Abstract
The application of dielectrophoretic field-flow fractionation (depFFF) to the isolation of circulating tumor cells (CTCs) from clinical blood specimens was studied using simulated cell mixtures of three different cultured tumor cell types with peripheral blood. The depFFF method can not only exploit intrinsic tumor cell properties so that labeling is unnecessary but can also deliver unmodified, viable tumor cells for culture and/or all types of molecular analysis. We investigated tumor cell recovery efficiency as a function of cell loading for a 25 mm wide × 300 mm long depFFF chamber. More than 90% of tumor cells were recovered for small samples but a larger chamber will be required if similarly high recovery efficiencies are to be realized for 10 mL blood specimens used for CTC analysis in clinics. We show that the factor limiting isolation efficiency is cell-cell dielectric interactions and that isolation protocols should be completed within ~15 minutes in order to avoid changes in cell dielectric properties associated with ion leakage.
Keywords: dielectrophoresis, field-flow fractionation, tumor, circulating tumor cells
Introduction
Many clinical problems demand isolation of one cell type from another in dense cell suspensions, an example of which is the detection of rare circulating tumor cells (CTC’s) in the peripheral blood of cancer patients. This is of importance to the prognosis and treatment of breast, prostate, ovarian, colon, and other cancers [1–7]. CTC concentrations found in the peripheral blood of patients varies in relation to the stage of the disease but is always very low compared to background blood cell count. It is inferred that a concentration above 5 CTC’s per 7.5 mL peripheral blood correlates to worsening outcome in breast cancer patients [3]. Methods for detecting CTCs include magnetic-labeled antibodies used in conjunction with conventional magnetically-activated cell separation (MACS) [2,8,9] and quadrupole magnetic sorting [10], fluorescence-activated cell sorting (FACS) [11] and PCR [8,12–14]. More recently, automated scanning fluorescence has been successfully applied [15] and microchip approaches have been developed that can filter cells by size using microfabricated screens [8, 12], or immobilize cells using antibodies attached to microposts [16]. Many of these methods do not provide unaltered, viable cells for molecular analysis or culture. Alternative methods that do not depend on antigens and can maintain cell function and viability following isolation are desirable.
We reported success in isolating viable cultured breast tumor cells from peripheral blood using dielectrophoresis (DEP)[17, 18], an electric method that can be used to manipulate cells in accordance with their phenotype and membrane capacitance without the need for labeling or modification [19–22]. We succeeded in isolating cultured leukemia cells from blood [23] and cultured breast tumor cells from CD34+ hemopoietic stem cells [24]. Other groups have used DEP to manipulate and sort tumor cells using DEP methods [25–31]. However, prior studies used small electrode arrays, limiting the number of cells that could be processed to at most a few hundred thousand. In the present research work we tested a larger DEP arrayand improved methods to process much higher cell numbers and we report data for the isolation of cultured MDA-MB-435, MDA-MB-468 and MDA-MB-231 tumor cells from mixtures with up to 30 million peripheral blood mononuclear cells (PBMN) cells, a clinically relevant specimen size for detecting CTCs. After isolation, tumor cells were successfully returned to growth in culture to demonstrate functional integrity and viability. Separation time, eluate flow rate, and cell loading concentration influenced the efficiency of DEP isolation. The impacts of these factors are analyzed and equations accounting for them are introduced to aid in the design of DEP-based cell separators.
Experimental
Cell samples
MDA-MB-435, MDA-MB-468 and MDA-MB-231 cells [32] were maintained in exponential growth in MEM (Cellgro, Mediatech, Monassas, VA) supplemented with 10% fetal bovine serum (Atlanta Biochemicals, Atlanta, GA). To sustain consistent cell dielectric properties, cells were harvested at about 65% confluence in exponential growth two days after seeding using trypsin-EDTA (Cellgro). Cell viability, determined by trypan blue dye exclusion, was at least 98% following harvest.
Peripheral blood was collected from volunteers into vacutainers containing EDTA anticoagulant (BD Biosciences, San Jose, CA) or purchased from Gulf Coast Regional Blood Bank (Houston, TX). Blood was diluted with RPMI, layered onto Histopaque-1077 density separation medium (H8889, Sigma-Aldrich, St. Louis, MO), and centrifuged at 300g for 20 mins. The PBMN cell layer was collected, washed in RPMI, centrifuged again, spiked with harvested tumor cells at concentrations between 10 and 1000 tumor cells per million PBMN cells, and suspended in a DEP running buffer (used as the eluate in all our experiments) composed of 9.5% ultrapure sucrose (S7903, Sigma-Aldrich, St. Louis, MO), 0.3% dextrose (Fisher D-16) and 0.1% Pluronic F68 (P1300, Sigma-Aldrich, St. Louis, MO) titrated to a conductivity of 30 mS.m−1 with KCl with the aid of a conductivity meter (EC Meter 19101-00, Cole-Parmer Instrument Co, Vernon Hills, IL). The running buffer had an osmolarity of 320 mOs.L−1 and a pH of 7 and experiments were conducted at room temperature (22 °C).
DEP separator
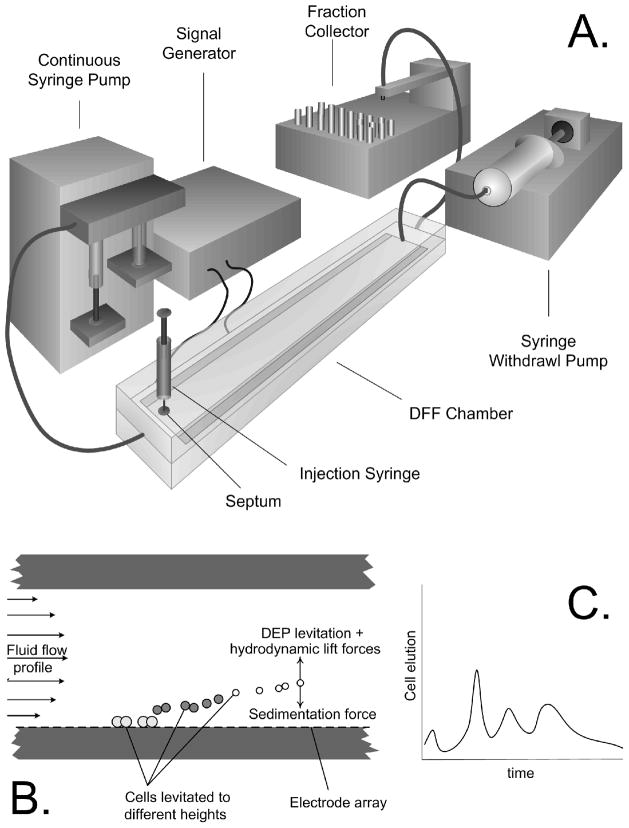
Cell fractionation was accomplished with the arrangement shown in Fig 1. The depFFF chamber contained a flow channel 0.6 mm high × 25 mm wide × 300 mm long having a floor lined with interdigitated 50 micron wide gold-on-copper electrodes spaced 50 microns apart on a Kapton substrate. 3000 electrode elements ran widthwise across the chamber with alternate elements being connected to two bus lines. To provide adequate current-carrying capacity, these bus lines were held in contact with heavy, gold-plated bus bars in the chamber top that were energized by a signal generator comprising a microcomputer-controlled (BX25, Basic X, NetMedia Inc, Tucson, AZ) digitally synthesized sine wave chip (AD9854, Analog Devices, Norwood, MA) and a video power operational amplifier output stage (PA09, Apex Microtechnology, Tucson, AZ) that could deliver sinusoidal signals in the frequency range 1 kHz to 2 MHz and up to 10 V peak-peak at a maximum current of 3 A RMS. The applied AC signals created inhomogeneous electrical fields between neighboring electrode elements and exerted dielectrophoretic forces on cells near the chamber floor. By setting signal properties appropriately, it was possible to alter the elution characteristics of the cells in accordance with their dielectric characteristics.
Figure 1.
A. The DEP flow fractionation (depFFF) configuration used in this study. Cells were injected into the chamber by syringe and allowed to settle before flow was initiated from the gear pump and an ac electrical signal was applied to the electrode to influence the cell elution characteristics. B. Dielectrophoretic, sedimentation and hydrodynamic forces combined to influence the cell position in the hydrodynamic flow profile. C. As a result, cells having different properties eluted at different times from the chamber.
The cell concentration of samples loaded into the depFFF chamber was varied in different experiments. Samples of between 0.25mL and 4.5 mL in volume, corresponding to between 5% and 90% of the chamber volume, and containing between 1 × 105.mL−1 and 5 × 106.mL−1 were studied. Most experiments were conducted at a ratio of 1 tumor cell per 103 PBMN cells to allow distribution, viability and growth potential of tumor cells in the collected fractions to be determined in a statistically relevant way. However, in one set of experiments, the ratio was varied from 1 tumor cell per 103 PBMN cells to 1 tumor cell per 105 PBMN cells to verify that fractionation behavior was independent of the tumor cell content of the samples.
Samples were injected into the separation chamber using a syringe via a septum. Cells were allowed to settle for 10 minutes in the chamber with the electric field turned off, allowing cells to accumulate on the chamber floor while avoiding dipole-dipole interactions between cells that could cause cell aggregation. The electrodes were then energized with a 2.8 V peak-peak AC signal at 60 kHz. Simultaneously, flow of DEP running buffer (see above for properties) was initiated, in different experiments, at 1.5, 2.5, 4.5, 6, 8, 10 or 12 mL.min−1 from a gear pump (Ismatec, Glattbrugg, Switzerland). Total current drawn by the electrode array was about 2.8 A RMS, corresponding to a power density of 40 mW.cm−2 over the electrode array lining the chamber floor. Eluant was gently heated by this power input as it passed through the chamber. The temperature difference between inlet and outlet ports was found by thermocouple to be < 5°C at 1.5 mL.min−1 flow rate and <0.5°C at 12 mL.min−1.
Eluant from the chamber passed through a laser forward scatter particle counter (PC 2400 D, ChemTrac Systems, Norcross, GA), which counted and profiled emerging cells by size. Cell counts were uploaded to a computer and analyzed by custom software. The eluant emerging from the counter was aliquotted by a fraction collector (CF-1, Spectra/Chrom, Spectrum Chromatography, Houston, TX) for cytological analysis and growth experiments.
Cell Isolation
The separation principle used to isolate tumor cells was dielectrophoretic field flow fractionation (see Discussion). Because of the cell dielectric properties, electric fields at or below 60 kHz are expected to pull tumor cells towards the microelectrode array on the chamber floor and repel blood cells in the 30 mS.m−1 eluant used for our experiments. Tumor cells were therefore expected to experience steric retardation that would slow their transit through the chamber while blood cells would be carried away by eluant (see Discussion). During cell fractionation, the laser counter-sizer was used to monitor elution of PBMN cells and, once their count rate fell to a negligible level, the DEP frequency was adjusted to 15 kHz. Because of their dielectric properties, tumor cells experienced repulsive DEP force from the electrode at this frequency and they were levitated and expelled rapidly from the chamber. In some experiments, eluant flow rate was held constant throughout the experiment until both blood and tumor cells had finished eluting. Alternatively, once the PBMNCs had eluted and the DEP frequency had been adjusted to 15 kHz, flow rate was increased to 50 mL.min−1 to provide a 20 mL burst of DEP buffer that flushed the tumor cells from the chamber in just a few seconds.
Assessment of cells following isolation
Cell membrane barrier function was assessed in collected fractions by trypan blue dye exclusion. Slides of the cell fractions were prepared using a cytocentrifuge (Cytopro-7620, Wescor, Logan, UT) and the cells were stained (Hema 3 Stain Set, Fisher Scientific, USA) and counted to determine cell recovery efficiency and the ratio of PBMN cells to tumor cells in each fraction.
In some experiments, tumor cells were labeled with dye (CellTracker Green C7025, Invitrogen, Carlsbad, CA) prior to mixing with PBMNCs. Following depFFF separation, collected fractions were examined by flow cytometry (excitation 488 nm, emission 520 nm, CyFlow, Partec, Munster, Germany). Because they were labeled with cell tracker dye, viable tumor cells were brightly fluorescent, and gating analysis of cytometric scattergrams allowed the ratios of tumor to blood cells to be measured accurately. Because the entire fractions were counted and the viability of the tumor cells was verified, this method was most accurate for assessing cell ratios and collection efficiencies.
In experiments to test growth potential of collected tumor cells, the DEP chamber and counter/sizer were sterilized with 70% ethanol and rinsed with sterile eluant. Running buffer was sterilized by passing it through an in-line 0.2 μm-filter just before it entered the depFFF chamber, and fraction collection was conducted in sterile tubes in a sterile enclosure. The PBMN: tumor cell ratio was calculated for each fraction via cytological examination of slides and part of each fraction was returned to growth medium and cultured in 12-well plates. Cell attachment was assessed at 24 hours and cell counts were followed for several days to determine growth potential.
Results
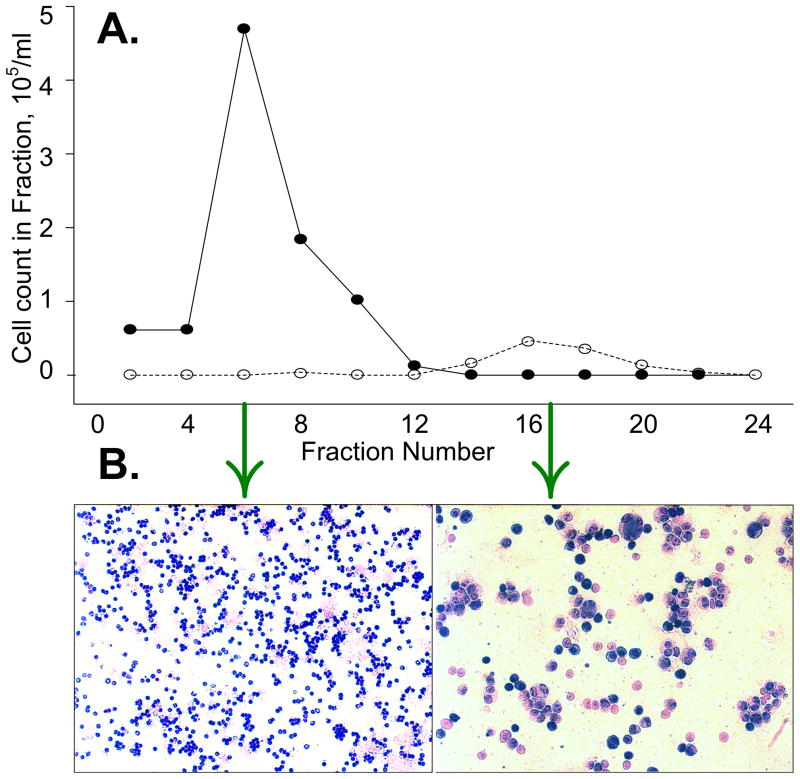
The distributions of PBMN and MDA-435 cells in fractions eluted from a sample containing 106 PBMN cells and 103 MDA435 tumor cells per mL at 10% chamber loading are shown in Figure 2A. Tumor cells were depleted from the fractions when the applied DEP frequency was 60 kHz and were concentrated in later fractions following a frequency shift to 15 kHz. The appearance of slides made from fractions collected during the PBMN and tumor cell elution phases is shown in Figure 2B. The tumor: PBMN cell concentration ratio was enriched more than 2000-fold in the example shown.
Figure 2.
A. The relative proportion of peripheral blood (●) and cancer cells (❍) varied in the different fractions. The applied DEP frequency was 60 kHz for fractions 1 to 12, then it was switched to 15 kHz for fractions 13 to 24. B. Wright staining reveals the high ratio of tumor cells collected in fraction 17 relative to fraction 6.
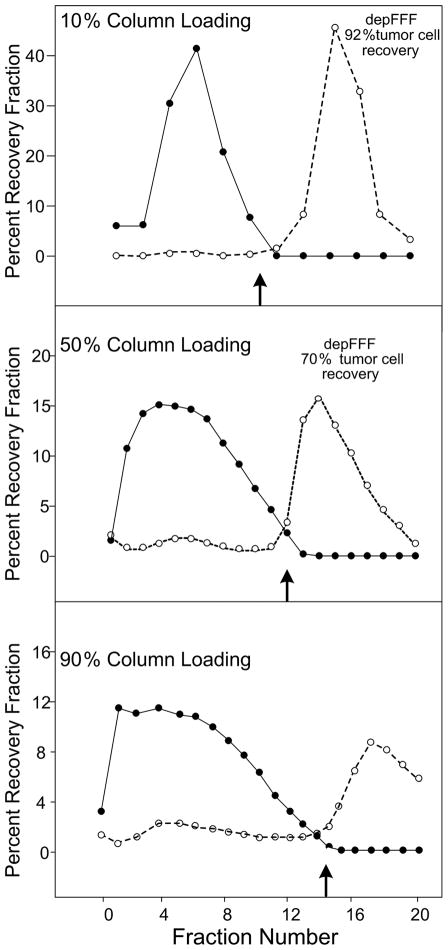
Cell isolation efficiency and recovery was a function of the volume of sample loaded into the chamber, the concentration of cells in the sample, and the method by which the tumor cells were recovered from the chamber. Figure 3 shows fractograms and cell recovery for samples that filled 10%, 50% and 90% of the chamber volume. Tumor cell recovery and puritydecreased approximately 10-fold as the chamber loading was increased from 10% to 90% as expected for a chromatographic method like depFFF.
Figure 3.
Elution profiles of PBMN (●) and cancer cells (❍) as a function of the percentage of the depFFF volume that was loaded with sample prior to cell settling. The DEP frequency was switched from 60 kHz to 15 kHz at the fractions shown by the small arrows. Better recovery of tumor cells occurred if the cells remaining in the depFFF were flushed rapidly from the chamber after the DEP frequency was changed to 15 kHz (Flush recovery).
Initially, a flow rate of 1.5 mL.min−1 was used for depFFF experiments and cell separations took 40–50 minutes to complete. Tumor cells exhibited decreasing responsiveness to the applied DEP signal after being immersed in the 30 mS.m−1 DEP running buffer for longer than ~30 minutes and could not be eluted from the chamber by adjusting the DEP frequency to 15 kHz at low eluate flow rates. Therefore, depFFF runs were hastened by using an initial flow rate of 4.5 mL.min−1 and by flushing the tumor cells from the chamber with a 20 mL burst of eluant at a 50 mL.min−1 flow rate once the PBMN cells had eluted and the frequency had been switched to 15 kHz. Under 90% column loading conditions, better recovery of tumor cells was achieved by this rapid flush method. With 10 minutes settling time and a 1.5 mL.min−1 flow rate, depFFF fractionation took about 55 minutes from start to finish while, at 4.5 mL.min−1 flow rate and rapid-flush recovery of the tumor cells, depFFF runs were completed in 20 minutes.
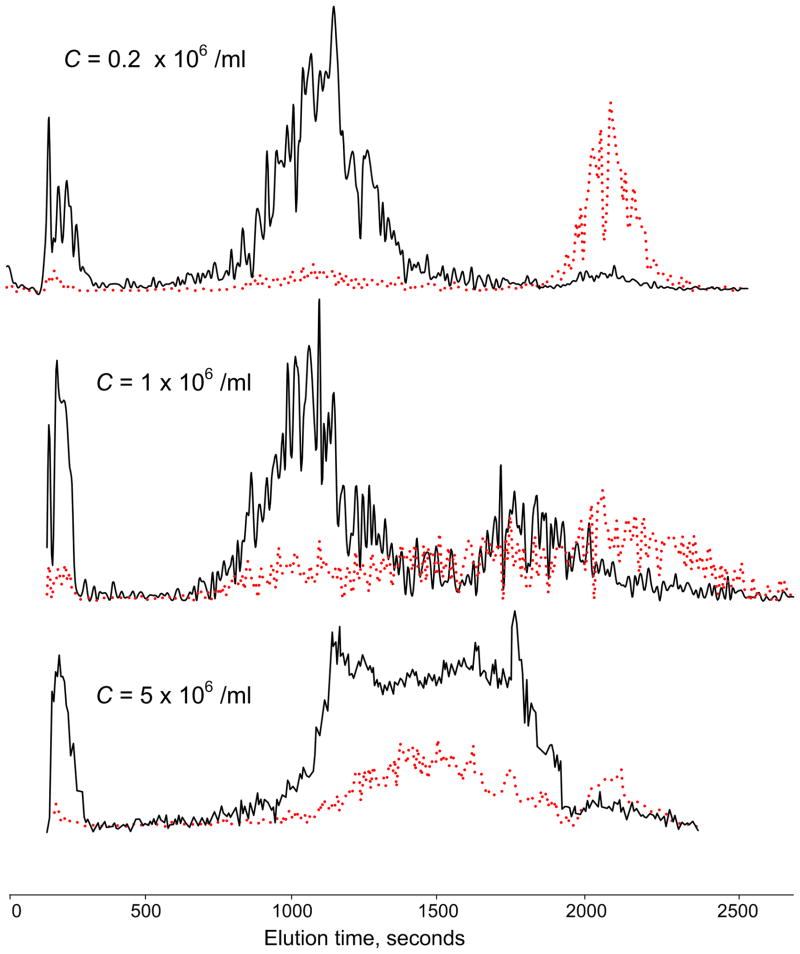
The effect of varying the concentration of PBMN cells in the samples at 20% chamber loading is shown in Figure 4. At concentrations below ~5 × 105 cells.ml−1, PBMN cells eluted at 60 kHz in a well-defined peak and few tumor cells emerged until the frequency was changed to 15 kHz. As the PBMN cell concentration was increased, the PBMN cell elution profile became more complex and more tumor cells co-eluted with them. At 5 × 106 PBMN cells.ml−1, almost 90% of the tumor cells co-eluted with the PBMN cells. Although the fractionation characteristics depended strongly on the PBMN cell concentration, it was independent of the tumor cell concentration in the tested range 10 to 103 tumor cells per 106 PBMN cells. Table 1 summarizes recovery efficiencies for MDA-435 cells as a function of the chamber loading volume, PBMN cell concentration, and tumor cell: blood cell ratio. The effect of increasing the total number of cells in the chamber, whether through higher sample volume or cell concentration, was to diminish isolation efficiency of the tumor cells. In the best case, with only 0.2 × 106 PBMN cells.ml−1 and 10% chamber loading (a total of 0.1 × 106 PBMN cells in the chamber), 92% of tumor cells were recovered. In the worst case, with a cell concentration of 5 × 106 PBMN cells.ml−1 and 90% chamber loading (a total of 23 × 106 PBMN cells in the chamber), only 10% of the tumor cells were recovered. Tumor cell collection efficiency was independent of tumor cell: blood cell ratio over the range examined.
Figure 4.
Elution profiles measured by the laser counter as a function of cell concentration loaded onto the depFFF chamber. PBMN cells (black) and tumor cells (red) were cleanly separated at a loading concentration C = 0.2 × 106 cells.ml−1, but elution peak broadening and co-elution of the PBMN and tumor cells occurred as the loading was increased.
Table 1.
depFFF tumor cell recovery was measured for tumor cells mixed into blood cell suspensions of different concentrations under 10%, 50% and 90% chamber loading conditions. Tumor cell recovery efficiency fell with increasing blood cell concentration and increasing chamber loading. The effect of changing the tumor: blood cell ratio was also measured with the blood cell concentration and chamber loading held constant. Tumor cell recovery efficiency did not depend on the tumor cell load at the tumor cell: blood cell ratios studied.
| Blood cell concentration (l | Tumor: normal cell ratio | chamber length loading | Total cell load | Tumor cell load | Recovery of tumor cells |
|---|---|---|---|---|---|
| 2.00E+05 | 1:1000 | 10% | 1.00E+05 | 100 | 92% |
| 2.00E+05 | 1:1000 | 50% | 5.00E+05 | 500 | 70% |
| 2.00E+05 | 1:1000 | 90% | 9.00E+05 | 900 | 40% |
| 1.00E+06 | 1:1000 | 10% | 5.00E+05 | 500 | 52% |
| 1.00E+06 | 1:1000 | 50% | 2.50E+06 | 2500 | 41% |
| 1.00E+06 | 1:1000 | 90% | 4.50E+06 | 4500 | 25% |
| 5.00E+06 | 1:1000 | 10% | 2.50E+06 | 2500 | 15% |
| 5.00E+06 | 1:1000 | 50% | 1.25E+07 | 12500 | 12% |
| 5.00E+06 | 1:1000 | 90% | 2.25E+07 | 22500 | 10% |
| 2.00E+06 | 1:1000 | 50% | 5.00E+06 | 5000 | 49% |
| 2.00E+06 | 1:400 | 50% | 5.00E+06 | 2000 | 39% |
| 2.00E+06 | 1:100 | 50% | 5.00E+06 | 500 | 44% |
| 2.00E+06 | 1:40 | 50% | 5.00E+06 | 200 | 42% |
| 2.00E+06 | 1:10 | 50% | 5.00E+06 | 56 | 48% |
The first three fractions in all runs had poor cell viability(well below 50%) as judged by trypan blue dye exclusion. Cytological examination revealed these fractions contained mostly ghosts and necrotic cells. Viability exceeded 90% for subsequent fractions but diminished for fractions that had been collected more than 30 minutes after cell loading. If long immersion times were avoided by rapidly flushing tumor cells from the depFFF chamber after the PBMN depletion phase, cells had a viability of ~90%, and more than 70% of these cells reattached to culture wells and resumed growth overnight.
While most experiments were conducted using MDA435 cells, we also studied the isolation characteristics of MDA468 and MDA231 cells at a dilution of 1 tumor cell per 103 PBMN cells and 20% column loading. Cell fractionation characteristics for tumor cells from these lines were similar to MDA435. This is consistent with the similarities in the dielectric properties and size of these cell types.
Discussion
Dielectric properties of cells
The cell dielectric measurements most familiar to biologists are electrophysiological determinations of cell total plasma membrane capacitance and conductance made with microelectrodes inserted through the plasma membrane [for example 33, 34]. The total cell capacitance reflects plasma membrane area, which depends both on cell size and features such as ruffles, folds and microvilli that contribute additional area. The total membrane area depends on cell transport phenomena [35] and metabolism [36] as well as processes such as apoptosis that lead to membrane shedding [37, 38]. Therefore total capacitance is a useful parameter for characterizing the gross morphology and physiological status of cells and has been used to quantify the heterogeneity of breast cancer cell lines [39]. Several groups including ours have determined membrane capacitance and conductance without the need to penetrate or contact the cells by inducing cell motion with alternating electric field gradients through dielectrophoresis (DEP) [40], by observing cell electrorotation induced by rotating AC electric fields [41], or by using impedance spectroscopy [39]. As well as establishing that there are significant differences in the membrane capacitance of different cell types [41], DEP has enabled cell membrane capacitance differences to be exploited to manipulate and isolate different cell types. For example, we have isolated breast cancer cells and leukemia cells from blood [23] and breast tumor cells from hemopoietic stem cells [24], and we have made use of changes in membrane properties to isolate malarially-parasitized erythrocytes from their uninfected counterparts [42] and to study drug and toxicant responses of cells [43]. DEP-based cell isolation can be achieved without the cell labeling that is the hallmark of magnetic activated cell sorting (MACS) and fluorescence activated cell sorting (FACS). Furthermore, cells isolated by DEP are viable and suitable for growth and molecular assays such as soluble protein, RT-PCR, and micro RNA determinations.
Dielectric Field-Flow Fractionation (depFFF)
In field-flow fractionation, one or more force fields are used to position particles in a fluid velocity gradient according to their physical properties and, as a result, the differentially positioned particles are carried at different velocities. Particles having different properties therefore transit the chamber at different speeds and become fractionated. We and others have shown that DEP, hydrodynamic lift (HDLF) and sedimentation forces can be used in concert to bring about the fractionation of cell mixtures (see figure 1B) [44]. We have also provided a theoretical basis for this methodology [43–45] which we call depFFF. The frequency of the applied electric field can be adjusted to cause either attractive or repulsive DEP forces to be imposed on cells in a mixture according to the cell membrane capacitance and conductance and the suspending medium conductivity, [46] (see below). If the DEP and sedimentation forces act in the same direction, cells are pulled to the electrodes where steric hindrance and the low flow rate that prevails close to the chamber floor slows their movement. Because cell size [47] and membrane flexibility[48] influence hydrodynamic lift forces that prevail close to the chamber floor, these parameters help determine the cell elution characteristics under these conditions and may be exploited to optimize separations. On the other hand, if the applied electric field frequency leads to repulsive DEP forces that oppose the sedimentation force on a given cell type, then that type is levitated into faster flow causing it to be transported more rapidly. In this case, cells are typically levitated several tens of μm above the chamber floor where hydrodynamic lift forces are small for the flow rates we studied.
The DEP force experienced by a cell in a medium of permittivity εs having an imposed inhomogeneous electric field E⃗ is given by
| (1) |
where r is the cell radius. fcm is the real part of the Claussius-Mossotti factor (CMF) which may be expressed in terms of the complex permittivities of the cell ( ) and suspending medium ( ) as
| (2) |
The FCM determines the relative strength and direction of the DEP force on a cell as a function of the applied field frequency. Benguigi and Lin [49] and Markx et al. [50] have discussed the relative contributions of cell and suspending medium conductivities and permittivities to the CMF. When the conductivity of the cell cytoplasm is much higher than, and the conductivity of the cell membrane is much less than, that of the suspending medium, the cell will exhibit negative DEP at low frequencies, positive DEP at higher frequencies, and no DEP at all at an intermediate DEP crossover frequency, f0. In this case, fcm can be conveniently approximated, in a frequency range close to f0 as
| (3) |
Our experiments exploit differences in f0 for the various cell types in our mixtures and it is convenient to write it using the expression of Jones & Kallio [51] as
| (4) |
where σs and σp are the conductivities, and εs and εp are the permittivities, of the suspending medium and cell, all respectively. According to the single shell dielectric model [52,53], and using expressions from Chan et al. [54] and Green & Morgan [55], the cell dielectric parameters in this frequency range are dominated by the properties of the plasma membrane and εp ≈ r·Cmem (where Cmem is the capacitance per unit area of the plasma membrane) and (where σm is depends on the transmembrane conductivity and Km on conductivity parallel to the membrane surface [55]).
For the viable cells examined here, εp ≫ εs and σp ≪ σs However, if the cell membrane barrier function collapses, the latter relationship, in particular, no longer pertains and f0 may alter by several orders of magnitude [38]. For this reason, depFFF makes the separation of viable and non-viable cells trivial while allowing cells having more subtle differences in membrane capacitance to be fractionated [24, 44]. While our previous experiments showed that cells can be isolated using depFFF, those experiments generally used small numbers of cells and small (<10 cm2) electrode arrays. Here we have applied the depFFF technique to larger numbers of cells on a 75 cm2 electrode array to establish whether the method can be scaled up for clinical applications.
Fractionation of cultured tumor cells from PBMN cells
Differences in cell size and membrane capacitance cause the breast tumor cell types we examined, including MDA435, MDA231, MDA-468, MDA-361 and MCF-7, to exhibit distinctly different fcm frequency responses from PBMN cell subpopulations [18,24,44] (see Figure 5). At the suspension conductivity of 0.03 S.m−1 used for the separations in this study, the f0 values for the tumor cells, granulocytes and lymphocytes were ~30 kHz, ~90 kHz, and ~140 kHz, respectively, and the DEP operating frequency of 60 kHz used for our separations led, from equation (3), to fcm values of +0.5, −0.22, and −0.39, respectively, for these cell types. Therefore, at 60 kHz the DEP and sedimentation forces acted cooperatively on the tumor cells and pulled them to the electrode array. At the same time, the DEP forces opposed and overcame the sedimentation forces on the blood cells, levitating them 20 to 30 microns above the electrode array. Consequently, the transit of tumor cells through the chamber was retarded, while the transit of blood cells was hastened, by the DEP forces.
Figure 5.
The real part of the Clausius-Mossotti factor fcm for the cancer cell lines studied and PBMN cells shown as a function of the frequency of the DEP voltage applied to the chamber electrodes. At 60 kHz, all breast tumor cell lines were attracted towards the electrodes by positive DEP forces and slowed by steric effects during separation while the PBMN cell types were levitated into the eluate flow stream by negative DEP forces and expelled quickly from the depFFF chamber. At 15 kHz, all cell types experienced repulsive DEP forces that promoted elution from the chamber.
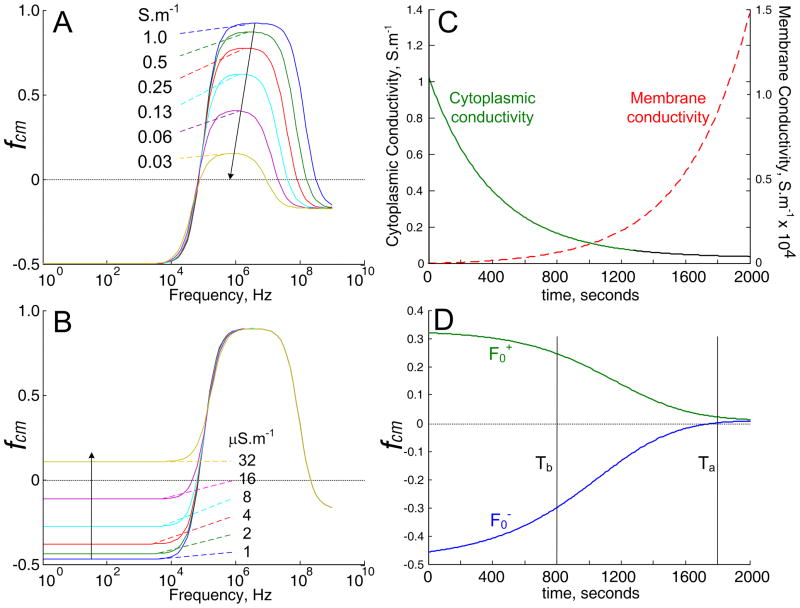
We found that the cells behaved according to these principles unless the flow rate used in separations was so slow that the cells had to be immersed in the low conductivity isotonic sucrose buffer for more than 40 minutes. The cells exhibited weakening positive and negative DEP responses after such long immersion times. A weakening attractive DEP force results if the cell cytoplasmic conductivity falls as shown by the single shell dielectric model simulation of fcm versus cytoplasmic conductivity (using MATLAB, The Mathworks, Natick, MA) in Figure (6A). It is reasonable to expect ions from the cytoplasm (initial conductivity ~ 1 S.m−1) to slowly leak through the plasma membrane into the 30 mS.m−1 sucrose suspending solution leading to reduced cytoplasmic conductivity and the decrease observed in attractive DEP forces at long immersion times. However, this effect cannot account for the decrease in repulsive DEP forces. To explain this, an increase in cell membrane conductivity is required, as simulated in Figure (6B). Interestingly, most cells did not stain with trypan blue after 40 minutes of immersion in sucrose buffer, showing that the cell membrane barrier function was intact. It is possible that ionic-strength-dependent channels opened as the cytoplasmic ions depleted by leakage. Such channels are present in most cell types [56], and these may also account for previous experiments with erythrocytes in which ionic-strength-dependent membrane conductivity was observed [57]. Opening of such channels would cause rapid loss of cytoplasmic ions to the suspending medium, therefore the positive and negative DEP responses might collapse suddenly when the channels triggered. Figure (6B) shows a simulation of the cytoplasmic and membrane conductivities, and Figure (6B) the resulting drop in attractive and repulsive DEP characteristics, when the leakage of ions and gating of channels are assumed to be slow. Even without rapid gating, these simulations predict that the cell DEP properties change little in the first 800 seconds of immersion in sucrose buffer, as we observed. Such time-dependency of cell dielectric properties needs to be taken into consideration when designing DEP separation experiments. Once the elution of PBMN cells was complete in our experiments, the DEP frequency was switched to 15 kHz. Because this was below the crossover frequency of the tumor cells, they experienced repulsive DEP under these conditions, were levitated into the flow stream, and eluted from the chamber. It was found that the most effective way to recover the tumor cells was with a rapid flush, which allowed longer immersion times in DEP buffer to be avoided.
Figure 6.
A and B. Simulations based on the single shell dielectric model of the frequency spectrum of the real part of the Claussius-Mossotti factor (fcm) that scales the DEP force for cells having a crossover frequency of 20 kHz. A. Effect on the fcm spectrum of decreasing the cytoplasmic conductivity to the values shown. B. Effect on the fcm spectrum of increasing cell membrane conductivity to the values shown. C. Simulations of the cytoplasmic and membrane conductivities versus time when the cytoplasmic ion concentration decays exponentially with a rate constant of 400 seconds and the membrane conductivity varies inversely with the cytoplasmic conductivity. D. Effect of the changing cytoplasmic and membrane conductivities shown in C on the fcm values for two cell types that initially exhibit positive and negative DEP responses. In this simulation, the two cell types are indistinguishable by DEP after about 2000 seconds because of the ion leakage and membrane conductivity increase.
The concentration of tumor cells found clinically in peripheral blood varies from below 1 per 106 PBMN cells to about 100 per 106 PBMN cells and it was important for us to investigate whether the depFFF method could recover tumor cells at such low levels. However, we wanted to obtain statistically significant results showing how the tumor cells were distributed among the collected fractions. Therefore, we used 1 tumor cell per 103 PBMN cells to derive cell recovery distributions for most experiments and then verified that the cell recovery efficiencies obtained for dilutions up to 1 tumor cell per 105 PBMN cells were consistent (see Table 1). Tumor cell isolation numbers were proportional to the number seeded into blood, showing that the isolation was independent of tumor cell load. This is reasonable given the very low tumor: PBMN ratio in all experiments and the correspondingly low probability that interactions occurred between tumor cells. It follows that our cell recovery data are pertinent to clinical rare tumor isolations and allow us to predict the expected tumor cell recovery characteristics.
While tumor cells were cleanly fractionated from PBMN cells under low sample loading conditions, increasing the cell concentration to >106 PBMN cells.ml−1 resulted in a broadening of the elution profile and an increasing proportion of tumor cells that co-eluted with PBMN cells. This loss of isolation efficiency can be understood in terms of cooperative dielectric effects between cells in close proximity. As their concentration increases, dielectric particles in an imposed electric field aggregate into chains in which the polarizability per particle increases many-fold as a result of multipole dielectric interactions [58]. Because these highly polarizable particle chains lie parallel to the electric field, the effective permittivity along different directions is modified differentially and the mixture exhibits dielectric anisotropy. Acrivos et al. [59] have shown that extremely high concentrations of plastic beads in an electric field go beyond chain formation and undergo an electrically-induced phase change in which the particle phase separates out completely from the suspending medium.
In our experiments, cells were injected as a homogeneous suspension but were given time prior to field application to settle to the bottom of the DEP chamber where they formed a layer equal in thickness to the cell diameter. If the layer was sufficiently concentrated, dipole and multipole interactions led to cell chain formation in this plane when the field was applied and the DEP analysis presented earlier, which assumes a homogeneous distribution of non-interacting cells, broke down. Jones [56] showed that cell-cell interactions become significant as the ratio of cell spacing δ to cell diameter d falls below ~5. The mean spacing of cells when a suspension of concentration X cells.m−3) settles onto the electrode plane from a chamber of height h is given by giving . Setting to avoid interactions, we find that the cell concentration cells.ml−1. For our chamber of height 0.6 mm, this suggests the cell concentration should be ≪2×106 cells.ml−1 for PBMN cells (diameter 8 μm) and ≪106 cells.ml−1 for MDA-435 cells (diameter 12 μm).
Figure 4 shows DEP elution profiles obtained for PBMN loading concentrations of 2 × 105 mL−1, 106 mL−1 and 5 ×06 mL−1. In agreement with the above predictions, loss of cell separation efficiency was apparent as the loading concentration approached 106 cells.ml−1. Furthermore, when cells were viewed through the clear acrylic top of the depFFF chamber with a long working distance microscope, aggregates of PBMN cells were seen entrapping tumor cells at high cell loading densities, confirming that field-induced dipole-dipole effects limited the separation efficiency. Based on these findings, a reasonable ceiling for cell loading when clean separations are desired may be taken as Cmax = 10−9/(d2h) corresponding to a maximum PBMNC loading concentration of 4 × 105 cells.ml−1 for our chamber.
Chromatographic fractionation methods like FFF are normally most efficient when the sample volume is small compared with the separation chamber volume and when the sample cell concentration is low. On the other hand, larger sample volumes and concentrations allow more cells to be processed, so operating conditions are normally chosen as a compromise that allows moderately large samples to be processed at adequate isolation efficiencies. Our dielectric analysis shows that the tumor cell recovery efficiencies in Table 1 could be improved if the size of the depFFF chamber were increased and the PBMN cell loading was reduced to attain the optimum loading ceiling.
The cultured breast tumor cell lines were considerably larger and had higher membrane capacitances than PBMN cells and these parameters contributed to the dielectric differences that allowed them to be separated from PBMN cells by depFFF. Although not studied here, differences in cell size and membrane flexibility also likely affected the cell separation characteristics by affecting the cell hydrodynamic lift properties. In assessing the applicability of depFFF to other cell isolation applications, it is important to take into account that a relationship exists between the fractionation efficiency, the sample size in terms of total cells, and the dielectric differences between the cells to be isolated. Fortunately, circulating tumor cells captured by MACS [2,5,10] and by microfluidic filters [8,12] showed significant size and morphological differences compared with PBMN cells, suggesting that CTCs in clinical specimens have similar morphological properties to the cells we used as surrogates in this study.
We found that recovered tumor cells could be returned to culture, proving they were not only intact and viable but also suitable for every type of post-separation analysis. The ability to isolate viable CTCs would not only allow a spectrum of molecular tests to be performed but would also permit primary cultures to be established from clinically derived CTCs. The depFFF apparatus is relatively simple, consisting of a small signal generator and a flow-through chamber, albeit one lined by an electrode. Therefore it could be operated in hospitals and clinics close to the point of care.
Conclusions
Our depFFF experiments expanded on earlier proof-of-concept findings and confirmed that tumor cells can be isolated from PBMN cells by DEP using clinically-relevant numbers of cells. We successfully isolated MDA435, MDA468 and MDA 231cells from blood samples containing up to 28 × 106 PBMN cells by exploiting cell morphological characteristics without the need for antibody or other labeling procedures. Tumor cell isolation efficiencies above 90% were achieved for small samples and fell with increasing cell loading to 10% for samples containing 23 × 106 PBMN cells. Our results suggest that the depFFF method may be applicable for isolating unlabeled CTCs from clinical samples, however in designing efficient DEP-based cell isolation protocols, a maximum loading density should be calculated using the criteria derived here. Larger chambers would be optimal for clinical samples. Experiment times should be chosen so that changes in the cytoplasmic and membrane conductivities do not confound cell separation. We showed that tumor cells recovered by DEP were viable, could be cultured, and were suitable for a wide spectrum of analyses.
Acknowledgments
This work was supported by NIH grant R33 CA88364-01. We are grateful to Dr. Jody Vykoukal for help with flow cytometry, to Dr. Chandra Das for preliminary studies on the blood cells, and to Sara Dholakia for editing.
References
- 1.Cristofanilli M. Cancer. 2005;103:877–880. doi: 10.1002/cncr.20838. [DOI] [PubMed] [Google Scholar]
- 2.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F, Jackson S, Gornet T, Christofanilli M, Pantel K. Clin Cancer Res. 2007;13:920–8. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 3.Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, Matera J, Allard WJ, Doyle GV, Terstappen LWWM. Clin Cancer Res. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 4.Fizazi K, Morat L, Chauveinc L, Prapotnich D, De Crevoisier R, Escudier B, Cathelineau X, Rozet F, Vallancien, Sabatier, Soria GLJ. Ann Oncol. 2007;18:518–21. doi: 10.1093/annonc/mdl419. [DOI] [PubMed] [Google Scholar]
- 5.Naoe M, Ogawa Y, Morita Omori J, Takeshita K, Shichiiyo T, Okumura T, Igarashi A, Yanaihara A, Iwamoto S, Fukagai T, Miyazaki, Yoshida AH. Cancer. 2007;109:1439–1445. doi: 10.1002/cncr.22543. [DOI] [PubMed] [Google Scholar]
- 6.Paterlini–Brechot P, Benali NL. Cancer Lett. 2007;253:180–204. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 7.Apostolaki S, Perraki M, Pallis A, Bozionelou V, Agelaki S, Kanellou P, Kotsakis A, Politaki E, Kalbakis K, Kalykaki A, Vamvakas L, Georgoulias V, Mavroudis D. Ann Oncol. 2007;18:851–858. doi: 10.1093/annonc/mdl502. [DOI] [PubMed] [Google Scholar]
- 8.Ring AE, Zabaglo L, Ormerod MG, Smith IE, Dowsett M. British J of Cancer. 2005;92:906–912. doi: 10.1038/sj.bjc.6602418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tveito S, Maelandsmo GM, Hoifodt HK, Rasmussen H, Fodstad O. Clin Exp Metastasis. 2007;24:317–27. doi: 10.1007/s10585-006-9052-8. [DOI] [PubMed] [Google Scholar]
- 10.Tong X, Yang L, Lang JC, Zborowski M, Chalmers JJ. Cytometry B Clin Cytom. 2007;72B:310–23. doi: 10.1002/cyto.b.20177. [DOI] [PubMed] [Google Scholar]
- 11.Damm-Welk C, Schieferstein J, Schwalm S, Reiter A, Woessmann W. Br J Haematol. 2007;138:459–66. doi: 10.1111/j.1365-2141.2007.06672.x. [DOI] [PubMed] [Google Scholar]
- 12.Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, Tai YC. J Chromatogr A. 2007;1162:154–61. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 13.Xi L, Nicastri DG, El-Hefnawy T, Hughes SJ, Luketich JD, Godfrey TE. Clin Chem. 2007;53:1206–15. doi: 10.1373/clinchem.2006.081828. [DOI] [PubMed] [Google Scholar]
- 14.Nakagawa T, Martinez SR, Goto Y, Koyanagi K, Kitago M, Shingai T, Elashoff DA, Ye X, Singer FR, Giuliano AE, Hoon DS. Clin Cancer Res. 2007;13:4105–10. doi: 10.1158/1078-0432.CCR-07-0419. [DOI] [PubMed] [Google Scholar]
- 15.Ntouroupi T, Ashraf S, McGregor S, Turney B, Seppo A, Kim Y, Wang X, Kilpatrick T, Tsipouras P, Tafas T, Bodmer W. Brit J Cancer. 2008 Aug 05;:1–7. doi: 10.1038/sj.bjc.6604545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagrath S, Sequist L, Maheswaran S, Bell D, Irimia D, Ulkus L, Smith M, Kwak E, Digumarthy S, Muzikansky A, Ryan P, Balis U, Tompkins R, Haber D, Toner M. Nature. 2007;450:1235–1241. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gascoyne PRC, Wang XB, Huang Y, Becker FF. IEEE transactions on industry applications. 1997;33:670–8. doi: 10.1109/28.585856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker FF, Wang X-B, Huang Y, Pethig R, Vykoukal J, Gascoyne PRC. Proc Natl Acad Sci. 1995;29:860–4. doi: 10.1073/pnas.92.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han A, Yang L, Frazier AB. Clin Cancer Res. 2007;12:139–43. doi: 10.1158/1078-0432.CCR-06-1346. [DOI] [PubMed] [Google Scholar]
- 20.Voldman J. Annu Rev Biomed Eng. 2006;8:425–54. doi: 10.1146/annurev.bioeng.8.061505.095739. [DOI] [PubMed] [Google Scholar]
- 21.Pethig R, Mark GH. Trends Biotechnol. 1997;15:426–32. doi: 10.1016/S0167-7799(97)01096-2. [DOI] [PubMed] [Google Scholar]
- 22.Das CM, Becker FF, Vernon S, Noshari J, Joyce C, Gascoyne PRC. Anal Chem. 2005;77:2708–19. doi: 10.1021/ac048196z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker FF, Wang XB, Huang Y, Pethig R, Vykoukal J. J Phys D Appl Phys. 1994;27:2659–62. [Google Scholar]
- 24.Huang Y, Yang J, Wang XB, Becker FF, Gascoyne PRC. J of Hematotherapy & Stem Cell Res. 1999;8:481–90. doi: 10.1089/152581699319939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng J, Sheldon EL, Wu L, Heller MJ, O’Connell JP. Anal Chem. 1998;70:2321–6. doi: 10.1021/ac971274g. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, Joo S, Duhon M, Heller M, Wallace B, Xu X. Anal Chem. 2002;74:3362–71. doi: 10.1021/ac011273v. [DOI] [PubMed] [Google Scholar]
- 27.Gambarim R, Borgatti M, Altomare L, Manaresi N, Medoro G, Romani A, Tartagni M, Guerrieri R. Cancer Res Treat. 2003;2:31–40. doi: 10.1177/153303460300200105. [DOI] [PubMed] [Google Scholar]
- 28.Altomare L, Borgatti M, Medoro G, Manaresi N, Tartagni M, Guerrieri R, Gambari R. Biotechnol Bioeng. 2003;82:474–9. doi: 10.1002/bit.10590. [DOI] [PubMed] [Google Scholar]
- 29.Broche LM, Bhadal N, Lewis MP, Porter S, Hughes MP, Labeed FH. Oral Oncol. 2007;43:199–203. doi: 10.1016/j.oraloncology.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 30.Cen EG, Dalton C, Li Y, Adamia S, Pilarski LM, Kaler KV. J Microbiol Methods. 2004;58:387–401. doi: 10.1016/j.mimet.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Hu X, Bessette PH, Qian J, Meinhart CD, Daugherty PS, Soh HT. Proc Natl Acad Sci USA. 2005;102:15757–61. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang RD, Fidler IJ, Price JE. Invasion Metastasis. 1991;204:204–15. [PubMed] [Google Scholar]
- 33.Bertrand CA, Laboisse CL, Hopfer U. Am J Physiol Cell Physiol. 1999;276:907–14. doi: 10.1152/ajpcell.1999.276.4.C907. [DOI] [PubMed] [Google Scholar]
- 34.Hallermann S, Pawlu C, Jonas P, Heckmann M. Proc Natl Acad Sci USA. 2003;100:8975–80. doi: 10.1073/pnas.1432836100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eliasson L, Ma X, Renstrom E, Barg S, Berggren PO, Galvanovskis J, Gromada J, Jing X, Lundquist I, Salehi A, Sewing S, Rorsman P. J Gen Physiol. 2003;121:181–97. doi: 10.1085/jgp.20028707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chowdhury HH, Grilc S, Zorec R. Ann NY Acad Sci. 2005;1048:281–6. doi: 10.1196/annals.1342.025. [DOI] [PubMed] [Google Scholar]
- 37.Huang C, Chen A, Guo M, Yu J. Biotechnol Lett. 2007;29:1307–13. doi: 10.1007/s10529-007-9414-6. [DOI] [PubMed] [Google Scholar]
- 38.Wang XJ, Becker FF, Gascoyne PRC. Biochim Biophys Acta. 2002;1564:412–20. doi: 10.1016/s0005-2736(02)00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han A, Yang L, Frazier AB. Clin Cancer Res. 2007;13:139–43. doi: 10.1158/1078-0432.CCR-06-1346. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y, Wang XB, Becker FF, Gascoyne PRC. Biochim Biophys Acta. 1996;1282:76–84. doi: 10.1016/0005-2736(96)00047-8. [DOI] [PubMed] [Google Scholar]
- 41.Gascoyne PRC, Mahidol C, Ruchirawat M, Satayavivad J, Watcharasit P, Becker FF. Lab on a Chip. 2002;2:70–75. doi: 10.1039/b110990c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gascoyne PRC, Mahidol C, Ruchirawat M, Satayavivad J, Watcharasit P, Becker FF. Lab on a chip. 2002;2:70–75. doi: 10.1039/b110990c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang XB, Huang Y, Becker FF, Gascoyne PRC. Biochim Biophys Acta. 1995;1243:185–94. doi: 10.1016/0304-4165(94)00146-o. [DOI] [PubMed] [Google Scholar]
- 44.Wang XB, Vykoukal J, Becker FF, Gascoyne PRC. J Biophy. 1998;74:2689–2701. doi: 10.1016/S0006-3495(98)77975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gascoyne PRC, Wang XB, Becker FF. Bioelectrochem Bioenergetics. 1995;36:115–25. [Google Scholar]
- 46.Gascoyne PRC, Wang XB, Huang Y, Becker FF. IEEE Industrial Application Society. 1997;33:670–678. doi: 10.1109/28.585856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams PS, Koch T, Giddings JC. Chem Eng Commun. 1992;111:121–147. [Google Scholar]
- 48.Tong X, Caldwell KD. J Chromatog B. 1995;674:39–47. doi: 10.1016/0378-4347(95)00297-0. [DOI] [PubMed] [Google Scholar]
- 49.Benguigui L, Lin IJ. J Appl Phys. 1984;56:3294–3297. [Google Scholar]
- 50.Markx GH, Huang Y, Zhou XF, Pethig R. Microbiology. 1994;140:585–591. [Google Scholar]
- 51.Jones B, Kallio GA. J Electrostatics. 1979;6:207–224. [Google Scholar]
- 52.Irimijiri A, Hanai T, Inouye A. J Theor Biol. 1979;78:251–269. doi: 10.1016/0022-5193(79)90268-6. [DOI] [PubMed] [Google Scholar]
- 53.Marszalek P, Zielinski JJ, Fikus M, Tsong TY. Biophys J. 1991;59:982–987. doi: 10.1016/S0006-3495(91)82312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan KL, Gascoyne PRC, Becker FF, Pethig R. Biophys Biochim Acta. 1997;1349:182–196. doi: 10.1016/s0005-2760(97)00092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Green NG, Morgan H. J Phys D: Appl Phys. 1997;30:L41–L84. [Google Scholar]
- 56.Biemens-Olehinkel E, Mahmood NABN, Poolman B. Proc Natl Acad Sci USA. 2006;103:10624–10629. doi: 10.1073/pnas.0603871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gascoyne PRC, Pethig R, Satayavivad J, Becker FF, Ruchirawat M. Biochimica et Biophysica Acta. 1997;1323:240–252. doi: 10.1016/s0005-2736(96)00191-5. [DOI] [PubMed] [Google Scholar]
- 58.Jones TB. Electromechanics of Particles. Chapters 6 and 7. Cambidge Univ. Press; New York: 1995. pp. 139–226. [Google Scholar]
- 59.Kumar A, Qiu Z, Acrivos A, Khusid B, Jacqmin D. Phys Rev. 2004:E69. doi: 10.1103/PhysRevE.69.021402. article 021402. [DOI] [PubMed] [Google Scholar]