Abstract
En masse cell migration is more relevant than single cell migration in physiological processes of tissue formation, such as embryogenesis, morphogenesis and wound healing. In these situations, cells are influenced by the proximity of other cells including interactions facilitated by substrate mechanics. Here we found that when fibroblasts migrated en masse over a hydrogel, they established a well-defined deformation field by traction forces and migrated along a trajectory defined by field gradients. The mechanics of the hydrogel determined the magnitude of the gradient. For materials stiff enough to withstand deformation related to cellular traction forces, such patterns did not form. Furthermore, migration patterns functioned poorly on very soft matrices where only a minimal traction gradient could be established. The largest degree of alignment and migration velocity occurred on the gels with the largest gradients. Granulation tissue formation in punch wounds of juvenile pigs was correlated strongly with the modulus of the implanted gel in agreement with in vitro en masse cell migration studies. These findings provide basic insight into the biomechanical influences on fibroblast movement in early wounds and relevant design criteria for development of tissue-engineered constructs that aim to stimulate en masse cell recruitment for rapid wound healing.
Keywords: wound healing, fibroblast, cell migration, biomechanics
Introduction
Tissue cell migration is the sine qua non of wound repair and regeneration (Singer and Clark, 1999). During the inflammatory phase of wound healing blood leukocytes sense chemical gradients emanating from the wound to which they respond by directed migration (chemotaxis) as single cells. In contrast, tissue cells often move en masse rather than singly. For example, in skin, epidermal cells quickly move across the wound space as a sheet in a tractor-tread fashion associated with dynamic formation and dissolution of cell junctions (Chometon et al., 2006; Vespa et al., 2005). Fibroblasts, which can respond to haptotactic (Carter, 1967; Raeber et al., 2008) as well as chemotactic signals, also move into the wound space en masse rather than as single cells during rapid granulation tissue formation (McClain et al., 1996), However, fibroblasts form relatively few cell junctions compared to epithelial and endothelial structures and these were only recently described (Morris et al., 2006). In addition to biochemical signals (Palecek et al., 1997), it has become increasingly clear that substrate mechanics is another key attribute that modulates tissue cell dynamics (Choquet et al., 1997; Discher et al., 2005; Georges and Janmey, 2005; Ghosh and Ingber, 2007; Lo et al., 2000; Pelham and Wang, 1997). Many groups have shown that cells can sense substrate stiffness through their adhesions to the extracellular matrix (ECM) and respond by altering their cytoskeletal organization and tension accordingly (Beningo and Wang, 2002; Galbraith and Sheetz, 1998; Geiger and Bershadsky, 2002; Ingber, 2003; Lo et al., 2000; Sheetz et al., 1998). The tension exerted onto the substrate via adhesion sites, called cellular traction forces, has been known as the mechanical reason for movement of a single cell (Oliver et al., 1999; Pan et al., 2009; Schmidt et al., 1993). More interestingly, because these traction forces can cause deformation on flexible substrates for distances large compared to the size of the cell, a mechanical means of communication between cells may occur over large distances and influence their en-masse behavior. The recent study of Reinhart-King et al. on paired endothelial cells on substrates with varying stiffness has clearly demonstrated that with appropriate substrate stiffness, cells can detect and respond to substrate strains created by neighboring cells (Reinhart-King et al., 2008) and Sen et al. applied a finite element analysis to study the length scale of the deformation at the interface of isolated cells and a gel substrate. (Sen et al., 2009). However, in most physiological processes, such as embryonic development, morphogenesis, and wound healing, en masse behavior of a large number of cells becomes more relevant (Clark 1996; Friedl and Gilmour, 2009; Rorth, 2007, 2009). In these situations where the cell density is high, the mechanism of communication between cells is more complicated, and can result in different migration behavior than for either single cells or a small clusters of cells. In this report we studied the dynamics of cells, plated at low density with those at high density, but placed on identical substrates and cultured in the same growth media. In this way we could directly compare the effect of substrate mechanics on the migration behavior of single cells with those subjected to influence from their neighbors. In contrast to previous in-vivo studies (McClain et al., 1996), where “en-masse” migration was defined in terms of the migration of multiple cell types within an extracellular matrix, here we focused on the en masse migration of one cell type out of an agarose droplet placed on a hydrogel substrate composed of crosslinked hyaluronic acid (HA) and fibronectin functional domains (FNfds).
By changing crosslink ratios, we varied the stiffness of substrates and studied their effects on en masse cell migration. Since the same substrates have been previously utilized to establish the correlation between substrate modulus and single cell migration (Ghosh et al., 2007), we could readily quantify the difference between en masse and single cell migration response to substrate stiffness. We then used an optimized version of the digital image speckle correlation (DISC) technique (Guan et al., 2004; Pan et al., 2009) to quantify displacement fields of cell ensembles as a function of substrate modulus, and demonstrated that a direct correlation could be established between en masse cell motion and displacement field topology. Since the HA/FNfds hydrogels used in vitro are bio-functional, biodegradable, and have moduli that could be varied in a range relevant to an early wound environment (Hinz, 2007), they were used in a porcine excisional wound model, where en masse fibroblast migration into wound site is critical for granulation tissue formation and subsequent healing (McClain et al., 1996). In agreement with in vitro findings, the percentage of granulation formed within a 4d porcine full-thickness excisional wound correlated positively with increasing modulus of the implant.
Results and Discussion
En masse cell migration as function of cell density
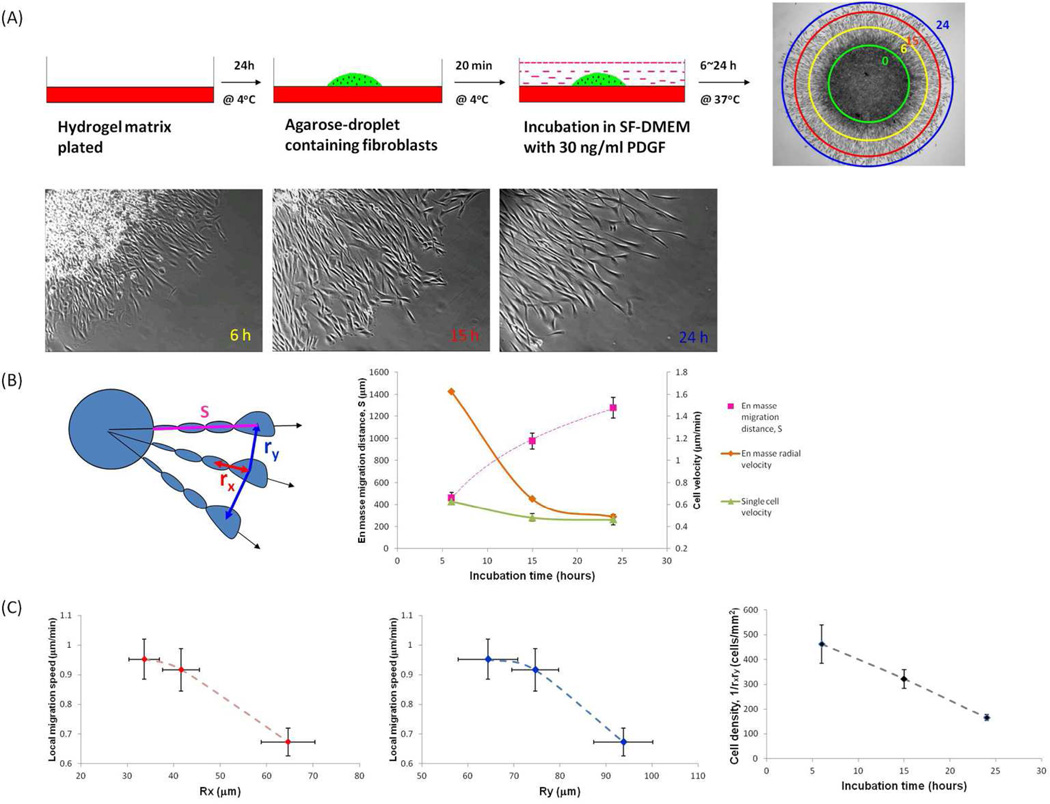
An agarose droplet loaded with adult human dermal fibroblasts (AHDF) was placed on a HA hydrogel functionalized with FNfds and crosslinked with PEGDA and incubated as shown in Figure 1A to allow cell attachment to the substrate. The outward migration of AHDF cells that emerged completely from the agarose and migrated on the hydrogel substrate was then monitored at 6,15, and 24h and the distance between the migration front, delineated by circular contours from the edge of the agarose droplet (Figure 1b) was recorded for each time. The shear modulus for this substrate was G’=4270 Pa, which was previously shown to support cell adhesion and actin stress fiber organization (Ghosh et al., 2007). As can be seen from the phase contrast images in Figure 1A, the AHDFs migrated en masse from the cell-laden agarose droplet in a radially outward manner that resulted in a continuous decreased in cell density with increasing distance from the droplet. Using time-lapse photography we recorded the cell movement at 15min intervals over 1h periods after 6, 15, and 24h incubations. Thus, we were able to obtain both cell velocity and regional cell density. In order to obtain single cell migration velocity, without interference from extraneous traction forces, cells were plated in the absence of agarose, at a very low density of ~500 cells/cm2 on HA hydrogel functionalized with FNfds hydrogels with the same stiffness (G’=4270 Pa) and observed following the same protocols. In the lower panel of Figure 1A we show high magnification phase contrast images corresponding to one quadrant of the outward migration pattern at the three incubation times. From the figures we see that the local cell density in the outermost ring of migrating cells at the leading edge is decreasing with increasing incubation time (Figure 1A). The average migration distance (S) from the droplet edge to the nucleus of a given cell at the migration front (Figure 1B schematic) was plotted as a function of incubation time, together with the derivative of the curve, which is the radial velocity function (Figure 1B graph). Single cell velocity was measured from the time-lapse images, as a function of incubation time (Figure 1B) and found that the velocity of leading cells during en masse outmigration decreased exponentially, approaching the single cell migration speeds that did not change significantly during the 15h incubation period.
Figure 1.
En masse cell migration decreased as cell density decreased. (A) Schematic of agarose droplet assay; low-magnification phase image obtained after 24h and rings indicating distance of migration at 0, 6, 15 and 24h; and high-magnification phase image of cell migration from agarose droplet at 6, 15 and 24h. (B) Schematic defines migration distance from edge of original droplet to nucleus of a given cell at migration front as S, and distance between nuclei of cells along the radial direction as rx, or along the circumference as ry. Graph shows average migration distance (S) as function of incubation time, together with derivative of curve, which is radial velocity function, compared to single cell migration velocity measured after the same incubation time. (C) Local migration speed of cells at the migration front as function of rx and ry, and local cell density (1/rxry) as function of incubation time. (B. en masse migration distance, mean±SD; single cell migration velocity, mean±SE, n=26–30; C mean±SE, n=12–18)
The local velocity of the leading cells was also measured as a function of the distance to the nearest neighbor cells along the edge (Figure 1C). The rx and ry show the distance from the nucleus of one cell to the nuclei of its neighboring cells along the radial directions and the circumference, respectively (Figure 1B schematic). The local density can then be approximately one cell per rxry unit area (1/rxry) and was plotted as a function of incubation time. The three panels of Figure 1C together showed that with longer incubation time, leading cells migrated slower with decreasing cell density. From these results it appears that local cell density is a primary driver controlling migration speed on the hydrogel surface.
En masse cell migration as function of substrate stiffness
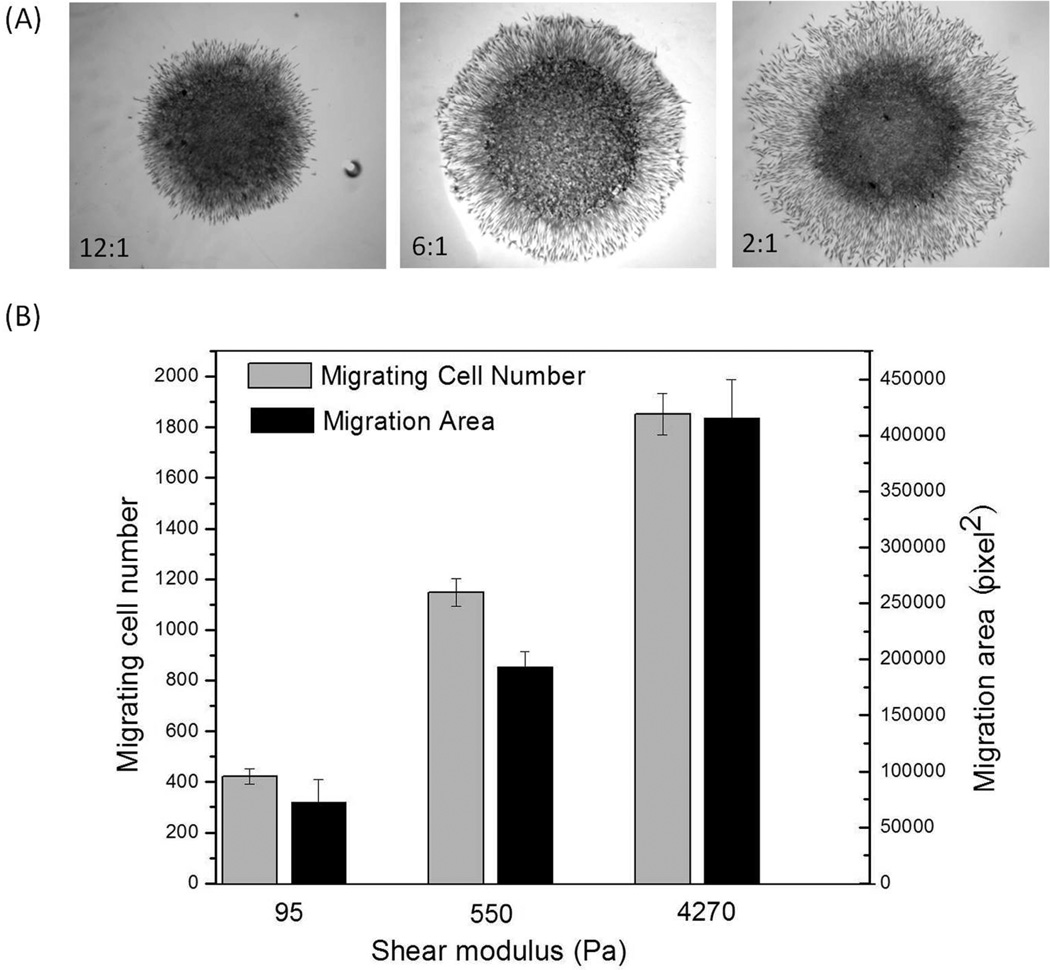
In addition to cell density, substrate stiffness also plays an important role in migration dynamics. It has previously been shown that the migration speed of single cells negatively correlates with the distribution and magnitude of substrate stiffness-dependent cell traction forces (Dembo et al., 1996; Dembo and Wang, 1999; Oliver et al., 1999; Pan et al., 2009). Here we show that, like single cells, en masse migrating cells also sense and respond to substrate elasticity although, contrary to single cells (Ghosh et al., 2007), en masse cell migration becomes faster on hydrogels of increasing stiffness (Figure 2A). When the average cell migration area and number of migrating cell from the radius and area of the corona around the droplet was measured as functions of shear moduli, G’ of the gels, it was confirmed quantitatively that en masse cell migration increased with increasing hydrogel moduli. A 5–10 fold increase in substrate shear modulus (G’) produced a 2–3 fold increase in both the number of cells which migrated out of the droplet and the area that they covered during the observation time. This result indicated that substrate mechanics alone can dramatically regulate en masse cell behavior in a different manner from single cells.
Figure 2.
En masse cell migration increased with increased substrate stiffness. (A) Phase images of en masse cell migration from agarose droplets onto hydrogels with different stiffness after 18 hours incubation. (B) Average en masse cell migration area and number of cells migrating out of the droplet as a function of the hydrogel stiffness. (A mean±SD, n-3–4)
Dynamic reciprocity between migrating cells and substrate deformation
Previously we have shown that isolated AHDF, which were adherent to fibronectin functional domains tethered to HA hydrogel, exerted traction forces whose magnitudes were related to substrate moduli. The substrate stiffness through the reactive cellular traction forces affected cell functions that were dependent on cellular adhesions, such as spreading, migration, and proliferation (Ghosh et al., 2007). However, substrate deformation extended distances that were much larger than the area occupied by individual cells. Thus, in addition to the more classical cell-cell communication systems such as cell-cell contact or paracrine gradients, local cell traction forces can create extended regions of substrate deformation that permit cells to communicate with each other by sensing the deformation gradients. This is especially relevant to understanding the influence of the substrate stiffness on the en masse cell migration where large deformation gradients are generated within the hydrogels and are in turn, sensed by the cells thereby altering both the direction and magnitude of resultant outward migration (Figure 3). In our system the leading edge of cells migrating out of the droplet, cell-cell adhesions are not apparent, even though they may be present inside the agarose droplet where the cells are at very high density. Furthermore, the migration speed decreases as the cells move further away from the droplet and their density decreases making cell contacts even less probable.
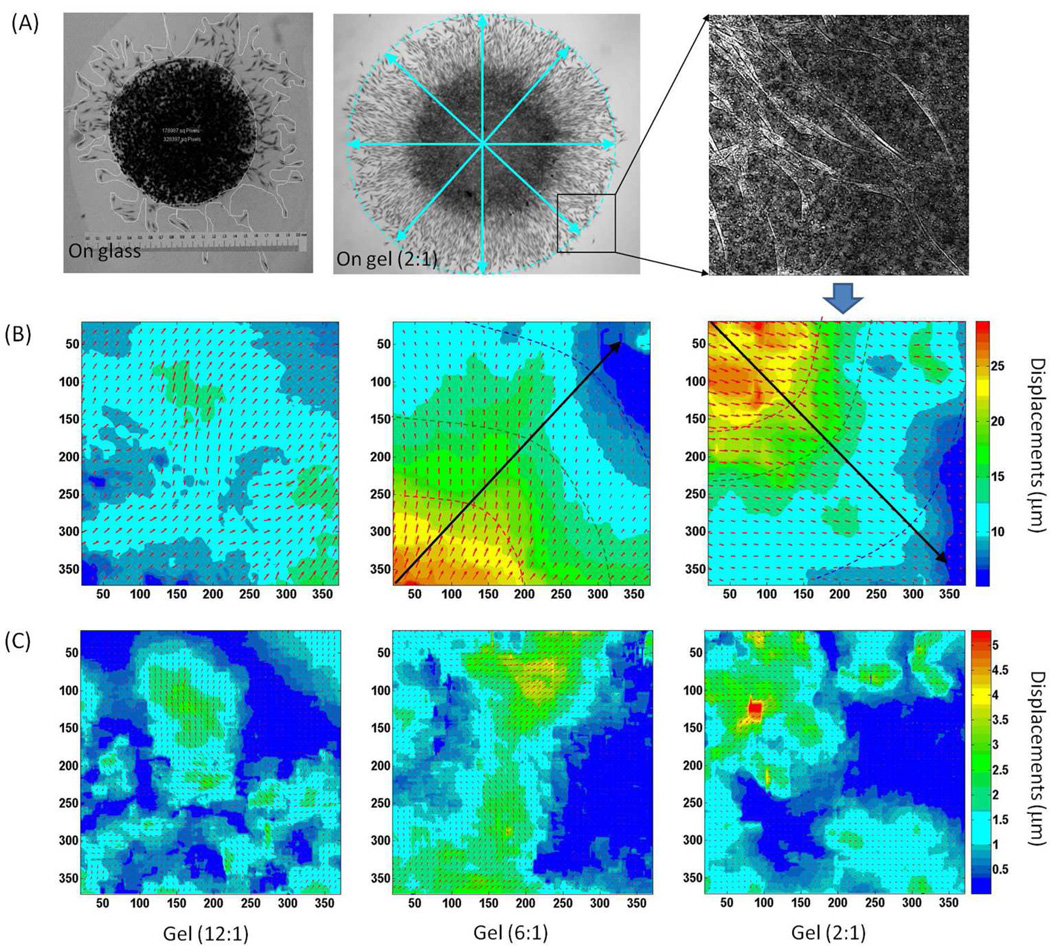
Figure 3.
Substrate displacement generated by en masse migrating cells. (A) Pattern of en masse cells migrating outward from a droplet placed on the stiffest hydrogel, together with the profile of en masse cells migrating out of a droplet placed on fibronectin-coated glass. (B) The local displacements around the edge of the droplets deposited on gels of different stiffness. The rays show the trajectories of the cells in the direction of decreasing deformation gradients. The colored dash curves approximate the equal deformation contours outwards the edge of the droplets. Since the deformation gradients can be estimated from the distance between the deformation contours, we can see that as en masse cells generate larger displacements on stiffer hydrogels (4270 Pa and 550 Pa) than on soft hydrogels (95 Pa), the deformation gradients decrease faster on the stiffest substrate (4270 Pa) than on the medium one (550 Pa). (C) Local displacements generated by the local cells at the migration front, which is calculated by removing the radial displacements from (B), showing that individual cells can generate larger displacements on stiffer substrate.
In Figure 3A, fibroblasts migrating out of an agarose droplet placed on a fibronectin-coated glass coverslip are shown together with cells migrating outward from an agarose droplet placed on the stiffest hydrogel (G’=4270 Pa). Compared to migration on gels, cell migration on glass was more disordered as cells did not follow each other in a radial pattern. Instead, cells appeared to leave the droplet, become physically isolated, and migrate in random directions. Furthermore, after some time they triangulated and ceased to migrate presumably secondary to strong adherence. In contrast, during en masse cell migration on hydrogels, the cells were elongated and appeared to follow each other. Although the migration patterns decrease cell density, only the distance between cells along the ry direction increased continuously. The distance along the rx direction remained fairly constant, with no significant change in the contacts between adjacent cells, even at the largest distances from the droplet. Random cell migration may explain the initial outward migration from the droplet, but it cannot account for the ray pattern formation. Rather the radial arrays might result directly from the hydrogel deformation fields around the edge of agarose droplets as measured by DISC (Figure 3B). In fact, cell migration rays appeared to follow deformation gradients, and form trajectories of cells in the direction of decreasing deformation gradients. Furthermore, the orderly progression of the cells out of the agarose droplet along rays appeared responsible for the larger number of cells that migrated out of the droplet on the gel surface relative to those on glass, i.e. directed migration versus random migration.
Hence deformation, in addition to modulus, becomes a key element in the migration pattern. Since the deformation patterns as well as the cellular response that creates them are both a function of the hydrogel moduli a system of dynamic reciprocity between cell traction forces and substrate mechanical properties is established. This interpretation does not rule out the possibility that the cells are also depositing extracellular matrix “tracks”, causing cells to emerge in interconnected streams, as previously proposed (Miron-Mendoza 2012) for corneal fibroblasts emerging from a cell laden button into a fibrin gel. However, in their case the migration was in a three-dimensional fibrin gel that most likely contained fibrin fibrils at different stages of formation. Therefore, it is not surprising that streamer morphology was due to remodeling fibers within the gel. In our case substrate deformation possibly modulates spatial deposition of newly synthesized ECM proteins that in turn imprints the oriented migratory patterns. Our results are consistent with the early observations of Paul Weiss whose work was reviewed recently (Grinnell 2010). Weis and co-workers first proposed that cell generated tension tracks within a fibrin gel along which cells migrated in tissue fragments (Weiss 1959). In this model the deformation fields, in combination with ECM proteins, established a pattern of “contact guidance” that resulted in coordinated en-mass migration and the formation of ordered hierarchial structures in tissue.
From the data we can also obtain the dependence of the deformation profile on the substrate modulus. Deformation contours on hydrogels of different stiffness, emanating outwards from the edge of the agarose droplets (colored dash curves in Figure 3B), can be mapped using the DISC analysis, and then the deformation gradients can be calculated from the distance between these curves. Not surprisingly, deformation gradients appeared dependent on the substrate modulus. The deformation contours were closer together on stiffer hydrogels, thus the deformation gradient increased with increasing hydrogel stiffness. In fact, a gradient was barely discerned on the softest gel. The solid lines (rays) originating at the droplet and drawn perpendicular to the contour lines delineate the direction of the deformation gradient (Figure 3B). A compelling interpretation of the migration pattern is that the cells can sense both the magnitude and the direction of the deformation gradient. Cell migration then follows the deformation gradient, which for an isolated droplet, results in the radial pattern of vectors pointing in the direction of decreasing deformation, orthogonal to the deformation contour.
In the case of the softest gel, the gradient is small and cells migrate least effectively out of the droplet. Even though the morphology of the cells on the softest gel is polarized and polarized cells are known to have the highest migration velocity, the lack of a well-defined gradient appears to prevent cells from finding a well-defined direction for migration. The displacements directly measured in Figure 3B were generated by all cells migrating outwards from the droplet. Removing radial displacements allowed measurement of local displacements generated by cells at the migration front (Figure 3C). Such measurements demonstrated that individual cells generate larger traction forces and displacements on stiffer substrate, a result consistent with previous single cell studies (Ghosh et al., 2007). In the previous work, the increased traction forces were responsible for reducing the single cell velocity of cells migrating in isolation. In the case of en-mass migration, it has been shown (Angelini, 2010, Trepat, 2009) that collective deformation induced by overlapping force fields of multiple cells, adds an additional factor in determining the cell migration velocity, hence differentiating cooperative migration from single cell migration. Our results show that the larger forces exerted by the cells on the stiffer substrates are also responsible for creating larger cooperative gradient force fields, which in turn are responsible for faster en mass migration.
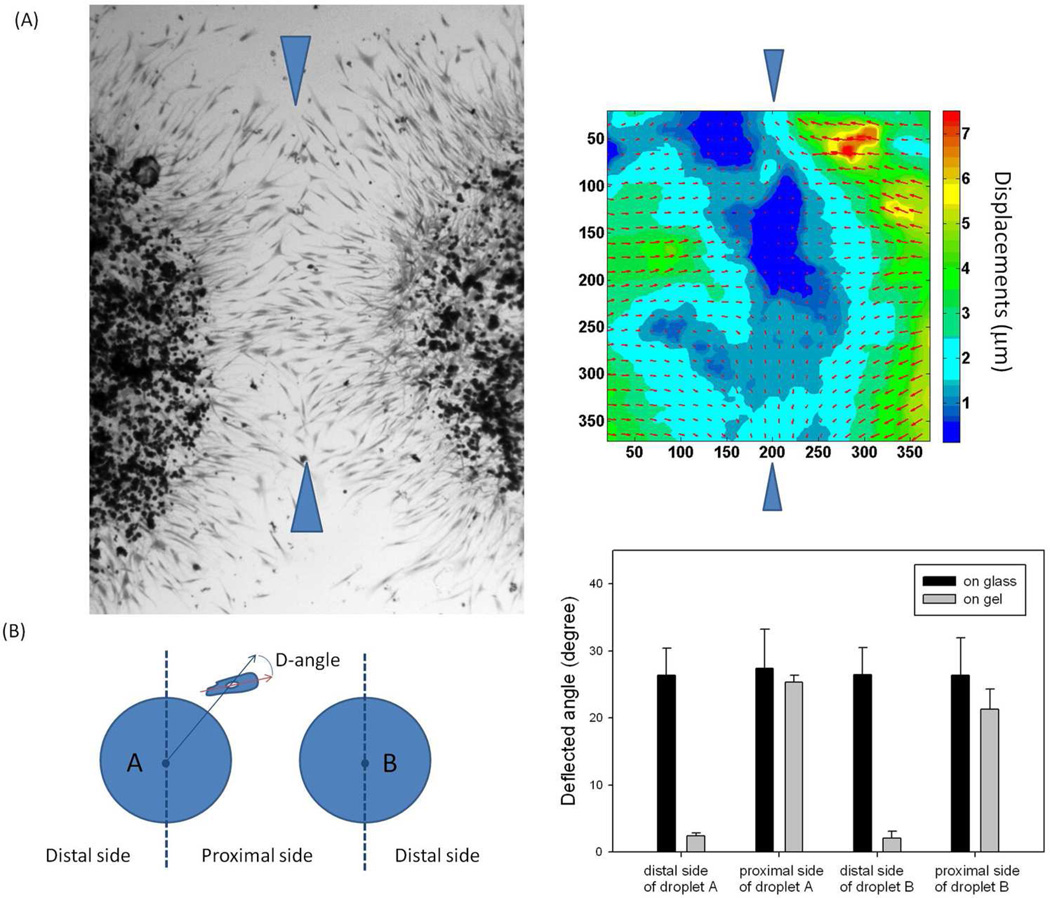
To test the deformation gradient model, we placed two agarose droplets loaded with fibroblasts in close proximity to each other and observed the patterns established as the cells began to migrate outward from both droplets simultaneously (Figure 4A). The cells initially migrated along radial rays, which put them on a trajectory towards each other. As they approached each other near the central line, the deformation gradient rotated by 90 degrees. The displacement map shows that the local displacements near the central line (between the two blue arrows) reach zero, which shift the deformation gradient about 90 degrees. This provides a clear biomechanical explanation for why cells in the middle tended to turn and migrate along the central line. A similar agarose arrangement was also made on glass, where such a phenomenon was not observed. The degree of correlation between the two droplets can be quantified by measuring the average deflected angles of lines drawn through the major axis of migrating cells from lines along the radii of the droplet (Figure 4B). Deflection angles in the region between droplets (proximal side) were compared to the deflection angles in the region on the opposite side of the droplet (distal side). The angles between rays and cells were small on the opposite side of the droplet indicating alignment, whereas the angles between rays and cells were large at the midway region between droplets showing the deflected trajectory correlated with shifted deformation gradients. The same analysis of cell migration from two agarose droplets placed on glass failed to show any deflection of cell migration trajectories at the midway region.
Figure 4.
Two-droplet model simulating the bidirectional migration during wound healing. (A) The pattern of en masse cells migrating outward from two agarose droplets loaded in close proximity to each other, and the local deformation generated in the middle. Since the local displacements near the central line (between the two blue arrows) reach zero, which shift the deformation gradient about 90 degrees, cells in the middle turn around and migrate along the central line. (B) The schematic defines the deflected angle (D-angle) as the angle between a given migrating cell along the edge and the radial direction that go through the cell nucleus. The degree of correlation between the two droplets is quantified by comparing the average D-angle on the proximal sides with that on the distal side. The D-angel on gel significantly increases on the proximal sides due to the influence of the neighboring droplet on the local displacements, while the D-angle is always large on glass since cells migrate randomly without the cue of deformation gradients. (B. mean + SD, 4 pairs of agarose droplets, more than 20 cells/droplet side were included in the measurement)
In vivo en masse cell migration: tissue repair as function of substrate stiffness
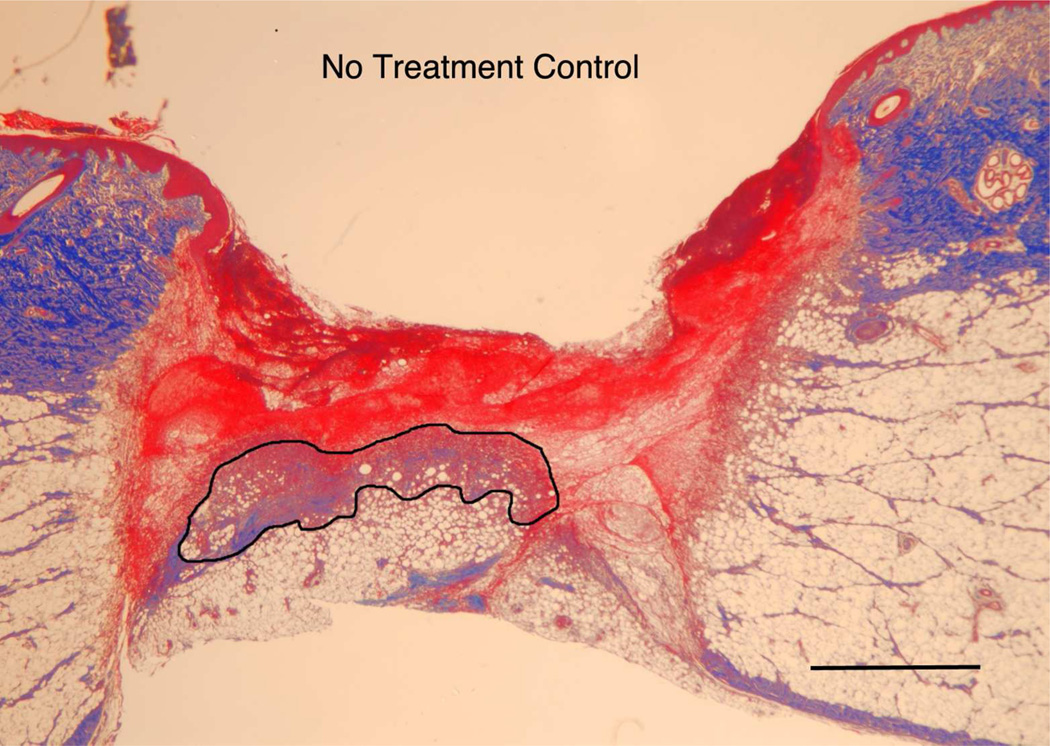
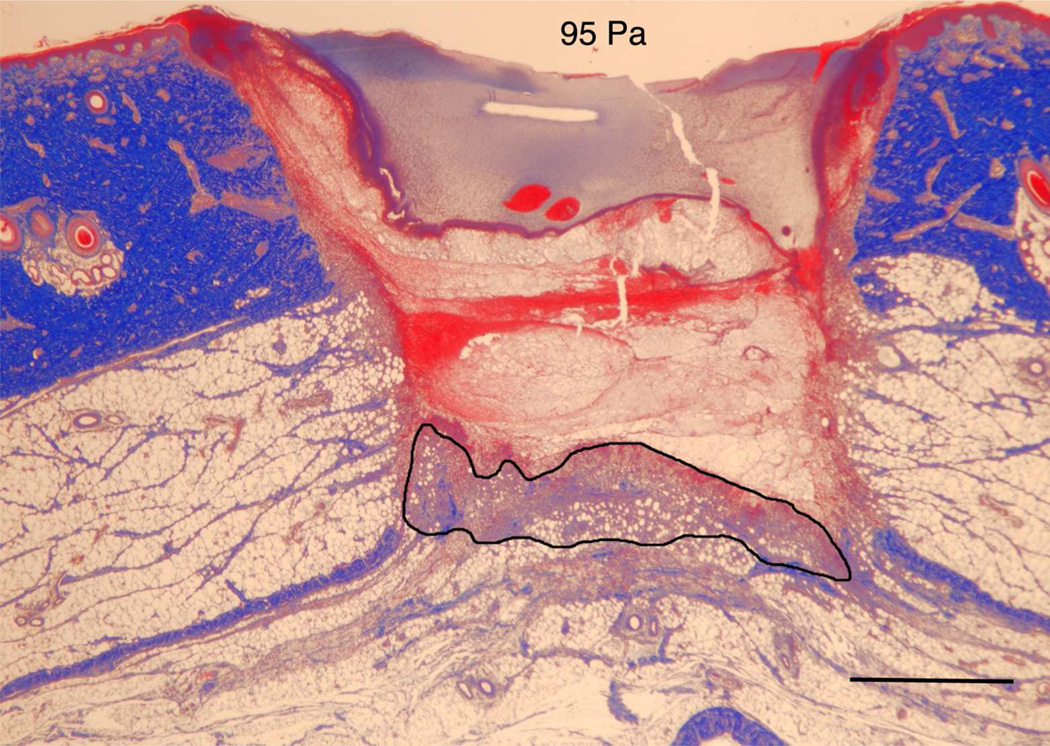
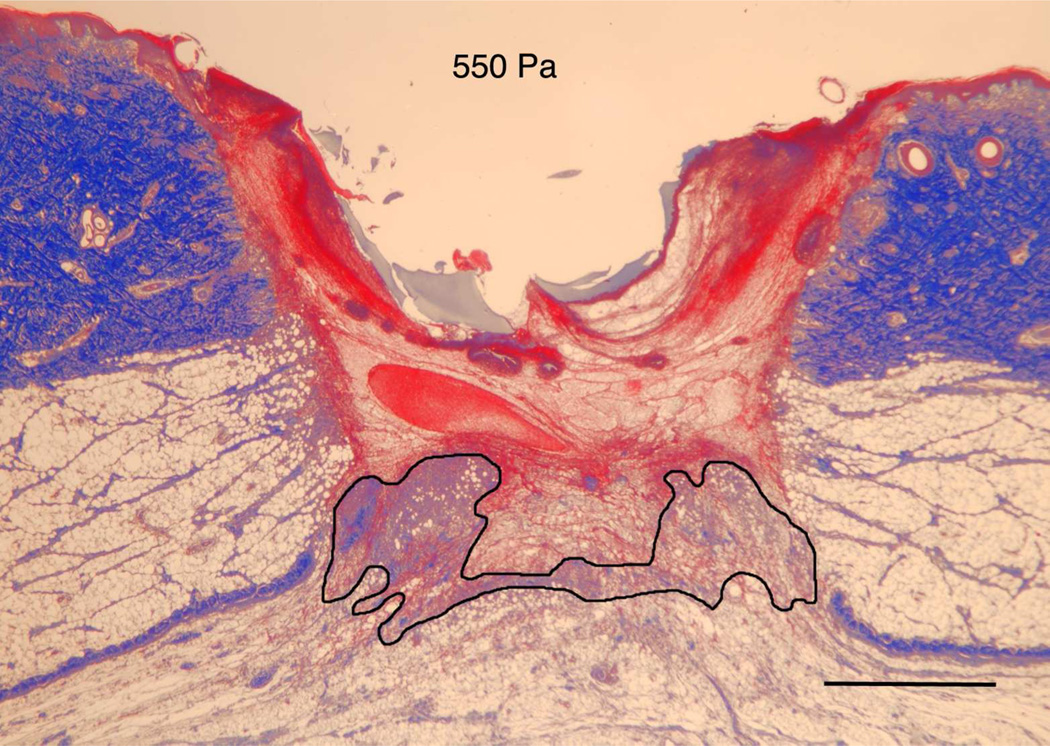
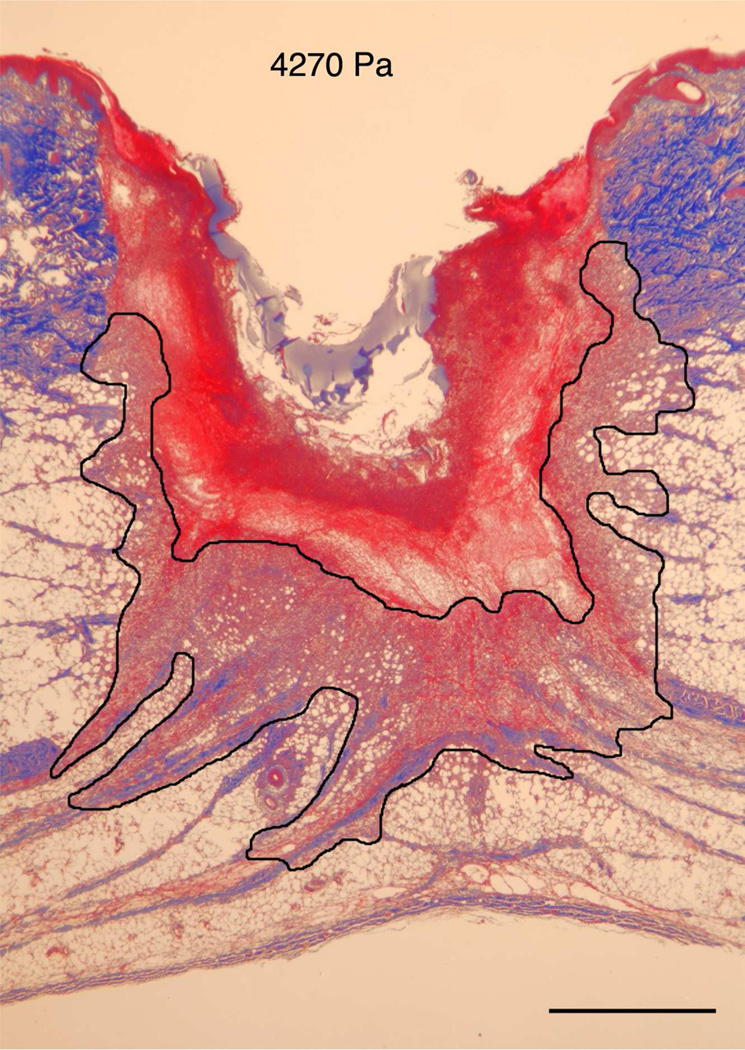
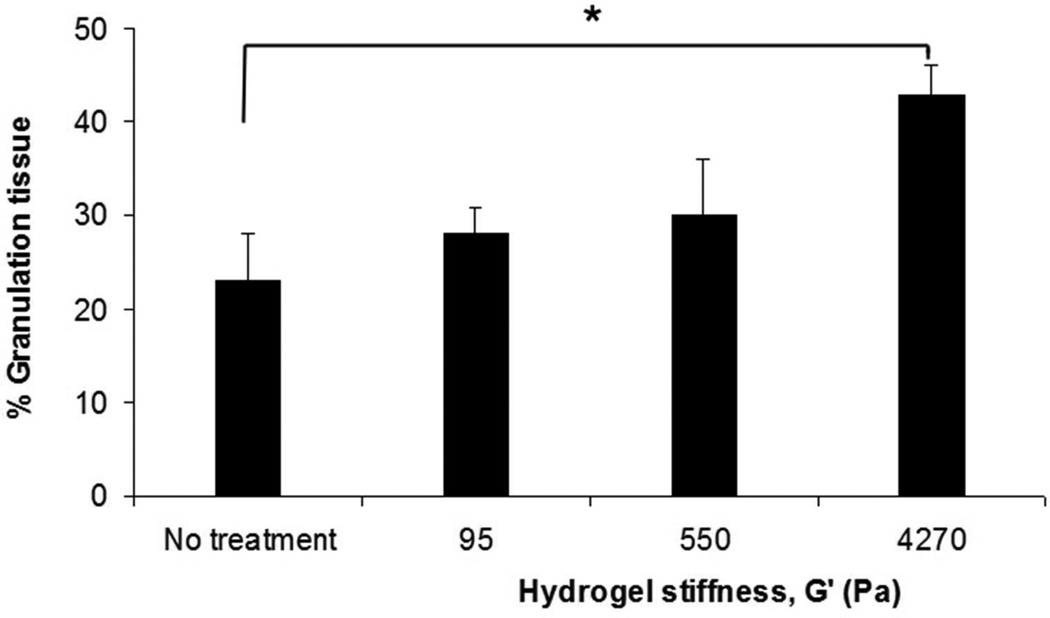
The in vitro model indicated that the modulus of the substrate can regulate both cell migration as well as cell orientation through the establishment of deformation gradient fields. Importantly, the substrate modulus had to be sufficiently high to maintain an adequate gradient, while simultaneously retaining the ability to deform in response to cell traction forces. In order to test this model in vivo, we used a porcine wound model. Full-thickness excisional wound sites were made on the back of healthy, juvenile Yorkshire pigs. HA/FNfds pre-gelling solutions with different crosslinking ratios were prepared and injected into the punch wound site where they gelled within 10 min after implantation. Four days later, the harvested wounds were processed and analyzed for the extent of granulation tissue formed (black line indicates boundary between granulation tissue and fibrin clot), as shown in Figure 5A. Morphometric analysis of cross sectional areas of such healing wounds demonstrated little to no difference in the degree of granulation tissue accumulation in wounds containing the softest gel relative to control wounds without gel (Figure 5B) However, a significant increase in granulation accumulation was observed for the stiffest gels, where we had previously demonstrated the optimal condition for cell migration in vitro.
Figure 5.
A) Masson Trichrome stained specimens showing granulation tissue regeneration in 4-day porcine excisional wounds as a function of hydrogel stiffness. Black lines encompass granulation tissue. No treatment: a partial rim of nascent pink granulation tissue appeared under the fibrin clot. 95 Pa hydrogel: a rim of granulation tissue underlies the fibrin clot. 550 Pa hydrogel: Increased granulation tissue appeared under the fibrin clot compared to no treatment control and 95 Pa hydrogel. 4270 Pa hydrogel: A remarkable increase in granulation tissue ingrowth (en masse fibroblast movement) appeared to be displacing the fibrin clot. B) Histogram of results. N=6. * indicates P < 0.05 by ANOVA and Tukey post-hoc analysis. Bars = 2mm
All together, we demonstrated the importance of deformation gradients in establishing the en masse migration patterns of human dermal fibroblast cells. We showed that the average en masse migration velocity of dermal fibroblasts is largest on stiffer hydrogel substrates, where they are also shown to exert the largest traction forces. We have also demonstrated that when cells migrate en masse on a compliant substrate, they establish a well-defined deformation field and migrate along a trajectory orthogonal to the field. In the case of cell migration outward from an agarose droplet, the trajectory assumed a radial pattern. The largest gradients occurred in the stiffer matrices (4270 Pa), where the largest degree of alignment occurred. For matrices that were too stiff and withstood deformation by the cellular traction forces, like glass, radial patterns did not form, consistent with the lack of a deformation field. Similarly, radial migration patterns functioned poorly on very soft matrices where only a minimal gradient could be established. When two droplets were placed adjacent to each other, a unique pattern was established only on the stiff but still compliant gels, indicating the importance of the gel modulus for cell organization. In vivo testing demonstrated that the largest degree of granulation tissue, following implantation of the gels in punch wounds of juvenile pigs, occurred for the stiffer gels, again illustrating the importance of substrate modulus in wound healing.
Materials and Methods
Preparation of hydrogel substrates
HA/FNfd hydrogel substrates were synthesized as reported previously (Ghosh et al., 2006). Briefly, the three major FNfds: FNIII8–11 (cell-binding domain, C), FNIII12V–15 (heparin-binding and variable domain, HV), and H, required for optimal fibroblast migration (Clark et al., 2003) were coupled to the crosslinker poly(ethylene glycol) diacrylate (PEGDA) (Nektar Therapeutics, Huntsville, AL) in PBS to form PEGDA-C, PEGDA-HV and PEGDA-H conjugates at 0.26 uM. Then these conjugates were mixed with 1.25%(w/v) thiol-functionalized HA (HA-DTPH) in serum free-DMEM (SF-DMEM, Sigma, St. Louis, MO) at volume ratio of 1:4 to obtain HA/FNfd that was allowed to gel in 35-mm tissue culture dishes. Final concentration of HA-DTPH was always 1% (w/v) in the hydrogels. The stiffness of these hydrogels was modulated by altering the concentration of PEGDA solutions from 4.5% to 1.5% and 0.75% (w/v) that resulted in crosslinking ratio from 2:1 to 6:1 and 12:1 and shear storage moduli of 4270 Pa, 550 Pa and 95 Pa, respectively, as measured by oscillatory shear rheometry (Ghosh et al., 2005).
Cell culture
Primary AHDFs obtained from a 31 years old Caucasian Female (CF-31, Clonetics, San Diego, CA) were used between passages 5 and 13. The cells were routinely cultured in Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% fetal bovine serum and 1% Penicillin, Streptomycin and L-glutamine, in a 37 °C, 5% CO2, 95% humidity incubator (Napco Scientific Company, Tualatin, OR). Additional experiments were also conducted using primary AHDF obtained from a 29 years old Caucasian Female, (CF-29, Clonetics, San Diego, CA) and AHDF obtained from surgical waste skin of an adult African American female with all identifiers removed. Since the results were qualitatively similar, only those obtained using the CF-31 cells are shown here.
Agarose droplet migration assay
After reaching 80–95% confluence, cells were detached using 0.05% trypsin, which was quenched with 10% serum in DMEM. After one rinse in 2%(w/v) bovine serum albumin (BSA) DMEM, cells were resuspended in 0.2% agarose SF-DMEM solution to obtain a final concentration of 1.5x107 cells/ml. Agarose droplets with cells (1.25µl) were placed on HA/FNfd hydrogels in 24-well dishes. Control samples, where the agarose droplet was plated on a circular fibronectin-coated glass coverslips were also placed in the 24-well dishes. After incubation at 4°C for 20min to allow the agarose droplets to set, SF-DMEM with 30ng/ml recombinant platelet derived growth factor-BB (PDGF) (ZymoGenetics, Seattle, WA) was added carefully. Then, the samples were incubated at 37°C for 18h.
After incubation, cells were fixed with 3.7% formaldehyde in PBS and then stained with filtered 0.1% crystal violet in 200 mM boric acid (pH 8.0) at 25°C for 10min. The cells were then rinsed with PBS. Samples were imaged using a Nikon SMX800 zoom stereomicroscope with a Diagnostic Instruments Spot RT camera Attachment (Sterling Heights, MI). Images were captured and analyzed with Spot 3.0 software. Migration area was determined by outlining the boundaries of the fibroblast migration and the agarose droplet. The area of migration (in square pixels) was then determined by the formula Total Area - Area of Droplet. The number of migrating cells in the migration area was quantified using NIH ImageJ. Each experimental group contained five replicates.
Time-lapse measurement of cell migration
AHDF in agarose droplets were seeded onto HA/FNfd hydrogel in 35mm cell culture dishes at ~500 /cm2. After incubation for 37°C for 6, 14, and 24h in serum-free DMEM with 30ng/ml PDGF. The maximum time was chosen to be less than the doubling time of the CF-31 AHDF cells on the stiffest substrate, previously reported (Ghosh 2007). Time-lapse phase images of cell migration were recorded every 15min for 60min with a MetaMorph®-operated CoolSNAP™ HQ camera (Universal Imaging Corporation, Downingtown, PA) attached to a Nikon Diaphot-TMD inverted microscope fitted with a 37°C stage incubator and a 10x objective lens. Migration speed was determined by tracking the distance covered by the center of a cell nucleus every 15min. The sample size n used was 5~10 cells/field×5 fields/replicate×3 replicates and 10~20 cells/field×3 fields/droplet×3 droplets, respectively, for single cells and en masse cells. Only single cells without cell-cell contact were chosen to measure single cell migration speed, and only cells migrating on the leading edge of droplet were tracked for evaluation of en masse cell migration.
Substrate deformation quantification using DISC
To measure substrate deformation generated by en masse cell migration, fluorescent beads (40 nm diameter, Molecular Probes, Eugene, OR) were sonicated and suspended uniformly at a concentration of 5%(w/v) in HA solution prior to gelation. Agarose droplets with cells were seeded onto HA/FNfd hydrogel in 35 mm cell culture dishes under the same conditions as cell migration experiments. After 6, 15 or 24h incubation at 37°C, distribution of fluorescent beads and cells in the field of interest were captured by Leica TCS SP2 laser scanning confocal microscope with a 20x water objective lens and a DIC lens, respectively, before and after separating cells from the substrate by the treatment with trypsin+EDTA. Using optimized DISC technique (Guan et al., 2004), the displacements of the substrate surface can be quantitatively determined by comparing the distribution change of the embedded beads. In order to highlight the local displacement generated by cells on the edge of droplets, radial displacements produced by the droplet were subtracted.
In vivo tissue repair with hydrogels of varying stiffness
Animal experiments were performed in accordance with Stony Brook University’s IACUC protocol. An established porcine full-thickness cutaneous wound model (McClain et al., 1996) was used to evaluate the effect of hydrogel stiffness on in vivo en masse cell migration. Full-thickness (8 mm) punch biopsy wounds were created on the back of female juvenile Yorkshire pigs weighing between 91–100 lbs (Archer Farms, VA). Sterile HA/FNfd pre-gelling solutions corresponding to 95, 550 and 4270 Pa stiffness (n=6) were then added to the wounds and allowed to set for 5 minutes. PDGF was added to all hydrogel samples at a final bulk density of 30 ng/ml. Six wounds received no hydrogels at all and were treated as controls. Following hydrogel gelation, wounds were dressed with Tegaderm® and allowed to heal for 4 days. Specimens from all wound sites were harvested after 4 days, immediately fixed in a 10% buffered formalin solution, embedded in paraffin, cut into 5 µm sections, mounted onto slides and stained with Masson trichrome (for type-I collagen) to delineate morphologic alterations.
Acknowledgements
We thank Dr. Xiangdong Ren for the cloning of FNfds, and Prof. Glenn D. Prestwich and Dr. Xiao Zheng Shu for providing HA-DTPH and sharing the chemistry to obtain cross-linked HA hydrogels. This work was supported by grants from the National Science Foundation MRSEC program and from NIH Merit Award AG 10143 to RAFC.
Footnotes
Conflict of Interest There is no conflict of interest among the authors
References
- Angelini TE, Hannezo E, Trepat X, Fredberg JJ, Weitz DA. Cell Migration Driven by Cooperative Substrate Deformation Patterns. Phys Rev Letts. 2010;104:168104. doi: 10.1103/PhysRevLett.104.168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beningo KA, Wang YL. Flexible substrata for the detection of cellular traction forces. Trends Cell Biol. 2002;12:79. doi: 10.1016/s0962-8924(01)02205-x. [DOI] [PubMed] [Google Scholar]
- Carter SB. Haptotaxis and methods of motility. Nature. 1967;213:256–260. doi: 10.1038/213256a0. [DOI] [PubMed] [Google Scholar]
- Chometon G, Zhang ZG, Rubinstein E, Boucheix C, Mauch C, Aumailley M. Dissociation of the complex between CD151 and laminin-binding integrins permits migration of epithelial cells. Experimental Cell Research. 2006;312:983–995. doi: 10.1016/j.yexcr.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrincytoskeleton linkages. Cell. 1997;88:39. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- Clark RAF. The Molecular and Celluar Biology of Wound Repair. Second Edition edn. New York and London: Plenum Press; 1996. [Google Scholar]
- Clark RAF, An JQ, Greiling D, Khan A, Schwarzbauer JE. Fibroblast migration on fibronectin requires three distinct functional domains. Journal of Investigative Dermatology. 2003;121:695–705. doi: 10.1046/j.1523-1747.2003.12484.x. [DOI] [PubMed] [Google Scholar]
- Dembo M, Oliver T, Ishihara A, Jacobson K. Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophys J. 1996;70:2008–2022. doi: 10.1016/S0006-3495(96)79767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76:2307. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher DE, Janmey P, Wang Y-l. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- Galbraith CG, Sheetz MP. Forces on adhesive contacts affect cell function. Curr Opin Cell Biol. 1998;10:566–571. doi: 10.1016/s0955-0674(98)80030-6. [DOI] [PubMed] [Google Scholar]
- Geiger B, Bershadsky A. Exploring the neighborhood: Adhesion-coupled cell mechanosensors. Cell. 2002;110:139. doi: 10.1016/s0092-8674(02)00831-0. [DOI] [PubMed] [Google Scholar]
- Georges PC, Janmey PA. Cell type-specific response to growth on soft materials. Journal of Applied Physiology. 2005;98:1547–1553. doi: 10.1152/japplphysiol.01121.2004. [DOI] [PubMed] [Google Scholar]
- Ghosh K, Ingber DE. Micromechanical control of cell and tissue development: Implications for tissue engineering. Adv Drug Deliv Rev. 2007;59:1306–1318. doi: 10.1016/j.addr.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Ghosh K, Pan Z, Guan E, Ge SR, Liu YJ, Nakamura T, et al. Cell adaptation to a physiologically relevant ECM mimic with different viscoelastic properties. Biomaterials. 2007;28:671–679. doi: 10.1016/j.biomaterials.2006.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh K, Ren XD, Shu XZ, Prestwich GD, Clark RAF. Fibronectin functional domains coupled to hyaluronan stimulate adult human dermal fibroblast responses critical for wound healing. Tissue Engineering. 2006;12:601–613. doi: 10.1089/ten.2006.12.601. [DOI] [PubMed] [Google Scholar]
- Ghosh K, Shu XZ, Mou R, Lombardi J, Prestwich GD, Rafailovich MH, et al. Rheological characterization of in situ cross-linkable hyaluronan hydrogels. Biomacromolecules. 2005;6:2857–2865. doi: 10.1021/bm050361c. [DOI] [PubMed] [Google Scholar]
- Goffin JM, Pittet P, Csucs G, Lussi JW, Meister JJ, Hinz B. Focal adhesion size controls tensiondependent recruitment of alpha-smooth muscle actin to stress fibers. J Cell Biol. 2006;172:259–268. doi: 10.1083/jcb.200506179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinnell Frederick, Matthew Petroll W. Cell Motility and Mechanics in Three-Dimensional Collagen Matrices. Annu. Rev. Cell Dev. Biol. 2010;26:335–361. doi: 10.1146/annurev.cellbio.042308.113318. [DOI] [PubMed] [Google Scholar]
- Guan E, Smilow S, Rafailovich M, Sokolov J. Determining the mechanical properties of rat skin with digital image speckle correlation. Dermatology. 2004;208:112–119. doi: 10.1159/000076483. [DOI] [PubMed] [Google Scholar]
- Hinz Boris. Formation and Function of the Myofibroblast during Tissue Repair. Journal of Investigative Dermatology. 2007;127:526–537. doi: 10.1038/sj.jid.5700613. [DOI] [PubMed] [Google Scholar]
- Ingber DE. Mechanosensation through integrins: Cells act locally but think globally. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2003;100:1472. doi: 10.1073/pnas.0530201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain SA, Simon M, Jones E, Nandi A, Gailit JO, Tonnesen MG, et al. Mesenchymal cell activation is the rate-limiting step of granulation tissue induction. American Journal of Pathology. 1996;149:1257–1270. [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Tawil A, Berkova Z, Wible L, Smith CW, Cunningham SA. Junctional Adhesion Molecules (JAMs) are differentially expressed in fibroblasts and co-localize with ZO-1 to adherens-like junctions. Cell Communication and Adhesion. 2006;13:233–247. doi: 10.1080/15419060600877978. [DOI] [PubMed] [Google Scholar]
- Oliver T, Dembo M, Jacobson K. Separation of propulsive and adhesive traction stresses in locomoting keratocytes. J Cell Biol. 1999;145:589–604. doi: 10.1083/jcb.145.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- Pan Z, Ghosh K, Liu YJ, Clark RAF, Rafailovich MH. Traction Stresses and Translational Distortion of the Nucleus During Fibroblast Migration on a Physiologically Relevant ECM Mimic. Biophys J. 2009;96:4286–4298. doi: 10.1016/j.bpj.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Wang YL. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proceedings Of The National Academy Of Sciences Of The United States Of America. 1997;94:13661. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeber GP, Lutolf MP, Hubbell JA. Part II: Fibroblasts preferentially migrate in the direction of principal strain. Biomechanics and Modeling in Mechanobiology. 2008;7:215–225. doi: 10.1007/s10237-007-0090-1. [DOI] [PubMed] [Google Scholar]
- Reinhart-King CA, Dembo M, Hammer DA. Cell-Cell Mechanical Communication through Compliant Substrates. Biophys J. 2008;95:6044–6051. doi: 10.1529/biophysj.107.127662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. Collective guidance of collective cell migration. Trends Cell Biol. 2007;17:575–579. doi: 10.1016/j.tcb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Rorth P. Collective Cell Migration. Annu Rev Cell Dev Biol. 2009;25:407–429. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- Schmidt CE, Horwitz AF, Lauffenburger DA, Sheetz MP. Integrin Cytoskeletal Interactions in Migrating Fibroblasts Are Dynamic, Asymmetric, and Regulated. J Cell Biol. 1993;123:977–991. doi: 10.1083/jcb.123.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Engler AJ, Discher DE. Matrix Strains Induced by Cells: Computing How Far Cells Can Feel. Cell Mol Bioeng. 2009;2:39–48. doi: 10.1007/s12195-009-0052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz MP, Felsenfeld DP, Galbraith CG. Cell migration: Regulation of force on extracellular-matrixintegrin complexes. Trends Cell Biol. 1998;8:51–54. doi: 10.1016/s0962-8924(98)80005-6. [DOI] [PubMed] [Google Scholar]
- Singer AJ, Clark RAF. Mechanisms of disease - Cutaneous wound healing. N Engl J Med. 1999;341:738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- Trepat1 X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, Fredberg J. Physical forces during collective cell migration. Nature Physics. 2009;5:426–430. [Google Scholar]
- Vespa A, D'Souza SJA, Dagnino L. A novel role for integrin-linked kinase in epithelial sheet morphogenesis. Molecular Biology of the Cell. 2005;16:4084–4095. doi: 10.1091/mbc.E05-02-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P. Cellular dynamics. Rev. Mod. Phys. 1959;31:11–20. [Google Scholar]