Abstract
Rhynchocyon udzungwensis is a recently described and poorly understood sengi (giant elephant-shrew) endemic to two small montane forests in Southern Tanzania, and surrounded in lower forests by R. cirnei reichardi. In this study, we investigate the molecular genetic relationship between R. udzungwensis and R. c. reichardi, and the possible role that shifting species distributions in response to climate fluctuations may have played in shaping their evolutionary history. Rhynchocyon udzungwensis and R. c. reichardi individuals were sampled from five localities for genetic analyses. Three mitochondrial and two nuclear loci were used to construct species trees for delimitation and to determine whether introgression was detectable either from ancient or ongoing hybridization. All species-tree results show R. udzungwensis and R. c. reichardi as distinct lineages, though mtDNA shows evidence of introgression in some populations. Nuclear loci of each species were monophyletic, implying introgression is exclusively historical. Because we found evidence of introgression, we used distribution data and species distribution modelling for present, glacial, and interglacial climate cycles to predict how shifting species distributions may have facilitated hybridization in some populations. Though interpretations are affected by the limited range of these species, a likely scenario is that the mtDNA introgression found in eastern mid-elevation populations was facilitated by low numbers of R. udzungwensis that expanded into lowland heavily occupied R. c. reichardi areas during interglacial climate cycles. These results imply that relationships within the genus Rhynchocyon may be confounded by porous species boundaries and introgression, even if species are not currently sympatric.
Introduction
When a new species is described to science, understanding its genetic and morphological distinctness, relationship to congeneric taxa, the permeability of species boundaries, and overall evolutionary history are necessary to place the lineage in the grand scheme of evolution. While many initial investigations document morphological and sometimes molecular relationships to related lineages, considering broader aspects of evolution including non-linear species divergence and interspecific interactions may provide a more accurate estimate of evolutionary history and uniqueness. The prevalence of introgression is non-trivial, as ∼10% of animal species appear to have hybridization and introgression in their evolutionary past [1]. In this study, we investigate the evolutionary history and genetic distinctness of the newly described grey-faced sengi or elephant-shrews (Rhynchocyon udzungwensis Rathbun and Rovero 2008) and its relationship to the parapatric chequered sengi (Rhynchocyon cirnei reichardi Peters 1847) in the Udzungwa Mountains of Tanzania. Evolutionary relationships within the genus Rhynchocyon are poorly understood, and the relationship of the newest member to other East African species and subspecies is of great interest to conservation efforts for preserving unique lineages in the face of small population sizes and human degradation of habitat.
The grey-faced sengi, R. udzungwensis, was described in 2008 [2] as the largest member of a poorly known group of African mammals of ancient and intriguing evolutionary history. Sengis are an ancient monophyletic order (Macroscelidea) of 18 extant species, endemic to Africa [3], [4]. Based largely on molecular data, sengis belonging to the super-cohort Afrotheria, which also includes elephants, hyraxes, sea cows (Paenungulata), tenrecs and golden moles (Afrosoricida), and the aardvark (Tubulidentata) [5]–[7]. The sengi order consists of two subfamilies: the forest-dwelling giant sengis (subfamily Rhynchocyoninae, single genus Rhynchocyon, species = 4), and the soft-furred, arid adapted sengis (subfamily Macroscelidinae, three genera: Macroscelides (species = 2), Petrodromus (species = 1), and Elephantulus (species = 11)). While a large-scale phylogeny of most sengis has been completed [8], little is known about the taxonomy and phylogenetic relationships of the giant sengis (Rhynchocyon) [3], [9]. Corbet & Hanks [10] recognized three Rhynchocyon species based on morphological differences and allopatric distribution (four are now recognized with the discovery of R. udzungwensis). They also described six subspecies within R. cirnei, where R. cirnei reichardi occurs in the Udzungwa Mountains and the highlands of southwestern Tanzania and northern Malawi. This taxonomy, however, is based nearly exclusively on pelage patterns and is debatable in some cases [3], [11]. While sufficient sampling for a robust phylogeny of Rhynchocyon is currently beyond our grasp because giant sengis are elusive and sufficient samples are not available, we now have adequate material to investigate the relationship between two parapatric lineages: the newly described R. udzungwensis and the more widespread R. cirnei reichardi.
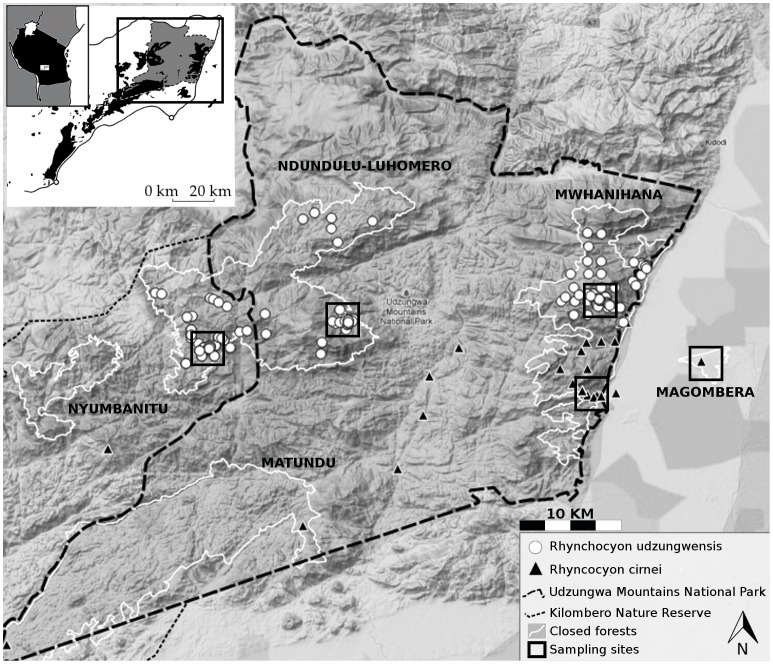
Rhynchocyon udzungwensis has an extremely small range (ca. 390 km2) limited to two forests in the northern Udzungwa Mountains in Tanzania: the western Ndundulu-Luhomero forest and the eastern Mwanihana forest, which are separated by a ∼25 km gap of lower elevation and drier woodland [2]. The range of R. udzungwensis is nearly encompassed by the range of R. cirnei reichardi, and the two taxa present partial overlap in elevation (R. udzungwensis occurring between 350 and 2300 meters elevation and R. c. reichardi between 290 and 1800 meters in the Udzungwa Mountains [2], [10]). The distributions of the two taxa meet only along the central part of the eastern Mwanihana forest ([12]; Fig. 1), with no indication of physical barriers, abrupt habitat changes, or steep rises. While there is no evidence of intermediate pelage colouration between these two taxa that might suggest hybridization (based on camera-trapping images, sightings, and voucher specimens), the physical proximity of these congeneric lineages, coupled with small population sizes estimated for R. udzungwensis, led us to include the relationship with R. c. reichardi when investigating R. udzungwensis’ evolutionary history.
Figure 1. Map of the north-eastern portion of the Udzungwa Mountains, south-central Tanzania with sampling localities.
Distribution records of the grey-faced sengi, Rhynchocyon udzungwensis, shown as white dots. The occurrence of the checkered sengi, R. cirnei reichardi, shown as black triangles. The western portion of the distribution of R. udzungwensis is in Ndundulu/Luhomero forest, and the eastern portion is in Mwanihana forest. Rhynchocyon c. reichardi’s distribution extends into the Magombera forest on the eastern plain. The five sampling sites for molecular analysis of the two species are also shown. The background layer is a topographic map (dark is lower elevation). The inset shows the location of the Udzungwa Mountains in Tanzania (left) and of the mapped area within the Udzungwa Mountains (right). Insert reprinted from [75] under a CC BY license, with permission from A. Marshall, original copyright 2005.
We began this study with a simple investigation of genetic distinction between the two morphological lineages, and then added additional molecular data, species tree analyses, and current and historical species distribution modelling (SDM; see [13] for a review) in order to provide a scenario that could accommodate non-bifurcating molecular data. In this way, we can determine how historical scenarios of contact, isolation, and introgression have shaped evolution for rare and isolated montane fauna, and better understand evolutionary dynamics in a biodiversity hotspot. Species with stable, allopatric distributions are not expected to be prone to hybridization, but species with historic range shifts tracking fluctuating climatic conditions may have experienced temporary sympatry and associated genetic introgression [14]–[17]. While climate-driven range shifts have been documented in a wide variety of settings, the most pronounced shifts are expected in montane species, as climatic similarity may correlate with distant montane areas rather than nearby lowlands [18], [19]. In particular, by using SDM through time, we are able to assess the potential of overlap between sengis during both current and past time periods to accommodate the possibility of low-level historical and/or current introgression [20].
Materials and Methods
Ethics Statement
All field procedures involving live animals met the standards for the ethical and humane treatment of animals of the American Society of Mammalogists and the 2000 American Veterinary Medical Association guidelines [21], [22], and all precautions to minimize pain, distress or suffering by trapped animals were taken. Vouchered animals were euthanized using chloroform, as approved by the Guidelines of the American Society of Mammalogists for the use of wild mammals in research [21]. All fieldwork was performed under research/collecting permit number 2008-311-ER-2008-150 to SR and permit number 2010-270-ER-2009-49 to FR, issued by the Tanzania Wildlife Research Institute. Research and collection permits were also issued by the Tanzania Commission for Science and Technology and by National Parks. Materials were exported under Tanzania Wildlife Division/CITES Office export permits #57189 and #58267 to SR.
Tissue Sampling
We collected tissues for molecular analysis from 31 specimens in Tanzania from 2006–2009: R. udzungwensis (n = 22, localities = 3), R. cirnei reichardi (n = 8, localities = 2), and one outgroup sengi, black-and-rufous sengi (Rhynchocyon petersi, n = 1, locality = riparian forest on the north side of the Rufiji river, 150 km east of the Udzungwa Mountains) (Fig. 1). For live animals to be released, tissue collection consisted of a small amount of hair along with a tiny piece of auricle. The ear was then disinfected with betadine and the animal was monitored for a few minutes to ensure no adverse reaction. Dead specimens and specimens destined for voucher had fresh muscle, liver, or kidney tissues preserved in 95% ethanol. Tissue samples for genetic analysis are stored at the Foundation Edmund Mach in Trento, Italy. A complete list of sampled specimens and sampling localities can be found in Table S1.
We trapped sengis with nylon twin snares [2] and double-door live traps (Tomahawk Live Traps, 24×6×6 inch) that were set where sengi were likely to travel. Capture and handling of specimens followed the Animal Care and Use Committee of the American Society of Mammalogists Guidelines. Voucher specimens were collected (R. udzungwensis: n = 8 (of which 4 were specimens taken for the description and of these, 3 were given to other museums), R. cirnei reichardi: n = 5 (including one given to California Academy of Sciences) and were prepared as study skins and skulls.
Genetic Analyses
Genomic DNA was extracted with the DNeasy Tissue kit (Qiagen, Valencia, USA) following manufacturer’s instructions. We used PCR to amplify fragments of three mitochondrial loci: NADH dehydrogenase 2 (ND2: Met-1 and Trp-2 [23]), the hypervariable 5′ end of the control region (D-loop: L0-F [24] and E3 [25]) and the ribosomal gene 12S (12S –F & 12S-R, [26]) totalling 1756 bp. For the mtDNA markers we used 10 ng of DNA and PCR conditions were as follows: 94°C for 10 min, followed by 35 cycles of 94°C for 1 min, 55°C for 45 sec, and 72°C for 1 min, with a final extension of 72°C for 10 min. To avoid comparing paralogs, we tested for the presence of numts (for details see Text S1). Starting from 50 ng of extracted DNA, we also amplified portions of two nuclear protein-coding genes, von Willebrand factor (vWF; 1100 bp) and Enamelin (ENAM; 2598 bp), using primers and PCR conditions as in [27]. During both DNA extraction and amplification, blank samples were inserted to check for contamination. All sequence data are deposited in GenBank (Accession Nos. KF202138–KF202289).
Sequencing was performed with the same set of primers used for PCR, using Big Dye terminator cycle sequencing with an ABI 3730×l. Sequences were edited and assembled with Sequencher 4.8 (Gene Codes Corporation). All sequences aligned unambiguously, but we additionally evaluated alignments in MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle; [28]. Heterozygous genotypes in the nuclear gene data were resolved using PHASE 2.1.1 [29], [30], under the default options of 100 iterations, 1 thinning interval, 100 burn-in iterations, and confidence probability thresholds of 0.90. One nuclear allele from each individual was chosen at random to represent the individual in species-tree analyses.
We concatenated all three mitochondrial loci to form a single mitochondrial dataset for phylogenetic analyses except where specified otherwise. Models of sequence evolution for each locus were selected using the corrected Akaike information criterion (AICc) as implemented in jModelTest [31] (Text S1). We assessed the performance of all Bayesian analyses (convergence and stationarity) with the program Tracer v. 1.5 [32]. Only runs with adequate mixing and ESS above 200 were considered for final analyses. Rhynchocyon petersi was included as an outgroup in initial analyses and removed when inclusion failed to alter relationships between R. c. reichardi and R. udzungwensis.
Single gene tree analysis
Each locus was individually analysed using Bayesian inference in MrBayes, [33]. All analyses were run for 200 million Markov chain Monte Carlo (MCMC) generations, sampling every 1000th tree. Half of the resulting trees were discarded as burnin. Maximum clade credibility trees were calculated in TreeAnnotator from Beast v. 1.6.2 [32].
Species tree construction
We employed several methods for estimating species trees with different assumptions on ILS (Incomplete Lineage Sorting) and horizontal gene transfer HGT to accommodate the possibility that introgression and HGT may be a substantial part of the evolutionary lineages of at least some populations within these species. Three species tree methods, BEST (Bayesian Estimation of Species Trees: [34], [35], *BEAST (Bayesian Evolutionary Analysis Sampling Trees: [36], and BP&P (Bayesian Phylogenetics & Phylogeography: [37], [38] have been shown to accurately estimate phylogenetic (species tree) relationships in spite of gene tree heterogeneity. They assume, however, that ILS is the only cause of gene tree-species tree discordance. To complement these assumptions with a model that is agnostic to the causes of gene tree heterogeneity, we also analysed our dataset in BUCKy (Bayesian Untangling of Concordance Knots, Bayesian concordance analysis: [39]–[41], which can be used to reject ILS as sufficient to explain gene tree discordance. If ILS cannot be rejected as the cause of mismatch between our datasets, then the species-trees produced from *BEAST and BEST should be good estimates of the true species tree without invoking HGT to explain discordance of genetrees. If BUCKy rejects ILS as sufficient to explain genetree discordance, the individual gene trees can be assessed to determine likely transgenic events [42].
BEST v2.3.1 is a hierarchical coalescent model implemented in a two-step MCMC algorithm. The first step estimates the posterior probability distributions of gene trees for each locus in MrBayes, and the second step uses these probabilities to estimate the posterior probability distribution of species trees. In our BEST analysis, we obtained Bayesian posterior probabilities from 100 million MCMC cycles with a sample frequency of 1,000 and a burn-in period of 25 million generations, with a relatively flat prior for θ (α = 3, β = 0.03) as employed in [43], [44]. Models of molecular evolution for each locus were defined from the highest scoring models in jModeltest available in BEST (mtDNA: nst = 2, haploid, Invgamma; ENAM: nst = 2, diploid, Invgamma; vWF: nst = 2, diploid, Equal).
The program *BEAST v. 1.6.2 was used to jointly estimate the posterior distributions of species trees and contained gene trees using coalescent models. The *BEAST analysis was conducted under a strict molecular clock model (no loci violated a strict clock assumption, data not shown) using the SRD06 model of sequence evolution [45] and a random starting tree. As no estimated mutation rates or calibrated phylogenies are available for this clade, relative evolutionary rates were estimated by setting the mean clock rate of mtDNA to 1.0 and allowing all other loci to be estimated in relation to this rate. The final analysis was run for 100 million generations, sampling every 1,000th generation. A maximum clade credibility tree was generated using the program Tree Annotator v.1.6.2 provided in the BEAST package, with a burn-in of 1,000 (10%).
We used the hyperresolved (population level) species tree generated in *BEAST as the user-specified “guide tree” for species tree estimation in the program BPP, v. 2.0. Population size parameters (qs) and the age of the root of the species tree (t0) were assigned gamma priors (α = 2, β = 1000), while all other divergence time parameters are assigned the Dirichlet prior [38]. Each analysis was run twice to confirm consistency between runs. Analyses were run for 150,000 generations, with the first 50,000 discarded as burn-in and a sampling frequency of 5. The species delimitation algorithms 0 and 1 were both run for this dataset (species delimitation: 1 0 5, reported). A variety of algorithms, a’s, e’s, and m’s were explored (algorithm 0, e = 5 & 20. algorithm 1, α = 1 & 2, m = 0.5, 1, 2) with similar results (Text S1). All five loci (3 mtDNA and 2 nuDNA) were allowed to vary independently in this analysis. Finetune variables were adjusted in the control file for each run so that the acceptance proportions were contained in the interval (0.15, 0.7).
BUCKy v.1.4.0 was used to estimate a primary concordance tree and a population tree from genetrees estimated in MrBayes. Analyses were run for 2 million generations with four different heating chains after a 200,000 generation burn-in. Two extremes of α were explored (α = 0.5, α = 10) with no change to our results. To determine whether HGT appears to play a major role within this region, we compare the Population Tree (PT) and the Primary Concordance Tree (PCT). If ILS is the sole source of gene tree conflict, the two trees should be in tight concordance [46]. If, however the Population Tree and Primary Concordance Tree are in disagreement, this is evidence of hybridization [42].
Species Distribution Models (SDMs)
Occurrence data
Data on the presence of R. udzungwensis and R. cirnei reichardi were collected from live traps and camera traps [12], museum records, and personal communications (Fig. 1 and Text S1) for a total of 32 and 172 localities for grey-faced and chequered sengi, respectively. To reduce taxonomic misidentification of sengis used in the modelling effort, only data from Tanzania were used to avoid uncertainty of identification and distribution of R. c. hendersoni in relation to R. c. reichardi [11]. Similarly, samples of R. c. reichardi from the western border of Lake Tanganyika were eliminated from our analyses due to uncertainty regarding the affinity with southern Tanzanian samples. Though the recently discovered Rhynchocyon in the northern coastal forests of Kenya [47] has pelage colouration and patterning very similar to R. udzungwensis, we have not included it in our analyses because it has not been adequately sampled or described.
Environmental data
Climate layers were acquired from the WorldClim data set [48]. Three time points representing the recent climate extremes of glacial and interglacial cycles were assessed: current, Last Glacial Maximum (LGM –21 kya, Community Climate System Model: [49], and Last Interglacial (LIG –120 kya: [50]. While these specific time points (21 kya and 120 kya) may not be directly related to range shifts or genetic events concerning these species, by representing well documented dry and wet extremes known to have impacted forest habitat and extent in East Africa, we can approximate the range of distributions experienced by these species. We ran an initial correlation assessment of Bioclim layers at each time point (calculated using ENMtools, v 1.3, [51] to attempt to reduce the number of auto-correlated variables which have the potential of over-fitting models of distribution predictions. As these correlations were not stable between time points or between species, all 19 variables were retained for analyses to avoid under-fitting the model. All layers were clipped to both the extent of Tanzanian boundaries and to a rectangle encompassing the area of species overlap in the Udzungwa Mountains (Latitude: −6° to −9.25°, Longitude: 35° to 37.25°) in ArcGIS (ESRI v.10.1).
Species Distribution Models (SDMs)
Species distribution models were created with MaxEnt version 3.3.3 [52]. MaxEnt was chosen because of its ability to make accurate predictions of species distributions on small and spatially limited datasets [53], [54]. All MaxEnt runs were trained on the current climate conditions with a convergence threshold of 0.00001, 1,000 iterations, 25% model testing, logistic output, and the 10% training presence threshold for outputs to create binary models of habitat suitability. Outputs were then projected onto climate models of the LGM and LIG. ENMtools was used to evaluate the similarity of the environmental niche occupied by the two lineages using the Niche Overlap, Niche Identity, and Background functions (200 randomized pseudoreplicates). Background comparisons were completed on using both the distributions envelope of each species (minus the other’s range) from which to generate random points, and the larger shared distribution of the limited area rectangle. The hypothesis of identical climatic niche is rejected when the empirically observed values of identity (D and I statistic: [55], [56] respectively) are significantly lower than the range of pseudoreplicate values [51]. Range overlap through time was calculated in ArcMap.
Results
Molecular Analysis
Gene trees
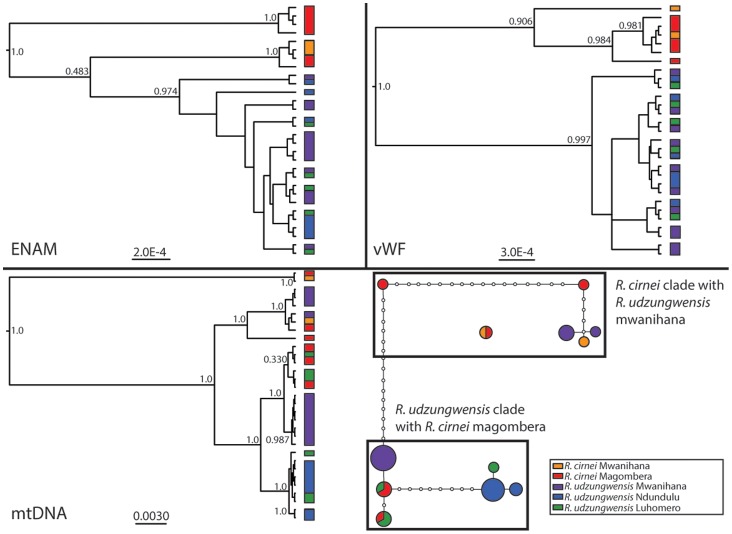
Nuclear loci were in agreement concerning the monophyly of R. udzungwensis. Rhynchocyon c. reichardi has strong support of monophyly for vWF, but an uncertain relationship to the R. udzungwensis clade in ENAM. Neither species showed intraspecific population structure for nuclear loci (Fig. 2, top). The concatenated mitochondrial loci, however, showed substantial paraphyly of the two species, particularly in regards to R. c. reichardi from Magombera and R. udzungwensis from Mwanihana forest, which both possessed mtDNA haplotypes from both major clades (Fig. 2, bottom). Dating divergence events between these lineages is currently impossible without a full phylogeny and fossil representation, but future work will help establish the timing of separation of these species and the approximate timing of genetic transfer between species.
Figure 2. Gene Trees and Haplotype networks.
Bayesian Gene trees of two nuclear loci (top: ENAM and vWF) and concatenated mtDNA (bottom left: ND2, D-loop region, and the ribosomal gene 12S) constructed in MrBayes. Posterior probabilities displayed for major nodes. Branch lengths proportional to genetic distance. (Bottom Right) Haplotype network of concatenated mtDNA. Each node represents one base pair change.
Species trees
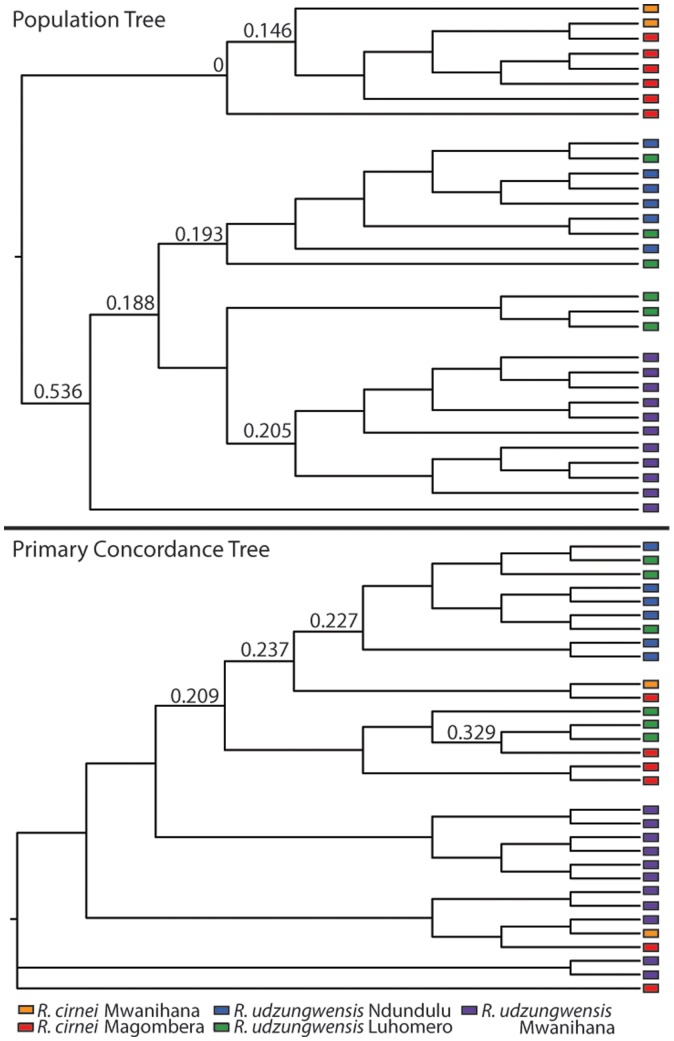
Species tree methods were in agreement concerning the monophyly of the species and phylogeographic relationships within species, with strong support (posterior = 1.00 for all three methods) for the basal split between the two species (Fig. 3). The BUCKy analysis, which was run with individuals as the terminals instead of populations as in the other species trees (e.g., [44]), showed monophyly in the Population Tree, yet extreme paraphyly in the Primary Concordance Tree (Fig. 4). As the CFs (concordance factors, which measure the genomic support of each clade) of the PCT were low, it is likely that many alternative topologies are possible for each node, consistent with Horizontal Gene Transfer.
Figure 3. Highest probability species tree selected by all three methods (*BEAST, BPP, and BEST).
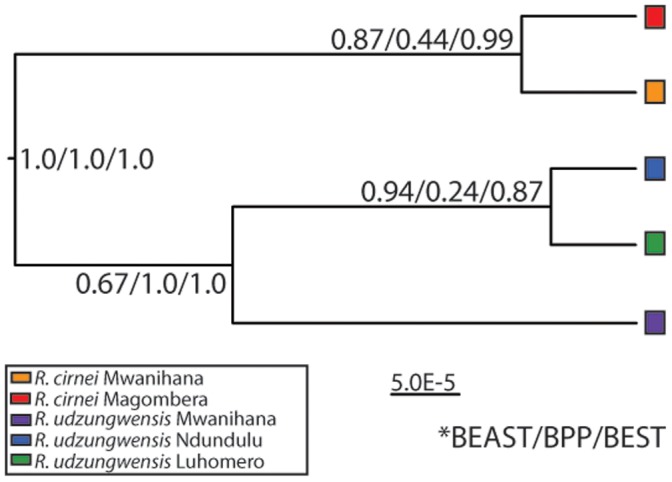
Posterior probabilities shown for each node. Both species are monophyletic for all species tree reconstructions. Branch lengths proportional to genetic distance.
Figure 4. Bayesian concordance analysis in BUCKy: Population Tree (top), Primary concordance tree (bottom).
Posterior mean concordance factors at major nodes are displayed above branches. Branches with concordance factors below 0.2 are not shown except in major nodes. The population tree shows monophyly for the two species, while the primary concordance tree shows admixture between the species and poorly supported relationships implying some level of admixture.
Species Distribution Models
Niche overlap between species
Geographic scale did not significantly impact the current distribution predictions or niche similarity estimates for these species, as models based on the full extent of Tanzania and models generated from just the rectangular region around the Udzungwa Mountains and Southern Highlands yielded similar results (Fig. S1). Projections into past climate conditions were less stable between the two scales, so results are interpreted from the limited distribution rectangle with insights from the larger scale. The AUC (Area Under the Curve) in the rectangle encompassing the Udzungwa Mountains was 0.901 for R. c. reichardi, and 0.994 for R. udzungwensis. The niches of the two species (R. udzungwensis and R. c. reichardi) are significantly more dissimilar than chance by all similarity measures (P<0.001; Fig. S2, S3). As expected given their separate distributions and elevation separation, all models confirmed that they currently occupy different niches despite their close spatial distribution.
Distributions and shifts
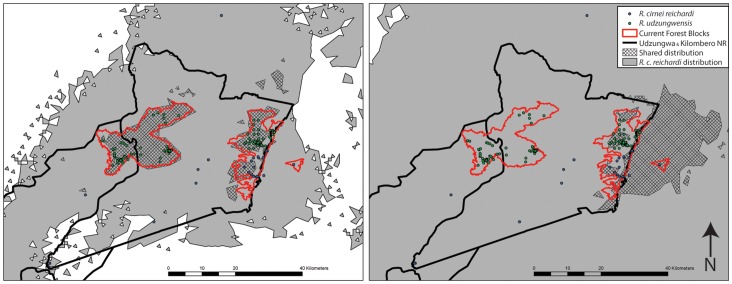
Under current climate conditions, R. c. reichardi has a much larger and more continuous distribution in the region of interest than does R. udzungwensis (Fig. 5, left), which appears to be limited to the two extant forest blocks. The distributions during past wet climate cycles (exemplified by the LIG) show increased suitability throughout the region for R. c. reichardi. Instead of expanding in a similar manner to R. c. reichardi, R. udzungwensis appears to experience a shift in range towards eastern, lower-elevation forests. Instead of the current distribution with one population in a western high elevation forest (Ndundulu-Luhomero) and one population in a central mid-elevation forest (Mwanihana), the LIG distribution consists of populations in the central mid-elevation (Mwanihana) and the eastern lower-elevation forest of Magombera (currently only occupied by R. c. reichardi) (Fig. 5, right). The distribution of both species during dry climate cycles (exemplified by the LGM) was likely greatly reduced, as our climate models fail to predict any substantial areas of suitable habitat within this region for either species.
Figure 5. Species Distribution Models during current climate scenario (left) and wetter Last Interglacial (right; LIG).
Current sampling points used in model construction are shown. The limited distribution of R. udzungwensis is entirely contained within the suitable area for R. c. reichardi at both time points. During the LIG, the ranges for both species were predicted to occur at lower elevations in the eastern areas of the Udzungwa Mountains foothills. During this time, R. udzungwensis shifted its entire distribution eastward resulting in a loss occupancy in the western Ndundulu/Luhomero forest, a reduced range in the Mwanihana forest, and a relatively extensive distribution east of Mwanihana in the Kilombero Valley including the Magombera forest (see Fig. 1 for place names).
Discussion
Species-tree methods confirm the genetic distinctness of the two species with very high support, validating the morphological description of R. udzungwensis [2]. This is a first, important result given the complex phylogeny of the genus. However, the discrepancy between the BUCKy population tree and primary concordance tree, and the clear introgression of mtDNA haplotypes despite nuclear monophyly imply that ancient hybridization occurred between these two taxa. The absence of current admixture inferred from monophyletic nuclear loci is supported by the fact that despite the close proximity of R. c. reichardi and R. udzungwensis populations in the Udzungwa Mountains, no morphologically intermediate individuals (presumed hybrids) have been found. Given the spatial proximity of these species, apparent mitochondrial introgression, and the narrow habitat requirements of R. udzungwensis, four related questions arise: (1) Why is there no evidence of nuclear gene introgression; (2) Why do some populations of R. udzungwensis exhibit ancient hybridization while others do not; (3) Why is there no evidence of current hybridization between the two species; and (4) Why do SDM models predict a near complete lack of suitable habitat in this area during the LGM?
Mitochondrial Introgression
The phenomenon of extensive mitochondrial introgression without a comparable nuclear signal is relatively common in phylogenetic studies (e.g., [57]–[60], exemplified by a stark disagreement between resolved nuclear genes and admixed mitochondrial loci. Given the ∼4-fold increase in mutation rate for mtDNA vs. nuclear [61], [62], these patterns are unlikely to be a result of incomplete lineage sorting [63], particularly when two morphologically and genetically distinct lineages share exact mitochondrial haplotypes. Though data are extremely limited within this sengis, no examples of incomplete lineage sorting of mtDNA have been found [4], [64].
The signal of mitochondrial introgression can persist between species, despite loss of a nuclear signal when two potentially hybridizing species meet in circumstances of disparate abundance. In these uneven encounters, normally choosy females of the less abundant species may accept heterospecific mates if conspecifics mates are rare [58]. In these instances, though both nuclear and mitochondrial genes from the rare species introgress into the common species, mitochondrial introgression invariably exceeds nuclear. This is particularly apparent when the proportion of immigrants is small [58].
Introgressed and Pure Populations
Historical uneven encounters could be expected in some populations of R. udzungwensis and R. c reichardi, while others may have remained isolated from introgression throughout history. Evidence from this study suggests that some populations have undergone introgression while others have not: (1) mtDNA of R. udzungwensis from the Luhomero forests appears introgressed into R. c. reichardi individuals from Magombera forest, (2) apparent mtDNA haplotypes from R. c. reichardi appear in R. udzungwensis individuals from Mwanihana, and (3) R. udzungwensis from Ndundulu appear isolated and free from admixture despite the fact that the Ndundulu forests are equally surrounded by R. c. reichardi as the other fragments occupied by R. udzungwensis.
The spatial distribution of habitat during both current and LIG climate conditions, and the unique nature of the Ndundulu forests offer possible explanations for spatially structured introgression. The Ndundulu forest appears to act as an ark of diversity for a number of forest-restricted species, providing stable habitat through time for its unusually divergent occupants (e.g., Udzungwa forest partridge (Xenoperdix udzungwensis, [65]–[67]), kipunji (Rungwecebus kipunji, [68], [69], and rufous-winged sunbird (Cinnyris rufipennis, [70]). The reason that distinct and relictual species are preserved in the Ndundulu forest is not understood, but perhaps various environmental aspects filter generalist species from invading and displacing or intermingling with local lineages.
Together, the current parapatric distribution, population-specific mtDNA introgression, and the predicted historical ranges of these species during interglacial cycles support a scenario of hybridization with R. c. reichardi through uneven encounters. As species ranges shifted in response to glacial climate cycles, rare colonists of displaced R. udzungwensis may have encountered abundant and established R. c. reichardi in the newly accessible flood plain forest of which Magombera is today a remnant (Fig. 5) leading to a transfer of mtDNA haplotypes into R. c. reichardi populations [71]. Likewise, R. c. reichardi may have expanded into the northern Mwanihana forests as R. udzungwensis’ range contracted, leading to the mtDNA introgression from R. c. reichardi into R. udzungwensis. These scenarios of predicted relative abundance (i.e., “rare colonists” and “abundant established”) reflect stable range distributions of one species (based on current and LIG models) vs. newly occupied areas for the other. With the current data, much of this interpretation is speculative, but fits with known models of interactions. Future studies incorporating larger sampling of the diversity within R. c. reichardi and R. cirnei as a whole will help to clarify these hypotheses.
Current Genetic Isolation
SDM show that the current R. udzungwensis range is completely within the predicted, and much broader, range of R. c. reichardi, yet no on-going hybridization appears to be taking place. In situations such as this, where one species is predicted to occupy the range of another yet is distinctly absent from that area, competition coupled with reinforcement for potentially hybridizing species is typically invoked. It is likely that the larger-bodied R. udzungwensis is able to outcompete R. c. reichardi at the higher/harsher elevations that it currently occupies, but R. c. reichardi is better able to utilize resources in the available landscape surrounding the R. udzungwensis refugia. In this case, gradual historic climate change may explain the trend towards reduced introgression at the present by reinforcing the concentration of R. udzungwensis in its wetter, highland refugia if it cannot compete in the drier mid-elevation areas occupied by R. c. reichardi. Though East Africa experienced many climate fluctuations in the past 5 my, oscillating from wet to dry climate conditions with associated forest expansions and contractions, there has been a general drying trend from 1.86 (+/−0.44 Ma) to present (ODP 721/722 dust flux record, [72]. From this point on, the range of R. udzungwensis would have concentrated in the highland forests of the Udzungwa Mountains tops instead of the expanded eastern range with R. c. reichardi. This scenario is difficult to verify without absolute dating of the larger phylogeny and the proposed introgression event (an ongoing effort of these authors and others), but is likely the levels of divergence and other phylogenetic and phylogeographic patterns in East Africa (discussed in [73]).
SDM Model Predictions
Species distribution modelling is an invaluable tool for many aspects of evolutionary and conservation biology, but there are limitations for range-restricted species that tend to have little latitude in their environmental envelope [74]. The modelling approach in this study, MaxEnt based on bioclimatic variables, did not predict any areas of stable habitat (using a threshold cut-off of suitability) within the Udzungwa Mountains region for either sengi in this study during the LGM, a result that seems unlikely given our molecular findings. Though this may imply that populations in this area were nearly extinct, as should be investigated with a larger population genetics dataset, it is also possible that the distribution requirements of sengis are either not adequately predicted by these models or that the reconstructions of climate during the LGM may be inadequate for this region. Future improvements in both models and sengi distribution requirements will aid in our understanding of these dynamics: (1) Models of past and future climate continue to evolve, particularly in terms of sensitivity to the under-represented African tropics. These advancements will greatly enhance the understanding of climatic effects on diversity within African lineages as a whole. (2) As more ecological information becomes available for sengis including microhabitat requirements, physiological limits, and potential competitors, we may be able to provide a more sensitive model of distributions. These traits, however, have limited ability to be predicted into past and future climates.
Advancing our Understanding of Sengis
This study is the first known case of introgression in sengis, a result that was only identifiable using multi-locus data from multiple populations. Previous investigations into relationships of co-distributed sengis failed to detect introgression, but lacked multi-locus data [4], [8]. A multi-locus perspective will be necessary in studies of sengis, including phylogenetic analyses, to adequately interpret interspecific relationships.
In the current study, we are able to see a clear signal of introgression using multiple individuals from multiple localities. In future studies, increased sample sizes sufficient to investigate demographic events (such as expansions and contractions) and estimate population diversity across ranges would both verify proposed demographic processes and ground our estimates of the extent of introgression between species.
This study demonstrates that multi-locus, population-level sampling including introgression analyses is imperative for any attempts at investigating the broader relationships and evolutionary history of Rhynchocyon. Within this, all subspecies, pelage variants, and allopatric populations should be considered for the fullest understanding of evolution and diversification within this poorly understood genus. It also highlights how comparisons with neighboring and closely related species highlight the uniqueness and complex history of a new and vulnerable giant sengi.
Supporting Information
ENM predictions of Tanzania vs. reduced rectangle. Current predictions for R. c. reichardi (left) and R. udzungwensis (right) with Udzungwa NP and Kilombero Forest reserve outlined in black for reference.
(TIF)
Niche Identity comparisons between R. udzungwensis and R. c. reichardi.
(TIF)
Similarity of R. udzungwensis and R. c. reichardi niche to backgrounds. Top of each set: Shared background of entire evaluated rectangle. Bottom of each set: compared to random points in the threshold occurrence envelope of the other species. Arrows represent actual similarity.
(TIF)
Collection and identification of specimens used in this study.
(XLS)
Supplementary information regarding jModelTest, BPP, exclusion of 12S numts, niche comparisons between R. udzungwensis and R. c. reichardi.
(DOCX)
Acknowledgments
Several field assistants, especially Ruben Mwakisoma, helped data collection invaluably. Barbara Crestanello provided sequencing assistance and Matteo Girardi undertook a great deal of the lab work. Galen Rathbun was an advisor during initiation of the project, provided access to his sengi distributional database, and greatly improved the clarity of the manuscript. Aaron Iemma helped making the map in Fig. 1. We are grateful to the Tanzania Wildlife Research Institute, Tanzania Commission for Science and Technology, Forestry and Beekeeping Division and Tanzania National Parks for research permits.
Funding Statement
This research was supported by grants from National Geographic’s Committee for Research and Exploration (grant 8497-08 to F.R.), Trento Autonomous Province, Trento Science Museum and the Critical Ecosystem Partnership Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mallet J (2005) Hybridization as an invasion of the genome. Trends Ecol Evol 20: 229–237 doi:10.1016/j.tree.2005.02.010 [DOI] [PubMed] [Google Scholar]
- 2. Rovero F, Rathbun GB, Perkin A, Jones T, Ribble DO, et al. (2008) A new species of giant sengi or elephant-shrew (genus Rhynchocyon) highlights the exceptional biodiversity of the Udzungwa Mountains of Tanzania. J Zool 274: 126–133 doi:10.1111/j.1469-7998.2007.00363.x [Google Scholar]
- 3. Rathbun GB (2009) Why is there discordant diversity in sengi (Mammalia: Afrotheria: Macroscelidea) taxonomy and ecology? Afr J Ecol 47: 1–13 doi:10.1111/j.1365-2028.2009.01102.x [Google Scholar]
- 4. Dumbacher JP, Rathbun GB, Smit HA, Eiseb SJ (2012) Phylogeny and Taxonomy of the Round-Eared Sengis or Elephant-Shrews, Genus Macroscelides (Mammalia, Afrotheria, Macroscelidea). PLoS ONE 7: e32410 doi:10.1371/journal.pone.0032410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Springer MS, Cleven GC, Madsen O, de Jong WW, Waddell VG, et al. (1997) Endemic African mammals shake the phylogenetic tree. Nature 388: 61–64 doi:10.1038/40386 [DOI] [PubMed] [Google Scholar]
- 6. Stanhope MJ, Waddell VG, Madsen O, de Jong W, Hedges SB, et al. (1998) Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals. PNAS 95: 9967–9972 doi:10.1073/pnas.95.17.9967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seiffert ER (2007) A new estimate of afrotherian phylogeny based on simultaneous analysis of genomic, morphological, and fossil evidence. BMC Evolutionary Biology 7: 224 doi:10.1186/1471-2148-7-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smit HA, Jansen van Vuuren B, O’Brien PCM, Ferguson-Smith M, Yang F, et al. (2011) Phylogenetic relationships of elephant-shrews (Afrotheria, Macroscelididae). J Zool 284: 133–143 doi:10.1111/j.1469-7998.2011.00790.x [Google Scholar]
- 9.Coals PGR, Rathbun GB (in press) The taxonomic status of giant sengis (genus Rhynchocyon) in Mozambique. JE Afr Nat Hist.
- 10. Corbet GB, Hanks J (1968) A revision of the elephant-shrews, Family Macroscelididae. Bull Br Mus Nat Hist Zool 16: 47–111. [Google Scholar]
- 11.Kingdon J (1974) East African mammals; An atlas of evolution in Africa. New York, USA: Academic Press.
- 12.Rovero F, Collett C, Ricci S, Martin E, Spitale D (in press) Distribution, occupancy and habitat associations of the grey-faced sengi (Rhynchocyon udzungwensis) as revealed by camera traps. J Mammal.
- 13. Guisan A, Zimmermann NE (2000) Predictive habitat distribution models in ecology. Ecol Model 135: 147–186 doi:10.1016/S0304-3800(00)00354-9 [Google Scholar]
- 14. Parmesan C (2006) Ecological and Evolutionary Responses to Recent Climate Change. Annu Rev Ecol Evol Syst 37: 637–669. [Google Scholar]
- 15. Alves PC, Melo-Ferreira J, Freitas H, Boursot P (2008) The Ubiquitous Mountain Hare Mitochondria: Multiple Introgressive Hybridization in Hares, Genus Lepus . Phil Trans R Soc B 363: 2831–2839 doi:10.1098/rstb.2008.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mallet J, Meyer A, Nosil P, Feder JL (2009) Space, sympatry and speciation. J Evol Biol 22: 2332–2341 doi:10.1111/j.1420-9101.2009.01816.x [DOI] [PubMed] [Google Scholar]
- 17. Garroway CJ, Bowman J, Cascaden TJ, Holloway GL, Mahan CG, et al. (2010) Climate change induced hybridization in flying squirrels. Glob Change Biol 16: 113–121 doi:10.1111/j.1365-2486.2009.01948.x [Google Scholar]
- 18. Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58: 247–276 doi:10.1006/bijl.1996.0035 [Google Scholar]
- 19. Guralnick R (2007) Differential effects of past climate warming on mountain and flatland species distributions: a multispecies North American mammal assessment. Global Ecol Biogeogr 16: 14–23 doi:10.1111/j.1466-8238.2006.00260.x [Google Scholar]
- 20. Nyari AS, Peterson AT, Rathbun GB (2010) Late Pleistocene potential distribution of the North African sengi or elephant-shrew Elephantulus rozeti (Mammalia: Macroscelidea). Afr Zool 45: 330–339 doi:10.3377/004.045.0212 [Google Scholar]
- 21. Sikes RS, Gannon WL (2011) Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J Mammal 92: 235–253 doi:10.1644/10-MAMM-F-355.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.AVMA (2000) AVMA guidelines on euthanasia. American Veterinary Medical Association. Available: http://www.avma.org/issues/animal_welfare/euthanasia.pdf.
- 23. Olson LE, Goodman SM, Yoder AD (2004) Illumination of cryptic species boundaries in long-tailed shrew tenrecs (Mammalia: Tenrecidae; Microgale), with new insights into geographic variation and distributional constraints. Biol J Linn Soc 83: 1–22 doi:10.1111/j.1095-8312.2004.00366.x [Google Scholar]
- 24. Douzery E, Randi E (1997) The mitochondrial control region of Cervidae: evolutionary patterns and phylogenetic content. Mol Biol Evol 14: 1154–1166. [DOI] [PubMed] [Google Scholar]
- 25. Huchon D, Delsuc F, Catzeflis FM, Douzery EJP (1999) Armadillos exhibit less genetic polymorphism in North America than in South America: nuclear and mitochondrial data confirm a founder effect in Dasypus novemcinctus (Xenarthra). Mol Ecol 8: 1743–1748 doi:10.1046/j.1365-294x.1999.00768.x [DOI] [PubMed] [Google Scholar]
- 26. Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, et al. (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. PNAS 86: 6196–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Douady CJ, Catzeflis F, Raman J, Springer MS, Stanhope MJ (2003) The Sahara as a vicariant agent, and the role of Miocene climatic events, in the diversification of the mammalian order Macroscelidea (elephant-shrews). PNAS 100: 8325–8330 doi:10.1073/pnas.0832467100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Edgar RC (2004) MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucl Acids Res 32: 1792–1797 doi:10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stephens M, Scheet P (2005) Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet 76: 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 32. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 34. Liu L, Pearl DK (2007) Species trees from gene trees: reconstructing Bayesian posterior distributions of a species phylogeny using estimated gene tree distributions. Syst Biol 56: 504–514 doi:10.1080/10635150701429982 [DOI] [PubMed] [Google Scholar]
- 35. Liu L (2008) BEST: Bayesian estimation of species trees under the coalescent model. Bioinformatics 24: 2542–2543 doi:10.1093/bioinformatics/btn484 [DOI] [PubMed] [Google Scholar]
- 36. Heled J, Drummond AJ (2010) Bayesian inference of species trees from multilocus data. Mol Biol Evol 27: 570–580 doi:10.1093/molbev/msp274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rannala B, Yang Z (2003) Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164: 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang Z, Rannala B (2010) Bayesian species delimitation using multilocus sequence data. PNAS 107: 9264–9269 doi:10.1073/pnas.0913022107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ané C, Larget B, Baum DA, Smith SD, Rokas A (2007) Bayesian estimation of concordance among gene trees. Mol Biol Evol 24: 412–426 doi:10.1093/molbev/msl170 [DOI] [PubMed] [Google Scholar]
- 40. Ané C, Dewey CN, Kotha SK, Larget BR (2010) Gene tree reconciliation: new developments in Bayesian concordance analysis with BUCKy. Bioinformatics 26: 2910–2911 doi:10.1093/bioinformatics/btq539 [DOI] [PubMed] [Google Scholar]
- 41. Larget BR, Kotha SK, Dewey CN, Ané C (2010) BUCKy: Gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics 26: 2910–2911 doi:10.1093/bioinformatics/btq539 [DOI] [PubMed] [Google Scholar]
- 42. Chung Y, Ané C (2011) Comparing two Bayesian methods for gene tree/species tree reconstruction: simulations with incomplete lineage sorting and horizontal gene transfer. Syst Biol 60: 261–275 doi:10.1093/sysbio/syr003 [DOI] [PubMed] [Google Scholar]
- 43. Leaché AD (2009) Species tree discordance traces to phylogeographic clade boundaries in North American fence lizards (Sceloporus). Syst Biol 58: 547–559 doi:10.1093/sysbio/syp057 [DOI] [PubMed] [Google Scholar]
- 44. Lee JY, Joseph L, Edwards SV (2012) A species tree for the Australo-Papuan Fairy-wrens and Allies (Aves: Maluridae). Syst Biol 61: 253–271 doi:10.1093/sysbio/syr101 [DOI] [PubMed] [Google Scholar]
- 45. Shapiro B, Rambaut A, Drummond AJ (2006) Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol 23: 7–9 doi:10.1093/molbev/msj021 [DOI] [PubMed] [Google Scholar]
- 46. Degnan JH, DeGiorgio M, Bryant D, Rosenberg NA (2009) Properties of consensus methods for inferring species trees from gene trees. Syst Biol 58: 35–54 doi:10.1093/sysbio/syp008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Andanje S, Agwanda BR, Ngaruiya GW, Amin R, Rathbun GB (2010) Sengi (elephant-shrew) observations from northern coastal Kenya. JE Afr Nat Hist 99: 1–8 doi:10.2982/028.099.0101 [Google Scholar]
- 48. Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25: 1965–1978 doi:10.1002/joc.1276 [Google Scholar]
- 49. Collins WD, Bitz CM, Blackmon ML, Bonan GB, Bretherton CS, et al. (2006) The community climate system model version 3 (CCSM3). J Climate 19: 2122–2143. [Google Scholar]
- 50.Otto-Bliesner BL, Marshall SJ, Overpeck JT, Miller GH, Hu A, et al.. (2006) Simulating Warm-Arctic Climate and Ice Sheet Sensitivity for the Last Interglaciation. Science: 1751–1753. [DOI] [PubMed]
- 51. Warren DL, Glor RE, Turelli M (2010) ENMTools: a toolbox for comparative studies of environmental niche models. Ecography 33: 607–611 doi:10.1111/j.1600-0587.2009.06142.x [Google Scholar]
- 52.Phillips SJ, Dudík M, Schapire RE (2004) A maximum entropy approach to species distribution modeling. Proceedings of the twenty-first international conference on Machine learning. ACM New York, NY, USA. 472–486.
- 53. Hernandez PA, Graham CH, Master LL, Albert DL (2006) The effect of sample size and species characteristics on performance of different species distribution modeling methods. Ecography 29: 773–785 doi:10.1111/j.0906-7590.2006.04700.x [Google Scholar]
- 54. Hernandez PA, Franke I, Herzog SK, Pacheco V, Paniagua L, et al. (2008) Predicting species distributions in poorly-studied landscapes. Biodivers Conserv 17: 1353–1366 doi:10.1007/s10531-007-9314-z [Google Scholar]
- 55. Schoener TW (1968) The Anolis lizards of Bimini: resource partitioning in a complex fauna. Ecology 49: 704–726. [Google Scholar]
- 56. Warren DL, Glor RE, Turelli M (2008) Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62: 2868–2883 doi:10.1111/j.1558-5646.2008.00482.x [DOI] [PubMed] [Google Scholar]
- 57. Funk DJ, Omland KE (2003) Species-Level Paraphyly and Polyphyly: Frequency, Causes, and Consequences, with Insights from Animal Mitochondrial DNA. Annu Rev Ecol Evol Syst 34: 397–423 doi:10.1146/annurev.ecolsys.34.011802.132421 [Google Scholar]
- 58. Chan KMA, Levin SA (2005) Leaky prezygotic isolation and porous genomes: rapid introgression of maternally inherited DNA. Evolution 59: 720–729 doi:10.1554/04-534 [PubMed] [Google Scholar]
- 59. Linnen CR, Farrell BD (2008) Comparison of methods for species-tree inference in the sawfly genus Neodiprion (Hymenoptera: Diprionidae). Syst Biol 57: 876–890 doi:10.1080/10635150802580949 [DOI] [PubMed] [Google Scholar]
- 60. Spinks PQ, Shaffer HB (2009) Conflicting mitochondrial and nuclear phylogenies for the widely disjunct Emys (Testudines: Emydidae) species complex, and what they tell us about biogeography and hybridization. Syst Biol 58: 1–20 doi:10.1093/sysbio/syp005 [DOI] [PubMed] [Google Scholar]
- 61. Moore WS (1995) Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees. Evolution 49: 718–726. [DOI] [PubMed] [Google Scholar]
- 62. Palumbi SR, Cipriano F, Hare MP (2001) Predicting nuclear gene coalescence from mitochondrial data: the three-times rule. Evolution 55: 859–868 doi:[];10.1554/0014-3820(2001)055[0859:PNGCFM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 63. Hudson RR, Coyne JA (2002) Mathematical consequences of the genealogical species concept. Evolution 56: 1557–1565 doi:[];10.1554/0014-3820(2002)056[1557:MCOTGS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 64. Smit HA, Robinson TJ, Van Vuuren BJ (2007) Coalescence methods reveal the impact of vicariance on the spatial genetic structure of Elephantulus edwardii (Afrotheria, Macroscelidea). Mol Ecol 16: 2680–2692 doi:10.1111/j.1365-294X.2007.03334.x [DOI] [PubMed] [Google Scholar]
- 65. Dinesen L, Lehmberg T, Svendsen TO, Hansen LA, Fjeldsaå J (1994) A new genus and species of perdicine bird (Phasianidae, Perdicini) from Tanzania; a relict form with Indo-Malayan affinities. Ibis 136: 3–11. [Google Scholar]
- 66. Fjeldså J, Kiure J (2003) A new population of the Udzungwa forest partridge. Bull Br Ornithol Club 123: 52–56. [Google Scholar]
- 67. Bowie RCK, Fjeldså J (2005) Genetic and morphological evidence for two species in the Udzungwa Forest Partridge Xenoperdix udzungwensis . JE Afr Nat Hist 94: 191–201. [Google Scholar]
- 68. Davenport TRB, Stanley WT, Sargis EJ, De Luca DW, Mpunga NE, et al. (2006) A new genus of African monkey, Rungwecebus: morphology, ecology, and molecular phylogenetics. Science 312: 1378–1381 doi:10.1126/science.1125631 [DOI] [PubMed] [Google Scholar]
- 69. Roberts TE, Davenport TRB, Hildebrandt KBP, Jones T, Stanley WT, et al. (2010) The biogeography of introgression in the critically endangered African monkey Rungwecebus kipunji . Biol Lett 6: 233–237 doi:10.1098/rsbl.2009.0741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dinesen L, Lehmberg T, Svendsen JO, Hansen LA (1993) Range extension and other notes on some restricted-range forest birds from West Kilombero in the Udzungwa Mountains, Tanzania. Scopus 17: 48–59. [Google Scholar]
- 71. McPeek MA, Gavrilets S (2006) The evolution of female mating preferences: differentiation from species with promiscuous males can promote speciation. Evolution 60: 1967–1980 doi:10.1554/06-184.1 [PubMed] [Google Scholar]
- 72. Trauth MH, Larrasoaña JC, Mudelsee M (2009) Trends, rhythms and events in Plio-Pleistocene African climate. Quat Sci Rev 28: 399–411 doi:10.1016/j.quascirev.2008.11.003 [Google Scholar]
- 73.Lawson LP (2010) The discordance of diversification: evolution in the tropical-montane frogs of the Eastern Arc Mountains of Tanzania. Mol Ecol 19, 4046–4060. doi: 10.1111/j.1365-294X.2010.04788.x. [DOI] [PubMed]
- 74. Graham CH, VanDerWal J, Phillips SJ, Moritz C, Williams SE (2010) Dynamic refugia and species persistence: tracking spatial shifts in habitat through time. Ecography 33: 1062–1069 doi:10.1111/j.1600-0587.2010.06430.x [Google Scholar]
- 75.Marshall AR, Topp-Jørgensen JE, Brink H, Fanning E (2005) Monkey abundance and social structure in two high elevation forest reserves in the Udzungwa Mountains of Tanzania. Int J Primatol 26(1), 127–145. doi: 10.1007/s10764-005-0011-z.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ENM predictions of Tanzania vs. reduced rectangle. Current predictions for R. c. reichardi (left) and R. udzungwensis (right) with Udzungwa NP and Kilombero Forest reserve outlined in black for reference.
(TIF)
Niche Identity comparisons between R. udzungwensis and R. c. reichardi.
(TIF)
Similarity of R. udzungwensis and R. c. reichardi niche to backgrounds. Top of each set: Shared background of entire evaluated rectangle. Bottom of each set: compared to random points in the threshold occurrence envelope of the other species. Arrows represent actual similarity.
(TIF)
Collection and identification of specimens used in this study.
(XLS)
Supplementary information regarding jModelTest, BPP, exclusion of 12S numts, niche comparisons between R. udzungwensis and R. c. reichardi.
(DOCX)