl and 750 mM sorbitol (data not shown). Additionally, the parental and ΔkdpFABC strain (data not shown) grew consistently and identically in high osmotic conditions regardless of K+ concentration, indicating that Ktr-mediated K+ uptake is a major regulator of cellular osmotic pressure.
Ktr mutants have increased membrane potential
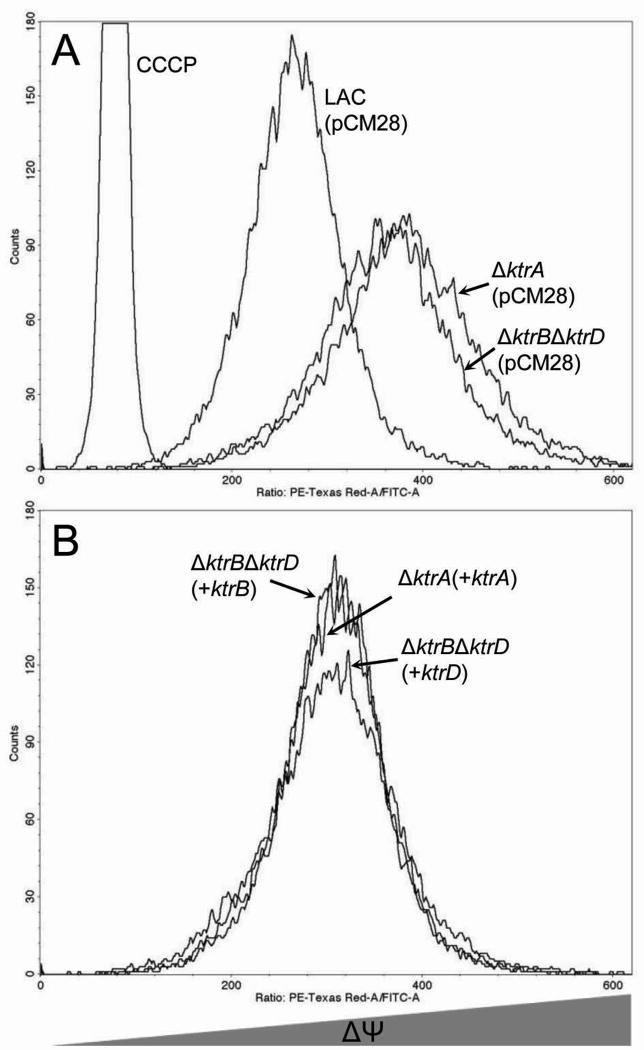
Efficient uptake of K+ is essential for regulation of the electrical potential across the bacterial cytoplasmic membrane (ΔΨ) (Bakker & Mangerich, 1981). As protons are translocated out of the cell, creating a proton motive force, the electro-chemical ion gradient is held constant, primarily as a result of K+ transport. To investigate the role of the Ktr system in regulation of S. aureus ΔΨ, we utilized flow cytometry to assess the fluorescence of cells stained with the carbocyanine dye, diethyloxacarbocyanine (DiOC2(3)) (Novo et al., 2000, Novo et al., 1999). As shown in figure 5A, the ΔktrA and ΔktrBΔktrD strains stained with DiOC2(3), produced an increased red:green fluorescence ratio compared to a similarly stained parental strain, demonstrating that the electrical potential across the plasma membrane of cells lacking a functional Ktr system is higher relative to wild-type cells. This hyperpolarization was restored to wildtype levels by complementation of the ΔktrA strain in trans with ktrA or complementation of ΔktrBΔktrD with either ktrB or ktrD (Fig. 5B). These data demonstrate that K+ uptake mediated by the Ktr system plays an important role in the regulation of ΔΨ in S. aureus.
Fig. 5.
Ktr-mediated regulation of membrane potential. The cyanine dye DiOC2(3) was utilized to determine the role of the Ktr system in regulation of S. aureus resting electrical membrane potential.
A. The ΔktrA and ΔktrBΔktrD mutant strains were hyperpolarized compared to wild-type.
B. Hyperpolarization was decreased to near wild-type levels with pCM28-based expression of ktrA in the ΔktrA mutant and either ktrB or ktrD in the ΔktrBΔktrD strain.
Results are representative of three independent experiments.
Ktr is required for antimicrobial resistance
Mediating cellular transmembrane electrical potential has been proposed as a mechanism of bacterial defense to the effects of cationic antimicrobial peptides (Yeaman & Yount, 2003, Hancock, 1997, Yeaman et al., 1998) and is known to play a role in aminoglycoside resistance in S. aureus (Mates et al., 1982). To determine the role of the Ktr system in antibiotic resistance, we compared the effective concentration of several aminoglycoside antibiotics, cationic antimicrobials, and immune cationic antimicrobial peptides against wild-type LAC and its isogenic ktrA mutant strain.
The ΔktrA strain exhibited markedly lower gentamicin and tobramycin minimum inhibitory concentrations (MIC) than the parental strain in 0.5 mM KCl (Table 2). Supplementation of the medium to 10 mM KCl partially restored the MIC of the ktrA mutant. Expression of ktrA from a plasmid fully restored the MIC of the ΔktrA strain to wild-type levels in all conditions tested. MIC values of 4-, 16-, and 4-fold lower than wild-type were observed when the ktrA mutant was exposed to cationic antimicrobial polymyxin B, epithelial cathelicidin LL-37 and human neutrophil peptide 1 (HNP-1), respectively, in 0.5 mM KCl. We also determined the effects of two ionophores, gramicidin and valinomycin, which form monovalent cation-permeable channels in cell membranes. Gramicidin had a marked effect on the ktrA mutant in 0.5 mM KCl with a MIC less than the limit of detection (<1 μg/mL) compared to the parental strain, which was greater than the highest concentration tested (>64 μg/mL). In 10 mM KCl, gramicidin had a moderate effect on the ktrA mutant strain, with a MIC of 32 μg/mL, while the MIC of the parental strain remained >64 μg/mL. Interestingly, the highly K+-selective ionophore valinomycin had no measurable effect on the parental or ΔktrA strains in low K+ conditions (0.5 mM KCl), but had a substantial inhibitory effect in the presence 10 mM KCl where the mutant exhibited a 2-fold lower MIC. Additionally, we found the ΔktrA strain to have a minor sensitivity to vancomycin (2-fold lower MIC than wildtype), a cationic antibiotic that acts on the cell wall.
Table 2.
Contribution of Ktr to antimicrobial resistance.
| Antibiotic | MIC Values | |||||
|---|---|---|---|---|---|---|
| 0.5 mM KCl | 10 mM KCl | |||||
| LAC (pCM28) | ΔktrA (pCM28) | ΔktrA (+ktrA) | LAC (pCM28) | ΔktrA (pCM28) | ΔktrA (+ktrA) | |
| Gentamicin | 0.5 | <0.0625 | 0.5 | 0.5 | 0.25 | 0.5 |
| Tobramycin | 1 | 0.125 | 1 | 1 | 0.5 | 1 |
| Polymyxin B | 32 | 8 | 32 | 32 | 16 | 32 |
| LL-37 | 256 | 16 | 256 | 512 | 64 | 512 |
| HNP-1 | 6 | 1.5 | 6 | 6 | 3 | 6 |
| Vancomycin | 2 | 1 | 2 | 2 | 1 | 2 |
| Gramicidin | >64 | <1 | >64 | >64 | 32 | >64 |
| Valinomycin | >64 | >64 | >64 | 4 | 2 | 4 |
MIC values reported are the concentration of antibiotic in which no growth was seen following 20 hrs of static incubation at 37°C.
Results are representative of at least two independent experiments.
To examine the effect of pH on the antimicrobial susceptibility of the ΔktrA strain, MIC assays were also performed in acidic, neutral, and alkaline conditions (Table S3). Consistent with its role in growth under these conditions, the gentamicin, polymyxin B, and HNP-1 MICs for the ΔktrA mutant gradually increased as the pH became less alkaline. However, the determined MIC values for the ΔktrA mutant were consistently less than that observed for the wild-type strain, regardless of the pH of the medium.
Ktr contributes to fitness during infection
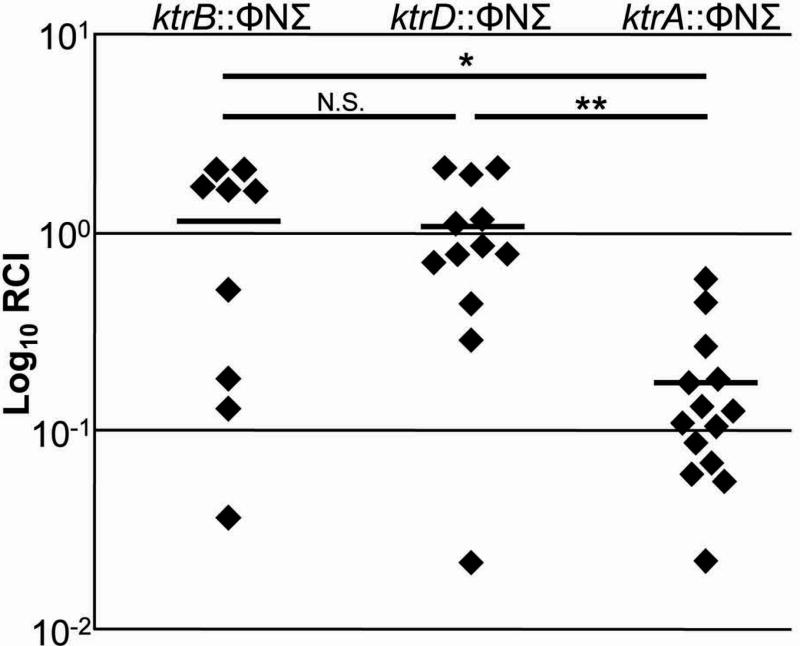
The ability of bacterial pathogens to grow within an animal host depends on their capacity to adapt to and survive within a hostile environment, one that contains limited available nutrients, osmotic stress, and an onslaught of antimicrobial immune components. To determine if the Ktr system contributes to cellular fitness during a S. aureus infection, we employed a competitive index model by infecting mice with a 1:1 mixture of ktrA, ktrB, or ktrD mutant and wild-type S. aureus cells. The ktrA::ΦNΣ, ktrB::ΦNΣ, and ktrD::ΦNΣ mutants from the Nebraska Transposon Mutant Library (Fey et al., 2013) were utilized for their post-infection selection due to the presence of a selectable erythromycin resistance gene within the transposon. After 9 days, bacterial burden within the kidneys was determined and the relative competitive index (RCI) was calculated. As shown in figure 6, the ktrA mutant was significantly outcompeted by JE2 (p=0.0001), demonstrating that the presence of a functional Ktr system confers a fitness advantage to S. aureus during infection. In contrast, the ktrB and ktrD mutants were able to compete effectively with the JE2 strain, indicating that these genes are individually dispensable for bacterial fitness within an infection. The RCI of the ktrA::ΦNΣ mutant was significantly less than that of the ktrB::ΦNΣ and ktrD::ΦNΣ mutants and is consistent with the lack of an observable phenotype in the other functional assays described above.
Fig. 6.
Ktr contributes for fitness in a mouse bacteremia model. C57BL/6 mice were infected with ∼3×106 CFU of ∼50% S. aureus JE2 and ∼50% ktrB::ΦNΣ (n=5 mice), ktrD::ΦNΣ (n=6 mice), or ktrA::ΦNΣ (n=7 mice). After 9 days, both kidneys were individually homogenized and bacterial burden of JE2 and ΦNΣ mutants were determined through viable cell count on TSA and TSA+Erm, respectively. RCI values were calculated by dividing the percentage of the ΦNΣ strain within each kidney by percentage of the ΦNΣ strain within the initial inoculum and plotted as an individual data point. Horizontal lines represents the RCI mean. The probability that significant differences in RCI are noted between ktrB::ΦNΣ and ktrA::ΦNΣ (*p=0.0128), ktrD::ΦNΣ and ktrA::ΦNΣ (**p=0.008), but not ktrB::ΦNΣ and ktrD::ΦNΣ (p=0.9716).
Discussion
Transport mechanisms of the essential mineral nutrient K+ have been well-described in a variety of bacterial species; however the specific mechanisms of its uptake in the Gram-positive pathogen, Staphylococcus aureus, have yet to be revealed. The data generated by this study demonstrate that S. aureus utilizes a unique Ktr K+ transport system in which two separate ion-conducting proteins, KtrB and KtrD, are regulated by a single protein, KtrA. This type of Ktr system has yet to be described and its occurrence appears to be conserved among many staphylococcal species (Table 1). We have shown that KtrA and either KtrB or KtrD are required for normal growth in medium containing less than 10 mM K+, and expression of either ktrB or ktrD from a plasmid fully complemented the growth defect of the ΔktrBΔktrD mutant strain (Fig. 2). Deletion of either ktrB or ktrD alone had no effect on K+-limited growth (Fig. S4). Furthermore, KtrA appears to have very tight regulation over KtrB and KtrD activity as the ΔktrA and ΔktrBΔktrD strains had identical growth characteristics. These results reveal that, while encoding two separate channel proteins is largely conserved within staphylococcal species, only one channel protein is necessary and sufficient for full Ktr function in S. aureus. However, any functional differences between the two K+ channel proteins were indistinguishable as the growth rates of the ktrB and ktrD mutants were identical to the wild-type strain, and the ΔktrBΔktrD phenotypes produced were equally complementable with either ktrB and ktrD, despite the fact that KtrB and KtrD share only 55% overall amino acid identity (data not shown). Moreover, with consideration that KtrA appears to regulate the activity of each of these proteins to a similar extent, it was unexpected that KtrB and KtrD share only 53% identity in the 15 amino acids described as essential for Ktr regulatory protein binding (Hanelt et al., 2010) (data not shown). Thus, additional work will be required to identify functional differences that may distinguish these channel proteins.
Although mutations rendering the S. aureus Ktr system nonfunctional resulted in an extended lag phase when grown in a K+-limited medium (Figs. 2B and C), the mutant strains were ultimately able to grow at the same rate and yield as the parental strain. These results indicate that, as K+-deplete stationary-phase cells are diluted from overnight cultures into fresh culture media, the Ktr system is responsible for the initial uptake of K+. Lacking a functional Ktr system, the cells appear to remain K+-deplete and are unable to begin growth at the same time as the wild-type strain. However, the Ktr mutant strains were eventually able to resume apparently normal growth after an amount of time that was dependent on the K+ concentration within the medium. Based on these results, we propose that the S. aureus Ktr system is a major effector in the initial acquisition of K+, enabling the cells to effectively reach a threshold intracellular K+ concentration that is growth permissive. While we have demonstrated a clear role for the Ktr system in low K+ growth, it is also apparent that S. aureus utilizes a second K+ transport mechanism in the absence of a functional Ktr system. Importantly, repeating the low K+ growth experiments with ΔktrA cells collected after growth recovery under these conditions resulted in a similar growth delay, indicating that the observed growth was not due to a spontaneous mutation that permitted growth in K+-limiting conditions (data not shown). Instead, the delayed growth of the ΔktrA mutant in 0.1 and 0.5 mM KCl appeared to be related to Kdp-ATPase system (Fig. 3B and C), despite having previously been characterized as having a negligible role in K+ uptake in S. aureus (Xue et al., 2011). Growth defects of ΔktrAΔkdpFABC were fully complementable by expression of ktrA in both conditions, confirming the role of the Kdp system in K+ acquisition in the absence of Ktr. Thus, it is clear that the Ktr system is the major K+ uptake system in S. aureus, and we propose that Kdp functions as K+-scavenger, secondarily to Ktr. Interestingly, the KdpDE two-component system has been shown to regulate the expression of the KdpFABC K+ transport system in Escherichia coli (Heermann & Jung, 2010, Voelkner et al., 1993, Polarek et al., 1992). Under K+ limitation or high osmolarity, the histidine kinase, KdpD, phosphorylates the response regulator, KdpE, increasing its affinity for the kdpFABC promoter and triggering transcription of this operon (Laimins et al., 1981, Epstein, 1992). However, KdpE functions to repress the transcription of kdpFABC in S. aureus (Xue et al., 2011), and this is likely due to the ability of the Ktr system to satisfy the majority of the K+ requirements in S. aureus.
K+ uptake in the face of acidic stress is important for compensation of H+ extrusion and resulting alkalinization of the bacterial cytoplasm (Bakker & Mangerich, 1981, Booth, 1985). Thus, it was surprising to find that the S. aureus Ktr system was essential for growth under alkaline stress, conditions in which efflux of K+ and/or Na+ typically occur (Padan et al., 2005, Krulwich et al., 2009), but not at a neutral or acidic pH (Fig. 1). Given that Ktr systems are characteristically dependent on inward Na+ gradients for K+ uptake activity (Matsuda et al., 2004, Tholema et al., 1999, Zulkifli et al., 2010), it is tempting to speculate that the S. aureus Ktr system may function differently due to the requirement for Ktr under alkaline growth but not in neutral or acidic conditions. Further mechanistic studies will be required to elucidate the K+/Na+ transport activity of the S. aureus Ktr system. Interestingly, the Ktr system of Enterococcus hirae has also previously been shown to be important for alkaline growth (Murata et al., 1996, Kawano et al., 2000). The E. hirae KtrB-like channel protein (NtpJ) is encoded as the last gene within the Ntp Na+-ATPase operon, implicating the Ktr system in Na+ cycling. While the ktr genes of S. aureus are not associated with any known Na+ efflux genes, it remains possible that cellular Na+ efflux activity is associated with Ktr activity. Notably, the ΔktrAΔkdpFABC mutant displayed similar growth profiles as the ΔktrA single mutant in 0.5 mM KCl at pH 5.21 and 7.02 (data not shown), indicating that a third, yet uncharacterized K+ transport system is required for low K+ growth in these conditions.
S. aureus is considered a highly osmotolerant organism (Scott, 1953). When cultured in medium of high osmolarity, other bacterial species are known to import K+ to help maintain the turgor pressure necessary for growth (Csonka, 1989). However, only the contribution of glycine betaine and proline have been examined in detail in S. aureus osmoadaptation (Pourkomailian & Booth, 1992, Townsend & Wilkinson, 1992). We observed an inability of the ΔktrA mutant cells to grow in 0.5 mM KCl medium containing 400 mM NaCl (Fig. 4A) suggesting that the Ktr-mediated import of K+ is also important for overcoming the strong osmotic pressure of NaCl in S. aureus. Growth in the presence of 400 mM NaCl could only be partially rescued by supplementation of the medium with 10 mM KCl (Fig. 4B) indicating that the Ktr system is required for osmotolerance in S. aureus at K+ concentrations beyond 10 mM. Since previous studies have demonstrated that Ktr systems in other organisms are energized by K+/Na+ symport (Matsuda et al., 2004, Tholema et al., 1999), we addressed the possibility that the abundant Na+ present in the growth medium was complicating the interpretation of these results. Thus, we performed similar experiments using high concentrations of the non-metabolic compound sorbitol. Supplemented with 0.5 mM K+ and 750 mM sorbitol, a 12.5-hour lag phase was observed before the ΔktrA mutant was able to resume growth (Fig. 4C). Once again, supplementing this medium with 10 mM KCl fully restored growth (Fig. 4D). We conclude from these data that the Ktr system plays an important role in the ability of S. aureus to overcome high osmotic pressures by facilitating the uptake of K+. The ability to adapt to osmotic fluctuations and grow in high osmotic conditions is essential for bacterial pathogens to infect. Within the intestinal lumen, for example, the osmolarity is very high (around 300 mM NaCl) and the blood stream has an osmolarity equivalent to about 150 mM NaCl.
Because of the role of K+ in the maintenance of the trans-membrane electrical potential (ΔΨ) (Bakker & Mangerich, 1981), we also investigated the potential effects of the ktr genes on ΔΨ. As protons are translocated out of bacterial cells, forming a proton motive force, a charge gradient develops across the membrane. Importantly, the import of K+ ions accompanies the export of protons such that the charge differential across the membrane is maintained. Thus, the inability to efficiently import K+ often accompanies alterations in ΔΨ (Bakker & Mangerich, 1981). Consistent with this, the ΔktrA and ΔktrBΔktrD mutations resulted in a hyperpolarized cell membrane (Fig. 5A) compared to the parental and complemented strains (Fig. 5B), indicative of an inability to maintain a homeostatic ion balance. One consequence of an altered ΔΨ is that it could affect antibiotic susceptibility. For example, ΔΨ in S. aureus has been shown to be positively correlated with the uptake of the aminoglycoside antibiotic gentamicin (Mates et al., 1982). Consistent with this was the finding that mutations affecting the Ktr system altered susceptibility to the effects of gentamicin and tobramycin (Table 2). Regulation of ΔΨ is also known to influence resistance to the activity of cationic antimicrobial peptides that target the bacterial lipid bilayer which are a major part of the biological defense arsenal against bacterial pathogens (Peschel & Sahl, 2006) by electrically repelling their entry (Hancock, 1997, Yeaman & Yount, 2003, Yeaman & Yount, 2007, Yount et al., 2006). Our studies demonstrated that the ktrA mutant was more susceptible than wild-type to the poly-cationic antibiotic polymyxin B, human cathelicidin LL-37, and human neutrophil peptide-1 (HNP-1) (Table 2). Likewise, susceptibility to the widely used antibiotic, vancomycin, was found to be effected by the ktrA mutation, although to a lesser extent. These data are consistent with a model proposing that tight regulation of ΔΨ mediated by K+ uptake is important to inhibit the insertion of cationic antimicrobials into the membrane (Parra-Lopez et al., 1994).
An alternative hypothesis is that K+ uptake is required for the reacquisition of K+ lost during antibiotic treatment (Stumpe & Bakker, 1997). The markedly lower MIC of the ΔktrA strain to the cationic ionophore gramicidin, which eliminated ion gradients (Katsu et al., 1986), suggests that the Ktr system may be required to restore intracellular K+ levels. However, treatment with the neutrally-charged ionophore, valinomycin (Pressman, 1976), demonstrates that cells with a non-functional Ktr system exhibit similar susceptibility to this antibiotic as wild-type cells. These results indicate that the decreased MIC of the ktrA mutant to gramicidin is likely due to its cationic nature (Kelkar & Chattopadhyay, 2007) rather than an inability to acquire K+. This is in agreement with previous findings showing S. aureus wild-type and ΔktrA strains had a comparable, two-fold MIC difference to the neutrally-charged ionophore, nigericin (Corrigan et al., 2013).
Based on the results of this study, we propose that the S. aureus Ktr system functions to maintain ion homeostasis by facilitating K+ uptake, thereby counterbalancing proton extrusion and neutralizing intracellular hydroxyl anions. Mediating membrane polarization (inside negative), the Ktr system plays a significant role in resisting the actions of a wide variety of antimicrobial agents including cationic antibiotics and antimicrobial peptides, potentially by preventing their membrane insertion and/or uptake. Considering that membrane energetics may serve as a selective mechanism for immune cationic peptide action, targeting these molecules to the invading bacteria (Yeaman & Yount, 2003, Yount & Yeaman, 2013), tight regulation of ΔΨ likely plays an important role in pathogen survival during infection. Furthermore, it has recently been hypothesized that K+sequestration from a common limited supply may be a critical host defense mechanism in resistance to bacterial infection (Freeman et al., 2013). Recent work has indicated the potential for K+ transport in S. aureus pathogenesis (Xue et al., 2011, Zhao et al., 2010), however, these studies focused on the less-effective Kdp system. The ability of the S. aureus Ktr system to mediate host antimicrobial peptide resistance lead us to propose that Ktr contributes significantly to cellular fitness during an infection. Indeed, interruption of ktrA resulted in a significant out-competition by wild-type in a mouse model of bacteremia (Fig. 6), indicating that the combination of limited K+ availability, osmotic stress, and/or the effects of immune cationic antimicrobials are factors that S. aureus must overcome to survive within a host.
Experimental Procedures
Bacterial strains and plasmids used
Escherichia coli DH5α was used for cloning and was grown at 37°C in LB medium supplemented with ampicillin (100 μg ml-1) or spectinomycin (50 μg ml-1) as needed for selection. Oligonucleotides (Supplementary Table 2) were synthesized by Integrated DNA Technologies (Coralville, IA) and PCR was performed with Mastercycler proS (Eppendorf, Hamburg, Germany) using KOD DNA polymerase (Novagen, Madison, WI). Amplified DNA fragments were confirmed by separation on a 1% agarose gel and extracting the desired band when required. DNA was recovered using the DNA Clean and Concentrator-5 Kit (Zymo Research, Orange, CA). Klenow fragment, DNA ligase, and restriction enzymes were obtained from New England Biolabs (Beverly, MA). Plasmids were purified using the Wizard Plus SV Miniprep DNA purification system (Promega Corporation, Madison, WI), and the nucleotide sequences of the plasmids generated were determined at the High-Throughput DNA Sequencing and Genotyping Core Facility at the University of Nebraska Medical Center (Omaha, NE) and analyzed using Vector NTI (Invitrogen, Carlsbad, CA) to ensure the absence of unintended mutations.
The Staphylococcus aureus strains used in this study were derived from a USA300 strain isolated from a skin and soft tissue infection in a detainee from the Los Angeles County Jail (LAC) (Voyich et al., 2005). Plasmids and oligonucleotides used in the manipulation of these strains (Supplementary Table 1) were generated based on the sequence of the highly related S. aureus strain FPR_3757 (NC_007793.1). Chromosomal changes in S. aureus were engineered by allelic exchange using the temperature-sensitive E. coli-S. aureus shuttle vector, pCL52.2 (Sau et al., 1997) or pJB38 (Bose et al., 2013), containing ∼1000 bp of upstream and downstream DNA from the gene of interest. Primers were designed to add restriction enzyme recognition sites to the 3′ and 5′ ends of the upstream and downstream products and fragments amplified from the chromosome of S. aureus LAC. Plasmids were generated in E. coli and transferred to the highly-transformable, restriction-deficient S. aureus strain RN4220 (Kreiswirth et al., 1983) by electroporation. Transduction into LAC was performed using Φ11 propagated on plasmid-containing RN4220 cells. Allelic exchange was carried out as previously described (Bose et al., 2012). Complementation fragments were cloned into pCM28 and included 500 bp upstream of the gene of interest to contain the native promoter.
Transposon insertion mutants were acquired from the Nebraska Transposon Mutant Library (Fey et al., 2013) and the transposon insertions were transduced into a fresh wild-type (JE2) strain using Φ11 selected by plating on tryptic soy agar (TSA) medium containing erythromycin (5 μg ml-1). Confirmation of the mutations was achieved by PCR amplification of DNA surrounding the gene harboring the transposon and pulse-field gel electrophoresis was used to confirm the genetic background prior to experimentation.
BLAST analysis
The Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov) was used to compare the amino acid sequences of Bacillus subtilis strain 168 KtrA (NP_390987), KtrB (NP_390988), KtrC (NP_389334), and KtrD (NP_389233) to the Staphylococcus sp. strains listed in Table 1. An Expected (E) value <10-10 and Query coverage >90% was required for protein homology consideration. Table S2 lists the locus and accession numbers for each protein listed. Amino acid alignments were performed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Growth conditions
Cells from overnight cultures were washed twice with K-CDM and diluted to OD600 = 0.05 in 200 μL of K-CDM supplemented with 0, 0.1, 0.5, or 10 mM KCl in wells of a 96-well cell culture plate (Corning Incorporated, Corning, NY). Growth was monitored by acquiring OD600 measurements every 30 or 60 minutes with incubation at 37°C and continuous orbital shaking using a TECAN Infinite 200 PRO (Tecan Group Ltd, Männedorf, Switzerland). For osmotolerance assays, cells were cultured in 0.5 and 10 mM KCl as described above except with the addition of either 400 mM NaCl or with 750 mM sorbitol.
Prior to the experiments, S. aureus strains harboring pCM28 and pCM28-derived plasmids were streaked for isolation on TSA medium containing chloramphenicol (10 μg ml-1) then grown overnight in a chemically-defined medium (CDM) as previously described (Hussain et al., 1991) except for the addition of 0.25% glucose and 10 mM KCl, the absence of KH2PO4, and a final pH of ∼8.4. For experiments testing growth in the presence of various K+ levels, KCl was added to the desired concentration by dilution from a 1M KCl stock solution. It should be noted that the K+-deficient medium in which no KH2PO4 or KCl was added (K-CDM) was not completely free of K+ due to trace contamination of commercially available reagents. Compensatory molar amounts of LiCl were added from a 1M stock so that the total ionic strength of the medium contributed from K+ and Li+ together was 10 mM at all times. Na2HPO4 was also eliminated during the making of the CDM and added, unless otherwise stated, to 142 mM (10 g/L). pH adjustments were made with addition of either HCl or Triethanolamine. All reagents were purchased from Sigma Aldrich (St. Louis, MO), EMD Chemical Inc (Gibbstown, NJ), or Becton, Dickinson, and Company (Sparks, MD).
Measurement of membrane potential
The resting membrane potential was determined using the carbocyanine dye 3,3′-diethyloxa-carbocyaninie iodide (DiOC2(3)) (BacLight B34950). Cells from overnight cultures were washed twice with K-CDM and diluted to OD600 = 0.1 in 3 mL of K-CDM plus 0.1 mM KCl in a 14 mL polystyrene culture tube (Becton, Dickinson, and Company, Sparks, MD). Cultures were then grown at 37°C with 250 rpm shaking until an OD600 of 2.0 was reached. 1-mL cell samples were centrifuged and resuspended in 1 mL K+-free phosphate-buffered saline. 0.5 mL of the cell suspension was added to a 14 mL flow cytometry tube and subsequently stained with 30 μM DiOC2(3) for 30 minutes. As a negative control, one sample was treated with 30 μM CCCP to depolarize the cells. 1.2-1.3 × 104 stained bacteria were assayed in a FACSAria flow cytometer (Becton-Dickinson) using a laser emitting at 488 nm and fluorescence was collected in the green (fluorescein) and red (Texas Red) channels. DiOC2(3) exhibits green fluorescence in all bacterial cells, but shifts to red as the dye molecules self-associate at higher cytosolic concentrations caused by a larger ΔΨ. The ratio of red to green fluorescence intensity was used as a size-independent indicator of membrane potential and plotted on a ratiometric histogram.
Determination of antibiotic MIC
Broth microdilution assays were performed in a 96-well plate using serial 2-fold dilutions of antibiotics in K-CDM supplemented with either 0.5 or 10 mM KCl. Susceptibility levels were defined based on the Clinical and Laboratory Standards Institute supplement M07-A9. MIC values reported are the concentration of antibiotic in which no growth was seen following 20 hrs of static incubation at 37°C.
Antibiotics were purchased from Sigma Aldrich (St. Louis, MO), Human Neutrophil Peptite-1 (NHP-1) was purchased from Peptides International (Louisville, KY), and LL-37 was synthesized at the University of Nebraska Medical Center by standard solid phase methods (AAPPTEC Apex 396 synthesizer) on a pre-loaded Ser Wang resin using the Fmoc (9-fluorenylmethoxy-carbonyl) method of orthogonal synthesis with HBTU [2-(1H-benzotriazol-1-yl)-1,1,3,3-tetra-methyluronium hexafluorophospate]-activated esters of the amino acids. LL-37 was characterized by confirmation of molecular mass using Electrospray mass spectrometry; Mcalc =4493, (M+3H)3+ =1498.25, (M+4H)4+ = 1123.92, (M+5H)5+ = 899.31, (M+6H)6+ =749.59, (M+7H)7+ = 642.66.
Mouse bacteremia model
Prior to inoculation, C57BL/6 mice were anesthetized using ketamine and xylazine. 100 μL containing approximately 3 × 106 CFU with 50% S. aureus JE2 and 50% ktrB::ΦNΣ, ktrD::ΦNΣ, or ktrA::ΦNΣ inoculated via retro-orbital injection. On day 9 following inoculation, the animals were sacrificed and the kidneys were excised, homogenized, and subsequently dilution plated on TSA and TSA+Erm (5 μg/mL). Only those kidneys containing greater than 100 CFU/g of tissue were statistically analyzed. Relative competitive index (RCI) values were calculated by dividing the percentage of mutant cells collected from the kidney by the percentage of mutant cells in the original inoculum.
To determine if significant out-competition occurred, pair-wise comparisons were conducted using the Wilcoxon signed-rank test method. Additionally, a two-sided Wilcoxon rank sum test was used to pair-wise compare two RCI values.
Supplementary Material
Acknowledgments
We would like to thank Dr. Sam Sanderson for the synthesis of LL-37 and Kari Nelson for aid in preparation of this manuscript. This work is supported by the National Institutes of Health grants PO1AI83211 (K.W.B.), RO1AI038901 (K.W.B.), and UNMC Graduate Student Assistantship/Fellowship (C.M.G.).
References
- Albright RA, Ibar JL, Kim CU, Gruner SM, Morais-Cabral JH. The RCK domain of the KtrAB K+ transporter: multiple conformations of an octameric ring. Cell. 2006;126:1147–1159. doi: 10.1016/j.cell.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Alkhuder K, Meibom KL, Dubail I, Dupuis M, Charbit A. Identification of trkH, encoding a potassium uptake protein required for Francisella tularensis systemic dissemination in mice. PloS one. 2010;5:e8966. doi: 10.1371/journal.pone.0008966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker EP. Alkali cation transport systems in prokaryotes. CRC Press; Boca Raton: 1993. p. 448. [Google Scholar]
- Bakker EP, Mangerich WE. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. Journal of bacteriology. 1981;147:820–826. doi: 10.1128/jb.147.3.820-826.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth IR. Regulation of cytoplasmic pH in bacteria. Microbiological reviews. 1985;49:359–378. doi: 10.1128/mr.49.4.359-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Fey PD, Bayles KW. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Applied and environmental microbiology. 2013;79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Lehman MK, Fey PD, Bayles KW. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PloS one. 2012;7:e42244. doi: 10.1371/journal.pone.0042244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corratge-Faillie C, Jabnoune M, Zimmermann S, Very AA, Fizames C, Sentenac H. Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family. Cellular and molecular life sciences : CMLS. 2010;67:2511–2532. doi: 10.1007/s00018-010-0317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Grundling A. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiological reviews. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Epstein W. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol Rev. 1986;39:73–78. [Google Scholar]
- Epstein W. Kdp, a bacterial P-type ATPase whose expression and activity are regulated by turgor pressure. Acta physiologica Scandinavica Supplementum. 1992;607:193–199. [PubMed] [Google Scholar]
- Epstein W. The roles and regulation of potassium in bacteria. Progress in nucleic acid research and molecular biology. 2003;75:293–320. doi: 10.1016/s0079-6603(03)75008-9. [DOI] [PubMed] [Google Scholar]
- Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. A genetic resource for rapid and comprehensive phenotype screening of nonessential. Staphylococcus aureus genes mBio. 2013;4:e00537–00512. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman ZN, Dorus S, Waterfield NR. The KdpD/KdpE two-component system: integrating K(+) homeostasis and virulence. PLoS pathogens. 2013;9:e1003201. doi: 10.1371/journal.ppat.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RE. Peptide antibiotics. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- Hanelt I, Lochte S, Sundermann L, Elbers K, Vor der Bruggen M, Bakker EP. Gain of function mutations in membrane region M2C2 of KtrB open a gate controlling K+ transport by the KtrAB system from Vibrio alginolyticus. The Journal of biological chemistry. 2010;285:10318–10327. doi: 10.1074/jbc.M109.089870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heermann R, Jung K. The complexity of the ‘simple’ two-component system KdpD/KdpE in Escherichia coli. FEMS microbiology letters. 2010;304:97–106. doi: 10.1111/j.1574-6968.2010.01906.x. [DOI] [PubMed] [Google Scholar]
- Holtmann G, Bakker EP, Uozumi N, Bremer E. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. Journal of bacteriology. 2003;185:1289–1298. doi: 10.1128/JB.185.4.1289-1298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Hastings JG, White PJ. A chemically defined medium for slime production by coagulase-negative staphylococci. Journal of medical microbiology. 1991;34:143–147. doi: 10.1099/00222615-34-3-143. [DOI] [PubMed] [Google Scholar]
- Kawano M, Abuki R, Igarashi K, Kakinuma Y. Evidence for Na(+) influx via the NtpJ protein of the KtrII K(+) uptake system in Enterococcus hirae. Journal of bacteriology. 2000;182:2507–2512. doi: 10.1128/jb.182.9.2507-2512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar DA, Chattopadhyay A. The gramicidin ion channel: a model membrane protein. Biochimica et biophysica acta. 2007;1768:2011–2025. doi: 10.1016/j.bbamem.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Kitko RD, Wilks JC, Garduque GM, Slonczewski JL. Osmolytes contribute to pH homeostasis of Escherichia coli. PloS one. 2010;5:e10078. doi: 10.1371/journal.pone.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Krulwich TA, Hicks DB, Ito M. Cation/proton antiporter complements of bacteria: why so large and diverse? Molecular microbiology. 2009;74:257–260. doi: 10.1111/j.1365-2958.2009.06842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laermann V, Cudic E, Kipschull K, Zimmann P, Altendorf K. The sensor kinase KdpD of Escherichia coli senses external K(+) Molecular microbiology. 2013;88:1194–1204. doi: 10.1111/mmi.12251. [DOI] [PubMed] [Google Scholar]
- Laimins LA, Rhoads DB, Epstein W. Osmotic control of kdp operon expression in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:464–468. doi: 10.1073/pnas.78.1.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Miller C. Silver as a probe of pore-forming residues in a potassium channel. Science. 1995;268:304–307. doi: 10.1126/science.7716526. [DOI] [PubMed] [Google Scholar]
- Mason KM, Bruggeman ME, Munson RS, Bakaletz LO. The non-typeable Haemophilus influenzae Sap transporter provides a mechanism of antimicrobial peptide resistance and SapD-dependent potassium acquisition. Molecular microbiology. 2006;62:1357–1372. doi: 10.1111/j.1365-2958.2006.05460.x. [DOI] [PubMed] [Google Scholar]
- Mates SM, Eisenberg ES, Mandel LJ, Patel L, Kaback HR, Miller MH. Membrane potential and gentamicin uptake in Staphylococcus aureus. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:6693–6697. doi: 10.1073/pnas.79.21.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Kobayashi H, Katoh H, Ogawa T, Futatsugi L, Nakamura T, Bakker EP, Uozumi N. Na+-dependent K+ uptake Ktr system from the cyanobacterium Synechocystis sp. PCC 6803 and its role in the early phases of cell adaptation to hyperosmotic shock. The Journal of biological chemistry. 2004;279:54952–54962. doi: 10.1074/jbc.M407268200. [DOI] [PubMed] [Google Scholar]
- Murata T, Takase K, Yamato I, Igarashi K, Kakinuma Y. The ntpJ gene in the Enterococcus hirae ntp operon encodes a component of KtrII potassium transport system functionally independent of vacuolar Na+-ATPase. The Journal of biological chemistry. 1996;271:10042–10047. doi: 10.1074/jbc.271.17.10042. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yamamuro N, Stumpe S, Unemoto T, Bakker EP. Cloning of the trkAH gene cluster and characterization of the Trk K(+)-uptake system of Vibrio alginolyticus. Microbiology. 1998;144(Pt 8):2281–2289. doi: 10.1099/00221287-144-8-2281. [DOI] [PubMed] [Google Scholar]
- Novo D, Perlmutter NG, Hunt RH, Shapiro HM. Accurate flow cytometric membrane potential measurement in bacteria using diethyloxacarbocyanine and a ratiometric technique. Cytometry. 1999;35:55–63. doi: 10.1002/(sici)1097-0320(19990101)35:1<55::aid-cyto8>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Novo DJ, Perlmutter NG, Hunt RH, Shapiro HM. Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrobial agents and chemotherapy. 2000;44:827–834. doi: 10.1128/aac.44.4.827-834.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padan E, Bibi E, Ito M, Krulwich TA. Alkaline pH homeostasis in bacteria: new insights. Biochimica et biophysica acta. 2005;1717:67–88. doi: 10.1016/j.bbamem.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Lopez C, Lin R, Aspedon A, Groisman EA. A Salmonella protein that is required for resistance to antimicrobial peptides and transport of potassium. The EMBO journal. 1994;13:3964–3972. doi: 10.1002/j.1460-2075.1994.tb06712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nature reviews Microbiology. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- Polarek JW, Williams G, Epstein W. The products of the kdpDE operon are required for expression of the Kdp ATPase of Escherichia coli. Journal of bacteriology. 1992;174:2145–2151. doi: 10.1128/jb.174.7.2145-2151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourkomailian B, Booth IR. Glycine betaine transport by Staphylococcus aureus: evidence for two transport systems and for their possible roles in osmoregulation. Journal of general microbiology. 1992;138:2515–2518. doi: 10.1099/00221287-138-12-2515. [DOI] [PubMed] [Google Scholar]
- Pressman BC. Biological applications of ionophores. Annual review of biochemistry. 1976;45:501–530. doi: 10.1146/annurev.bi.45.070176.002441. [DOI] [PubMed] [Google Scholar]
- Roosild TP, Castronovo S, Miller S, Li C, Rasmussen T, Bartlett W, Gunasekera B, Choe S, Booth IR. KTN (RCK) domains regulate K+ channels and transporters by controlling the dimer-hinge conformation. Structure. 2009;17:893–903. doi: 10.1016/j.str.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosild TP, Miller S, Booth IR, Choe S. A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch. Cell. 2002;109:781–791. doi: 10.1016/s0092-8674(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Sau S, Sun J, Lee CY. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. Journal of bacteriology. 1997;179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WJ. Water relations of Staphylococcus aureus at 30 degrees C. Australian journal of biological sciences. 1953;6:549–564. [PubMed] [Google Scholar]
- Stingl K, Brandt S, Uhlemann EM, Schmid R, Altendorf K, Zeilinger C, Ecobichon C, Labigne A, Bakker EP, de Reuse H. Channel-mediated potassium uptake in Helicobacter pylori is essential for gastric colonization. The EMBO journal. 2007;26:232–241. doi: 10.1038/sj.emboj.7601471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpe S, Bakker EP. Requirement of a large K+-uptake capacity and of extracytoplasmic protease activity for protamine resistance of Escherichia coli. Archives of microbiology. 1997;167:126–136. [PubMed] [Google Scholar]
- Su J, Gong H, Lai J, Main A, Lu S. The potassium transporter Trk and external potassium modulate Salmonella enterica protein secretion and virulence. Infection and immunity. 2009;77:667–675. doi: 10.1128/IAI.01027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholema N, Bakker EP, Suzuki A, Nakamura T. Change to alanine of one out of four selectivity filter glycines in KtrB causes a two orders of magnitude decrease in the affinities for both K+ and Na+ of the Na+ dependent K+ uptake system KtrAB from Vibrio alginolyticus. FEBS letters. 1999;450:217–220. doi: 10.1016/s0014-5793(99)00504-9. [DOI] [PubMed] [Google Scholar]
- Tholema N, Vor der Bruggen M, Maser P, Nakamura T, Schroeder JI, Kobayashi H, Uozumi N, Bakker EP. All four putative selectivity filter glycine residues in KtrB are essential for high affinity and selective K+ uptake by the KtrAB system from Vibrio alginolyticus. The Journal of biological chemistry. 2005;280:41146–41154. doi: 10.1074/jbc.M507647200. [DOI] [PubMed] [Google Scholar]
- Townsend DE, Wilkinson BJ. Proline transport in Staphylococcus aureus: a high-affinity system and a low-affinity system involved in osmoregulation. Journal of bacteriology. 1992;174:2702–2710. doi: 10.1128/jb.174.8.2702-2710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkner P, Puppe W, Altendorf K. Characterization of the KdpD protein, the sensor kinase of the K(+)-translocating Kdp system of Escherichia coli. European journal of biochemistry/FEBS. 1993;217:1019–1026. doi: 10.1111/j.1432-1033.1993.tb18333.x. [DOI] [PubMed] [Google Scholar]
- Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Said-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. Journal of immunology. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- Walderhaug MO, Polarek JW, Voelkner P, Daniel JM, Hesse JE, Altendorf K, Epstein W. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. Journal of bacteriology. 1992;174:2152–2159. doi: 10.1128/jb.174.7.2152-2159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore AM, Chudek JA, Reed RH. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. Journal of general microbiology. 1990;136:2527–2535. doi: 10.1099/00221287-136-12-2527. [DOI] [PubMed] [Google Scholar]
- Xue T, You Y, Hong D, Sun H, Sun B. The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and Agr signaling to infection programming. Infection and immunity. 2011;79:2154–2167. doi: 10.1128/IAI.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman MR, Bayer AS, Koo SP, Foss W, Sullam PM. Platelet microbicidal proteins and neutrophil defensin disrupt the Staphylococcus aureus cytoplasmic membrane by distinct mechanisms of action. The Journal of clinical investigation. 1998;101:178–187. doi: 10.1172/JCI562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacological reviews. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- Yeaman MR, Yount NY. Unifying themes in host defence effector polypeptides. Nature reviews Microbiology. 2007;5:727–740. doi: 10.1038/nrmicro1744. [DOI] [PubMed] [Google Scholar]
- Yount NY, Bayer AS, Xiong YQ, Yeaman MR. Advances in antimicrobial peptide immunobiology. Biopolymers. 2006;84:435–458. doi: 10.1002/bip.20543. [DOI] [PubMed] [Google Scholar]
- Yount NY, Yeaman MR. Peptide antimicrobials: cell wall as a bacterial target. Annals of the New York Academy of Sciences. 2013;1277:127–138. doi: 10.1111/nyas.12005. [DOI] [PubMed] [Google Scholar]
- Zhao L, Xue T, Shang F, Sun H, Sun B. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infection and immunity. 2010;78:3506–3515. doi: 10.1128/IAI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- Zulkifli L, Akai M, Yoshikawa A, Shimojima M, Ohta H, Guy HR, Uozumi N. The KtrA and KtrE subunits are required for Na+-dependent K+ uptake by KtrB across the plasma membrane in Synechocystis sp. strain PCC 6803. Journal of bacteriology. 2010;192:5063–5070. doi: 10.1128/JB.00569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulkifli L, Uozumi N. Mutation of His-157 in the second pore loop drastically reduces the activity of the Synechocystis Ktr-type transporter. Journal of bacteriology. 2006;188:7985–7987. doi: 10.1128/JB.00886-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.