Abstract
Although dielectrophoresis (DEP) has great potential for addressing clinical cell isolation problems based on cell dielectric differences, a biological basis for predicting the DEP behavior of cells has been lacking. Here, the dielectric properties of the NCI-60 panel of tumor cell types have been measured by dielectrophoretic (DEP) field-flow fractionation, correlated with the exterior morphologies of the cells during growth, and compared with the dielectric and morphological characteristics of the subpopulations of peripheral blood. In agreement with earlier findings, cell total capacitance varied with both cell size and plasma membrane folding and the dielectric properties of the NCI-60 cell types in suspension reflected the plasma membrane area and volume of the cells at their growth sites. Therefore, the behavior of cells in DEP-based manipulations is largely determined by their exterior morphological characteristics prior to release into suspension. As a consequence, DEP is able to discriminate between cells of similar size having different morphological origins, offering a significant advantage over size-based filtering for isolating circulating tumor cells, for example. The findings provide a framework for anticipating cell dielectric behavior on the basis of structure-function relationships and suggest that DEP should be widely applicable as a surface marker-independent method for sorting cells.
Keywords: Cell exterior morphology, Cell dielectric properties, Cell membrane, Dielectrophoresis, Cell isolation, Circulating tumor cells
1 Introduction
Dielectrophoresis (DEP) has been widely used for the determination of cell dielectric characteristics and is applicable to the manipulation, separation and isolation of target cells from mixtures in suspension. Recently, the applications of DEP have been reviewed comprehensively by Pethig [1]. DEP methods have been shown to have potential for identifying and isolating stem cells [2], platelets [3], white blood cells [4],pancreatic b-cells [5], osteoblasts [6, 7], prostate tumor initiating cells[8], oral cancer cells[9, 10], melanoma[11], colorectal cancer cells[12], and circulating tumor cells (CTCs) from blood[13-17]. Despite the potential applications based on these findings, the dielectric differences that enable discrimination between different cell types do not have a well-founded biological basis that can act as a predictor of cell behavior in DEP separation applications. This stands in contrast to cell isolation methods such as antibody-based approaches that target cell-surface proteins that mark specific cell types in terms of identified structure-function relationships. Without a similar understanding, the reliability of DEP as a mode of cell discrimination is hard to anticipate. For example, the extent to which DEP will be applicable to the important problem of CTC isolation in different types of cancer is unclear. In this article, we apply DEP-field field flow fractionation (DEP-FFF) as an analytical tool to characterize the dielectric properties of the entire NCI-60 panel of cells [18-22]. This set of cell types is representative of a wide range of cancers derived from different organs and includes those of diverse morphological types. We show that the dielectric properties of different cell types in suspension, including the subpopulations of peripheral blood cells, correlate closely with cell morphological characteristics that derive directly from structure-function relationships appropriate to cell type and site of origin. We thereby establish a framework for predicting the behavior of cells in DEP applications.
2 Materials and Methods
2.1 Cell culture
NCI-60 cells were cultured to 50-70% confluence in RPMI (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS) (GIBCO, Grand Island, NY). Just prior to harvest, cells were photographed using phase contrast microscopy. Adherent cultures were harvested by rinsing with calcium- and magnesium-free Hank's buffered saline solution and incubated at 37°C for 5 minutes with Trypsin/EDTA followed by sharp tapping, and neutralization with RPMI+10% fetal bovine serum (FBS). Non-adherent cell cultures were spun down from culture and suspended directly in RPMI + 10% FBS. Cells were counted to ensure >98% viability by trypan blue dye exclusion and suspended at ~106 cell/mL in RPMI + 10% FBS in conical tubes for DEP analysis. In supplementary experiments, non-adherent cultures were exposed to trypsin/EDTA and their properties in subsequent experiments were compared to those of untrypsinized cells to verify that trypsinization did not impact the data upon which conclusions of this study are based.
Cell line identities were confirmed by short tandem repeat DNA fingerprinting in the University of Texas M.D. Anderson Cell Core facility.
2.2 Dielectrophoretic field-flow fractionation (DEP-FFF)
DEP-FFF is a chromatographic method in which cell elution times reflect the positions of cells in a hydrodynamic flow profile under the control of sedimentation, DEP and hydrodynamic lift forces, FSED, FDEP and FHDL, respectively. To characterize the biophysical properties of each cell type in the NCI-60 panel, elution profiles were measured using the apparatus and methods described earlier under three conditions chosen to allow FSED, FDEP and FHDL to be disentangled from one another [23-25]. Cell elution times were measured using a laser light-scatter detector (PC2400D, ChemTrac Systems, Norcross, GA). Cell size reported for a cell by laser light scattering can be considered to be a mean value averaged over its surface. Cell radius values used in the later DEP analysis were taken to be the average of the cell sizes reported for the eluting cell populations.
For our analysis, the DEP-FFF buffer consisted of an aqueous solution of 9.5% sucrose (S7903, Sigma-Aldich, St Louis, MO), 0.1 mg/mL dextrose (S73418-1, Fisher, Fair Lawn, NJ), 0.1% pluronic F68 (P1300, Sigma-Aldich, St Louis, MO), 0.1% bovine serum albumin (A7906, Sigma-Aldich, St Louis, MO), 1 mM phosphate buffer pH 7.0, 0.1 mM CaAcetate, 0.5 mM MgAcetate and 100 units/mL catalase (C30, Sigma-Aldich, St Louis, MO) adjusted to a conductivity of 30 mS/m with KCl.
2.3 Cell exterior morphology
Previous work on harvested cells in suspension established that cell plasma membrane surface area, as estimated from electron micrographs of the fixed cells, was a key determining factor of their dielectric properties [26, 27]. The NCI-60 panel includes cell lines that grow in culture with strikingly different exterior morphologies including those of spherical, lymphoblastic-like, epithelial-like, fibroblastic-like, dendritic-like and mixed appearances. Therefore, this panel is ideal for testing whether the exterior morphology of cells prior to detachment from their growth environment is also a significant determinant of their dielectric behavior. Forty of the NCI-60 lines were photographed just prior to harvest and the variety of cell morphologies observed is illustrated in Fig. 1. The complexity of the exterior morphological diversity, as well as the distribution of morphologies for cells within each photograph, made it impractical to use an image analysis algorithm to score the cell morphological properties. Instead, we defined a visual scoring system based on easily identified external morphological characteristics that contributed to enhanced surface area relative to smooth spherical cells. These characteristics were cell flattening (spreading) on the surface of the culture flask (F), cell elongation or the presence of long dendritic projections (P), and cell membrane roughening associated with ruffles, folds and microvilli that contributed to specularity of the cell surface and margins (R). To provide a scoring guide that related each characteristic to cell membrane area, Matlab was used to generate bodies of identical volume for which these morphological characteristics contributed a surface area 1.5, 2.0 and 3.0 times that of a smooth sphere (see Fig. 2). Each author analyzed the cell photographs independently and scored each morphological characteristic on a scale of 1 to 3. A membrane area morphological score M = (F + P + R)/3 was then calculated for each photograph to estimate the extent to which cell exterior morphology increased cell surface area compared with perfectly smooth, spherical cells. On this scale, M = 1 corresponds to a smooth, round cell with no spreading or surface projections while M = 3 corresponds to a highly flattened cell with extensive projections and striking surface roughness.
Figure 1.
Micrographs showing typical morphological variations amongst the NCI-60 panel of cultured cell lines and illustrating the surface spreading, projections and roughness features used in this work as indicators of increased cell surface area compared with a perfectly smooth, spherical cell. MOLT4 (an acute lymphoblastic leukemia) shows a spherical exterior morphology with little cell flattening, no projections and relatively smooth cell surface appearance. This is as close to a smooth, spherical reference cell type of minimal surface area as was found in the NCI-60 panel. NCI-H226 (a non-small lung cancer) exhibits cell flattening resulting from cell spreading on the surface of the culture flask with projections and membrane surface roughness also apparent in some cells. SKMEL28 (a melanoma) demonstrates marked dendritic projections with minor flattening and occasional cell surface roughness. HCT116 (a colon cancer) shows marked surface roughness, with minor flattening and projections. In general, cell lines in the NCI-60 panel show combinations of these morphological traits that contribute to different, increased cell surface areas compared with smooth, spherical spheres.
Figure 2.
Simulated cell surfaces showing how exterior morphological differences can contribute to an increased cell membrane area compared with the smooth reference sphere having identical volume shown at top left. Cells having surface areas 1.5, 2.0 and 3.0 times that of the reference sphere are shown. In the left hand panel, the effect of cell flattening is illustrated using oblate spheroids as models. The circular bodies simulate the appearance of cells viewed from above while the gauge lines show how thick the model cells would be if viewed from the side. In the center panel, cells having dendritic projections are modeled as Gaussian probability curves that have been rotated in space to create solid bodies. Surface areas and volumes can be easily solved explicitly for these shapes. In the right hand panel, cell surface roughness is simulated by adding noise at three scale lengths (0.1 × radius, 0.4 × radius and 1.6 × radius) to represent microvilli, ruffles and folds on the cell surface. These simulations were made in MATLAB using distortions of the sphere function and the surface areas were computed by summing the areas of all facets on the resulting bodies.
3 Results and Discussion
Density ρ, hydrodynamic flexibility parameters Φ and crossover frequency fco were derived by measuring DEP-FFF elution profiles for each of the NCI-60 cell types and analyzing these data as detailed earlier [23, 24, 28]. The parameters obtained from our analysis together with the visual cell morphological analysis are shown in Table 1. Results for blood cells were taken from the literature where the dielectric data were measured by electrorotation (ROT) or by DEP crossover frequency methods (DCO). A number of cell types in Table 1 were measured by ROT and/or DCO as well as by DEP-FFF and good agreement in derived cell dielectric properties was found for these different methods.
Table 1.
Dielectric and exterior morphological parameters for the NCI-60 panel and blood cells.
| Cell Type | Method | Tissue Type | r, μm | fco, kHz | Cmem, mF.m−2 | Ctot, pF | Folding, ϕ | M=(F+P+R)/3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Basophils | DCO [31] | Blood | 3.58 | ± | 0.03 | 169 | ± | 18.8 | 11.2 | ± | 1.3 | 1.8 | ± | 0.2 | 1.24 | ± | 0.14 | 1.38 |
| B-Lymphocytes | ROT [49] | Blood | 3.29 | ± | 0.03 | 163 | ± | 45.3 | 12.6 | ± | 3.5 | 1.7 | ± | 0.5 | 1.40 | ± | 0.39 | 1.25 |
| DCO [31] | 3.09 | ± | 0.22 | 221 | ± | 17.8 | 9.9 | ± | 0.8 | 1.2 | ± | 0.1 | 1.10 | ± | 0.09 | |||
| CD34+ HSC | ROT [50] | Blood | 3.50 | ± | 0.05 | 189 | ± | 27.8 | 10.2 | ± | 1.5 | 1.6 | ± | 0.2 | 1.13 | ± | 0.17 | 1.25 |
| Eosinophils | DCO [31] | Blood | 4.19 | ± | 0.07 | 172 | ± | 7.3 | 9.4 | ± | 0.4 | 2.1 | ± | 0.1 | 1.04 | ± | 0.04 | 1.13 |
| Erythrocytes | ROT [51] | Blood | 3.10 | ± | 0.02 | 218 | ± | 21.8 | 10 | ± | 1.0 | 1.2 | ± | 0.1 | 1.11 | ± | 0.11 | 1.31 |
| DCO [51] | 3.10 | ± | 0.02 | 185 | ± | 23.5 | 11.8 | ± | 1.5 | 1.4 | ± | 0.2 | 1.31 | ± | 0.17 | |||
| ROT [14] | 2.8 | ± | 0.1 | 268 | ± | 23.8 | 9 | ± | 0.8 | 0.9 | ± | 0.1 | 1.00 | ± | 0.09 | |||
| Granulocytes (mixed) | ROT [49] | Blood | 4.71 | ± | 0.23 | 130 | ± | 37.9 | 11 | ± | 3.2 | 3.1 | ± | 0.9 | 1.22 | ± | 0.36 | |
| DFF | 4.70 | ± | 0.23 | 95 | ± | 13.9 | 15.1 | ± | 2.2 | 4.2 | ± | 0.6 | 1.68 | ± | 0.24 | |||
| Lymphocytes-mitototic | ROT | Blood | 4.54 | ± | 0.63 | 92 | ± | 17.8 | 16.1 | ± | 3.1 | 4.2 | ± | 0.8 | 1.79 | ± | 0.34 | 1.80 |
| Monocytes | ROT [49] | Blood | 4.63 | ± | 0.36 | 95 | ± | 26.8 | 15.3 | ± | 4.3 | 4.1 | ± | 1.2 | 1.70 | ± | 0.48 | 1.50 |
| DCO [31] | 4.21 | ± | 0.05 | 113 | ± | 6.4 | 14.2 | ± | 0.8 | 3.2 | ± | 0.2 | 1.58 | ± | 0.09 | |||
| Neutrophils | DCO [31] | Blood | 4.06 | ± | 0.06 | 170 | ± | 1.2 | 9.8 | ± | 0.1 | 2.0 | ± | 0.0 | 1.09 | ± | 0.01 | 1.13 |
| T-Lymphocytes | ROT [14] | Blood | 3.5 | ± | 0.2 | 176 | ± | 17.6 | 11 | ± | 1.1 | 1.7 | ± | 0.2 | 1.22 | ± | 0.12 | 1.38 |
| DCO [31] | 3.40 | ± | 0.08 | 149 | ± | 20.2 | 13.3 | ± | 1.8 | 1.9 | ± | 0.3 | 1.48 | ± | 0.20 | |||
| ROT [33] | 3.04 | ± | 0.26 | 184 | ± | 22.8 | 12.1 | ± | 1.5 | 1.4 | ± | 0.2 | 1.34 | ± | 0.17 | |||
| DFF | 3.40 | ± | 1.29 | 155 | ± | 35.2 | 12.8 | ± | 2.9 | 1.9 | ± | 0.4 | 1.42 | ± | 0.32 | |||
| BT-549 | DFF | Breast | 10.36 | ± | 3.53 | 45 | ± | 15.7 | 14.4 | ± | 5.0 | 20 | ± | 6.8 | 1.6 | ± | 0.56 | 2.00 |
| HS 578T | DFF | Breast | 8.88 | ± | 3.63 | 40.9 | ± | 12.4 | 18.5 | ± | 5.6 | 19 | ± | 5.6 | 2.05 | ± | 0.62 | |
| MCF7 | DFF | Breast | 9.08 | ± | 3.71 | 33.4 | ± | 5.4 | 22.2 | ± | 3.6 | 23 | ± | 3.8 | 2.46 | ± | 0.40 | |
| MDA-MB-231/ATTC | ROT [14] | Breast | 6.20 | ± | 0.58 | 42 | ± | 6.8 | 25.9 | ± | 4.2 | 12.5 | ± | 2.0 | 2.88 | ± | 0.47 | |
| DFF | 7.57 | ± | 2.58 | 32.3 | ± | 4.4 | 27.5 | ± | 3.7 | 20 | ± | 2.7 | 3.06 | ± | 0.41 | |||
| MDA-MB-435 | ROT [50] | Breast | 7.69 | ± | 2.92 | 38 | ± | 11.8 | 23 | ± | 7.1 | 17.1 | ± | 5.3 | 2.56 | ± | 0.79 | |
| ROT [14] | 7.70 | ± | 0.72 | 34 | ± | 5.5 | 26 | ± | 4.2 | 19.4 | ± | 3.1 | 2.89 | ± | 0.47 | |||
| DFF | 7.69 | ± | 2.92 | 32 | ± | 3.7 | 27.3 | ± | 3.2 | 20 | ± | 2.4 | 3.03 | ± | 0.35 | |||
| NCI/ADR-RES | DFF | Breast | 8.78 | ± | 3.28 | 28.4 | ± | 3.4 | 26.9 | ± | 3.2 | 26 | ± | 3.1 | 2.99 | ± | 0.36 | |
| SF-268 | DFF | CNS | 7.53 | ± | 2.77 | 32.4 | ± | 7.4 | 27.5 | ± | 6.3 | 20 | ± | 4.5 | 3.06 | ± | 0.70 | 2.18 |
| SF-295 | DFF | CNS | 10.02 | ± | 3.05 | 28 | ± | 3.2 | 24 | ± | 2.8 | 30 | ± | 3.5 | 2.67 | ± | 0.31 | 2.16 |
| SF-539 | DFF | CNS | 10.45 | ± | 3.35 | 25.6 | ± | 4.1 | 25.2 | ± | 4.0 | 35 | ± | 5.5 | 2.8 | ± | 0.44 | |
| SNB-19 | DFF | CNS | 5.83 | ± | 2.83 | 42.2 | ± | 14.2 | 27.4 | ± | 9.2 | 12 | ± | 3.9 | 3.05 | ± | 1.02 | |
| SNB-75 | DFF | CNS | 8.70 | ± | 3.20 | 26 | ± | 3.4 | 29.7 | ± | 3.9 | 29 | ± | 3.7 | 3.3 | ± | 0.43 | 2.24 |
| U251 | DFF | CNS | 7.67 | ± | 3.12 | 24.5 | ± | 4.5 | 35.8 | ± | 6.5 | 27 | ± | 4.9 | 3.97 | ± | 0.73 | 2.66 |
| COLO 205 | DFF | Colon | 9.39 | ± | 3.51 | 44.1 | ± | 7.2 | 16.3 | ± | 2.7 | 18 | ± | 3.0 | 1.81 | ± | 0.30 | |
| HCC-2998 | DFF | Colon | 8.45 | ± | 4.15 | 52.9 | ± | 8.8 | 15.1 | ± | 2.5 | 14 | ± | 2.3 | 1.67 | ± | 0.28 | 1.96 |
| HCT-116 | DFF | Colon | 8.84 | ± | 3.97 | 34.4 | ± | 6.1 | 22.1 | ± | 3.9 | 22 | ± | 3.9 | 2.46 | ± | 0.44 | 2.11 |
| HCT-15 | DFF | Colon | 7.49 | ± | 3.40 | 51.6 | ± | 9.7 | 17.3 | ± | 3.2 | 12 | ± | 2.3 | 1.92 | ± | 0.36 | 2.08 |
| HT-29 | DFF | Colon | 11.66 | ± | 4.43 | 45 | ± | 7.5 | 12.9 | ± | 2.2 | 22 | ± | 3.7 | 1.43 | ± | 0.24 | |
| KM12 | DFF | Colon | 9.52 | ± | 3.50 | 43 | ± | 7.1 | 16.4 | ± | 2.7 | 19 | ± | 3.1 | 1.83 | ± | 0.30 | |
| SW-620 | DFF | Colon | 6.36 | ± | 2.33 | 59.4 | ± | 10.0 | 17.8 | ± | 3.0 | 9.1 | ± | 1.5 | 1.98 | ± | 0.33 | 1.58 |
| 786-0 | DFF | Kidney | 8.97 | ± | 3.16 | 31.7 | ± | 4.5 | 23.6 | ± | 3.3 | 24 | ± | 3.4 | 2.63 | ± | 0.37 | 1.91 |
| A498 | DFF | Kidney | 10.29 | ± | 3.46 | 24.6 | ± | 3.0 | 26.5 | ± | 3.2 | 36 | ± | 4.3 | 2.94 | ± | 0.35 | 2.17 |
| ACHN | DFF | Kidney | 7.58 | ± | 3.03 | 32.1 | ± | 3.8 | 27.7 | ± | 3.3 | 20 | ± | 2.4 | 3.07 | ± | 0.36 | 2.05 |
| CAKI-1 | DFF | Kidney | 8.82 | ± | 3.60 | 25.2 | ± | 5.0 | 30.2 | ± | 6.0 | 30 | ± | 5.9 | 3.36 | ± | 0.67 | 2.53 |
| RXF 393 | DFF | Kidney | 9.97 | ± | 3.43 | 34.1 | ± | 6.6 | 19.8 | ± | 3.8 | 25 | ± | 4.8 | 2.2 | ± | 0.43 | 1.82 |
| SN-12C | DFF | Kidney | 9.64 | ± | 3.28 | 34 | ± | 5.7 | 20.4 | ± | 3.4 | 24 | ± | 4.0 | 2.26 | ± | 0.38 | |
| TK-10 | DFF | Kidney | 8.51 | ± | 3.06 | 33.8 | ± | 3.8 | 23.3 | ± | 2.6 | 21 | ± | 2.4 | 2.59 | ± | 0.29 | 2.11 |
| UO-31 | DFF | Kidney | 6.86 | ± | 1.62 | 34.1 | ± | 4.4 | 28.7 | ± | 3.7 | 17 | ± | 2.2 | 3.19 | ± | 0.42 | 2.31 |
| CCRF-CEM | DFF | Leukemia | 4.45 | ± | 1.69 | 88.2 | ± | 15.1 | 17.1 | ± | 2.9 | 4.3 | ± | 0.7 | 1.9 | ± | 0.33 | 1.22 |
| HL-60 | DFF | Leukemia | 8.42 | ± | 3.20 | 69 | ± | 11.5 | 11.6 | ± | 1.9 | 10.3 | ± | 1.7 | 1.29 | ± | 0.21 | |
| K-562 | DFF | Leukemia | 5.87 | ± | 2.48 | 54.9 | ± | 13.8 | 20.9 | ± | 5.2 | 9.1 | ± | 2.3 | 2.32 | ± | 0.58 | 1.28 |
| MOLT-4 | DFF | Leukemia | 5.56 | ± | 2.07 | 102 | ± | 14.7 | 11.8 | ± | 1.7 | 4.6 | ± | 0.7 | 1.31 | ± | 0.19 | 1.06 |
| RPMI-8226 | DFF | Leukemia | 7.67 | ± | 2.85 | 51.5 | ± | 12.0 | 17 | ± | 4.0 | 13 | ± | 2.9 | 1.89 | ± | 0.44 | 1.38 |
| SR | DFF | Leukemia | 6.81 | ± | 2.83 | 79.2 | ± | 20.5 | 12.5 | ± | 3.2 | 7.3 | ± | 1.9 | 1.39 | ± | 0.36 | 1.18 |
| LOX IMVI | DFF | Melanoma | 9.38 | ± | 3.49 | 53.2 | ± | 16.4 | 13.5 | ± | 4.2 | 15 | ± | 4.6 | 1.5 | ± | 0.46 | 2.13 |
| M14 | DFF | Melanoma | 7.70 | ± | 3.22 | 36.3 | ± | 4.8 | 24.1 | ± | 3.2 | 18 | ± | 2.4 | 2.68 | ± | 0.35 | 1.61 |
| MALME-3M | DFF | Melanoma | 9.29 | ± | 3.39 | 31 | ± | 3.4 | 23.3 | ± | 2.5 | 26 | ± | 2.8 | 2.59 | ± | 0.28 | 2.19 |
| SK-MEL-2 | DFF | Melanoma | 8.52 | ± | 3.20 | 33.6 | ± | 4.2 | 23.5 | ± | 2.9 | 22 | ± | 2.7 | 2.61 | ± | 0.32 | 2.11 |
| SK-MEL-28 | DFF | Melanoma | 8.82 | ± | 4.01 | 29.3 | ± | 3.5 | 26.1 | ± | 3.1 | 26 | ± | 3.1 | 2.9 | ± | 0.35 | 2.39 |
| UACC-257 | DFF | Melanoma | 7.76 | ± | 3.46 | 38.7 | ± | 4.9 | 22.5 | ± | 2.8 | 17 | ± | 2.1 | 2.5 | ± | 0.31 | |
| UACC-62 | DFF | Melanoma | 8.27 | ± | 3.86 | 37.8 | ± | 4.5 | 21.6 | ± | 2.6 | 19 | ± | 2.2 | 2.4 | ± | 0.28 | 2.33 |
| A549/ATCC | DFF | NSCL | 9.31 | ± | 2.99 | 29.4 | ± | 3.3 | 24.6 | ± | 2.7 | 27 | ± | 3.0 | 2.73 | ± | 0.30 | 2.06 |
| EKVX | DFF | NSCL | 10.74 | ± | 3.15 | 30.4 | ± | 4.2 | 20.6 | ± | 2.8 | 30 | ± | 4.1 | 2.29 | ± | 0.31 | 1.74 |
| HOP-62 | DFF | NSCL | 9.84 | ± | 3.84 | 33 | ± | 11.3 | 20.7 | ± | 7.1 | 25 | ± | 8.7 | 2.29 | ± | 0.78 | 1.97 |
| HOP-92 | DFF | NSCL | 10.68 | ± | 3.33 | 30.8 | ± | 3.0 | 20.5 | ± | 2.0 | 30 | ± | 2.9 | 2.27 | ± | 0.22 | 1.86 |
| NCI-H226 | DFF | NSCL | 8.57 | ± | 3.58 | 23.4 | ± | 3.1 | 33.6 | ± | 4.4 | 31 | ± | 4.1 | 3.73 | ± | 0.49 | 2.58 |
| NCI-H23 | DFF | NSCL | 8.94 | ± | 3.41 | 59.1 | ± | 18.2 | 12.8 | ± | 3.9 | 13 | ± | 3.9 | 1.42 | ± | 0.44 | |
| NCI-H322M | DFF | NSCL | 9.37 | ± | 3.12 | 26.5 | ± | 5.9 | 27.1 | ± | 6.1 | 30 | ± | 6.7 | 3.01 | ± | 0.67 | 2.69 |
| NCI-H460 | DFF | NSCL | 9.85 | ± | 3.08 | 38.2 | ± | 5.4 | 17.9 | ± | 2.5 | 22 | ± | 3.1 | 1.99 | ± | 0.28 | 1.61 |
| NCI-H522 | DFF | NSCL | 8.51 | ± | 3.76 | 53.4 | ± | 16.5 | 14.8 | ± | 4.6 | 14 | ± | 4.2 | 1.64 | ± | 0.51 | 2.27 |
| IGR-OV1 | DFF | Ovarian | 9.90 | ± | 3.56 | 40.4 | ± | 6.7 | 16.8 | ± | 2.8 | 21 | ± | 3.5 | 1.86 | ± | 0.31 | 1.61 |
| OVCAR-3 | DFF | Ovarian | 10.37 | ± | 3.94 | 27 | ± | 4.5 | 24.1 | ± | 4.0 | 33 | ± | 5.4 | 2.68 | ± | 0.45 | |
| OVCAR-4 | DFF | Ovarian | 10.21 | ± | 3.79 | 35.6 | ± | 5.2 | 18.5 | ± | 2.7 | 24 | ± | 3.6 | 2.06 | ± | 0.30 | 1.72 |
| OVCAR-5 | DFF | Ovarian | 8.49 | ± | 3.21 | 38.3 | ± | 5.3 | 20.7 | ± | 2.8 | 19 | ± | 2.6 | 2.3 | ± | 0.32 | 2.32 |
| OVCAR-8 | DFF | Ovarian | 7.63 | ± | 3.26 | 24.1 | ± | 3.7 | 36.3 | ± | 5.6 | 27 | ± | 4.2 | 4.04 | ± | 0.63 | 2.71 |
| SKOV-3 | DFF | Ovarian | 10.41 | ± | 3.96 | 33 | ± | 5.5 | 19.7 | ± | 3.3 | 27 | ± | 4.5 | 2.19 | ± | 0.37 | |
| DU-145 | DFF | Prostate | 8.28 | ± | 3.01 | 40.5 | ± | 5.7 | 20.1 | ± | 2.8 | 17 | ± | 2.4 | 2.23 | ± | 0.31 | 1.69 |
| PC-3 | DFF | Prostate | 11.42 | ± | 4.34 | 26 | ± | 4.4 | 22.4 | ± | 3.7 | 37 | ± | 6.1 | 2.49 | ± | 0.42 | |
For the NCI-60 cells, dielectric parameters were derived from DEP-FFF [23-25]. Some literature values were obtained by electrorotation (ROT) or DEP crossover (DCO) methods, which give comparable results. Morphological parameters were scored visually as described in the text. Blood cell morphological parameters were scored from The Clinical Hematology Atlas[52]. (CNS=Central nervous system; NSCL=Non-small cell lung).
The magnitude and direction of the DEP force imposed on an intact cell suspended in a medium having a conductivity much lower than that of its cytoplasm by an inhomogeneous alternating AC electric field of frequency f may be approximated as
| (1) |
where r is the cell radius, ∇E2 is the gradient of the mean squared field strength, εsε0 is the electric permittivity of the suspending medium, and fCO is the cell crossover frequency, which is proportional to the rate at which the plasma membrane capacitance can be charged in the ionic milieu of the suspending medium in response to an applied electric field. The DEP force is null and changes direction when the applied electric field frequency is fCO. At DEP frequencies well below 1 MHz, a spherical mammalian cell under these conditions has a crossover frequency that may be approximated by the expression
| (2) |
where σs is the medium conductivity [27, 29, 30]. We and others have shown that Cmem, the capacitance per unit area of the cell plasma membrane, varies substantially between different cell types[31], cells in different states of differentiation[26, 27, 32], in different stages of the cell cycle[33, 34] and following exposure of cells to apoptosis-inducing agents[35, 36] and to toxicants[36, 37]. By correlating cell dielectric measurements with membrane features revealed by electron microscopy, we showed early on that these differences in Cmem were related to differences in membrane area associated with cell size and with surface features such as ruffles, folds and microvilli [26, 33]. This finding was consistent with the interpretation of electrophysiological measurements of cell capacitance where Ctot is used to infer the total area of the cell plasma membrane[38]. Smooth plasma membrane has a capacitance of C0 ≈ 9 mF.m−2 [39] but features such as membrane folds, ruffles and microvilli cause the area to increase by an effective folding factor ϕ ≥ 1 compared to that of a smooth, spherical cell. Then ϕ is the ratio of the actual cell membrane surface area to that of an ideal, smooth sphere of the same radius and a measure of how wrinkled the cell surface is in suspension. The effective capacitance per unit area of the cell may then be written Cmem = ϕ C0 and the total capacitance of the cell is Ctot = 4π r2ϕ C0 [26]. It follows that our early work drew attention to two cell variables that determine DEP properties of viable cells in suspension, namely the radius, r, which is different for different cell types, and the membrane folding factor, ϕ, which is also known to vary from one cell type to another independently of radius. The goal of this article is to examine the biological basis for these cell differences to better understand the applicability of DEP to research and clinical problems. To accomplish this, we employed the NCI-60 panel of cell types[20, 22], which represents a diverse range of cancers from different organs, of primary versus metastatic origin, of epithelial versus mesenchymal character, and of vastly different doubling times, metabolic rates, and degrees of differentiation.
Using Eqn. 2, we can express the cell membrane folding factor and total capacitance in terms of the DEP crossover frequency as
| (3a) |
| (3b) |
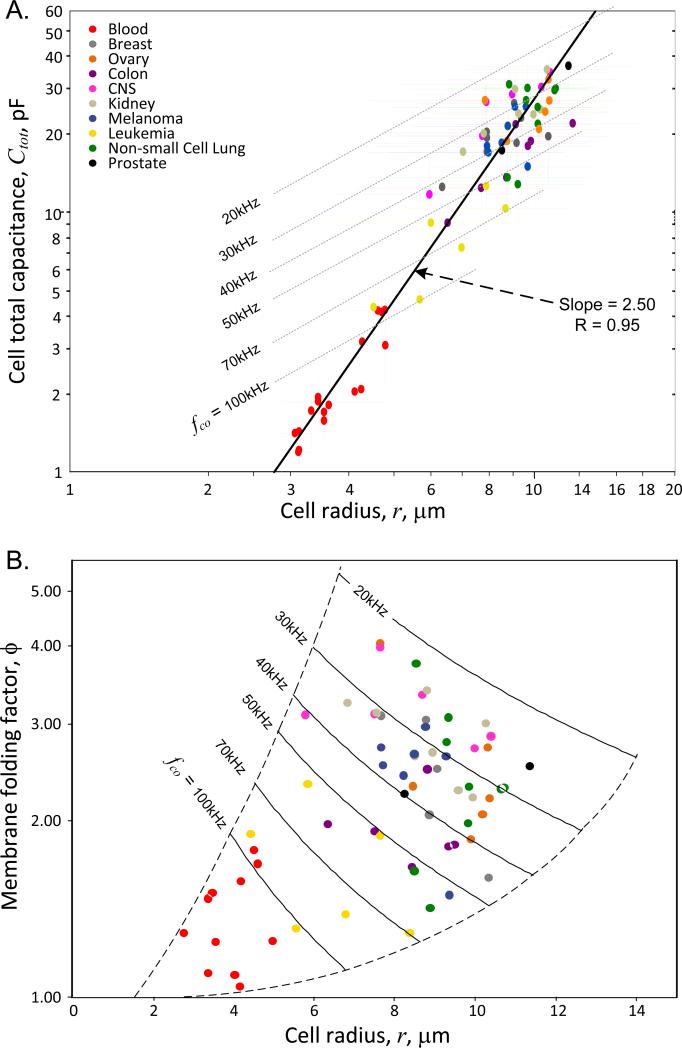
The DEP crossover frequencies and corresponding cell capacitances and membrane folding factors calculated according to these expressions for the NCI-60 cell lines and for the cell subpopulations of peripheral blood are given in Table 1. The relationships between these derived parameters and cell size are plotted in Fig. 3. The slope of the regression line in Fig. 3A shows that the cell total capacitance tends to vary as r2.5, indicating that the cell plasma membrane area bears a fractal dimensional relationship to cell size. Earlier, we found a similar fractal relationship for a different set of 26 cell lines [23] and also showed that the dielectric frequency spectrum of single cells is consistent with fractal membrane morphology [40]. Fractal relationships are common when transport through surfaces supports biological functions that are distributed through volumes such as in the alveoli of lungs [41, 42] and the capillary beds of organs [42]. The plasma membrane surface area of a single cell must similarly provide transmembrane flux to support homeostasis within the entire cell volume, requiring the surface to volume ratio to deviate from that of a smooth sphere and leading to the fractal relationship seen in Fig. 3A between cell membrane area (as manifested in cell capacitance) and cell size. Nevertheless, it is also apparent from Fig. 3A that membrane capacitance values for any given cell size span a wide range (see dashed upper and lower bounds), indicating that cell radius is not the sole driver of membrane area.
Figure 3.
(A) Dependency of total capacitance on cell radius, r, for the NCI-60 and blood cell types. Three broad clusters comprising the blood cell types (red), the leukemia cell types (yellow), and all other cell types are apparent. The slope of this log-log plot shows that cell capacitance varies as r2.5, consistent with a fractal relationship between cell surface area and cell volume. (B) Cell capacitance data for the NCI-60 and blood cell types expressed as the dependency of the plasma membrane folding factor, ϕ, on cell radius. Loci corresponding to constant DEP crossover frequencies based on Eqn. 2 are shown. The plasma membrane folding factor tends to increase with increasing cell size.
While the exterior morphology of each cell type in the NCI-60 panel may be different, we can state in general that any cell has a plasma membrane area A and a volume V that may deviate substantially from the corresponding parameters of a smooth sphere. If the cell has an exterior morphology that deviates significantly from that of a sphere then the area A needed to enclose the cell volume will be significantly greater than that needed to enclose a spherical cell of the same volume V. Once the cell is released into suspension during harvest or shedding from its tissue of origin, however, the anchorage points that maintain its conformation at the site of origin will be broken and tension in the cytoskeleton will tends to round the cell into a (wrinkled) sphere. If the cell membrane area and volume are conserved during this transition, the radius of the rounded cell will become to conserve cell volume and the plasma membrane will assume a folding factor ϕ = A/(4π · r2) to conserve cell membrane area. Eliminating r between these expressions gives
| (4) |
We can then rewrite the cell crossover frequency according to Eqn. 2 in a novel way in terms of the cell volume and area as
| (5) |
As already indicated, our previous studies established that the crossover frequency of cells in suspension is inversely proportional to the total cell surface area including that associated with membrane-rich features such as microvilli, ruffles and folds [2, 26, 43]. Eqn. 4 and 5 go a step further by suggesting that these membrane-rich surface features can be understood in terms of conservation of cell plasma membrane area during release into suspension. Eqn. 5 shows that, in general, the crossover frequency of any cell can be defined in terms of the morphological length parameter .
The rich variety of cell morphologies within the NCI-60 panel allowed us to examine the relationships in Eqn. 4 and 5. The M scores were obtained by examining the cell exterior morphologies prior to release into suspension in order to provide a visual estimate of the relative membrane areas of the cells at their growth sites in comparison to smooth spheres (M = 1) of similar volume. We stained some of the cell lines in culture with CellMask orange fluorescent plasma membrane stain (C10045, Molecular Probes, Life Technologies) and green-fluorescent calcein-AM viable cell stain (L-3224, Molecular Probes, Life Technologies) and this allowed us to visualize by fluorescent microscopy the fate of the cell membranes and the cytoplasmic contents during harvest. Cells were found to retain essentially all of their membrane while showing no obvious leakage of cytoplasmic fluorescence during release, suggesting that the cell membrane area and cell volume were conserved. Under these circumstances, the membrane folding factors ϕ derived from DEP-FFF should correlate with the M scores. Fig. 4, shows that the expected correlation between ϕ and M exists, demonstrating clearly that the DEP properties of the various cell types depended not only on cell radius after release into suspension but also on the exterior morphology of the cell types at their site of origin prior to release.
Figure 4.
Relationship between the membrane folding factors of the NCI-60 cell types as measured by DEP-FFF in cell suspension and the cell exterior morphologies in culture just prior to harvest as judged by visual estimates of the combination of spreading, elongation and surface roughness (see text and Fig. 1 and 2). Blood cell types are plotted based on DEP-derived folding factors with estimates of their morphological scores based on microographs in the literature.
The results in Fig. 3 and 4 may be considered in terms of the source of the different cell types. Blood cells are usually in the resting G0 phase of the growth cycle and are adapted to travelling through small capillary vessels of the circulatory system while being suspended in nutrient-rich plasma. Their small size, spherical, relatively smooth membrane morphology and small membrane folding factors represent structures that are appropriate for these functions. Peripheral blood cell subpopulations (red circles) lay clustered nearest to the origins of Fig. 4 and 3B and these cell characteristics lead to crossover frequencies mostly in excess of 100 kHz in our DEP eluate buffer of 30 mS.m−1 conductivity. Leukemia cells (yellow circles) are similarly suited to travelling through capillary vessels while being suspended in nutrient-rich plasma. However, these cells have a tendency to be outside the resting G0 phase of the cell division cycle and exhibit a slightly larger size and membrane morphological complexity. As a result, these cells tend to have slightly lower crossover frequencies than the normal, resting peripheral blood cells in Fig. 3. Finally, solid cancer cell types are adapted to close coordination, spreading and contact with neighboring cells and retain varying degrees of the morphological characteristics of differentiated tissue. Fig. 4 and 3b show that while the membrane morphological complexities of cells of solid cancer origin vary over a wide range, they are much greater than the variations seen in normal blood cell types. The larger size and greater membrane folding of solid tumor types result in crossover frequencies that are much lower than those of the blood cell subpopulations.
These results show that the dielectric properties of suspended cells depend on membrane morphology at the cells’ native growth sites. This finding has great significance to the application of DEP to cell separation and isolation problems because it allows the properties of cells to be anticipated in terms of cell structure-function relationships. We showed earlier that cancer cells have lower crossover frequencies than peripheral blood cells and the data presented here reinforces this by demonstrating that all solid tumor types in the NCI-60 panel also exhibit DEP crossover frequencies that are distinct from those of peripheral blood cell subpopulations and would permit them to be isolated from blood.
Our results also show that, while cell size clearly does contribute significantly to dielectric differences between different cell types soon after harvest, cell membrane morphology can also vary significantly among cells of the same size. For example, in Fig. 3A cells of radius close to 8.7 μm include a leukemia with a cell capacitance of 11.7 pF, prostate, ovary and kidney lines having capacitances near 20 pF, and non-small cell lung, breast and central nervous system cancers having capacitances near 30 pF - roughly a three-fold range in cell capacitance among cells of similar size. These different clusters of cell types may be distinguished and isolated from one another by DEP.
This disconnect between cell size and cell dielectric properties has potential importance for the isolation of circulating tumor cells. Tumor cells that have been released and remain in suspension undergo remodeling whereby membrane and cytoplasm are shed over a period of hours without loss of cell viability [23]. This process eventually leads the cancer cells to reach the same diameter as monocytes or granulocytes and causes CTCs to exhibit dramatic pleomorphism. Yet, because of the fundamentally different initial exterior morphology characteristics of cancer and blood cell types, the capacitance of the cancer cells remains higher than that of blood cells during shedding so that the cancer cells can still be isolated from blood by DEP[23] even when their size is comparable to blood cells. Small, remodeled cancer cells of this type cannot be isolated from blood by size-based filtering.
Our results provide a biological rationale for the plasma membrane-associated dielectric differences between different cell types and suggest the possibility of additional analysis. The membrane morphology at the site of origin of cells must reflect their gene expression profiles and future work may allow cell dielectric differences to be related to specific patterns of gene expression. For example, cell size is known to be affected by the mTOR pathway[44] and a variety of cell adhesion[45] and cytoskeletal[46] proteins are known to influence cell adhesion and spreading. The current clinical method[47, 48] of isolating CTCs employs EpCAM cell surface protein that is associated with cell-cell adhesion at the site of origin of CTCs of epithelial phenotype. Even though DEP does not depend on cell surface markers, its ability to discriminate between cell types nevertheless appears to build broadly on this concept of cell properties associated with the site of origin.
4 Concluding remarks
We have presented dielectric data for the NCI-60 panel of cells and compared them with similar data for normal peripheral blood cells. The results confirm our earlier findings that the dielectric properties of cells in suspension correlate not only with size but also with membrane morphology. We show further that these cell properties in suspension can be explained by the conservation of cell volume and membrane area during the release of cells into suspension from their sites of origin. Our findings thereby provide a rationale for anticipating the dielectric properties of cells in suspension based on their native structure-function relationships. This allows us to understand earlier results, and additional ones presented here, showing that cancer cells have consistently lower DEP crossover frequencies than peripheral blood cells. As a consequence of the conservation of cell properties, DEP is able to discriminate between cells of similar size having different morphological origins and is therefore able to isolate cancer cells from blood cells of the same size. This suggests that DEP offers significant advantages over size-based filtering for isolating circulating tumor cells.
Acknowledgements
We thank Tom Anderson for design, fabrication and engineering in creating and maintaining the DEP-FFF instrumentation used in this study. We are also grateful to Patrick Girardet for undertaking experiments to track the fate of cell plasma membranes during harvest og cultured cells. NCI-60 cells are under Materials Transfer Agreement and graciously contributed by Dr. Gordon B. Mills. This work was supported by grant RP100934 from the Cancer Prevention and Research Institute of Texas (CPRIT) and funding from the Kleberg Center for Molecular Markers. KSH is also supported by a Stand Up to Cancer Dream Team Translational Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0209). STR DNA fingerprinting was done by the Cancer Center Support Grant-funded Characterized Cell Line core, NCI # CA016672.
Abbreviations
- CTC
circulating tumor cell
- FFF
field-flow fractionation
- PBMN
peripheral blood mononuclear cell
Footnotes
Conflict of Interest
PG and FFB qualify to receive royalty distributions from patents assigned to the Regents of the University of Texas and licensed for commercial development related to DEP-FFF technology.
References
- 1.Pethig R. Biomicrofluidics. 2010:4. doi: 10.1063/1.3456626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pethig R, Menachery A, Pells S, De Sousa P. J Biomed Biotechnol. 2010:182581. doi: 10.1155/2010/182581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piacentini N, Mernier G, Tornay R, Renaud P. Biomicrofluidics. 2011:5. doi: 10.1063/1.3640045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borgatti M, Altomare L, Baruffa M, Fabbri E, et al. International Journal of Molecular Medicine. 2005;15:913–920. [PubMed] [Google Scholar]
- 5.Oeinck M, Riehemann K, Fuchs H, Schnekenburger J. Pancreas. 2009;38:1034–1034. [Google Scholar]
- 6.Thomas RS, Mitchell PD, Oreffo RO, Morgan H. Biomicrofluidics. 2010:4. doi: 10.1063/1.3406951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou H, Syms RRA, Mellon S, Tanner KE. Micro and Nano Technology. 2009:60–61. 63–67. [Google Scholar]
- 8.Salmanzadeh A, Romero L, Shafiee H, Gallo-Villanueva RC, et al. Lab on a Chip. 2012;12:182–189. doi: 10.1039/c1lc20701f. [DOI] [PubMed] [Google Scholar]
- 9.Broche LM, Bhadal N, Lewis MP, Porter S, et al. Oral Oncology. 2007;43:199–203. doi: 10.1016/j.oraloncology.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Mulhall HJ, Labeed FH, Kazmi B, Costea DE, et al. Anal Bioanal Chem. 2011;401:2455–2463. doi: 10.1007/s00216-011-5337-0. [DOI] [PubMed] [Google Scholar]
- 11.Sabuncu AC, Liu JA, Beebe SJ, Beskok A. Biomicrofluidics. 2010;4 doi: 10.1063/1.3447702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang F, Yang XM, Jiang H, Bulkhaults P, et al. Biomicrofluidics. 2010;4 doi: 10.1063/1.3279786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker FF, Wang XB, Huang Y, Pethig R, et al. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:860–864. doi: 10.1073/pnas.92.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gascoyne PR, Wang XB, Huang Y, Becker FF. IEEE Trans Ind Appl. 1997;33:670–678. doi: 10.1109/28.585856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon HS, Kwon K, Kim SI, Han H, et al. Lab Chip. 2011;11:1118–1125. doi: 10.1039/c0lc00345j. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Lu J, Marchenko SA, Monuki ES, et al. Electrophoresis. 2009;30:782–791. doi: 10.1002/elps.200800637. [DOI] [PubMed] [Google Scholar]
- 17.Pratt ED, Huang C, Hawkins BG, Gleghorn JP, Kirby BJ. Chemical Engineering Science. 2011;66:1508–1522. doi: 10.1016/j.ces.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokilde R, Kaczkowski B, Podolska A, Cirera S, et al. Mol Cancer Ther. 2010;10:375–384. doi: 10.1158/1535-7163.MCT-10-0605. [DOI] [PubMed] [Google Scholar]
- 19.Pfister TD, Reinhold WC, Agama K, Gupta S, et al. Mol Cancer Ther. 2009;8:1878–1884. doi: 10.1158/1535-7163.MCT-09-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenzi PL, Reinhold WC, Varma S, Hutchinson AA, et al. Mol Cancer Ther. 2009;8:713–724. doi: 10.1158/1535-7163.MCT-08-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shankavaram UT, Reinhold WC, Nishizuka S, Major S, et al. Mol Cancer Ther. 2007;6:820–832. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 22.Roschke AV, Tonon G, Gehlhaus KS, McTyre N, et al. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- 23.Shim S, Gascoyne P, Noshari J, Hale KS. Integrative Biology. 2011;3:850–862. doi: 10.1039/c1ib00032b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gascoyne PRC. Field-Flow Fractionation in Biopolymer Analysis. Springer-Verlag; 2012. Isolation and Characterization of Cells by Dielectrophoretic Field-Flow Fractionation. [Google Scholar]
- 25.Shim S, Stemke-Hale K, Noshari J, Becker F, Gascoyne P. Biomicrofluidics. 2013;7 doi: 10.1063/1.4774307. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang XB, Huang Y, Gascoyne PR, Becker FF, et al. Biochimica et Biophysica Acta. 1994;1193:330–344. doi: 10.1016/0005-2736(94)90170-8. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Wang XB, Becker FF, Gascoyne PR. Biochimica et Biophysica Acta. 1996;1282:76–84. doi: 10.1016/0005-2736(96)00047-8. [DOI] [PubMed] [Google Scholar]
- 28.Gascoyne PR. Anal Chem. 2009;81:8878–8885. doi: 10.1021/ac901470z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marszalek P, Zielinsky JJ, Fikus M, Tsong TY. Biophys J. 1991;59:982–987. doi: 10.1016/S0006-3495(91)82312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones TB, Kallio GA. Journal of Electrostatics. 1979;6:18. [Google Scholar]
- 31.Vykoukal DM, Gascoyne PRC, Vykoukal J. Integrative Biology:Quantitative Biosciences from Nano to Macro. 2009;1:477–484. doi: 10.1039/b906137a. [DOI] [PubMed] [Google Scholar]
- 32.Cristofanilli M, De Gasperis G, Zhang L, Hung M-C, et al. Clinical Cancer Research. 2002;8:615–619. [PubMed] [Google Scholar]
- 33.Huang Y, Wang XB, Gascoyne PR, Becker FF. Biochimica et Biophysica Acta. 1999;1417:51–62. doi: 10.1016/s0005-2736(98)00253-3. [DOI] [PubMed] [Google Scholar]
- 34.Kim U, Shu C-W, Dane KY, Daugherty PS, et al. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20708–20712. doi: 10.1073/pnas.0708760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Becker FF, Gascoyne PR. Biochim Biophys Acta. 2002;1564:412–420. doi: 10.1016/s0005-2736(02)00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pui-ock S, Ruchirawat M, Gascoyne P. Analytical Chemistry. 2008;80:7727–7734. doi: 10.1021/ac801095p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ratanachoo K, Gascoyne PRC, Ruchirawat M. Biochimica et Biophysica Acta. 2002;1564:449–458. doi: 10.1016/s0005-2736(02)00494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chowdhury HH, Zorec R. Archives of physiology and biochemistry. 2012;118:121–127. doi: 10.3109/13813455.2012.677453. [DOI] [PubMed] [Google Scholar]
- 39.Pethig R, Kell DB. Phys Med Biol. 1987;32:933–970. doi: 10.1088/0031-9155/32/8/001. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Becker FF, Gascoyne PRC. Chaos. 2011;20:043133. doi: 10.1063/1.3526972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tinajero JP, Robledo RF, Lantz RC, Sobonya RE, et al. Res Commun Mol Pathol Pharmacol. 1997;95:275–285. [PubMed] [Google Scholar]
- 42.Lorthois S, Cassot F. Journal of theoretical biology. 2010;262:614–633. doi: 10.1016/j.jtbi.2009.10.037. [DOI] [PubMed] [Google Scholar]
- 43.Gascoyne PR, Pethig R, Burt JP, Becker FF. Biochimica et Biophysica Acta. 1993;1149:119–126. doi: 10.1016/0005-2736(93)90032-u. [DOI] [PubMed] [Google Scholar]
- 44.Hall MN, Raff M, Thomas G. Cell growth: Control of Cell Size. Cold Spring Harbor Laboratory Press; Woodbury, New York: 2004. [Google Scholar]
- 45.Tominaga T, Barber DL. Mol Biol Cell. 1998;9:2287–2303. doi: 10.1091/mbc.9.8.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maurin B, Canadas P, Baudriller H, Montcourrier P, Bettache N. Journal of biomechanics. 2008;41:2036–2041. doi: 10.1016/j.jbiomech.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 47.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, et al. J Clin Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 48.Budd GT, Cristofanilli M, Ellis MJ, Stopeck A, et al. Clin Cancer Res. 2006;12:6403–6409. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- 49.Yang J, Huang Y, Wang X, Wang XB, et al. Biophysical Journal. 1999;76:3307–3314. doi: 10.1016/S0006-3495(99)77483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Y, Yang J, Wang XB, Becker FF, Gascoyne PR. Journal of Hematotherapy & Stem Cell Research. 1999;8:481–490. doi: 10.1089/152581699319939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gascoyne P, Pethig R, Satayavivad J, Becker FF, Ruchirawat M. Biochimica et Biophysica Acta. 1997;1323:240–252. doi: 10.1016/s0005-2736(96)00191-5. [DOI] [PubMed] [Google Scholar]
- 52.Carr J, Rodak B. Clinical Hematology Atlas. 3rd Edition Elsevier (W.B. Saunders); 2008. [Google Scholar]