Abstract
Mass spectrometric approaches have been fundamental to the identification of metabolites associated with steroid hormones, yet this topic has not been reviewed in depth in recent years. To this end, and given the increasing relevance of liquid chromatrography-mass spectrometry (LC-MS) studies on steroid hormones and their metabolites, the present review addresses this subject. This review provides a timely summary of the use of various mass spectrometry-based analytical techniques during the evaluation of steroidal biomarkers in a range of human disease settings. The sensitivity and specificity of these technologies are clearly providing valuable new insights into breast cancer and cardiovascular disease.
We aim to contribute to an enhanced understanding of steroid metabolism and how it can be profiled by LC-MS techniques.
Keywords: Mass spectrometry, LC-MS, estradiol metabolites, cholesterol metabolites, cancer biomarkers
Mass spectrometry overview
Mass spectrometry has an important history in the identification of drug metabolites and has recently emerged as the foremost technology in endogenous metabolite research [1], given its proven success in several drug metabolite analysis and pharmacokinetic studies [2–12]. In fact, the recent rise of the «metabolomics era» stems from the enhanced ability to perform faster, more accurate and comprehensive metabolite analyses, along with the need to understand intracellular biochemical events towards identification of both disease and pharmaceutical biomarkers [1].
Metabolite analyses have been typically carried out by means of liquid or gas chromatography hyphenated with mass spectrometry (LC-MS or GC-MS, respectively), or inclusively high field proton nuclear magnetic resonance (NMR). The primary advantage of NMR in metabolite analysis is its ability to measure analytes in biofluids quickly and accurately, without the need of initial processing or separation [13–15]. Over recent years, improvements have included higher spectral resolution, lower instrument cost, and the addition of stop-flow chromatography on fractions of samples. Yet, the major weakness of NMR is that it has a poor dynamic range (100–1000) that results in only the major components being observed [1]. High-resolution capillary GC-MS has also been a landmark in metabolite research and disease diagnosis, as it enables identification of key small molecules, such as fatty acids, amino acids and organic acids, in biofluids, particularly in urine and blood [16–18]. This technique has been influential in providing diagnostic information for many inherited diseases, such as numerous metabolic disorders, disorders of the metabolism of amino acids [19–22], bile acids [23–24] and steroids [25–27]. Nevertheless, GC-MS techniques have limited applicability to metabolite profiling, as they usually require (i) convoluted sample preparation including metabolite extraction and subsequent derivatization to volatile adducts, (ii) long analysis times, and (iii) ideal size and type of molecules to be analyzable; in other words, non-volatile, highly polar and/or large molecules cannot be studied by GC-MS [1]. In this context, LC-MS present several advantages over NMR or GC-MS techniques in metabolite profiling, namely greater sensitivity and dynamic range. Therefore, LC-MS techniques will be overviewed in greater detail.
Competing MS technologies
LC-MS with an electrospray ionization interface (LC-ESI-MS) has become a popular choice for metabolite analysis and studies for new biomarkers [18,28]. This technique is advantageous over GC-MS in that sample preparation and analysis are relatively simple, providing access to metabolites of higher structural diversity. ESI offers many advantages over other ionization techniques, for example, the ability to analyze low- and high-molecular weight compounds, excellent quantitative capabilities and reproducibility, high sensitivity, simple sample preparation, amenability to automation, soft ionization and absence of matrix [29]. The utility of ESI lies in its ability to generate gas-phase ions directly from the liquid phase, which establishes the technique as a convenient mass-analysis platform for both LC and direct flow injection analysis (FIA), especially when combined with tandem mass spectrometry (MS/MS) [1]. While previous LC separation of the diverse molecules present in biofluids can reduce ESI ion suppression [30–32], making LC-MS especially attractive in the initial stages of metabolite research, it also delays data acquisition and analysis. Therefore, for ESI-MS quantitation of a known biomarker, extraction combined with flow injection analysis (FIA) is the method of choice, as the extracted sample is directly injected into the mass spectrometer, without prior chromatographic separation [33]. Altogether, ESI-MS techniques result in a selective approach that allows for both qualitative and quantitative metabolite analysis, while sensitivities in the pg/mL range can be readily achieved [34]. Still, a challenge in metabolite profiling is that potential biomarkers may be present in the biofluid in even lower abundances, thus requiring specially sensitive techniques, like nano-LC-ESI-MS; this technique is performed at flow rates (~200 nL·min−1) much lower than those in standard LC-ESI-MS (~300 L·min−1), which produces ions with less evaporation, thus enabling detection of highly diluted species. This improves the sensitivity and ultimately offers a greater dynamic range in metabolite discovery [30–36].
Finally, though atmospheric-pressure chemical or photo-ionization mass spectrometry (APCI-MS or APPI-MS, respectively) are not widely used in metabolite-profiling studies, they have been employed in the analysis of more easily ionizable molecules, such as phospholipids, to produce molecular and fragment ions complementary to those obtained by ESI with collision-induced dissociation (CID). APCI-MS provides a dynamic range higher than ESI-MS and is considered robust, easy to operate and relatively tolerant to higher buffer concentrations. Yet, it is a mass-sensitive rather than concentration-sensitive technique, so no sensitivity gain can be reached with smaller columns or lower flow rates.
A summary of the main characteristics of the techniques outlined above is provided in Table 1.
Table 1.
Summary of the major characteristics of LC-MS, FIA, GC-MS and NMR techniques used in metabolite-profiling studies.
| Technique Characteristics | LC-MS | FIA | GC-MS | NMR |
|---|---|---|---|---|
| Sample preparation | extraction | extraction | Extraction and chemical modification | Typically none |
| Chromatographic Separation | Medium-resolution separation | No separation | High-resolution separation | No separation |
| Sensitivity | Millimolar to nanomolar | Millimolar to micromolar | Millimolar to nanomolar | Millimolar to high micromolar |
| Dynamic range | 106 | 104 | 106 | 103 |
| Speed | Slow (5–90 min) | Rapid (1 to 5 min) | Slow (~30 min) | Rapid (1 to 5 min) |
| Quantitative accuracy | ±10 % | ±10 % | ±10 % | ±10 % |
| Significant advantages | Soft ionization Large mass range | Data in one spectrum fast | High resolution ESI-MS library available | No sample preparation |
| Significant disadvantages | Speed of analysis | Signal suppression from multiple components | Significant sample preparation with chemical modification; Slow analysis; Harsh ionization; Limited number of molecules can be analyzed | Poor sensitivity and dynamic range Some chemical classes are not detected |
Another critical parameter in MS-based metabolite studies is the mass analyzer, a central piece in the performance of any mass spectrometer. Among the most commonly used are the quadrupole, the quadrupole ion trap, the time-of-flight (TOF) reflectron, and the Fourier transform ion cyclotron resonance (FTMS) analyzer.
Quadrupole is presently the most common type of mass analyzers; quadrupoles tolerate relatively high pressures, have the capability of analyzing up to an m/z of 4000 and are relatively low cost instruments. Yet, a triple-quadrupole is required if tandem mass analysis is to be performed; the three quadrupoles are placed in series, and each of them has a separate function: the first (Q1) is used to scan across the full m/z range and select an ion of interest; the second (Q2), also known as the collision cell, focuses and transmits the ions while introducing a collision gas (argon or helium) into the flight path of the selected ion; the third (Q3) serves to analyze the fragment ions generated in the collision cell (Q2) [29].
Quadrupole ion trap analyzers are also useful in tandem MS analysis, as a single ion species can be isolated by ejecting all others from the trap, enabling the isolated species to be further fragmented by collisional activation (CID); a key advantage of quadrupole ion traps is that multiple CID experiments can be performed quickly without requiring multiple analyzers. Other advantages include their ability to trap and accumulate ions to provide a better signal-to-noise ratio and their mass range up to ~4000 m/z. Yet, quadrupole ion traps are unable to perform high-sensitivity triple quadrupole-type precursor-ion scanning and neutral loss scanning experiments; also, the upper limit on the ratio between precursor m/z and the lowest trapped fragment ion is about 0.3 (also known as the “one-third rule”) and their dynamic range is limited, as when too many ions are in the trap, space charge effects diminish the performance of the ion-trap analyzer [29].
The linear time-of-flight (TOF) is the simplest mass analyzer with a virtually unlimited mass range. It has gained wide use due to its fast scanning capabilities (milliseconds), good mass range (up to m/z ~10000), and an accuracy in the order of 5 ppm. Quadrupole-TOF mass analyzers combine the stability of a quadrupole analyzer with high efficiency, sensitivity, and accuracy of time-of-flight reflectron mass analyzer and are typically coupled to ESI sources. The quadrupole can act as a simple quadrupole analyzer to scan across a specified m/z range. Quadrupole-TOF exploits the quadrupole’s ability to select a particular ion and the ability of TOF-MS to achieve simultaneous and accurate measurements of ions across the full mass range. They offer significantly higher sensitivity and accuracy of tandem quadrupole instruments when acquiring full-fragment mass spectra.
FTMS offers high resolution, the ability to perform multiple collision experiments (MSn), and high-accuracy fragment masses (often at the part-per-million level). It is now becoming more common to couple ultra high resolution (> 105) FTMS to a wide variety of ionization sources, including MALDI, ESI, APCI and EI (electron impact ionization). Quadrupole-FTMS and quadrupole linear ion-trap FTMS mass analyzers that have recently been introduced are typically coupled to ESI sources. The quadrupole FTMS combines the stability of a quadrupole analyzer with high accuracy of FTMS. A specified m/z range can be scanned by using the quadrupole, which can also be used to selectively isolate a precursor ion. This ion can be directed into the collision cell or the FTMS, and the resultant precursor and fragment ions can then be analyzed by the FTMS [29].
MS instrumentation related before will be important to understand some conclusions in the next MS biological studies, and we think that this instrumental review clarifies some researchers.
MS studies on estradiol metabolites as potential cancer biomarkers
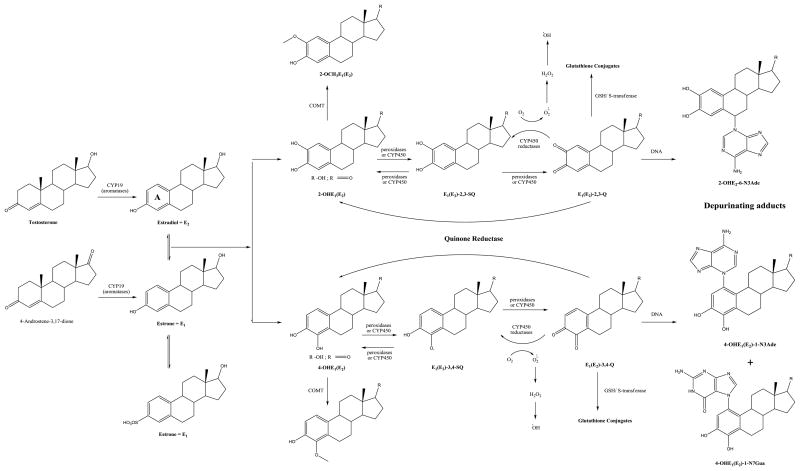
Metabolism of estrogens is characterized by two major pathways, respectively started by hydroxylation in ring A of molecule (Figure 1) to produce the 2- and 4-catechol estrogens, and hydroxylation at the 16α position (Fig. 1). In the catechol pathway, the metabolism involves further oxidation to semiquinones and quinones, including formation of the catechol estrogen-3,4-quinones (E-3,4-Q), the major carcinogenic metabolites of estrogens. These electrophilic compounds react with DNA to form the depurinating adducts 4-hydroxyestrone(estradiol)-1-N3Adenine (4-OHE1(E2)-1-N3Ade) and 4-hydroxyestrone- (estradiol)-1-N7Guanine (4-OHE1(E2)-1-N7Gua) [1]. Oxidation of catechol estrogens to their quinones is homeostatically controlled which minimizes formation of the quinones and their apurinic adducts. When the homeostasis is disrupted, excessive amount of catechol estrogen quinones are formed and the resulting increase in depurinating DNA adducts can lead to initiation of carcinogenesis [37]. Cavalieri and co-workers conducted several studies using ESI-MS and LC-MS/MS techniques to prove the formation of the aforementioned DNA adducts and their relevance in the initiation of carcinogenesis. Moreover, they used those techniques to study the effect of specific antioxidants, such as N-acetylcysteine and resveratrol (Resv), as well as of different enzymes, on estrogen metabolism [38].
Fig. 1.
Biosynthesis and metabolic activation of estrogens E1 and E2. One of the major pathways of E1 and E2 leads to 2- and 4-catechol derivatives, which are further, oxidized to yield the corresponding reactive quinones; these can react with DNA to form depurinating DNA adducts. In the deactivation pathway, which operates in parallel, the catechol derivatives are methylated to form methoxy catechol estrogens. In addition, the quinones are reduced by quinone reductase, as well as conjugated to GSH and thus rendered harmless. A shift in the apparent balance between activating and deactivating pathways towards formation of depurinating DNA adducts could lead to initiation of breast cancer (adapted from [38].)
2-OHE1(E2) – 2-hydroxyestrone(estradiol); 4-OHE1(E2) - 4-hydroxyestrone(estradiol); 2-OCH3E1(E2) – 2-methoxyestrone(estradiol); 4-OCH3E1(E2)- 4-methoxyestrone(estradiol); E1(E2)-2,3-SQ- Estrone(estradiol)-2,3-semiquinone; E1(E2)-2,3-Q – Estrone(estradiol)-2,3-quinone; E1(E2)-3,4-SQ - Estrone(estradiol)-3,4-semiquinone; E1(E2)-3,4-Q Estrone(estradiol)-3,4-quinones; 2-OHE2-6-N3Ade- 2-hydroxyestradiol-6-N3Adenine; 4-OHE1(E2)-1-N3Ade- 4-hydroxyestrone(estradiol)-1-N3Adenine; 4-OHE1(E2)-1-N7Gua- 4-hydroxyestrone(estradiol)-1-N7Guanine.
Gaikwad and colleagues studied the evidence for reduction of the carcinogenic ortho-quinones mediated by NRH quinone oxidoreductase 2 (NQO2). These authors showed for the first time that NQO2 catalyzes the reduction of electrophilic estrogen quinones and thereby may act as a detoxification enzyme. Binding of E2-3,4-Q with NQO2 was confirmed by ESI-MS, and further corroborated by analyzing the NQO2-E2-3,4-Q complex by matrix-assisted laser desorption/ionization mass spectrometry with a TOF analyzer (MALDI-TOF). Both UV and LC-MS/MS assays unequivocally corroborate the reduction of estrogen ortho-quinones by NQO2, indicating that this could be a novel target for prevention of breast cancer initiation [39].
In a separate report, Gaikwad et al. analyzed urine samples from 46 healthy control women, 17 women with breast cancer and 12 women in high risk of acquiring breast cancer. After partial purification of urine samples by solid phase extraction (SPE), 40 estrogen-related compounds were identified and quantitated by ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). Each metabolite was detected and identified based on unique parameters including mass retention time and ionization mode (positive or negative). The authors proposed that levels of depurinating DNA adducts to their respective estrogen metabolites and conjugates could provide an invaluable biomarker allowing differentiation between women at high risk of developing breast cancer, women with breast cancer and healthy women [40].
The potential function of various cytochrome CYP450 enzymes in oxidizing catechol estrogens to quinones was identified by Zhang et al. These investigators used different human CYP isoforms with the aim of oxidizing the catechol estrogens 2-OHE2 and 4-OHE2 to their respective estrogen quinones, which then react with DNA to form depurinating adducts in vitro. The reaction products were analyzed by UPLC-MS/MS. These experiments demonstrated that CYP isoforms are able to oxidize catechol estrogens to their respective quinones, which can further react with proteins, GSH and DNA, the latter resulting in depurinating adducts that can lead to mutagenesis [41]. Pruthi et al. conducted a study to determine whether the ratio of estrogen DNA adducts to their metabolites and conjugates in serum differed between women with early-onset breast cancer and those with average or high risk of developing breast cancer (serum samples from women at average risk (n=63) or high risk (n=80) using Gail model). The goal of this study was to investigate the imbalance of estrogen metabolism in serum expressed as estrogen-DNA adduct ratio to examine its potential as a biomarker for increased breast cancer risk [42]. Serum samples from women diagnosed with early breast cancer were analyzed by UPLC-MS/MS, allowing observation that level of depurinating estrogen-DNA adducts where significantly higher in women at high risk for developing breast cancer than in women at average risk. These findings suggest that serum estrogen-DNA adducts are potential biomarkers not only for determining the risk for developing breast cancer, but also for monitoring the effects of therapy. Yet, these findings are quite recent and require further studies towards unequivocal validation [42]. Zahid et al. reported a further study of benzene catechol (1,2-dihydroxybenzene, CAT) and N-acetyldopamine (NADA), which is itself a catechol. Benzene is metabolized to phenol in the liver by cytochrome CYP2E1. Other metabolites include CAT and hydroxyquinone (1,4-dihydroxybenzene). Oxidation of CAT and hydroxyquinone is catalyzed by peroxidases, forming quinones that can exert myelotoxic effects, and produce stable and depurinating DNA adducts. In this study, those authors analyzed reactions of catechol quinones such as the leukemogenic benzene quinone (CAT-Q) and N-acetyldopamine quinone (NADA-Q) with dG or DNA, using ESI-MS and UPLC-MS. They concluded that catechol quinones of natural and synthetic estrogens, benzene, naphthalene and dopamine react with DNA through an 1,4-Michael addition to form predominantly depurinating N3Ade and N7Gua adducts. With all of these compounds, the N3Ade adduct depurinates instantaneously from DNA, whereas the N7Gua adduct depurinates slowly, with a half-time of a few hours. For that reason, Zahid et al. proposed that common features may lead to the initiation of cancer and neurodegenerative diseases [43].
The effects of N-acetylcysteine N-AcCys on the metabolism of two cell lines MCF-10F (a normal human breast epithelial cell line) and E6 (a normal mouse breast epithelial cell line) were studied by Zahid et al. [43]. The cells were treated with 4-OHE2 or E2-3,4-Q, after which analysis using HPLC-EDC (electro-chemical detection) and UPLC-MS/MS demonstrated that N-AcCys inhibits the formation of depurinating adducts in an apparently similar way for both cell lines, despite originating from different mammal species. Therefore, it seems that, by blocking formation of estrogen-DNA adducts, N-AcCys could prevent the initiation of cancer by estrogens [44].
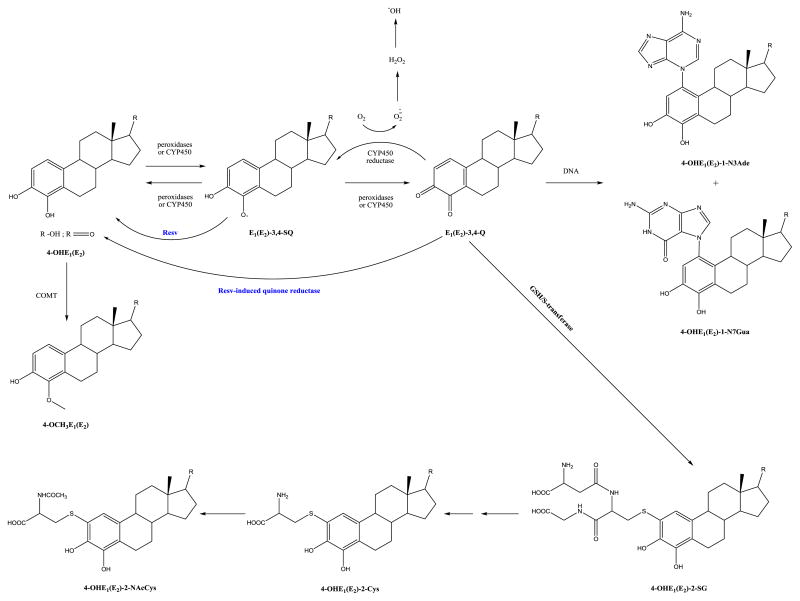
The same authors conducted another study using MCF-10F cells to evaluate the prevention of estrogen-DNA adduct formation by resveratrol (Resv), as Resv acts as both an antioxidant and an inducer of the phase II enzyme NQO1. The effects of Resv on the metabolism of estrogens was assessed by treatment of MCF-10F cells with 4-OHE2 or E2-3,4-Q, where UPLC-MS/MS was employed to analyze the estrogen-DNA adducts formed and determine the activity of NQO1 and catechol-O-methyl transferase (COMT). It was found that Resv decreased the formation of depurinating estrogen-DNA adducts from 4-OHE2 or E2-3,4-Q and increased formation of methoxycatechol estrogens; DNA adducts were not detectable, which indicates that Resv may be effective in preventing estrogen-mediated carcinogenesis, by estrogen by blocking multiple sites in the estrogen genotoxicity pathway, as shown in Fig. 2 [45].
Fig. 2.
Mechanism by which Resv is proposed to prevent estrogen-initiated breast cancer (reproduced from [45]).
Gaikwad and colleagues demonstrated the possibility of circumventing the problem of nonenzymatic reduction of estrogen quinone by NAD(P)H, given that NQO1 catalyzes the reduction of estrogen quinones. They performed mass spectrometric binding studies involving E2-3,4-Q or menadione with NQO1 which support the hypothesis of formation of an enzyme-substrate complex. Two different strategies were employed to ascertain the NQO1 activity in estrogen quinone reduction: first, the ping-pong mechanism of NQO1 catalysis was utilized to overcome the problem of nonenzymatic reduction of the substrate by NAD(P)H; second, tetrahydrophilic acid, which has a reducing potential lower than NAD(P)H was used as an alternative cofactor. Both strategies confirmed the reduction of E2-3,4-Q by NQO1, according to UV or LC/MS-MS analysis of the assay mixtures [10]. In view of this, the authors concluded that E2-3,4-Q is a substrate for NQO1, and NQO1 has a significant role in deactivation of estrogen ortho-quinone that might lead to initiation of cancer after formation of depurinating DNA adducts. These conclusions have broad implications for development of potential inducers of NQO1 that could ultimately prevent estrogen-initiated carcinogenesis [46].
In another study by Zahid et al., natural antioxidants, such as N-AcCys, melatonin, reduced lipoic acid and Resv, were investigated for their ability to prevent the reaction of E2-3,4-Q with DNA [43]. DNA was incubated with E2-3,4-Q or lactoperoxidase-activated 4-OHE2 in the presence of one of such antioxidants; after precipitation of DNA and centrifugation, supernatants were analyzed by LC-MS/MS and it was found that Resv and melatonin did not affect the formation of depurinating adducts when E2-3,4-Q was reacted with DNA [11]. On the other hand, N-AcCys and lipoic acid showed a significant inhibition of the formation of depurinating adducts by E2-3.4-Q. In the case of lactoperoxidase-activated 4-OHE2 reaction with DNA, Resv achieved the highest level of inhibition, N-AcCys and reduced lipoic acid produced moderate inhibition, whereas melatonin had the lower inhibitory capability. Therefore, this LC-MS/MS study provided demonstration that all four antioxidants inhibited formation of adducts involved in malignant transformation of mammary epithelial cells, identifying them as potential chemo-preventing agents of cancer initiation, particularly breast and prostate cancer [47].
Gaikwad et al. also reported a study where the aim was to investigate urine biomarkers of risk in the molecular etiology of breast cancer. To this end, urine samples from 40 healthy control women, 40 high risk women (Gail Model scores were 1.67%–11.7%) and 40 women newly diagnosed with breast cancer where analyzed by UPLC-MS/MS for quantitation of estrogen metabolites such as conjugates and depurinating DNA adducts [38]. At the outset, they confirmed that relatively high levels of depurinating estrogen-DNA adducts were present in women at high risk for breast cancer or diagnosed as already diseased. Estrogen metabolism was shifted from protective methoxylation and conjugation pathways in the healthy individuals towards activating pathways leading to formation of depurinating DNA adducts in women at high risk or with breast cancer. These results support the hypothesis that breast cancer is initiated by mutations derived from depurination of estrogen-DNA adducts. Therefore, relative levels of depurinating estrogen-DNA adducts could become biomarkers for early detection of breast cancer risk and aid in prevention [38].
A recent analysis of urine samples from premenopausal women during the luteal phase showed that nine of the steroidal estrogens (E1, E2, 16α-OHE1, E3, 16-ketoE2, 2-OHE1, 2-OHE2, 2-MeOE1, 4-OHE1) represent more than 90% of the measured urinary estrogen metabolites [48]. This was corroborated by an analogous study involving 10 premenopausal women also during luteal phase, where the same nine compounds represented 89% of all measured urinary estrogen metabolites [49]. At a much larger scale, Franke et al. analyzed steroids in 232 urine samples from 78 premenopausal women, using a benchtop orbitrap LC-MS and a single quadrupole GC-MS; while GC-MS allows the measurement of a wide range of steroids, including non-polar analytes that elude detection by LC-MS, orbitrap-based LC-MS is more sensitive, faster, cheaper and allows post-data acquisition reinterrogation of analytes not targeted a priori [50]. In this combined LC-MS/GC-MS study, sixteen steroidal estrogens, including oxidized metabolites, could be detected; the LC-MS/GC-MS Spearman rank correlation coefficients (ρ) for the relative concentrations of major estrogens E1, E2, E3, 16α-OHE1 and 2-OHE1 were very high (ρ~ 0.72 to 0.91), and absolute concentrations as determined by both techniques were also in agreement (below 5% difference for E1, E2, E3, 16α-OHE1). LC-MS allowed reinterrogation of the acquired data due to the orbitrap technology, which permitted post-analysis quantitation of progesterone, cortisol, and cortisone with an LC-MS/GC-MS ρ between 0.80 and 0.84, and differences between absolute concentrations below 7% (n=13).
Mass spectrometry techniques have also been used in the survey of potential biomarkers of bladder cancer caused by the parasitic flatworm Schistosoma haematobium; this eukaryotic pathogen infects millions of people mostly in the rural regions of sub-Saharan Africa, and is associated with high incidence of bladder cancer, although why this happens remains uncertain [51]. Since it has long been known that schistosomes have estradiol receptors, Botelho et al. conducted a series of studies on sera from S. haematobium infected persons, as potentially carcinogenic antigens/components from these parasites may be useful to decipher schistosome-associated oncogenesis [52]; thus, those authors readily identified and quantitated the sex hormones estradiol, testosterone and luteinizing stimulating hormone (LH) and found that, in all cases, serum levels of estradiol were remarkably high as compared to those in sera from non-infected persons; the other hormones were not dissimilar from normal levels [52]. In view of this, the authors hypothesized that the levels of estradiol observed in infected patients were of schistosome origin, and therefore analyzed the estradiol content in extracts of worms, to find significant expression of estradiol-related molecules [52,53]. The same authors employed LC-ESI-MS to investigate the possible presence of additional undisclosed estrogenic molecules in worm extracts and sera of schistosome-infected persons [53,54]; novel estrogen-related molecules were identified in both worm tissues and sera of infected individuals, but not in the plasma of a healthy donor, and their structures suggest that they were formed upon reaction of estrogen quinone with DNA (Fig. 1). Therefore, given the aforementioned carcinogenic potential of this estrogen adduct, the estrogen-related molecules found in extracts of S. haematobium may be the link between schistosomiasis and squamous cell carcinoma of bladder [54].
MS methods in the analysis of cholesterol, bile acids and related metabolites
Estrogens have not been the only steroids targeted by MS-based metabolomics studies: androgens, corticoids and other steroids such as bile acids, vitamin D, cardiac steroids and cholesterol itself exhibit physiologically relevant activities, and therefore are frequently monitored in diverse biological samples [55]. Due to the metabolic versatility of steroid molecules, extremely complex mixtures are often encountered, making their analysis quite demanding and requiring chromatographic separation prior to MS analysis [52]. Both GC-MS and LC-MS have been used in the study of steroids and their metabolites, but LC-MS is considered as the most promising analytical method for determination of steroids and steroid conjugates, due to its sensitivity, specificity and versatility [55,56]. Cholesterol is undeniably the most emblematic of all steroids; it is a constituent of cell membranes and a bioprecursor of bile acids and steroid hormones. Cholesterol is also one of the major risk factors for arteriosclerotic diseases, such as hypertension and cerebrovascular disease. Thus, the remainder of this review will focus on literature reports about MS-based studies on cholesterol, bile acids and derivatives.
Kock et al. developed an LC-isotope dilution-MS procedure to quantify total cholesterol in serum, without the need of an in-line derivatization method, using [25,26,27-13C] cholesterol as the internal standard [56]; a particle-beam interface was used for coupling the LC and the MS [20], whereas alkaline hydrolysis followed by extraction with cyclohexane were employed for sample preparation; results obtained correlated well with those produced by the stable isotope dilution GC-MS method, but some interference by other steroids in the cholesterol quantitation by the LC-MS approach was observed [55,56].
Griffiths et al. developed prototypic LC-MS/MS methods for prenatal diagnosis of Smith-Lemli-Opitz syndrome (SLOS), a severe disorder in cholesterol synthesis that is classically diagnosed prenatally by GC-MS analysis of sterol in the amniotic fluids [57]; in this work, the 3β-hydroxysterols from amniotic fluids were oxidized with cholesterol oxidase to their corresponding 3-ketones, which were then derivatized with Girard P (GP) hydrazine in a “one-pot” reaction; the GP-hydrazones formed were analyzed by LC-ESI-MS/MS, and results provided proof of concept on the potential application of this approach, once optimized, in the prenatal diagnosis of SLOS [57]. Marbel et al. developed a novel LC-MS/MS method for quantitative determination of 4β-hydroxycholesterol after analyte extraction from plasma with hexane and extract purification by normal-phase SPE followed by 4β-hydroxycholesterol isolation from cholesterol and endogenous isobaric plasma oxysterols by reversed-phase HPLC; detection was achieved by APPI-MS/MS in the positive mode, using toluene as dopant [58]. This LC-APPI-MS/MS method allowed quantitation of 4B-hydroxycholesterol in human plasma at relevant physiological levels (10.0–250 nM), which makes it highly suited for the clinics, given the method’s high sensitivity, selectivity, accuracy and relative short analysis time [58].
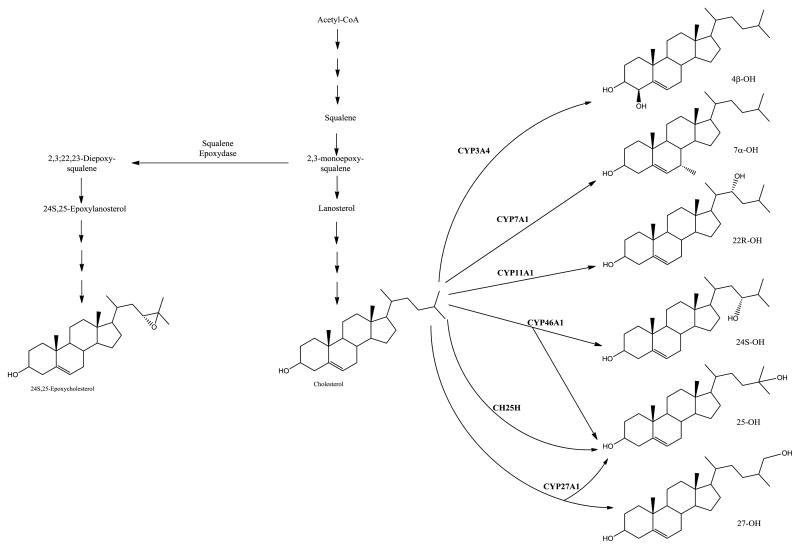
Oxysterols, which are mostly oxygenated forms of cholesterol in mammals (Fig. 3) recently found to be biologically active per se [59], have also been the focus of some methodological studies based on MS techniques. For example, Honda and co-authors [60] described a highly sensitive and specific method, based on a stable isotope dilution LC-MS/MS technique, for the quantitation of the oxysterol 7α-hydroxy-4-cholesten-3-one (C4), which has been used as a biomarker for bile acid biosynthesis [24]; C4 was extracted from human serum by a salting-out procedure, derivatized into its picolinyl ester and then isolated by SPE [60] for subsequent quantitation by LC-ESI-MS/MS; the method provided highly reliable and reproducible results for quantiation of C4 in sera, especially from small volumes of blood samples [60]. Application of this method was then expanded by the same authors for the quantitation, in serum or rat liver microsomes, of numerous key regulatory oxysterols [61], and also for the analysis of serum sterol profiles [62]. These works allowed confirming that derivatization of (i) dihydroxy- and epoxysterols [61], and (ii) neutral sterols [62] into the respective picolinyl esters enabled fast LC-ESI-MS/MS quantitation with high sensitivity and reproducibility. Also, it allowed to identify, in addition to cholesterol, 19 cholesterol precursors, cholestanol, campresterol, sitosterol, and sitostanol [62]. Overall, this method seems to be potentially useful in lipid metabolism studies or in the clinics for (i) diagnosis of cholesterol/oxysterol metabolism-related disorders, or (ii) quantitation of serum biomarkers for the synthesis/absorption of cholesterol.
Fig. 3.
Biosynthetic pathways for key regulatory oxysterols. Hydroxycholesterols are synthesized from cholesterol, whereas 24S,25-epoxycholesterol is derived from a shunt in the cholesterol biosynthetic pathway. CH25H, cholesterol 25-hydroxylase; 4β-OH, 4β-hydroxycholesterol; 7α-OH, 7α-hydroxycholesterol; 22R-OH, 22R-hydroxycholesterol; 24S-OH, 24S-hydroxycholesterol; 25-OH, 25-hydroxycholesterol and 27-OH, 27-hydroxycholesterol (reproduced from (61).)
An alternative method for the quantitation of C4 in serum, without the need for chemical derivatization, was described by Lövgren-Sandblom [63]; this method was used successfully in studies on sera from healthy fasting volunteers, and results obtained were consistent with the possibility that part of 7α-hydroxy-3-oxo-4-cholestenoic acid in blood originates from extrahepatic C4; therefore, the authors hypothesized that the rate of production of C4 in liver is directly reflected by levels of α-hydroxy-3-oxo-4-cholestenoic acid in blood, which is consistent with use of the latter as a marker for cholesterol 7α-hydroxylase activity [63].
Oxysterols occur in mammalian brain at ng/g- μg/g levels, while cholesterol is present at mg/g levels, which makes oxysterol analysis in brain rather challenging. Karu et al. developed a nano-LC-MS/MS method for the analysis of oxysterols in brain, based on SPE of the oxysterol fraction followed by and an oxidation derivatization protocol and then nano-flow-LC-MSn analysis [64]. According to these authors, while the oxidation derivatization method improved detection limits, nano-LC-MSn provided separation of isomers and allowed for accurate oxysterol quantification [28]; in fact, they were able to identify 13 discrete oxysterols in rat brain, including 24S-hydroxycholesterol, 24S-25-epoxycholesterol and 7α,26-dihydroxycholest-4-en-3-one [64]. The same research group developed a novel LC-MSn methodology for the identification of cholesterol metabolites in rat brain with high sensitivity, i.e., at the low pg level [65]; the method includes derivatization to enhance ionization, exact mass analysis at high resolution to identify potential metabolites, and MSn (n=3) to allow their structural characterization; this provided confirmation of 24S-hydroxycholesterol as a major oxysterol in rat brain, and identification of other formerly undisclosed oxysterols in brain, such as 24,25-, 24,27-, 25,27-, 6,24-, 7α-25-, and 7α-27-dihydroxycholesterols. Additionally, two molecules linked to protein amyloidogenesis, 3β-hydroxy-5-oxo-5,6-seccholestan-6-al and its aldol, were also identified in the same study [65].
An LC-MS/MS approach to determine 24S-hydroxycholesterol (24SOHChol) separately from 25-hydroxycholesterol in plasma was described by DeBarber and colleagues, as blood levels of 24SOHChol are a practical measure of cholesterol efflux from human brain; the method was found be accurate and free of interference by endogenous 25-hydroxycholesterol interference, with the advantage of involving simplified sample work-up and analysis [66]. Development of highly sensitive and accurate methods for the analysis of 24SOHChol is extremely relevant also due to the fact that this oxysterol, as well as 27-hydroxycholesterol, are under investigation as potential biomarkers associated with neurodegenerative disorders such as Alzheimer’s disease and multiple sclerosis [67]. In this connection, Griffiths et al. developed a new LC-MS approach using charge-tagging and high resolution MS providing identification in plasma of several oxysterols and downstream metabolites such as, 7α-, 24S-, and 27-hydroxycholesterol, the cholestenetriol 7α-27-dihydroxycholesterol, and 3β-hydroxycholest-5-en-27-oic acid and its metabolite 3β-7α-dihydroxycholest-5-en-27-oic acid [67].
The formation of bile acids and bile alcohols is of major importance for the maintenance of cholesterol homeostasis, as besides their functions in lipid absorption, bile acids/alcohols are regulatory molecules for a number of metabolic processes. Their effects are structure-dependent, and numerous metabolic conversions result in a complex mixture of biologically active and inactive forms. MS is the basic detection technique for the analysis of bile acids/alcohols in biological media, usually after at least an LC separation step. Capillary LC-ESI-MS normally provides the highest sensitivity, but depending on the nature of the bile acid/alcohol mixture and the range of concentrations, discrete sample preparation sequences, ranging from simple extractions to complex group separations and derivatizations, are applicable [68].
Ikegawa et al. developed an LC-ESI-MS method for simultaneous individual determination of different bile acid 3-sulfates in human urine; the urine sample was subjected to SPE followed by ion-exchange chromatography on a lipophilic gel, and then submitted to LC-ESI-MS analysis; the 3-sulfates were characterized by an abundant pseudo-molecular ion [M-H]− along with a doubly charged ion [M-2H]2−, whose ratio was markedly influenced by an acidic component added to the LC mobile phase [69]. The application of this method to the analysis of the urine from a healthy volunteer allowed detection of chenodeoxycholic acid, deoxycholic acid and lithocholic acid 3-sulfate as glycine conjugates, with very small amounts of unconjugated and taurine-conjugated bile acid-3-sulfates [34]. In turn, analysis of urine from patients with obstructive jaundice led to identification of the glycine conjugates of chenodeoxycholic acid and cholic acid 3-sulfates, but not of lithocholic acid 3-sulfate [70]. Ikegawa’s group also developed a method for separation and determination of bile acid 24-glucuronides in urine using LC-ESI-MS, to provide more information about the metabolic profile of bile acids and potentially serve as diagnosis tool for hepatobiliary diseases; the extracted glucoronides were subjected to LC-ESI-MS analysis employing an 18O-labeled internal standard, and detected as intense peaks due to the deprotonated molecule [M-H]− and a fragment ion [M-H-176]− [69].
LC-ESI-MS studies by Goto et al. provided unprecedented identification of bile acid acyl galactosides in urine from healthy donors; the urine specimens were subjected to SPE followed by LC separation and alkalyne hydrolysis, after which cholic acid (CA) and deoxycholic acid (DCA) were identified as liberated bile acids, detected along with other unknown components [71,72]. To identify the latter, a portion of the alkaline hydrolysate was reacted with 1-phenyl-3-methyl-5-pyrazolone, which enabled detection of galactose derivatives by LC-ESI-MS; further analyses, using adequate controls, provided confirmation of the derivatives structures as being CA 24-galactoside and DCA 24-galactoside, whose biosynthesis in the human body was thus confirmed. Goto et al. have also developed a highly sensitive LC/ESI-MS/MS method, using selected reaction monitoring (SRM) analysis, for quantitation of bile acid derivatives in human urine; this provided simultaneous analysis of bile acid 3-sulfates, including nonamidated glycine-, and taurine-conjugated bile acid, cholic acid, chenodeoxycholic acid, deoxycholic acid, ursodeoxycholic acid and lithocholic acid, and first-time identification of 3β,12α-dihydroxy-5β-cholanoic acid 3-sulfate in human urine [72].
Last, but not least, Muto et al. have very recently described a new, simple and sensitive LC-ESI-MS/MS method for the identification and characterization of 39 conjugated and unconjugated bile acids, including Δ4-3-oxo- and Δ4,6-3-oxo-bile acids (markers for Δ4-3-oxo-steroid-5β-reductase deficiency) [73]. In this method, a concentrated desalted urine sample was diluted in ethanol and directly injected into the LC-ESI-MS/MS, with detection in the negative ion mode and quantitation by SRM. The remarkable performance of this new approach was confirmed by comparison with a previously validated GC-MS analysis method, using urine from two patients with genetically confirmed Δ4-3-oxo-steroid-5β-reductase deficiency and from a third patient with abnormally high levels of conjugated and unconjugated Δ4–3-oxo-bile acids [73].
Mass spectrometry to assess androgen status
The highly sensitive and specific LC-MS/MS methods have been shown to be superior to conventional immunoassays to assess low sex hormone concentrations. Haring et al. [74] determined age-specific reference ranges for LC-MS/MS-measured total testosterone and prohormone androstenedione (is the most common sex hormone precursor in both sexes and plays a crucial role in the biosynthesis of testosterone) as well as calculated free testosterone, in a large population-based sample of women aged 20–80 years. This LC-MS/MS method allows rapid, sensitive, and specific determination of serum sex hormone concentrations in women and is therefore suitable for routine clinical practice and research. Thus, the presented age-specific reference limits are particularly valuable to translate testosterone concentrations outside the reference range into clinical treatment and to establish appropriate cutoffs for clinical guidelines and epidemiological studies [74].
Reference ranges are essential for partitioning testosterone levels into low or normal and making the diagnosis of androgen deficiency. Bhasin et al. established reference ranges for total testosterone and free testosterone in a community-based sample of men, using using liquid chromatography tandem mass spectrometry [75]. Reference ranges generated in a community-based sample of men provide a rational basis for categorizing testosterone levels as low or normal. Men with low total testosterone or free testosterone by these criteria had higher prevalence of physical dysfunction, sexual dysfunction, and diabetes [75].
The association between aging-related testosterone deficiency and late-onset hypogonadism in men remains a controversial concept. Mass spectrometry was used to develop criteria for identifying late-onset hypogonadism in the general population on the basis of an association between symptoms and a low testosterone level [76]. Late-onset hypogonadism can be defined by the presence of at least three sexual symptoms associated with a total testosterone level of less than 11 nmol per liter (3.2 ng per milliliter) and a free testosterone level of less than 220 pmol per liter (64 pg per milliliter) [76].
Recently, testosterone is measured for the investigation of female hyperandrogenism and male hypogonadism [77]. LC-MS/MS is becoming the method of choice but comprehensive reference ranges are lacking. Testosterone was measured by tandem MS on 90 healthy women, 67 young healthy men and pregnant women (59 first trimester and 60 second trimester). The male, male calculated free, first trimester and second trimester testosterone reference ranges (derived using the antilog of mean ± 1.96 SD of log transformed data) were 10.6–31.9, 0.23–0.63, 0.6–4.9 and 0.9–4.9 nmol/L, respectively. The female testosterone upper reference range limit, derived non-parametrically from the 97.5th centile, was < 1.7 nmol/L [77].
Conclusion
The introduction of soft ionization methods in MS and the evolution of highly sensitive and versatile LC-MS/MS techniques has made possible the analysis of steroid hormones with small sample requirements, and simplified sample preparation. The latest reports in this field, as those reviewed here, demonstrate that MS-based techniques have a central role in steroid metabolomics, contributing not only to a deeper understanding of steroid metabolism, but also to the identification of clinically relevant disease biomarkers.
Acknowledgments
PG and NV thank Fundação para a Ciência e Tecnologia (FCT, Portugal) and FEDER (European Union) for funding through project grants CONC-REEQ/275/QUI and PEst-C/QUI/UI0081/2011. NV also thanks FCT for Post-Doc grant SFRH/BPD/48345/2008.
PJB received support from award R01CA155297 from the National Cancer Institute The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI or NIH.
Abbreviations
- APCI-MS
Atmospheric-pressure chemical ionization mass spectrometry
- APPI-MS
Atmospheric-pressure photoionization mass spectrometry
- CA
Cholic acid
- CAT
1,2-Dihydroxybenzene (benzene catechol)
- CAT-Q
1,2-Dihydroxybenzene – Quinone
- CID
Collision-induced dissociation
- COMT
Catechol-O-methyltransferase
- DCA
Deoxycholic acid
- ESI-MS
Electrospray ionizationp
- FIA
Flow injection analysis
- FTMS
Fourier transform mass spectrometry
- GC-MS
Gas chromatography-mass spectrometry
- GP
Girard P
- HPLC-EDC
High performance liquid chromatography-electro-chemical detection
- LC-MS
Liquid chromatography-mass spectrometry
- MALDI-TOF
Matrix-assisted laser desorption/ionization-time-of-flight
- N-AcCys
N-acetylcysteine
- NADA
N-acetyldopamine
- NADA-Q
N-acetyldopamine-quinone
- NQO-2
NRH quinone oxidoreductase 2
- Resv
Resveratrol
- SPE
Solid phase extration
- SLOS
Smith-Lemli-Opitz syndrome
- SRM
Selected reaction monitoring
- TOF
Time-of-flight
- UPLC-MS/MS
Ultra-performance liquid chromatography-tandem mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Want EJ, Cravatt BF, Siuzdak G. The expanding role of mass spectrometry in metabolite profiling and characterization. ChemBioChem. 2005;6:1941–51. doi: 10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- 2.Dear GJ, Plumb AR, Fraser IJ. The rapid identification of drug metabolites using capillary liquid chromatography coupled to an ion trap mass spectrometer. Rapid Commun Mass Spectrom. 1999;3:456–63. doi: 10.1002/(SICI)1097-0231(19990315)13:5<456::AID-RCM508>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 3.Zhang N, Fountain ST, Bi H, Rossi ST. Quantification and rapid metabolite identification in drug discovery using API Time-of-Flight LC/MS. Anal Chem. 2000;72:800–6. doi: 10.1021/ac9911701. [DOI] [PubMed] [Google Scholar]
- 4.Shockor JP, Holmes E. Metabonomic Applications in Toxicity Screening and Disease Diagnosis. Current Trop Med Chem. 2002;2:35–51. doi: 10.2174/1568026023394498. [DOI] [PubMed] [Google Scholar]
- 5.Plumb RS, Stumpf CL, Granger JH, Castro-Perez J, Haselden JN, Dear GJ. Use of liquid chromatography/time-of-flight mass spectrometry and multivariate statistical analysis shows promise for the detection of drug metabolites in biological fluids. Rapid Commun Mass Spectrom. 2003;17:2632–38. doi: 10.1002/rcm.1250. [DOI] [PubMed] [Google Scholar]
- 6.Tiller PR, Romanyshyn LA. Liquid chromatography/tandem mass spectrometric quantification with metabolite screening as a strategy to enhance the early drug discovery process. Rapid Commun Mass Spectrom. 2002;16:1225–39. doi: 10.1002/rcm.708. [DOI] [PubMed] [Google Scholar]
- 7.Deng Y, Wu JT, Zhang H, Olah TV. Quantitation of drug metabolites in the absence of pure metabolite standards by high-performance liquid chromatography coupled with a chemiluminescence nitrogen detector and mass spectrometer. Rapid Commun Mass Spectrom. 2004;18:1681–85. doi: 10.1002/rcm.1540. [DOI] [PubMed] [Google Scholar]
- 8.Maurer HH. Advances in analytical toxicology: Current role of liquid chromatography-mass spectrometry for drug quantification in blood and oral fluid. Anal Bioanal Chem. 2005;381:110–18. doi: 10.1007/s00216-004-2774-z. [DOI] [PubMed] [Google Scholar]
- 9.Staack RF, Varesio E, Hopfgartner G. The combination of liquid chromatography/tandem mass spectrometry and chip-based infusion for improved screening and characterization of drug metabolites. Rapid Commun Mass Spectrom. 2005;19:618–26. doi: 10.1002/rcm.1829. [DOI] [PubMed] [Google Scholar]
- 10.Liu DQ, Hop CE. Strategies for characterization of drug metabolites using liquid chromatography–tandem mass spectrometry in conjunction with chemical derivatization and on-line H/D exchange approaches. J Pharm Biomed Anal. 2005;37:1–18. doi: 10.1016/j.jpba.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Cravatt BF, Prospero-Garcia O, Siuzdak G, Gilula NB, Henriksen SJ, Boger DL, Lerner RA. Chemical characterization of a family of brain lipids that induce sleep. Science. 1995;268:1506–09. doi: 10.1126/science.7770779. [DOI] [PubMed] [Google Scholar]
- 12.Saghatelien A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43:14332–39. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discovery. 2002;1:153–61. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 14.Reo NV. NMR-Based Metabolomics. Drug Chem Toxicol. 2002;25:375–82. doi: 10.1081/dct-120014789. [DOI] [PubMed] [Google Scholar]
- 15.Griffin JL. Metabonomics: NMR spectroscopy and pattern recognition analysis of body fluids and tissues for characterisation of xenobiotic toxicity and disease diagnosis. Curr Opin Chem Biol. 2003;7:648–54. doi: 10.1016/j.cbpa.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Niwa T. Procedures for MS analysis of clinically relevant compounds. Clin Chim Acta. 1995;241–242:75–152. [PubMed] [Google Scholar]
- 17.Niwa T. Metabolic profiling with gas chromatography-mass spectrometry and its application to clinical medicine. J Chromatogr B Biomed Sci Appl. 1986;379:313–45. doi: 10.1016/s0378-4347(00)80688-x. [DOI] [PubMed] [Google Scholar]
- 18.Gelpi E. Biomedical and biochemical applications of liquid chromatography-mass spectrometry. J Chromatogr A. 1985;703:59–80. doi: 10.1016/0021-9673(94)01287-o. [DOI] [PubMed] [Google Scholar]
- 19.Deng C, Deng Y, Wang B, Yang X. as chromatography–mass spectrometry method for determination of phenylalanine and tyrosine in neonatal blood spots. J Chromatogr B. 2002;780:407–13. doi: 10.1016/s1570-0232(02)00632-3. [DOI] [PubMed] [Google Scholar]
- 20.Deng C, Shang C, Hu Y, Zhang X. Rapid diagnosis of phenylketonuria and other aminoacidemias by quantitative analysis of amino acids in neonatal blood spots by gas chromatography–mass spectrometry. J Chromatogr B. 2002;775:115–20. doi: 10.1016/s1570-0232(02)00283-0. [DOI] [PubMed] [Google Scholar]
- 21.Deng C, Deng Y. Diagnosis of maple syrup urine disease by determination of L-valine, L-isoleucine, L-leucine and L-phenylalanine in neonatal blood spots by gas chromatography mass–spectrometry. J Chromatogr B. 2003;792:261–68. doi: 10.1016/s1570-0232(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 22.Deng C, Li N, Zhang X. Rapid determination of amino acids in neonatal blood samples based on derivatization with isobutyl chloroformate followed by solid-phase microextraction and gas chromatography/mass spectrometry. Rapid Commun Mass Spectrom. 2004;18:2558–64. doi: 10.1002/rcm.1660. [DOI] [PubMed] [Google Scholar]
- 23.Sjövall J, Lawson AM, Stechell KD. Mass spectrometry of bile acids. Methods Enzymol. 1985;111:63–113. doi: 10.1016/s0076-6879(85)11006-2. [DOI] [PubMed] [Google Scholar]
- 24.Perwalz S, Mignault D, Tuchweber B, Yousef IM. Rapid and improved method for the determination of bile acids in human feces using MS. Lipids. 2002;37:1093–100. doi: 10.1007/s11745-002-1005-0. [DOI] [PubMed] [Google Scholar]
- 25.Shackleton CH. Profiling steroid hormones and urinary steroids. J Chromatogr. 1986;379:91–156. doi: 10.1016/s0378-4347(00)80683-0. [DOI] [PubMed] [Google Scholar]
- 26.Caufield MP, Lynn T, Gottschalk ME, Jones KL, Taylor NF, Malunowicz EM, Shackleton CH, Reitz RE, Fisher DA. The Diagnosis of Congenital Adrenal Hyperplasia in the Newborn by Gas Chromatography/Mass Spectrometry Analysis of Random Urine Specimens. J Clin Endocrinol Metabol. 2002;87:3682–90. doi: 10.1210/jcem.87.8.8712. [DOI] [PubMed] [Google Scholar]
- 27.Wudy SA, Hartmann MF. Gas Chromatography-Mass Spectrometry Profiling of Steroids in Times of Molecular Biology. Horm Metab Res. 2004;36:415–22. doi: 10.1055/s-2004-814565. [DOI] [PubMed] [Google Scholar]
- 28.Kushnir MM, Komaromy-Hiller G, Shushan B, Urry FM, Roberts WL. Analysis of dicarboxylic acids by tandem Mass Spectrometry. High-throughput quantitative measurement of methylmalonic acid in serum, plasma, and urine. Clin Chem. 2001;47:1993–2002. [PubMed] [Google Scholar]
- 29.Siuzdak G. The expanding role of mass spectrometry in Biotechnology. MCC Press; San Diego: 2003. [Google Scholar]
- 30.Chatman K, Hollenbeck T, Hagey L, Vallee M, Purdy R, Weiss F, Siuzdak G. Nanoelectrospray Mass Spectrometry and Precursor Ion Monitoring for Quantitative Steroid Analysis and Attomole Sensitivity. Anal Chem. 1999;71:2358–63. doi: 10.1021/ac9806411. [DOI] [PubMed] [Google Scholar]
- 31.Gangl ET, Annan MM, Spooner N, Vouros P. Reduction of Signal Suppression Effects in ESI-MS Using a Nanosplitting Device. Anal Chem. 2001;73:5635–44. doi: 10.1021/ac010501i. [DOI] [PubMed] [Google Scholar]
- 32.Gustavsson SA, Samskog J, Markides KE, Längström B. Studies of signal suppression in liquid chromatography-electrospray ionization mass spectrometry using volatile ion-pairing reagents. J Chromatogr A. 2001;937:41–7. doi: 10.1016/s0021-9673(01)01328-0. [DOI] [PubMed] [Google Scholar]
- 33.Chace DH. Mass Spectrometry in the Clinical Laboratory. Chem Rev. 2001;101:445–77. doi: 10.1021/cr990077+. [DOI] [PubMed] [Google Scholar]
- 34.Plumb RS, Warwick H, Higton D, Dear GJ, Mallett DN. Determination of 4-hydroxytamoxifen in mouse plasma in the pg/mL range by gradient capillary liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2001;15:297–303. doi: 10.1002/rcm.225. [DOI] [PubMed] [Google Scholar]
- 35.Griffiths WJ, Liu S, Yang Y, Purdy RH, Sjövall J. Nano-electrospray tandem mass spectrometry for the analysis of neurosteroid sulphate. Rapid Commun Mass Spectrom. 1999;13:1595–610. doi: 10.1002/(SICI)1097-0231(19990815)13:15<1595::AID-RCM681>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 36.Ablan J, Oosterkamp AJ, Gelpi E. Comparison of conventional, narrow-bore and capillary liquid chromatography/mass spectrometry for electrospray ionization mass spectrometry: practical considerations. J Mass Spectrom. 1999;34:244–54. [Google Scholar]
- 37.Cavalieri EL, Rogan EG. Unbalanced metabolism of endogenous estrogens in the etiology and prevention of human cancer. J Steroid Biochem Mol Biol. 2011;125:169–80. doi: 10.1016/j.jsbmb.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaikwad NW, Yang L, Pruthi S, Ingle JN, Sandhu N, Rogan EG, Cavalieri EL. Urine biomarkers of risk in the molecular etiology of breast cancer. Breast Cancer: Basic Clin Res. 2009;3:1–8. doi: 10.4137/bcbcr.s2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaikwad NW, Yang L, Rogan EC, Cavalieri EL. Evidence for NQO2-mediated reduction of the carcinogenic estrogen ortho-quinones. Free Radic Biol Med. 2009;46:253–62. doi: 10.1016/j.freeradbiomed.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaikwad NW, Yang L, Muti P, Meza JL, Pruthi S, Ingle JN, Rogan EG, Cavalieri EL. The molecular etiology of breast cancer: Evidence from biomarkers of risk. Int J Cancer. 2008;122:1949–57. doi: 10.1002/ijc.23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Gaikwad NW, Olson K, Zahid M, Cavalieri EL, Rogan EG. Cytochrome P450 isoforms catalyze formation of catechol estrogen quinones that can react with DNA. Metabolism. 2007;56:887–94. doi: 10.1016/j.metabol.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 42.Phruthi S, Yang L, Sandhu NP, Ingle JN, Beseler CL, Suman VJ, Cavalieri EL, Rogan EL. Evaluation of serum estrogen-DNA adducts as potential biomarkers for breast cancer risk. J Steroid Biochem Mol Biol. 2012;132:73–9. doi: 10.1016/j.jsbmb.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zahid M, Saeed M, Rogan EG, Cavalieri EL. Benzene and dopamine catechol quinones could inititate cancer or neurogenic disease. Free Radic Biol Med. 2010;48:318–24. doi: 10.1016/j.freeradbiomed.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zahid M, Saeed M, Ali MF, Rogan EG, Cavalieri EL. N-acetylcysteine blocks formation of cancer-initiating estrogen-DNA adducts in cells. Free Radic Biol Med. 2010;49:392–400. doi: 10.1016/j.freeradbiomed.2010.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zahid M, Gaikwad NW, Ali MF, Lu F, Saeed M, Yang L, Rogan EG, Cavalieri EL. Prevention of estrogen-DNA adduct formation in MCF-10F cells by resveratrol. Free Radic Biol Med. 2008;45:136–145. doi: 10.1016/j.freeradbiomed.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaikwad NW, Rogan EG, Cavalieri EL. Evidence from ESI-MS for NQO1-catalyzed reduction of estrogen ortho-quinones. Free Radic Biol Med. 2007;43:1289–98. doi: 10.1016/j.freeradbiomed.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zahid M, Gaikwad NW, Rogan EG, Cavalieri EL. Inhibition of depurinating estrogen-DNA adduct formation by natural compounds. Chem Res Toxicol. 207;20:1947–53. doi: 10.1021/tx700269s. [DOI] [PubMed] [Google Scholar]
- 48.Eliassen AH, Ziegler RG, Veenstra TD, Roman JM, Xu X, Hankinson SE. Reproducibility of fifteen urinary estrogens and estrogen metabolites over a 2- to 3-year period premenopausal women. Cancer Epidemiol Biomark Prev. 2009;18:2860–68. doi: 10.1158/1055-9965.EPI-09-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falk RT, Xu X, Keefer L, Veenstra TD, Ziegler RG. A liquid chromatography mass spectrometry method for the simultaneous measurement of 15 urinary estrogens and estrogen metabolites: assay reproducibility and interindividual variability. Cancer Epidemiol Biomark Prev. 2008;17:3411–18. doi: 10.1158/1055-9965.EPI-08-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franke A, Custer LJ, Morimoto Y, Nordt FJ, Maskarinec G. Analysis of urinary estrogens, their oxidized metabolites, and other endogenous steroids by benchtop orbitrap LCMS versus traditional quadropole GCMS. Anal Bioanal Chem. 2011;401:1319–30. doi: 10.1007/s00216-011-5164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Botelho MC, Machado JC, Costa JMC. Schistosoma haematobium and bladder cancer. Virulence. 2010;1:84–87. doi: 10.4161/viru.1.2.10487. [DOI] [PubMed] [Google Scholar]
- 52.Botelho MC, Machado JC, Brindley PJ, Costa JMC. Targeting molecular signaling pathways of Schistosoma haematobium infection in bladder cancer. Virulence. 2011;2:267–79. doi: 10.4161/viru.2.4.16734. [DOI] [PubMed] [Google Scholar]
- 53.Botelho MC, Crespo M, Almeida A, Vieira P, Delgado ML, Araujo L, Machado JC, Costa JMC. Schistosoma haematobium and Schistosomiasis mansoni: Production of an estradiol-related compound detected by elisa. Exp Parasitol. 2009;122:250–3. doi: 10.1016/j.exppara.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 54.Botelho MC, Soares R, Vale N, Ribeiro R, Camilo V, Almeida R, Medeiros R, Gomes P, Machado JC, Costa JMC. Schistosoma haematobium: Identification of new estrogenic molecules with estradiol antagonist activity and ability to inactivate estrogen receptor in mammalian cells. Exp Parasitol. 2010;126:526–35. doi: 10.1016/j.exppara.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 55.Shimada K, Mitamura K, Higashi T. Gas chromatography and high-performance liquid chromatography of natural steroids. J Chromatogr A. 2001;935:141–72. doi: 10.1016/s0021-9673(01)00943-8. [DOI] [PubMed] [Google Scholar]
- 56.Kock R, Delvoux B, Greiling H. Determination of total cholesterol in serum by liquid-chromatography-isotope dilution mass spectrometry. Clin Chem. 1997;43:1896–903. [PubMed] [Google Scholar]
- 57.Griffiths WJ, Wang Y, Karu K, Samuel E, McDonnell S, Hornshaw M, Shackleton C. Potential of sterol analysis by liquid chromatography-tandem mass spectrometry for the Prenatal diagnosis of Smith-Lemli-Opitz Syndrome. Clin Chem. 2008;54:1317–24. doi: 10.1373/clinchem.2007.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van de Merbel NC, Bronsema KJ, van Hout MWJ, Nilsson R, Sillén H. A validated liquid chromatography-tandem mass spectrometry method for the quantitative determination of 4β-hydroxycholesterol in human plasma. J Pharm Biomed Anal. 2011;55:1089–95. doi: 10.1016/j.jpba.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 59.Griffiths WJ, Wang Y. Analysis of oxysterols metabolomes. Biochim Biophys Acta. 2011;1811:784–99. doi: 10.1016/j.bbalip.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 60.Honda A, Yamashita K, Numazawa M, Ikegami T, Doy M, Matsuzaki Y, Miyazaki H. Highly sensitive quantification of 7a-hydroxy-4-cholesten-3-one in human serum by LC-ESI-MS/MS. J Lipid Res. 2007;48:458–64. doi: 10.1194/jlr.D600032-JLR200. [DOI] [PubMed] [Google Scholar]
- 61.Honda A, Yamashita K, Hara T, Ikegami T, Miyazaki H, Shirai M, Xu G, Numazawa M, Matsuzaki Y. High sensitive quantification of key regulatory oxysterols in biological samples by LC-ESI-MS/MS. J Lipid Res. 2009;50:350–57. doi: 10.1194/jlr.D800040-JLR200. [DOI] [PubMed] [Google Scholar]
- 62.Honda A, Yamashita K, Hara T, Miyazaki H, Shirai M, Ikegami T, Xu G, Numazawa M, Matsuzaki Y. Highly sensitive analysis of sterol profiles in human serum by LC-ESI-MS/MS. J Lipid Res. 2008;49:2063–73. doi: 10.1194/jlr.D800017-JLR200. [DOI] [PubMed] [Google Scholar]
- 63.Lövgren-Sandlom A, Heverin M, Larsson H, Lundström E, Wahren J, Diczfalusy U, Björkhem I. Novel LC-MS/MS method of assay of 7a-hydroxy-4-cholesten-3-one in human plasma. Evidence for a significant extrahepatic metabolism. J Chromatogr A. 2007;856:15–19. doi: 10.1016/j.jchromb.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 64.Karu K, Turton J, Wang Y, Griffiths WJ. Nano-liquid chromatography-tandem mass spectrometry analysis of oxysterols in brain: monitoring of cholesterol autoxidation. Chem Phys Lipid. 2011;164:411–24. doi: 10.1016/j.chemphyslip.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 65.Karu K, Hornshaw M, Woffendin G, Bodin K, Hamberg M, Alvelius G, Sjövall J, Turton J, Wang Y, Griffiths WJ. Liquid chromatography-mass spectrometry utilizing multi-stage fragmentation for the identification of oxysterols. J Lipid Res. 2007;48:976–87. doi: 10.1194/jlr.M600497-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeBarber AE, Lütjohann D, Merken SL, Steiner RD. Liquid chromatography-tandem mass spectrometry determination of plasma 24S-hydroxycholesterol with chromatographic separation of 25-hydroxycholesterol. Anal Biochem. 2008;381:151–53. doi: 10.1016/j.ab.2008.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Griffiths WJ, Hornshaw M, Woffending G, Baker SF, Lockhart A, Heidelberger S, Gustafsson M, Sjövall J, Wang Y. Discovering oxysterols in plasma: a window on the metabolome. J Proteome Res. 2008;7:3602–12. doi: 10.1021/pr8001639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Griffiths WJ, Sjövall J. Bile acids: analysis in biological fluids and tissues. J Lipid Res. 2010;51:23–41. doi: 10.1194/jlr.R001941-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ikegawa S, Okuyama H, Oohashi J, Murao N, Goto J. Separation and detection of bile acid 24-glucuronides in human urine by liquid chromatography combined with electrspray ionization mass spectrometry. Analyt Sci. 1999;15:625–31. [Google Scholar]
- 70.Ikegawa S, Yanagihara T, Murao N, Watanabe H, Goto J, Niwa T. Separatory determination of bile acid-3-sulfates by liquid chromatography/electrospray ionization mass spectrometry. J Mass Spectom. 1997;32:401–07. [Google Scholar]
- 71.Goto T, Shibata A, Sasaki D, Suzuki N, Hishinuma T, Kakiyama G, Iida T, Mano N, Goto J. Identification of a novel conjugate in human urine: bile acid acyl galactosides. Steroids. 2005;70:185–92. doi: 10.1016/j.steroids.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 72.Goto T, Myint KT, Sato K, Wada O, Kakiyama G, Iida T, Hishinuma T, Mano N, Goto J. LC/ESI-tandem mass spectrometric determination of bile acid 3-sulfates in human urine 3b-sulfooxy-12a-hydroxy-5b-cholanoic acid is an abundant nonamidated sulfate. J Chromatogr B. 2007;846:69–77. doi: 10.1016/j.jchromb.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 73.Muto A, Takei H, Unno A, Murai T, Kurosawa T, Ogawa S, Iida T, Ikegawa S, Mori J, Ohtake A, Hoshina T, Mizuochi T, Kimura A, Hofmann AF, Hagey LR, Nittono H. Detection of Δ4-3-oxo-steroid 5b-reductase deficiency by LC-ESI-MS/MS measurement of urinary bile acids. J Chromatogr B. 2012;900:24–31. doi: 10.1016/j.jchromb.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 74.Haring R, Hannemann A, John U, Radke D, Nauck M, Wallaschofski H, Owen L, Adaway J, Keevil BG, Brabant G. Age-specific reference ranges for serum testosterone and androstenedione concentrations in women measured by liquid chromatography-tandem mass spectrometry. J Clin Endocrinol Metab. 2012;97:408–415. doi: 10.1210/jc.2011-2134. [DOI] [PubMed] [Google Scholar]
- 75.Bhasin S, Pencina M, Jasuja GK, Travison TG, Coviello A, Orwoll E, Wang PY, Nielson C, Wu F, Tajar A, Labrie F, Vesper H, Zhang A, Ulloor J, Singh R, D’Agostino R, Vasan RS. Reference ranges for testosterone in men generated using liquid chromatography tandem mass spectrometry in a community-based sample of healthy nonobese young men in the Framingham Heart Study and applied to three geographically distinct cohorts. J Clin Endocrinol Metab. 2011;96:2430–2439. doi: 10.1210/jc.2010-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu FC, Tajar A, Beynon JM, Pye SR, Silman AJ, Finn JD, O’Neill TW, Bartfai G, Casanueva FF, Forti G, Giwercman A, Han TS, Kula K, Lean ME, Pendleton N, Punab M, Boonen S, Vanderschueren D, Labrie F, Huhtaniemi IT. Identification of late-onset hypogonadism in middle-aged and elderly men. N Engl J Med. 2010;363:123–135. doi: 10.1056/NEJMoa0911101. [DOI] [PubMed] [Google Scholar]
- 77.Neale SM, Hocking R, Biswas M, Turkes A, Rees D, Rees DA, Evans C. Adult testosterone and calculated free testosterone reference ranges by tandem mass spectrometry. Ann Clin Biochem. 2013 doi: 10.1258/acb.2012.012047. In press. [DOI] [PubMed] [Google Scholar]