Abstract
Increased O2•− and NO production is a key mechanism of mitochondrial dysfunction in myocardial ischemia/reperfusion injury. In complex II, oxidative impairment and enhanced tyrosine nitration of the 70 kDa FAD-binding protein occurs in the post-ischemic myocardium, and is thought to be mediated by peroxynitrite (OONO−) in vivo (Chen et al. (2008) J. Biol. Chem. 283 27991–28003). To gain the deeper insights into the redox protein thiols involved in OONO− –mediated oxidative post-translational modifications relevant in myocardial infarction, isolated myocardial complex II was subjected to in vitro protein nitration with OONO−. This resulted in site-specific nitration at the 70 kDa polypeptide and impairment of complex II-derived electron transfer activity. Under reducing conditions, the gel band of the 70 kDa polypeptide was subjected to in-gel trypsin/chymotrypsin digestion and then LC/MS/MS analysis. Nitration of Y56 and Y142 was previously reported. Further analysis revealed that C267, C476, and C537 are involved in OONO− –mediated S-sulfonation. To identify the disulfide formation mediated by OONO−, the nitrated complex II was alkylated with iodoacetamide. In-gel proteolytic digestion and LC/MS/MS analysis were carried out under non-reducing conditions. The MS/MS data were examined with MassMatrix program, indicating that three cysteine pairs, C306-C312, C439-C444, and C288-C575 were involved in OONO− –mediated disulfide formation. Immuno-spin trapping with anti-DMPO antibody and subsequent MS was used to define oxidative modification with protein radical formation. An OONO− –dependent DMPO adduct was detected, and further LC/MS/MS analysis indicated C288 and C655 were involved in DMPO-binding. These results offered a complete profile of OONO− –mediated oxidative modifications that may be relevant in the disease model of myocardial infarction.
Mitochondrial complex II (EC 1.3.5.1. succinate ubiquinone reductase, SQR) is a key membrane complex in the tricarboxylic acid cycle that catalyzes the oxidation of succinate to fumarate in the mitochondrial matrix. Succinate oxidation is coupled to reduction of ubiquinone at the mitochondrial inner membrane as one part of electron transport chain. Complex II mediates electron transfer from succinate to ubiquinone through the prosthetic groups of FAD, [2Fe-2S] (S1), [4Fe-4S] (S2), [3Fe-4S] (S3), and heme b. The enzyme is composed of a soluble succinate dehydrogenase hosting a 70 kDa FAD-binding protein, a 27 kDa iron-sulfur protein, and a membrane-anchoring protein fraction hosting two hydrophobic peptides (CybL/14 kDa and CybS/9 kDa) with heme b binding (1).
In the animal disease model of myocardial ischemia/reperfusion injury, oxidative impairment of the electron transfer activity of complex II is marked in the region of myocardial infarction (2). The injury of complex II is closely related to the mitochondrial dysfunction (loss of FAD-linked oxygen consumption or state 3 respiration) in the post-ischemic myocardium. Further evaluation of redox biochemistry of complex II indicated alternations of oxidative post-translational modification is marked in the post-ischemic myocardium, including deglutathiolation (loss of glutathione binding) and increase of protein tyrosine nitration at the 70 kDa polypeptide of complex II (2, 3).
Myocardial ischemia/reperfusion can provide a stimulus to alter NO metabolism. Enhancement of protein nitration in the myocardium is marked in the post-ischemic heart (4–7). The marked elevation of protein nitration has been hypothesized due to increased NO production and subsequent superoxide radical anion (O2•−) formation during ischemia/reperfusion (5–7). The above hypothesis has been evaluated in the post-ischemic myocardium of eNOS−/−, in which eNOS knock out resulted in the decline of oxygen consumption by mitochondria and reduction of protein nitration after myocardial infarction (6). Therefore, post-ischemic oxygen consumption mediated by eNOS-derived NO is linked to oxidative inactivation of electron transport chain including complex II injury. It is well known that NO traps O2•− to form peroxynitrite (OONO−) at a very fast rate (k ~ 109–1010 M−1s−1), thus lending support that OONO− formation mediates the enhancement of protein nitration of complex II and other proteins in the post-ischemic myocardium.
In the cellular models of cardiac myoblast H9c2 and endothelium, excess NO can stimulate overproduction of O2•− in mitochondria via the FAD-binding site of complex II. OONO− –mediated protein tyrosine nitration of complex II 70 kDa subunit has been reported in the post-hypoxic H9c2 and fully characterized in the isolated enzyme (3). The 70 kDa flavin subunit of complex II contains as many as 18 cysteinyl residues. It is one of the major components to host reactive/regulatory thiols, which are thought to have biological functions of antioxidant defense and redox signaling. It is logical to hypothesize that other important oxidative post-translational modifications involved in the redox thiols of 70 kDa subunit can also be mediated by the OONO− produced during myocardial ischemia and reperfusion. This study was thus undertaken to gain a deeper insight into OONO− –mediated oxidative modifications relevant in the myocardial infarction. In addition to OONO− –mediated protein tyrosine nitration, we have also detected oxidative modifications of specific cysteinyl residues, including S-sulfonation, disulfide bond formation, and protein radical intermediate resulting from in vitro OONO− –induced protein tyrosine nitration of complex II.
EXPERIMENTAL METHODS
Isolation of Cardiomyocytes
Adult ventricular myocytes were isolated from Sprague-Dawley rats (~ 300–350 g) by enzymatic dissociation according to the published procedure by Garg et al (8). Myocytes were placed in 100 mm petri dishes and incubated at 37 °C in a humidified 5% CO2 − 95% air mix prior to hypoxia/reoxygenation treatment.
In Vivo Myocardial Regional I/R Model
The procedure for the in vivo ischemia-reperfusion rat model was performed by the technique reported in previous publication (2). Sprague-Dawley rats (~ 300–350 g) were anesthetized with Nembutal administered intraperitoneally (80–100 mg/kg). The left anterior descending coronary artery (LAD) was then occluded. After 30 min of ischemia, the suture around the coronary artery was untied, allowing reperfusion to occur. At 24 hours post infarction, rats were then sacrificed, and the hearts excised and placed in PBS buffer. The infarct area (or risk region) was delineated by 2, 3, 5-triphenyltetrazolium chloride (TTC) staining.
Preparations of Mitochondrial Complex II
Complex II was isolated from succinate cytochrome c reductase (a supercomplex hosting complex II and complex III) by calcium phosphate-cellulose chromatography under non-reducing conditions according to the published method developed by Yu et al. (9, 10). Succinate was included in all buffers used in the purification procedure. Complex II-containing fractions obtained from the second calcium phosphate-cellulose column were concentrated by 43% ammonium sulfate saturation and centrifugation at 48,000 × g for 20 min (9). The precipitate obtained was dissolved in 50 mM Na/K phosphate, pH 7.8, containing 0.2% sodium cholate and 10% glycerol. The specific activity of purified complex II is ~ 15.2 μmol succinate oxidized or dichlorophenol indophenol (DCPIP) reduction/min/mg protein.
Analytical Methods
Optical spectra were measured on a Shimadzu 2401 UV/VIS recording spectrophotometer. The protein concentration of complex II was determined by the Lowry method using BSA as standard. The enzyme activity of complex II was assayed by measuring Q2-stimulated DCPIP reduction by succinate as described in the literature (9). To measure the electron transfer activity of complex II, an appropriate amount of complex II or myocardial tissue homogenate was added to an assay mixture (1.00 ml) containing 50 mM phosphate buffer, pH 7.4, 0.1 mM EDTA, 75 μM DCPIP, 50 μM Q2, and 20 mM succinate as developed by Hatefi et al. (11). The complex II activity was determined by measuring the decrease in absorbance at 600 nm, and confirmed by inhibition using thenoyl trifluroacetone (TTFA). The specific activity of complex II [nmol DCPIP reduction (or succinate oxidized)/min/mg of complex II] was calculated using a molar extinction coefficient ε600 nm = 21 mM−1cm−1.
Alkylation of Complex II with Iodoacetamide
The sample of nitrated complex II was subjected to alkylation with iodoacetamide (ICH2CONH2) to block free thiols on the surface of protein. The nitrated complex II (0.2 mg/ml) in reaction mixture was incubated with iodoacetamide (2 mM) at room temperature. After 1 h incubation, more iodoacetamide was added to the final concentration of 4 mM and the mixture was incubated at 4 °C for 8 h. The gel band of the 70 kDa subunit in the SDS-PAGE of nitrated complex II was subjected to in-gel digestion with trypsin or chymotrypsin or both, and followed by LC/MS/MS analysis.
In Gel Digestion and Capillary-Liquid Chromatography-Nanospray Tandem Mass Spectrometry (Nano-LC/MS/MS) Analysis
The protein separated by SDS-PAGE gels was digested with sequencing grade trypsin (Promega, Madison, WI), chymotrypsin (Roche, Indianapolis, IN) and a combination of both enzymes following the protocol described in previous studies (12). The peptides were extracted from the polyacrylamide with 50% acetonitrile and 5% formic acid several times and concentrated in a speed vac to ~ 25 μl for LC/MS/MS analysis. Nano LC/MS/MS was performed on a Thermo Finnigan LTQ mass spectrometer equipped with a nanospray source operated in positive ion mode. The LC system was an UltiMate™ Plus HPLC system from LC-Packings, a Dionex Co (Sunnyvale, CA). The sample was first cleaned on the trapping column (LC-Packings A Dionex Co, Sunnyvale, CA), with 50 mM acetic acid before eluting off onto the column (ProteoPep II C18, 5 cm X75 μm, New Objective, Inc. Woburn, MA) connected directly in the nanospray tip. Solvent A was water containing 50mM acetic acid and solvent B was acetonitrile. Peptides were eluted directly off the column into the LTQ system using a gradient of 2–80% B over 30 min with a flow rate of 300 nL/min. The MS/MS data was acquired based on the TopTen™ method-the analysis was programmed for a full scan recorded between 350–2000 Da, and an MS/MS scan to generate product ion spectra in consecutive instrument scans of the ten most abundant peak in the spectrum. The CID fragmentation energy was set to 35%. To exclude multiple MS/MS, dynamic exclusion is enabled with a repeat count of 30 s, exclusion duration of 350 s and a low mass width of 0.5 and high mass width of 1.50 Da.
Sequence information from the MS/MS data was processed using the MASCOT 2.0 active perl script with standard data processing parameters to form a peaklist (mgf file). Database searching was performed against SWISSPROT database using the MASCOT 2.0 (Matrix Science, Boston, MA) for protein identification. The data was also searched by MassMatrix (13), for the identification of post-translational modifications (S-sulfonation, tyrosine nitration, and DMPO adducts) and disulfide bonds linkage on 70 kDa subunit. The mass accuracy of the precursor ions was set to 1.8 Da to accommodate accidental selection of the C13 ion and the fragment mass accuracy was set to 0.8 Da. Possible hits were manually verified.
RESULTS AND DISCUSSION
Peroxynitrite Formation in the Mitochondria of Post-hypoxic Myocytes
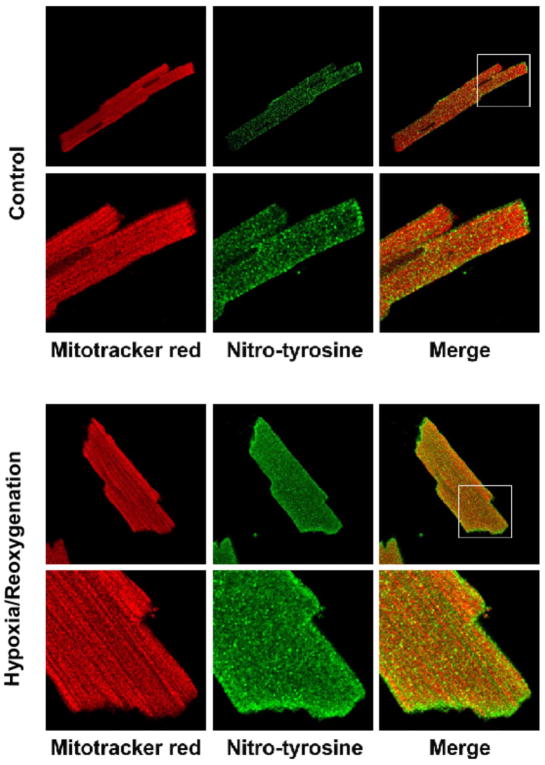
Formation of OONO− in the rat myocardium during ischemia/reperfusion (I/R) injury has been reported (7). The marker of OONO− detected in tissues or cells has been recognized as an indicator that NO and O2•− are overproduced, subsequently forming OONO− under the conditions of disease. In a recent study, Han et al. have reported nitrotyrosine staining in the mitochondria of endothelial cells under the conditions of I/R (14). We have conducted a similar experiment using adult rat ventricular myocytes obtained by enzymatic dissociation according to published procedures (8). Myocytes were subjected to the control or hypoxia (1 h)/reoxygenation (2 h) (H/RO) at 37 °C prior to immunofluorescence staining. To determine the effects of H/RO-mediated protein nitration, nitrotyrosine in myocytes was examined by immunofluorescence confocal microscopy. A basal level of nitrotyrosine staining was observed in myocytes under the control conditions. A small amount of nitrotyrosine was colocalized with the mitochondrial marker Mitotracker (Fig. 1, upper panel). Exposing myocytes to hypoxia followed by reoxygenation resulted in substantially more intense nitrotyrosine staining that showed significant localization of nitrotyrosine in mitochondria (Fig. 1, lower panel). The ratio of signal intensity obtained from anti-3-nitrotyrosine antibody that of Mitotracker Red staining is ~ 0.49 for the control myocyte and ~ 0.74 for post-hypoxic myocyte, indicating overproduction of NO and OONO− formation in mitochondria during I/R injury.
Fig. 1. Hypoxia/Reoxygenation (H/RO) increases 3-nitrotyrosine staining in the mitochondria of myocytes.
Cardiomyocytes were plated on laminin-coated coverslips in 24 well plates. Hypoxic treatment was accomplished by forming a layer of oil on the surface of the glucose-free medium and incubating for 1 h, after which reoxygenation was carried out by incubating in medium with glucose for 2 h. After treatment myocytes were loaded with MitoTracker® Red (250 nM for 15 min), fixed, and stained for nitrotyrosine using anti-nitrotyrosine polyclonal antibody (the secondary antibody was Alexa 488 conjugated). Fluorescence images were acquired with confocal microscopy and were merged in order to determine whether the increase in nitrotyrosine signal colocalized in mitochondria.
Protein Tyrosine Nitration at the 70 kDa Subunit of Complex II in Myocardial Tissue after Myocardial Infarction
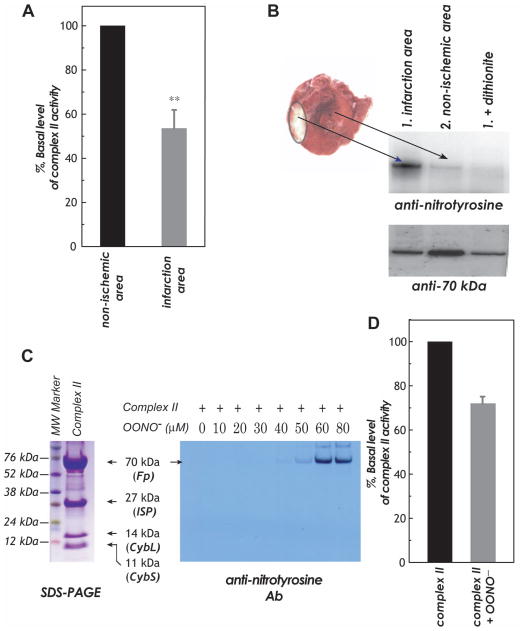
Protein nitration of the complex II 70 kDa FAD-binding polypeptide was evaluated in the disease model of myocardial infarction resulting from ischemia/reperfusion injury. Rats were subjected to 30 min of coronary ligation followed by 24 h reperfusion according to previous published procedure (2, 3). Tissue homogenates were prepared from non-ischemic region and infarct region according to published method (2). The electron transfer activity of complex II in myocardial tissue homogenates was measured by Q2-stimulated DCPIP reduction by succinate as described in the “Experimental Methods.” As indicated in Fig. 2A, complex II activity in the tissue homogenates of infarction area was decreased by 46.5 ± 8.4% (n = 6, p < 0.01) while compared to that of non-ischemic area. The immobilized polyclonal antibodies against 70 kDa was subsequently used to immunoprecipitate the 70 kDa FAD-binding subunit from the tissue homogenate of post-ischemic myocardium, and followed by immunoblotting with a polyclonal antibody against 3-nitrotyrosine. As indicated in the Fig. 2B, we have detected a weak signal of nitrotyrosine on the 70 kDa subunit of complex II from the non-ischemic region (Fig. 2B, upper panel, lane 1). The signal intensity of protein nitration on the 70 kDa subunit of complex II was enhanced on the infarct region of post-ischemic myocardium (upper panel, lane 2). The detected Western blot was abolished by pre-treatment of sample with dithionite due to reduction of 3-nitrotyrosine to 3-aminotyrosine (Fig. 2B, upper panel, lanes 3 and 4). The tissue homogenates were further immunoblotted with anti-70 kDa Ab to quantitate the protein loading of SDS-PAGE (Fig. 2B, lower panel). The ratio of signal intensity obtained from anti-3-nitrotyrosine Ab to that of anti-70 kDa Ab is ~ 1.21 for infarct region, and ~ 0.32 for non-ischemic region.
Fig. 2.
A and B, The rat heart model of in vivo myocardial ischemia/reperfusion and TCC staining of infarct region in the post-ischemic myocardium. Myocardial tissue homogenates from non-ischemic and infarct (risk) regions were subjected to analysis of complex II activity (in A) and immunoprecipitation with a polyclonal antibody against complex II 70 kDa and subsequently subjected to SDS-PAGE and immunoblotted with anti-3-nitrotyrosine (upper panel) and anti-70 kDa (lower panel) antibodies (in B). Note that 100% of the basal level of enzymatic activity (TTFA sensitive) is 60.3 nmole of DCPIP reduction/min/mg of protein in A. C, Right panel, isolated complex II was subjected to in vitro protein tyrosine nitration. Protein (1 μM, based on heme b) was incubated with various concentrations of OONO− (0–80 μM) at 37 °C for 1 h. Excess OONO− was removed by uric acid (1 mM). The OONO− –treated complex II was subjected to SDS-PAGE and then immunoblotting with anti-3-nitrotyrosine antibody. Left panel, SDS-PAGE of OONO− –treated complex II and stained by Coomassie blue. D, the OONO− –treated complex II was subjected to analysis of electron transfer activity. 100% of basal level of purified bovine complex II activity is 15.0 μmol of succinate oxidized/min/mg protein.
Peroxynitrite–mediated Protein Tyrosine Nitration at the 70 kDa Subunit of Isolated Complex II
It is widely accepted that protein nitration is the fingerprint of OONO− overproduction in vivo, and that protein nitration detected in the post-ischemic myocardium is mediated by OONO−. In vitro studies using the isolated complex II have the advantage to provide precise measurements and unequivocal results, which can complement the in vivo studies using post-ischemic myocadium. The isolated complex II (1 μM based on heme b) was subjected to in vitro protein nitration using various concentrations of OONO− (0–80 μM) treatments (3). The resulting OONO− –treated complex II was subjected to SDS-PAGE in the presence of β-mercaptoethanol (β-ME, Fig. 2C), and followed by immunoblotting with anti-3-nitrotyrosine Ab. It was observed the 70 kDa subunit of complex II was involved in site-specific protein nitration as indicated in the Fig. 2C (right panel). The detected Western blot signal was increased in proportion to the dosage of OONO− used, and the optimal signal intensity of protein nitration was observed at the dosage of 60 μM OONO− used. The electron transfer activity of complex II was decreased by 28.1 ± 1.9% resulting from OONO− treatment (Fig. 2D, 60 μM OONO− used, 37 °C for 1 h).
Peroxynitrite-mediated Cysteine S-Sulfonation of Complex II 70 kDa Subunit
To gain a deeper insight into the complex II-derived oxidative modifications mediated by OONO−, the protein band of the 70 kDa subunit of OONO− treated complex II [lane 2 in the SDS-PAGE of Fig. 2C (left panel) and complex II was treated with 60 μM of OONO−] was subjected to in-gel digestion with trypsin/chymotrypsin and analyzed by LC/MS/MS. With this technique, 93.4% of the amino acid sequence was identified in the MS/MS spectra. The coverage of amino acid sequence of the complex II 70 kDa subunit was indicated in the Fig 3A.
Fig. 3.
Amino acid sequence of the precursor of the complex II-70 kDa FAD-binding subunit. The regions labeled with bold represent the amino acid residues identified with LC/MS/MS under the reduced conditions in the presence of β-ME (A) and non-reduced conditions in the absence of β-ME (B). In (A), the cysteinyl residues involved in S-sulfonation are highlighted with gray and they are C267, C476, and C537. The cysteinyl residues involved in protein radical formation are underlined and they are C288 and C655. In (B), the cysteinyl residues involved in disulfide linkage are highlighted with gray and they are C306, C312, C439, C444, C288, and C575. The region labeled with a dotted underline is the N-terminal extension (aa 1–43), which acts as an import sequence and does not exist in the mature protein.
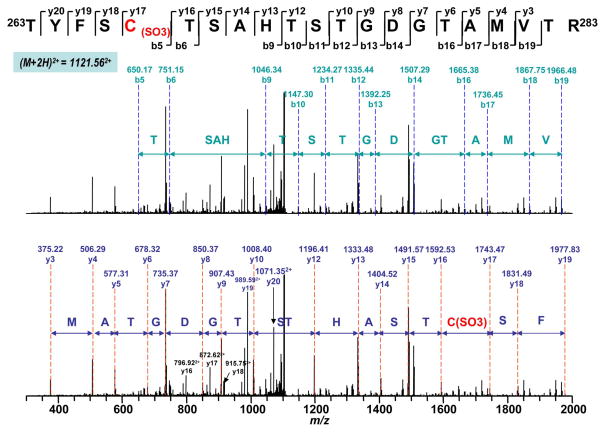
The MS/MS spectra obtained from nitrated complex II was examined for S-sulfonation (conversion of –SH to –SO3H) which occurs during cysteine oxidation. The mass spectra were examined for the mass shift of 48 Da caused by S-sulfonation. Detailed MS/MS analysis provided additional sequential information for the localization of S-sulfonation. For example, a mass shift of 48 Da was seen for doubly charged ion at m/z 1121.562+ (M + H = 2242.12), compared with the unmodified tryptic peptide 263TYFSC267TSAHTSTGDGTAMVTR283 (theoretical M + H = 2193.9539). Analysis of MS/MS spectrum of this peptide also revealed that a mass shift of 48 Da was observed in the fragment ions of b5-b6, b9-b14, b16-b19, and y17-y20 (Fig. 4), indicating the location of S-sulfonation was on C267. Likewise, mass shift of 48 Da was detected in the MS/MS spectra of doubly charged ions of tryptic peptides 467AC(CAM)468ALSIAESC476RPGDK481 and 529VGSVLQEGC537EKISSLYGDLR548 (m/z 813.352+ and m/z 1101.672+, Fig. S1–S2 and Table 1). Detailed MS/MS analysis provided additional information to sequence the two peptides and revealed S-sulfonation occurred on C476 and C537, respectively.
Fig. 4.
MS/MS of doubly protonated molecular ion of S-sulfonated peptide (263TYFSC267(SO3) TSAHTSTGDGTAMVTR283) (where Cys267 was sulfonated) of the 70 kDa subunit from peroxynitrite-treated complex II. The sequence-specific ions are labeled as y and b ions on the spectrum. Note that the same spectrum is shown in upper and lower panels.
Table 1.
Summary of the Peptide Sequence and Corresponding Oxidative Post-translational Modification (OPTM) Obtained from MS/MS Analysis
| Amnio Acid Residue in the 70 kDa | Theoretical m/z | Observed m/z | Peptide Sequence and OPTM | Remarks |
|---|---|---|---|---|
| Y56 | 982.15273+ | 982.903+ | 48VSDAISAQY56(NO2)PVVDHEFDAVVVGAGGAGLR76 | Reference 3 |
| Y142 | 1286.90523+ | 1286.763+ | 130GSDWLGDQDAIHYMTEQAPASVVELENY142(NO2)GMPFSR163 | Reference 3 |
| C267 | 1121.47302+ | 1121.562+ | 263TYFSC267(SO3)TSAHTSTGDGTAMVTR283 | |
| C476 | 813.36652+ | 813.352+ | 467AC(CAM)ALSIAESC476(SO3)RPGDK481 | O2•−–mediated S-sulfonation |
| C537 | 1101.04142+ | 1101.672+ | 529VGSVLQEGC537(SO3)EKISSLYGDLR548 | Glutathionylation (Reference 2) |
| C306/C312 | 802.38202+ | 802.422+ | 303GAGC306(SS)LITEG C312(SS)RGEGGIL319 | |
| C439/C444 | 941.10173+ | 941.433+ | 425HVNGQDQVVPGLYAC439(SS)GEAAC444(SS)ASVHGANR452 | |
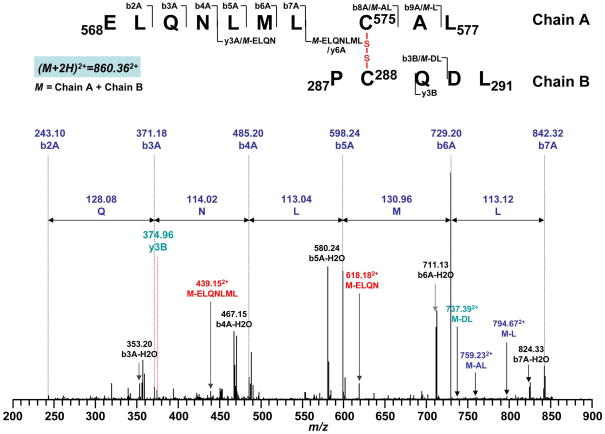
| C288/C575 | 860.40942+ | 860.362+ | 287PC288(SS)QDL291/568ELQNLMLC575(SS)AL577 | |
| C288 | 789.38712+ | 789.852+ | 284 AGLPC288(DMPO)QDL EFVQF296 | |
| C655 | 856.43782+ | 856.482+ | 649 TLN ETDC655(DMPO)ATVPPA IR663 | Glutathionylation (Reference 2) |
Based on the x-ray structure of mammalian complex II (pdb 1ZOY) (15), C267, C476, and C537 are surface-exposed and therefore are possibly involved in OONO− –mediated S-sulfonation. C267 (C224 in mature protein) is located in the hydrophilic pocket of β-barrel subdomain of large FAD binding domain. C476 (C433 in mature protein) is situated at the terminus of α-helix in the FAD-binding domain. C537 (C494 in mature protein) is located on the surface of three-helix bundle from the helical domain. All of them are surface-exposed (Fig. S3A), thus logically susceptible to OONO− –mediated oxidation. Furthermore, all these three cysteine residues are conserved in mammalian enzyme, but not conserved in bacterial enzyme. Therefore, we suggest that these reactive cysteines of mammalian complex II play the regulatory function in response to oxidant stress under the pathophysiological conditions, forming oxidized cysteines.
C267 is located in the part of FAD-binding domain (residues 10-273 in mature protein). The x-ray structure also reveals that C267 is near the riboflavin moiety of FAD (9.0 Å). Sulfonation of C267 likely induced a functional impact on the FAD, which subsequently impaired the electron transfer activity of complex II and mitochondrial function in vivo. Sulfonation of C476 and C537 was not likely exerting a significant impact on the electron transfer activity of complex II due to a much longer distance (23 Å and 28 Å) away from the FAD moiety.
Previous study indicated that superoxide generation is mediated by complex II in the presence of succinate (2). Oxidative attack of superoxide induced protein thiyl radical formation was detected by immuno-spin trapping (2). However, cysteine oxidation with S-sulfonation at C476 was also detected by LC/MS/MS (m/z 813.422+, spectrum not shown) under the conditions of superoxide generation by complex II. Therefore, C476 is the unique thiol susceptible to S-sulfonation induced by the attack of oxygen free radical(s).
C537 has been reported to be the secondary site (C90 is the primary site) involved in S-glutathionylation induced by GSH/GSSG in vitro (2). Loss of glutathione-binding or deglutathionylation has been detected at the 70 kDa subunit of complex II under the conditions of myocardial ischemia and reperfusion (2). Presumably, highly reductive conditions of ischemia triggers deglutathionylation of complex II. The detection of S-sulfonation at the C537 suggests the possibility that S-sulfonation follows deglutathionylation at the same cysteine residue of the complex II 70 kDa subunit under the conditions of ischemia (eq. 1) and reperfusion (eq. 2).
| (eq. 1) |
| (eq. 2) |
Peroxynitrite-mediated Cysteine Disulfide Formation of Complex II 70 kDa FAD-binding Subunit
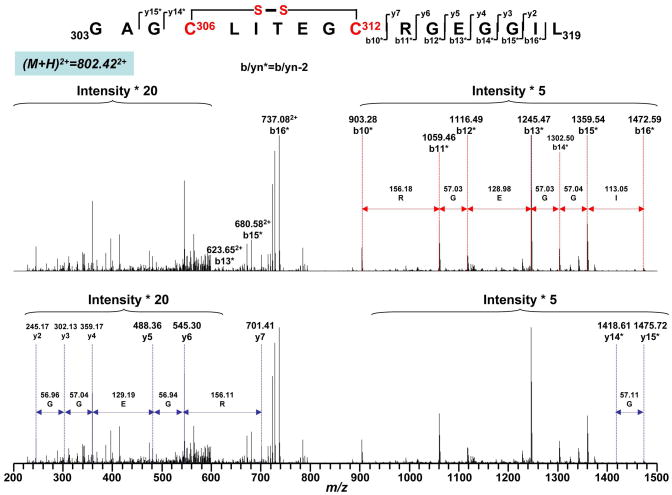
Oxidative post-translational modification with disulfide formation in the complex II 70 kDa subunit can occur under conditions of oxidative stress induced by OONO−. To test this hypothesis, iodoacetamide was added to the nitrated complex II to block free cysteines through carbamidomethylation (addition of –CH2CONH2) reaction and thus prevented the random disulfide bond formation. The alkylated and nitrated complex II was then subjected to SDS-PAGE under non-reducing conditions in the absence of β-ME. The protein band corresponding to 70 kDa subunit was subjected to in-gel proteolytic digestion and nano-LC/MS/MS analysis. 86.6% of the amino acid sequence was identified in the MS/MS spectra of the tryptic/chymotryptic peptides of the complex II 70 kDa subunit (Fig. 3B). The MS/MS data were examined by MassMatrix program (13) to identify disulfide bonds formed in this subunit. Possible hits were manually checked and three cysteine pairs, C306-C312 (m/z 802.652+, Fig. 5), C439-C444 (m/z 941.433+, Fig. S4) and C288-C575 (m/z 860.362+, Fig. 6), were verified as OONO− –mediated disulfide formation (as summarized in the Table 1). Disulfide bond formation in protein can be classified as inter-chain (disulfide bonds formed between two separate peptides) and intra-chain (disulfide bond formed within the same peptide) linkage. Fragmentation of intra-chain linkage generates b and y ions before disulfide bond linkage and corresponding y-2 and b-2 ions after disulfide bond linkage. For example, as shown in the Fig. 5, the doubly charged peptide at m/z 802.422+ was identified as peptide 303GAGC306LITEGC312RGEGGIL319 with disulfide bond linked between C306 and C312. Observed M+H (1603.83) was 2Da less then the theoretical M + H (1605.7723) of the peptide without disulfide bond linkage. Fragmentation ions observed in MS/MS spectrum were y2-y7 and y14-2 (y14*), y15-2 (y15*) and b10-2 (b10*) to b16-2 (b16*) where the loss of 2Da was caused by the formation of disulfide bond. These observations, especially the characteristic y-2 and b-2 fragmentation ions, confirmed the linkage between C306 and C312. Likewise, fragmentation of intra-chain linkage to generate y-2 and b-2 ions was also detected at the triply charged peptide with an m/z 941.433+, confirming the linkage between C439 and C444 (Fig. S 4).
Fig. 5.
Disulfide bond linkage between C306 and C312 as determined by MS/MS spectrum of tryptic/chymotryptic digests of the 70 kDa subunit of peroxynitrite-treated complex II. The sequence-specific ions are labeled as yn (n=2–7), yn* (yn-2, n=14–15), and bn* (bn-2, n=10–16) on the spectrum. Note that the same spectrum is shown in upper and lower panels.
Fig. 6.
Disulfide bond linkage between C288 and C575 as determined by MS/MS spectrum of tryptic/chymotryptic digests of the 70 kDa subunit of peroxynitrite-treated complex II. Chain A and Chain B represent peptides containing aa 568–577 and aa 287–291, respectively. A disulfide bond linking Chain A with Chain B is indicated in red. The sequence-specific ions are labeled as bnA (n = 2–9) for Chain A, y3B and b3B for Chain B on the spectrum. M indicates Chain A plus Chain B.
When two cysteine residues were linked through inter-chain linkage, each peptide chain may undergo fragmentation independently. Therefore, when one chain is fragmentized to create product ions, the other chain can be considered as a modification to cysteine residue on the first chain. As shown in the Fig. 6, doubly charge peak at m/z 860.362+ was identified as two cysteine-containing peptides 568ELANLMLC575AL577 and 287PC288QDL291 linked through C575 and C288. Fragmentation ions b2A-b7A from chain A and y3B from chain B were observed. Additional sequential information including M-L, M-AL, M-ELQN, M-ELQNLML (M = chain A + chain B) were assigned as b9A, b8A, y3A and y6A if considering chain B as a modification group to C575 while M-DL was assigned as b3B if considering chain A as a modification group to C288. Further experiments showed these disulfide bonds containing peptide ions subsequently disappeared under the reducing conditions in the presence of β-ME. In addition, these ions were not observed in the control experiment in which complex II was subjected to iodoacetamide alkylation without OONO− treatment. LC/MS/MS analysis revealed that C306, C312, C439, C444, C288 and C575 were carbamidomethylated as indicated in the Table 2. Therefore, these three disulfide bond linkages were formed specifically during the nitration of complex II.
Table 2.
Detected Fragments Containing Carbamoylmethylated/Alkylated (Addition of –CH2CONH2) Cysteines in the 70 kDa Polypeptide of Complex II Treated with Iodoacetamide.
| Theoretical m/z | Detected m/z | Enzyme | Fragment | Cys Detected |
|---|---|---|---|---|
| 770.40102+ | 770.532+ | trypsin | 234GVIALCIEDGSIHR247 | C239 |
| 513.93643+ | 513.843+ | trypsin | 234GVIALCIEDGSIHR247 | C239 |
| 1122.86173+ | 1122.913+ | trypsin | 284AGLPCQDLEFVQFHPTGIYGAGCLITEGCR 313 | C288, 306, 312 |
| 1184.92273+ | 1185.583+ | trypsin | 556GMVWNTDLVETLELQNLMLCALQTIYGAEAR586 | C575 |
| 860.41132+ | 860.202+ | chymotrypsin | 303GAGCLITEGCRGEGGIL319 | C306, 312 |
| 884.42703+ | 884.643+ | chymotrypsin | 303GAGCLITEGCRGEGGILINSQGERF327 | C306, 312 |
| 556.77762+ | 556.882+ | chymotrypsin | 575MICALQTIY581 | C575 |
| 770.40103+ | 770.693+ | Tryp-chymo | 234GVIALCIEDGSIHR247 | C239 |
| 543.75352+ | 543.742+ | Tryp-chymo | 239CIEDGSIHR247 | C239 |
| 762.36362+ | 762.362+ | Tryp-chymo | 288 AGLPCQDLEFVQF296 | C288 |
| 597.2737+ | 597.34+ | Tryp-chymo | 303GAGCLITEGCR313 | C306, 312 |
| 860.41132+ | 860.462+ | Tryp-chymo | 303GAGCLITEGCRGEGGIL319 | C306, 312 |
| 510.88943+ | 511.173+ | Tryp-chymo | 438ACGEAACASVHGANR452 | C439, 444 |
| 765.83042+ | 765.822+ | Tryp-chymo | 438ACGEAACASVHGANR452 | C439, 444 |
| 868.42331+ | 868.441+ | Tryp-chymo | 575CALQTIY581 | C575 |
Based on the x-ray structure (pdb 1ZOY), the distances of C306-C312, C439-C444 and C288-C575, are 13.4Å, 11.7Å and 6.8Å respectively in the native complex II. The cysteinyl pair of C288-C575 is within the reasonable distance (6.4Å (16)) required for disulfide formation. Therefore, it is possible that oxidant stress of complex II by OONO− induced some conformational changes to facilitate the formation of disulfide pairs C306-C312 and C439-C444. These conformational changes mediated by OONO− likely impaired the electron transfer activity of complex II, leading to mitochondrial dysfunction.
Peroxynitrite-mediated Protein Radical Formation at the 70 kDa Subunit of Complex II
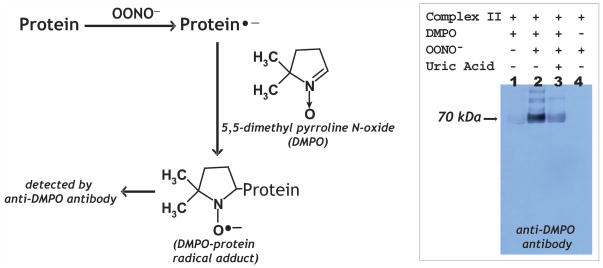
It has been reported that O2•− production by complex II induced self-inactivation through the mechanism in which O2•− may induce oxidative attack on the protein matrix of complex II, forming the protein radical (5). Similar mechanism of protein radical formation resulted from O2•− attack was observed in the flavoprotein subcomplex of complex I (17). OONO− –mediated one-electron oxidation of albumin in human plasma, forming a protein-derived thiyl radical, has been reported and implicated to be a cytotoxic mechanism of the OONO− (18). It is hypothesized that oxidative modification with protein radical formation can be induced with OONO− (Fig. 7, scheme). Immuno-spin trapping with anti-DMPO (DMPO, 5, 5-dimethyl pyrroline N-oxide) polyclonal antibody was used to define complex II-derived protein radical formation as described previously (17, 19, 20). Isolated complex II (1 μM based on heme b) was incubated with a nitrone spin trap, DMPO (100 mM), in PBS and the reaction was initiated by the addition of 60 μM OONO− at 37 °C. After 1 h incubation, the aliquots were subjected to SDS-PAGE and Western blot using an anti-DMPO antibody. The immobilized nitrone adduct of complex II was detected by immunoblotting (Fig. 7, right panel, lane 2) at the 70 kDa subunit of complex II, indicating the formation of complex II-derived protein radical(s). The detected Western blot signal was entirely dependent on OONO− (Fig. 7, right panel, lane 1) and DMPO (Fig. 7, right panel, lane 4). The detected signal of DMPO adduct could be inhibited by OONO− scavenger, uric acid (1 mM, Fig. 7, right panel, lane 3), and therefore confirming that the protein radical formation resulting from oxidative attack by OONO−.
Fig. 7. Left panel.
Schematic delineation of immuno-spin trapping of OONO−–induced protein radical with anti-DMPO polyclonal antibody. Right panel: Detection of the DMPO adduct of the complex II-derived protein radicals by Western blot using anti-DMPO nitrone adduct polyclonal antibody (lane 3). The reaction mixture contained complex II (dithiothretol treated, 1 μM heme b) and DMPO (100 mM) in PBS. OONO− (60 μM) was added to initiate the reaction. The reaction was allowed to incubate for 1 h at 37 °C, terminated by addition of uric acid (0.2 mM) and sample buffer containing 0.4% SDS and 1% β-ME, and then heated at 70 °C for 5 min. Aliquotes of 30 pmol of protein were subjected to SDS-PAGE and Western blot using anti-DMPO polyclonal antibody. Lane 1, OONO− was removed from the complete system. Lane 3, uric acid (0.2 mM) was pre-incubated with complex II and DMPO before the reaction was initiated by OONO−. Lane 4, DMPO was omitted from the complete system.
Involvement of Cys288 and Cys655 of the 70 kDa Subunit in the DMPO-binding Sites Determined by Mass Spectrometry
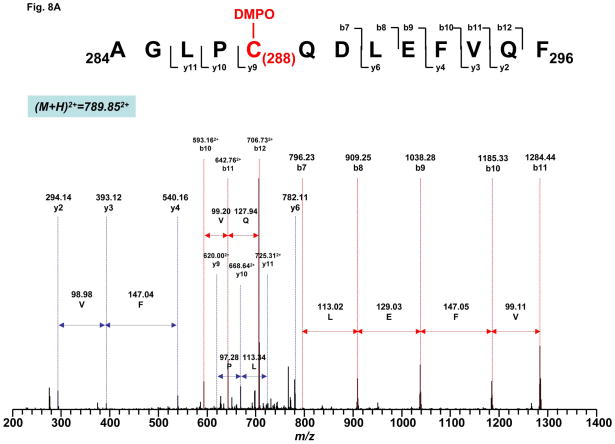
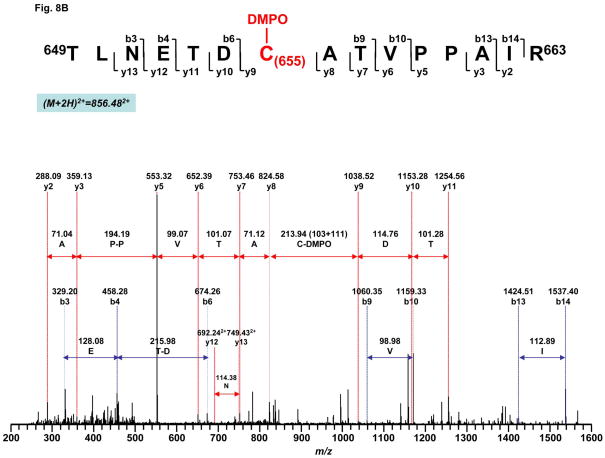
To further provide direct evidence for the molecular mechanism of the complex II-derived radical induced by OONO−, it is imperative to determine the location of DMPO binding. The DMPO nitrone adduct of complex II was subjected to SDS-PAGE under reducing conditions. The protein band at 70 kDa was cut out and subjected to in-gel digestion with trypsin and chymotrypsin, respectively, followed by nano-LC/MS/MS analysis. The resulting mass spectra contain ions that account for over 87% of amino acid sequence of 70 kDa polypeptide.
The binding of DMPO to cysteine residues causes a mass shift of 111 Da to the native intact peptide. Thus, the proteolytic peptides were investigated for the addition of 111 Da to their original intact sequence. This mass difference was observed for one chymotryptic peptide (284AGLPC288QDLEFVQF296, observed m/z 789.852+ v.s. theoretical m/z 733.85292+) and one tryptic peptide (649TLNETDC655ATVPPAIR663, observed m/z 856.482+ v.s. theoretical m/z 800.90362+). Further observation of fragmentation ions y9-y11 and b7 with mass shift of 111Da in the tandem mass spectrum of peptide 284AGLPC288QDLEFVQF296 (Fig. 8A) suggested that one DMPO molecule is covalently bound to C288 in this peptide. Similarly, observation of fragmentation ions y9-y13, b9-b10 and b13-b14 in the MS/MS spectrum of 649TLNETDC655ATVPPAIR663 confirmed that C655 was modified by the addition of DMPO (Fig. 8B).
Fig. 8. MS/MS spectra of the doubly protonated molecular ions of the DMPO-binding peptides.
A, 284AGLPC288QDLEFVQF296. B, 649TLNETDC655ATVPPAIR663. The sequence-specific ions are labeled as y and b ions on the spectra. The amino acid residues involved in DMPO binding are identified to be C288 and C655.
C288 is located in the large FAD-binding domain and the residue is surface exposed (Fig. S3B). C288 (C245 in mature protein) is also involved in the OONO− –mediated disulfide bond formation with C575 (C532 in mature protein) (Fig. 6). This distance between C288 and C575 is 6.8 Å. These results suggest that the protein radical intermediate at C288 can participate in the mechanism to facilitate OONO− –mediated disulfide formation between C288 and C575 (eqs. 3 and 4). Protein radical at C288 likely plays the catalytic role in the formation of disulfide bond with C575, which may indirectly contribute to OONO− –mediated complex II impairment.
| (eq. 3) |
| (eq. 4) |
C655 is located in the C-terminal domain and highly surface-exposed (Fig. S3B) based on x-ray structure. It is not likely that protein radical formation at C655 significantly affected on the moiety of FAD due to a longer distance (24 Å). However, protein thiyl radical is highly reactive species capable of addition even across carbon-carbon double bonds and possibly capable of initiating lipid peroxidation (21, 22), leading to mitochondrial dysfunction. C655 has been identified to be the other secondary site (C90 is the primary site) involved in S-glutathionylation induced by GSH/GSSG in vitro (2). C655 is also associated with the protein radical formation resulted from oxidative attack of O2•− (2). These data provide evidence of possible involvement of protein-derived thiyl radical in the reactive glutathionylating species that may contribute to S-glutathionylation at C655 in the presence of GSH (eq. 5).
| (eq. 5) |
CONCLUSIONS
Mitochondrial thiols are composed of protein thiols and GSH pool. The proteins of the mitochondrial electron transport chain are rich in protein thiols. Therefore, the protein thiols of ETC appear to play a critical role in controlling the redox state of mitochondria. Mitochondrial complex II is the major component of the ETC and hosts both structural thiols involved in the ligands of iron sulfur clusters and the reactive/or regulatory thiols that are thought to have biological functions of antioxidant and redox signaling. For example, the physiological role of complex II-derived thiols has been related to inhibiting respiration by nitrosative stress (23) and redox modification with S-glutathionylation during myocardial ischemia and reperfusion (2).
Despite of that in vivo relevance remains investigated, our data obtained from in vitro model of current study should provide deeper insights into the major mechanism of OONO− –mediated oxidative modifications. The current studies revealed C267, C476, C537, C306, C312, C439, C444, C288, C575, C655 (out of 18 cysteinyl residues available on the 70 kDa subunit) to be involved in oxidative modifications by OONO−. The residue of C288 is likely the primary reactive thiol in response to OONO− since C288 participates to the formation of protein radical and disulfide linkage (Table 1). OONO− –mediated oxidation of C537 (S-sulfonation) and C655 (protein thiyl radical) is related to the redox event of complex II-derived S-glutathionylation. It has been documented that S-glutathionylation of complex II 70 kDa subunit preserves enzymatic inactivation by OONO− (3). Therefore, S-glutathionylation of C537 and C655 should protect the residues from S-sulfonation and protein thiyl radical formation induced by OONO−. In contrast, deglutathionylation under highly reductive conditions (ischemia) facilitates the sulfonation and protein radical formation under oxidative conditions (reperfusion) (2). Disulfide formation of C306/C312 and C439/C444 should result from OONO− –induced conformational change in 70 kDa protein.
The milieu of the mitochondrial matrix is anoxic in the presence of the GSH/GSSG pool under normal physiological conditions (24). Analysis of redox compartmentation indicates that the relative redox states from most reductive to most oxidative are as follows: mitochondria > nuclei > endoplamic reticulum > extracellular space (24). Thus, it is expected that low oxygen tension in the mitochondrial environment should facilitate the free thiol state for most cysteines of complex II 70 kDa; mitochondrial thiols are the targets of oxidants such as OONO −. They are vulnerable to oxidation such as S-sulfonation, protein-thiyl radical, and disulfide formation. Recognition of this molecular event is valuable in understanding the fundamental basis of disease pathogenesis of myocardial infarction.
Acknowledgments
The authors thank Dr. Bradley E. Sturgeon (Department of Chemistry, Monmouth University, Monmouth, IL) for critical review of this manuscript.
Abbreviations
- Complex II
succinate ubiquinone reductase or SQR
- O2•−
superoxide anion radical
- OONO−
peroxynitrite
- DTT
dithiothretol
- ETC
electron transport chain
- DCPIP
dichlorophenol indophenol
- SMP
submitochondrial particles
- SDS-PAGE
SDS polyacrylamide gel electrophoresis
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- β-ME
beta mercaptoethanol
- H/RO
hypoxia/reoxygenation
- I/R
ischemia and reperfusion
Footnotes
This work was supported by National Institutes of Health Grant HL83237 (YRC).
SUPPORTING INFORMATION AVAILABLE
LC-MS/MS evidence of peroxnitrite-mediated S-sulfonation at the residues of C476 and C537 (Figs. S1 & S2) and intra-chain disulfide formation between C439 and C444 (Fig. S4). X-ray structure of mammalian complex II (1ZOY) showing the surface-exposed nature of specific cysteinyl residues (C267, C476, C537, C288, and C655) susceptible to oxidative modifications by peroxynitrite (Fig. S3). This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Lemos RS, Fernandes AS, Pereira MM, Gomes CM, Teixeira M. Quinol:fumarate oxidoreductases and succinate:quinone oxidoreductases: phylogenetic relationships, metal centres and membrane attachment. Biochim Biophys Acta. 2002;1553:158–170. doi: 10.1016/s0005-2728(01)00239-0. [DOI] [PubMed] [Google Scholar]
- 2.Chen YR, Chen CL, Pfeiffer DR, Zweier JL. Mitochondrial Complex II in the Post-ischemic Heart: OXIDATIVE INJURY AND THE ROLE OF PROTEIN S-GLUTATHIONYLATION. J Biol Chem. 2007;282:32640–32654. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 3.Chen CL, Chen J, Rawale S, Varadharaj S, Kaumaya PP, Zweier JL, Chen YR. Protein tyrosine nitration of the flavin subunit is associated with oxidative modification of mitochondrial complex II in the post-ischemic myocardium. J Biol Chem. 2008;283:27991–28003. doi: 10.1074/jbc.M802691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu B, Tewari AK, Zhang L, Green-Church KB, Zweier JL, Chen YR, He G. Proteomic analysis of protein tyrosine nitration after ischemia reperfusion injury: Mitochondria as the major target. Biochim Biophys Acta. 2009;1794:476–485. doi: 10.1016/j.bbapap.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao X, Chen YR, He G, Zhang A, Druhan LJ, Strauch AR, Zweier JL. Endothelial nitric oxide synthase (NOS3) knockout decreases NOS2 induction, limiting hyperoxygenation and conferring protection in the postischemic heart. Am J Physiol Heart Circ Physiol. 2007;292:H1541–1550. doi: 10.1152/ajpheart.00264.2006. [DOI] [PubMed] [Google Scholar]
- 6.Zhao X, He G, Chen YR, Pandian RP, Kuppusamy P, Zweier JL. Endothelium-derived nitric oxide regulates postischemic myocardial oxygenation and oxygen consumption by modulation of mitochondrial electron transport. Circulation. 2005;111:2966–2972. doi: 10.1161/CIRCULATIONAHA.104.527226. [DOI] [PubMed] [Google Scholar]
- 7.Wang P, Zweier JL. Measurement of nitric oxide and peroxynitrite generation in the postischemic heart. Evidence for peroxynitrite-mediated reperfusion injury. J Biol Chem. 1996;271:29223–29230. doi: 10.1074/jbc.271.46.29223. [DOI] [PubMed] [Google Scholar]
- 8.Garg V, Hu K. Protein kinase C isoform-dependent modulation of ATP-sensitive K+ channels in mitochondrial inner membrane. Am J Physiol Heart Circ Physiol. 2007;293:H322–332. doi: 10.1152/ajpheart.01035.2006. [DOI] [PubMed] [Google Scholar]
- 9.Lee GY, He DY, Yu L, Yu CA. Identification of the ubiquinone-binding domain in QPs1 of succinate-ubiquinone reductase. Journal of Biological Chemistry. 1995;270:6193–6198. doi: 10.1074/jbc.270.11.6193. [DOI] [PubMed] [Google Scholar]
- 10.Yu L, Yu CA. Quantitative resolution of succinate-cytochrome c reductase into succinate-ubiquinone and ubiquinol-cytochrome c reductases. J Biol Chem. 1982;257:2016–2021. [PubMed] [Google Scholar]
- 11.Hatefi Y. Preparation and properties of succinate: ubiquinone oxidoreductase (complex II) Methods Enzymol. 1978;53:21–27. doi: 10.1016/s0076-6879(78)53008-5. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Xu H, Chen CL, Green-Church KB, Freitas MA, Chen YR. Mass spectrometry profiles superoxide-induced intramolecular disulfide in the FMN-binding subunit of mitochondrial Complex I. J Am Soc Mass Spectrom. 2008;19:1875–1886. doi: 10.1016/j.jasms.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Z, Chen YR, Jones CI, 3rd, Meenakshisundaram G, Zweier JL, Alevriadou BR. Shear-induced reactive nitrogen species inhibit mitochondrial respiratory complex activities in cultured vascular endothelial cells. Am J Physiol Cell Physiol. 2007;292:C1103–1112. doi: 10.1152/ajpcell.00389.2006. [DOI] [PubMed] [Google Scholar]
- 15.Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 16.Seeger MA, von Ballmoos C, Eicher T, Brandstatter L, Verrey F, Diederichs K, Pos KM. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 17.Chen YR, Chen CL, Zhang L, Green-Church KB, Zweier JL. Superoxide generation from mitochondrial NADH dehydrogenase induces self-inactivation with specific protein radical formation. J Biol Chem. 2005;280:37339–37348. doi: 10.1074/jbc.M503936200. [DOI] [PubMed] [Google Scholar]
- 18.Vasquez-Vivar J, Santos AM, Junqueira VB, Augusto O. Peroxynitrite-mediated formation of free radicals in human plasma: EPR detection of ascorbyl, albumin-thiyl and uric acid-derived free radicals. Biochem J. 1996;314:869–876. doi: 10.1042/bj3140869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Detweiler CD, Deterding LJ, Tomer KB, Chignell CF, Germolec D, Mason RP. Immunological identification of the heart myoglobin radical formed by hydrogen peroxide. Free Radic Biol Med. 2002;33:364–369. doi: 10.1016/s0891-5849(02)00895-x. [DOI] [PubMed] [Google Scholar]
- 20.Chen YR, Chen CL, Liu X, Li H, Zweier JL, Mason RP. Involvement of protein radical, protein aggregation, and effects on NO metabolism in the hypochlorite-mediated oxidation of mitochondrial cytochrome c. Free Radic Biol Med. 2004;37:1591–1603. doi: 10.1016/j.freeradbiomed.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Schoneich C, Asmus KD, Dillinger U, von Bruchhausen F. Thiyl radical attack on polyunsaturated fatty acids: a possible route to lipid peroxidation. Biochem Biophys Res Commun. 1989;161:113–120. doi: 10.1016/0006-291x(89)91568-4. [DOI] [PubMed] [Google Scholar]
- 22.Schoneich C, Dillinger U, von Bruchhausen F, Asmus KD. Oxidation of polyunsaturated fatty acids and lipids through thiyl and sulfonyl radicals: reaction kinetics, and influence of oxygen and structure of thiyl radicals. Arch Biochem Biophys. 1992;292:456–467. doi: 10.1016/0003-9861(92)90016-p. [DOI] [PubMed] [Google Scholar]
- 23.Shiva S, Crawford JH, Ramachandran A, Ceaser EK, Hillson T, Brookes PS, Patel RP, Darley-Usmar VM. Mechanisms of the interaction of nitroxyl with mitochondria. Biochem J. 2004;379:359–366. doi: 10.1042/BJ20031758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu Rev Pharmacol Toxicol. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]