Abstract
S-Nitrosylation of protein cysteine residues is known to be an important mechanism for nitric oxide signaling. However, the detection of protein S-nitrosylation is still challenging due to technical limitations of current methods. This article provides a brief review on recent developments of methods, which directly target S-nitroso moieties for detection. We also describe in detail the protocol of an organophosphine-based biotin labeling of protein S-nitroso moieties.
Keywords: Nitric oxide, S-nitrosylation, S-nitrosothiol, protein labeling, phosphine, bioorthogonal reaction
1. Introduction
Nitric oxide (NO) is a cell signaling molecule involved in a number of physiological and pathophysiological processes. NO is synthesized by a family of enzymes known as nitric oxide synthases (NOS). Biological responses to NO are related to its in vivo reactions. NO can directly interact with some biomolecules such as heme centers and metalloproteins. NO can also undergo oxidation or other metabolic processes to form reactive nitrogen species (RNS). In particular, the reaction of RNS with protein cysteine residues (-SH) that results in S-nitrosylation (also known as S-nitrosation) has received a great deal of attention, because it represents an important post-translational modification (PTM) that transduces NO-dependent signals. To date, over 3,000 peptides and proteins have been characterized and studied as S-nitrosylation targets. In many cases S-nitrosylation is believed to regulate protein function and activity with consequences reflected both physiologically and pathologically.
Although significant progress has been made to understand the biological importance of protein S-nitrosylation, the detection of S-nitrosylation in complex biological sample is still a challenge [1–11]. The products of S-nitrosylation are S-nitrosothiols (SNO). From a chemistry point-of-view, all of protein SNO belong to primary SNO compounds, which are unstable compounds due to the reactive/unstable nature of the S-N bond. It is known that SNO can easily undergo photolytic decomposition to form NO and thiyl radicals (RS·). Cytosolic reducing agents such as ascorbate, glutathione, and reduced metals, especially Cu(I), can also break S-N bonds. One should be aware of these possible side reactions when attempting to measure SNO concentrations in certain biological samples.
In the past years, many methods have been developed for the detection of SNO. These methods can be classified into two categories: 1) indirect detection methods and 2) direct detection methods. Indirect methods are widely used and very popular in the field. By definition, these methods are not targeting the SNO adducts as a whole to generate the detection signals. Instead, they usually break the unstable S-N bonds and capture either the sulfur or the nitrogen part for detection. Representative methods in this category include 1) chemiluminescence methods to analyze NO radicals generated from SNO; 2) Saville assay to analyze nitrite (NO2−) generated from SNO; 3) fluorescence detection (using 4,5-diaminofluorescein, for example) to analyze NO+ generated from SNO; and 4) biotin-switch based methods to conjugate free thiols generated from SNO. These methods have made significant contributions to SNO research. In particular, a method reported two decades ago by Stamler and Loscalzo that involves the photolytic dissociation of the RS-NO bond followed by chemiluminescence, has been the landmark for quantitative analysis of protein S-nitrosylation [12–13]. The basis of this method stands on the ability of S-NO bond to undergo homolytic cleavage upon UV irradiation (~300–350 nm), to yield a thiyl radical (RS·) and NO. Then NO reacts with ozone (O3) and the light produced from the reaction can be detected by chemiluminescence. This method has been used to detect individual SNO-proteins following their purification [14–15]. A recent modification by Schoenfisch employs visible photolysis (500–550 nm) followed amperometric NO detection [16]. This strategy proved to be highly sensitive.
In this article, we will not review the progress in the category described above. Readers interested in indirect methods should read some review articles published recently [1–11]. We will focus our attention to review recent developments in direct methods for SNO detection in proteins. We will also provide detailed protocols of a direct biotinylation method developed by this laboratory.
2. Direct methods for the detection of protein S-nitrosylation: non-derivatization based methods
Direct methods target intact SNO moieties in samples. These methods should be able to selectively recognize SNO from complex biological systems and generate sufficient signals for detection. Non-derivatization based methods in this category include mass spectrometry and anti-SNO antibody based methods.
2.1 Mass spectrometry
Identification of protein SNO sites by mass spectrometry is usually challenging because of the lability of the SNO groups. Commonly employed mass ionization sources, such as electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI), under typical conditions can induce dissociation of the S-N bonds [17–19]. ESI is more suitable than MALDI for qualitative analysis because ESI conditions are milder and may preserve the labile SNO groups. It was reported that, under very strict ionization conditions, S-nitrosylated proteins can be analyzed by mass spectrometry. One example is the characterization of SNO-thioredoxin-1 by Wang et al [20]. In this example, thioredoxin-1 was subjected to S-nitrosylation, precipitation, digestion, and finally analyzed by LC/MS/MS using ESI quadrupole time of flight (QTOF) mass spectrometry. The authors stated that the optimization of voltage and collision energy was critical to achieve reliable signal/noise ratios. Buffer composition was also carefully controlled to minimize S-NO depletion. Although thioredoxin-1 was effectively analyzed by this technique, the application of it to other proteins will require extensive adjustment of experimental parameters. The stability of SNO in peptides is dependent upon the peptide sequence. Therefore the optimal voltage of ionization for any given peptide should be determined separately. In addition, analysis of mass results may not be trivial. Under some circumstances 3-nitroso (3-NO) tyrosine could be formed as the nitrosylation product, [21] and this could be misidentified as an SNO moiety. Therefore, control experiments must be performed to rule out possible false positive results regarding this post-translational modification. Up to date, the expansion of direct mass spectrometry-based methods for SNO proteomic studies is still limited and very challenging.
2.2 Anti-S-nitrosocysteine antibody: Immunohistochemical approach
Detection of protein S-nitrosylation using traditional methods like immunoprecipitation or Western blotting is not typically performed because the unstable S-N bonds may be broken during SDS-PAGE separation. Nevertheless the antibody can be used on non-reducing gels and to immunoprecipate SNO-proteins [22–23]. There has been some progress using anti-S-nitrosocysteine antibodies as a detection tool. Both monoclonal [24] and polyclonal antibodies [25] have been produced which bind the SNO functionality. This method consists of labeling SNO-protein with a primary antibody, anti-S-nitrosocysteine, followed by detection with a secondary antibody labeled with a chemical or biochemical reporter (horseradish peroxidase, 125I, biotinylated secondary antibody or fluorescein isothiocyanate). The anti-SNO antibody method has been applied for immunohistochemical detection of S-nitrosylated proteins in cells and in lung [25] and vascular [24,26] tissues. Both positive and negative controls are required and should be conducted on serial sections in which the test section is flanked by both a positive and a negative control.
3. Direct methods for the detection of protein S-nitrosylation: derivatization based methods
Methods in this category usually rely on chemical approaches using reagents which can directly react with SNO to form stable adducts. Reagents based on these reactions can be used to enrich SNO proteins and analyze SNO components.
3.1 Gold nanoparticles
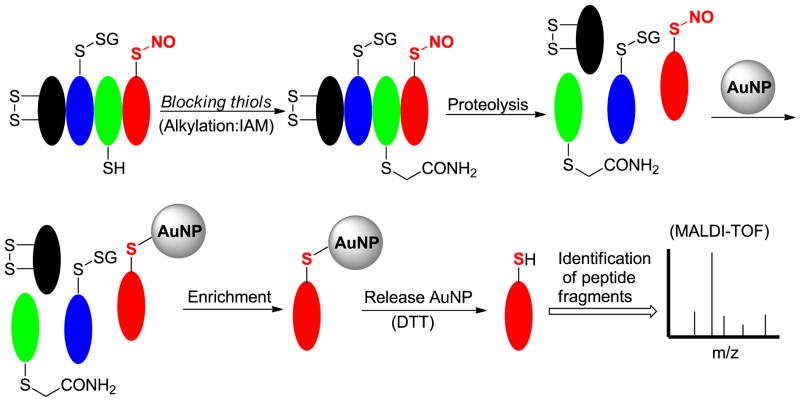
Some transition metals like gold have high affinity toward sulfur. Recent studies found that gold nanoparticles (AuNP) can react with SNO to form AuNP-protein thiolate conjugates [27]. Based on this reactivity, Mutus et al. developed a AuNP enrichment method for identifying protein SNO sites [28]. This method is described in Scheme 1. Free thiols in protein are first blocked by a SH-alkylation reagent-iodoacetamide (IAM). The protein is then proteolyzed and the digested fragments are treated with AuNP. SNO containing peptides are expected to react with AuNP to form AuNP-thiolate conjugates. The AuNP-bound peptides can then be harvested, released from AuNP surface, and finally identified by mass spectrometry.
Scheme 1.
AuNP enrichment method
(This color figure is for color reproduction on the Web, and in black-and-white in print)
This method is quite sensitive and easy to use. However, like the biotin switch method, this method is still a subtractive method. Since AuNP can also react with thiols to form AuNP-thiolate conjugates, all free thiols in protein have to be blocked. Given the fact that SNO concentrations are very low compared to free thiols in real biological systems, incomplete free thiol blocking could lead to false positive results. Another problem is that the target protein was subjected to proteolysis to generate SNO-peptides before AuNP was introduced. It is unclear how much SNO can be retained in peptide fragments. The environment/structure change on SNO adducts could dramatically affect the stability of SNO. Therefore some SNO peptides could decompose under the proteolysis step to form other products like disulfides. Finally, the authors stated that bulky disulfide containing peptides should not interact with AuNP, while less bulky disulfide peptides may react with AuNP to form Au-thiolate conjugates. This may also lead to false positive signals with some proteins.
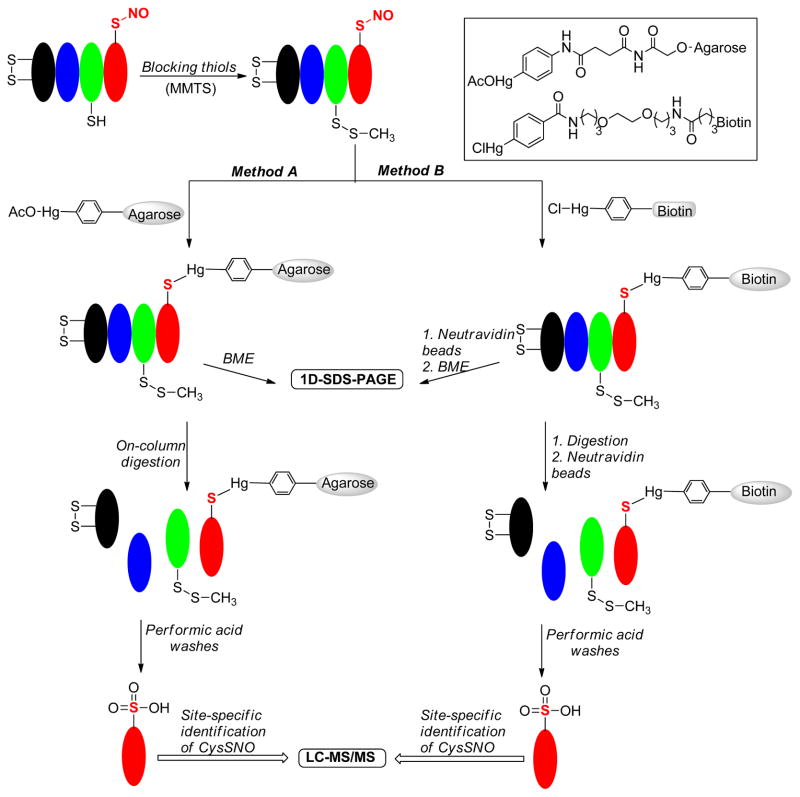
3.2 Organomercury resin capture (MRC)
Organomercury complexes are known to react with S-nitrosothiols to form stable mercury-thiolate conjugates [29]. Ischiropoulos et al applied this reaction in their proteomic studies of endogenous S-nitrosylation [30]. As shown in Scheme 2, this method consists of three steps: 1) free thiols are blocked with reagents like methyl methanethiosulfonate (MMTS); 2) SNO proteins are captured by organomercury reagents; 3) after enrichment, SNO proteins are released from the reagents and subjected to LC-MS/MS analysis. This method was adapted to both solid (method A) and solution phase (method B) sample enrichments by conjugating phenylmercury moiety with agarose beads or with a biotin tag. Performic acid was used to release bound protein or peptide fragments from the mercury surface. Interestingly, performic acid can oxidize the resulting thiols to sulfonic acid, producing a unique MS signature for site-specific identification.
Scheme 2.
MRC enrichment method
(This color figure is for color reproduction on the Web, and in black-and-white in print)
This mercury based method has been used to identify a large library of SNO proteins ranging from 15 to 270 kDa. In addition, the organo-mercury reagents seem to be highly specific for SNO and do not react with disulfides to form mercury-thiolate conjugates [31]. However, organo-mercurial can react with free thiols and therefore SH should be blocked. Further evaluation of the reactivity of organo-mercurial reagents against other Cys PTMs, especially persulfide (-S-SH) and polysulfides (-S-Sn-S-R) should be addressed. Regarding the operation of this assay, great caution has to be taken because organo-mercury compounds are highly toxic.
3.3 Organophosphine based SNO direct labeling
Bioorthogonal reactions of SNO are reactions which specifically target SNO moieties and convert unstable SNO to stable or detectable products. Such reactions should not react with other biological moieties, especially sulfur based derivatives such as thiol (-SH), disulfide (-S-S-), sulfenic acid (-S-OH), and perthiol (-S-SH). If suitable SNO bioorthogonal reactions can be developed, they should be useful for SNO detection. With this idea in mind, our group studied a series organophosphine based bioorthogonal reactions of SNO [4]. Previous research on the Staudinger ligation has demonstrated that triaryl substituted phosphines are mild reagents and can be used in living systems [32]. Such phosphines do not react with disulfides [32]. Interestingly, we found that triaryl-substituted phosphines showed high reactivity toward SNO [4]. They can rapidly react with SNO to form azaylide intermediates (Scheme 3). Upon manipulating the substituents on the aryl rings, the azaylide intermediates can undergo fast intramolecular reactions and be converted into different products: sulfenamides (via reductive ligation) [33], disulfide-phosphoimine adducts (via bis-ligation) [34], dehydroalanine adducts (via reductive elimination) [35], or simple disulfide conjugates (via the one-step disulfide formation) [36]. These potential bioorthogonal reactions of SNO are promising for the development of SNO detection methods. Among these four reactions, the one-pot disulfide formation has been applied to label protein SNO moieties [36]. Here we describe the detailed procedure of this method.
Scheme 3.
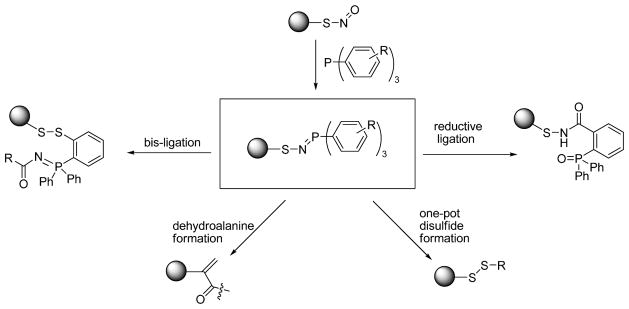
Phosphine based methods
4 Labeling protein SNO using the one-step disulfide formation strategy
4.1 Synthesis of phosphine substrate A
Biotin-linked phosphine A was used to label S-nitrosylated protein directly. This compound was prepared from three subunits S1, S2, and S3. S2 and S3 are commercially available. S1 was prepared starting from tetraethylene glycol using conventional organic chemistry (not shown here). The key synthetic sequence for A is shown in Scheme 4 and described below.
Scheme 4.
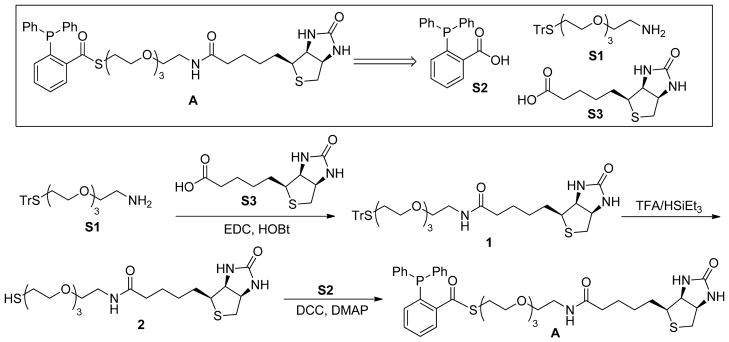
Synthesis of the biotin-phosphine substrate
Compound 1
S1 (728 mg, 1.61 mmol) and S3 (393 mg, 1.61 mmol) were dissolved in DMF (22 mL) and cooled to 0 °C. To this solution hydroxybenzotriazole (HOBt; 261 mg, 1.93 mmol) was added. After having stirred for 20 min, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC·HCl; 340 mg, 1.77 mmol) was added. The resulting mixture was warmed to room temperature and stirred overnight. The solvent was removed and the crude product was dissolved in CH2Cl2 (200 mL). The organic layer was washed with 10 % citric acid (100 mL), saturated NaHCO3 solution, and dried over anhydrous MgSO4. The crude product was purified by flash column chromatography (CH2Cl2/MeOH = 20/1). Compound 1 was obtained in 79% yield (860 mg). 1H NMR (300 MHz, CDCl3), δ (ppm): 7.38 (m, 6H), 7.16 (m, 9H), 6.70 (m, 1H), 6.59 (s, 1H), 5.56 (s, 1H), 4.40 (m, 1H), 4.21 (m, 1H), 3.51 (m, 8H), 3.38 (m, 4H), 3.24 (m, 2H), 3.03 (m, 1H), 2.81 (m, 1H), 2.65 (d, J = 12.6 Hz, 1H), 2.35 (td, J1 = 6.6 Hz, J2 = 0.9 Hz, 2H), 2.13 (t, J = 7.2Hz, 2H), 1.57 (m, 4H), 1.34 (m, 2H); 13C NMR (75 MHz, CDCl3), δ (ppm): 173.5, 164.3, 145.0, 129.8, 138.1, 126.9, 70.6, 70.5, 70.4, 70.3, 70.2, 69.9, 61.9, 60.4, 55.8, 40.8, 39.4, 36.2, 31.9, 28.3, 28.29, 25.8, 25.79.
Compound 2
Under an argon atmosphere, compound 1 (471 mg, 0.70 mmol) was dissolved in CH2Cl2 (10 mL) with trifluoroacetic acid (TFA, 0.4 mL). To this solution, triethylsilane (Et3SiH; 130 μL, 0.77 mmol) was added. The mixture was stirred for 2 h. After removal of the solvent, the resulting residue was purified by flash column chromatography (CH2Cl2/MeOH = 4/1). Compound 2 was obtained in 99% yield (303 mg). 1H NMR (300 MHz, CDCl3), δ (ppm): 6.48 (br, 2H), 4.47 (m, 1H), 4.27 (m, 1H), 3.54 (m, 15H), 3.08 (m, 1H), 2.83 (m, 1H), 2.64 (m, 3H), 2.15 (m, 2H), 1.63 (m, 5H), 1.38 (m, 2H); 13C NMR (75 MHz, CDCl3), δ (ppm): 73.1, 70.7, 70.4, 70.0, 62.2, 60.8, 55.8, 41.3, 39.4, 36.1, 36.1, 36.0, 28.4, 28.3, 25.6, 24.8.
Compound A
The final step for the synthesis of A was reported in [36].
4.2 Protein labeling
Incubation of protein nitrosothiols with compound A leads to the addition of biotin at the SNO site through the formation of a disulfide bond. The following approaches can be used to label cellular SNO proteins. To generate SNO proteins, cells in culture can be incubated with CysNO in Hanks Balanced Salt Solution buffered with 25 mM HEPES. The amount of S-nitrosylated proteins generated is dependent on the CysNO concentrations used. In general, the amount of SNO formed from 100–200 μM CysNO is easily detected by Western blot.
Fixation: S-nitrosylated cells can then be fixed to the culture substrate surface by treatment with −20 °C methanol for 15 minutes. This step allows the attachment of cells to the culture substrate surface so that excess blocking/labeling agents can easily be removed by rinsing.
Permeabilization/Blocking: fixed cells can then be treated with phosphate buffered saline containing 0.2% Triton X-100 containing 40 mM N-ethylmaleimide (NEM) for 1 hour at room temperature to permeabilize the membrane and alkylate (block) all free thiols. In the widely used Biotin Switch Assay, free thiol blocking is extremely critical, since any unblocked free thiols will be subsequently labeled and give signals. This false positive signal is considered a major pitfall of this assay. This is not a problem with our approach using compound A since free protein thiols will not be labeled. However it is important to add NEM to prevent possible trans-nitrosation from one thiol to another within or between proteins and to prevent possible loss of protein SNO due to trans-nitrosation reactions with small molecular weight cellular thiols (glutathione, cysteine, etc). Excess NEM can be removed by rinsing.
Biotinylation: labeling of SNO proteins is then accomplished by incubating cells with 3 mM compound A (in DMSO) for 30 minutes at room temperature. Excess reagent can be removed by rinsing. DMSO is necessary in this step to dissolve compound A in a reaction mixture compatible with cellular protein labeling.
Detection of the biotin conjugate: the added biotin label can be easily detected after SDS PAGE gel electrophoresis using readily available reagents. Prior to gel separation cellular proteins can be scraped into RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 1% SDS), sonicated briefly and centrifuged at 14,000 rpm for 10 minutes. The supernatant can be mixed with 2 X Laemmli loading buffer. It is important to note that neither DTT nor mercaptoethanol should be added to the loading buffer since thiol reducting reagents would reverse the disulfide labeling. Solubilized protein can then be separated on 10% SDS-PAGE gel and transferred to a PVDF membrane using standard approaches. The biotin label can then be detected by incubation with NeutrAvidin-HRP and visualized using enhanced chemiluminescence reagents (ECL plus reagents). To demonstrate equal protein loading, membranes may be stripped by treatment with 100 mM mercaptoethanol, 2% SDS, 62.5mM Tris–HCl, pH 6.7, at 50 °C for 30 min, and reprobed using a GAPDH antibody.
Control experiments
Two control experiments have been designed to address the specificity of the phosphine-biotin labeling approach:
DTT cleavage of the disulfide link: At the end of step 3 in the labeling method, DTT may be added after incubation with compound A. DTT will reduce the disulfide bond formed during the phosphine reaction with SNO and reverse labeling to confirm that any labeling seen in the absence of DTT treatment is due to biotin conjugation through a disulfide linkage.
Control labeling using compound B (Scheme 5): the phosphine-biotin substrate A contains a thioester functional group. One concern is that it may undergo hydrolysis or react with biological nucleophiles such as amines to generate biotin-linked thiols. These products may, in turn, react with SNO or even protein disulfide to give false positive results. To address this possibility, compound B (the preparation of B was reported in [36]) may be used as negative control. Compound B also contains the biotin-thioester functional group, but it lacks the phosphine function. If used in place of compound A, no biotin labeling of SNO proteins would be expected and serves to confirm specificity of this approach.
Scheme 5.
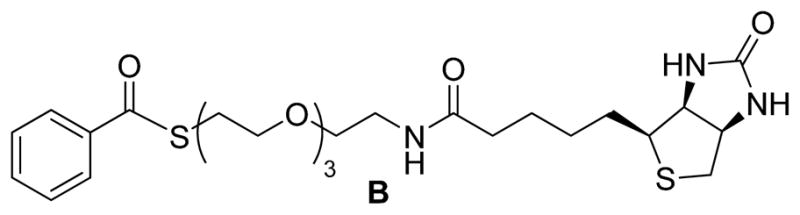
Structure of Compound B
This one-step phosphine based labeling method has recently been used by us to conjugate SNO proteins directly in COS7 cells which had been pretreated with CysNO [36]. Biotin conjugation was reversed by DTT and was not seen when compound B was substituted for compound A. These results demonstrated that the labeling was via a disulfide linkage and was specific for the reaction between compound A and S-nitrosothiols.
5 Summary and perspectives
Currently the detection of SNO in complex systems is still challenging despite many methods having been developed in the past years. Each method has its own advantages and disadvantages. In our opinion, future research in this field should focus on direct methods which target SNO, not the by-products from S-N bond cleavage. In this regard, organophosphine mediated bioorthogonal reactions are promising. The key step in these reactions is the formation of a sulfur linked azaylide intermediate, which is unique and only occurs with SNO moieties. Other biological sulfur adducts such as thiols (-SH), disulfide (-S-S-), sulfenic acid (-S-OH), perthiol (-S-SH), etc. should not form corresponding azaylide intermediates. Therefore, such reactions are selective for SNO. It is worth noting that the King group recently reported another phosphine-based approach for SNO labeling [37]. The King’s method uses tris(4,6-dimethyl-3-sulfonatophenyl-phosphine trisodium salt hydrate (TXPTS) to attack SNO. The resulted covalent S-alkylphosphonium (RS-PR3+) products can be identified by NMR and mass spectrometry. Both LMW S-nitrosothiols (such as CysSNO and GSNO) and SNO-proteins can be examined [37]. However, the products derived from the reaction, i.e., S-alkylphosphonium salts, showed instability under reducing conditions (DTT) and, also prone to hydrolysis. In addition, control experiments with GSSG and Cys disulfide showed that TXPTS can cleave –S-S-adducts to produce corresponding RS-PR3+ species in low yields (~3% and ~15%). Although they demonstrated that S-alkylphosphonium adduct of SNO peroxiredoxin mutant C165S alkyl hydroperoxide reductase C is apparently more stable [37], it is unclear if this is applicable for all other SNO proteins. Also, the reactivity of TXPTS against Cys disulfide linkage or S-glutathionylated Cys may differ within a protein, or in different proteins. Therefore careful control experiments should be performed to subtract any of these discrepancies.
The one-pot disulfide formation described in this article demonstrated that organophosphine mediated bioorthogonal reactions can be applied for protein SNO direct labeling. Control experiments have proved its selectivity. However, further optimization of this method is still needed. The following problems should be addressed in future studies: 1) significant organic solvents such as DMSO are required with current reagent, i.e. A. Reagents with better water solubility should be developed. 2) SNO labeling with the current reagent was carried out with artificial SNO concentrations. The efficiency of new water soluble reagents should be evaluated under biological SNO concentrations. 3) The current strategy converts SNO to disulfide conjugates, which cannot survive under some commonly used reductants such as DTT. Design of new tandem reactions that can further convert disulfide to more stable conjugates would be more useful. We recently showed one example of forming stable thioethers [38]. More strategies are to be discovered.
Highlights.
Recent developments of direct methods for protein SNO labeling are reviewed.
Detailed procedures for the preparation of the biotin-phosphine substrate are reported.
Detailed procedures of using the phosphine substrate to label protein SNO in cells are described.
Acknowledgments
This work was supported by NIH (R01GM088226 to M.X. and R01HL088559 to A.R.W.) and NSF (CHE-0844931 to M.X.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Foster MW. Methodologies for the characterization, identification and quantification of S-nitrosylated proteins. Biochim Biophys Acta. 2012;1820:675–683. doi: 10.1016/j.bbagen.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bechtold E, King SB. Chemical methods for the detection and labeling of S-nitrosothiols. Antioxid Redox Sign. 2012;17:981–991. doi: 10.1089/ars.2012.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seth D, Stamler JS. The SNO-proteome: causation and classifications. Curr Opin Chem Biol. 2011;15:129–136. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Xian M. Chemical methods to detect S-nitrosation. Curr Opin Chem Biol. 2011;15:32–37. doi: 10.1016/j.cbpa.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torta F, Elviri L, Bachi A. Direct and indirect methods for the analysis of S-nitrosylated peptides and proteins. Meth Enzymol. 2010;473:265–280. doi: 10.1016/S0076-6879(10)73014-7. [DOI] [PubMed] [Google Scholar]
- 6.Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein Snitrosylation with the biotin-switch technique. Free Rad Biol Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basu S, Wang X, Gladwin MT, Kim-Shapiro DB. Chemiluminescent detection of S-nitrosated proteins: comparison of tri-iodide, copper/CO/cysteine, and modified copper/cysteine methods. Methods Enzymol. 2008;440:137–156. doi: 10.1016/S0076-6879(07)00808-7. [DOI] [PubMed] [Google Scholar]
- 8.Palmer LA, Gaston B. S-Nitrosothiols assays that avoid the use of iodine. Methods Enzymol. 2008;440:157–176. doi: 10.1016/S0076-6879(07)00809-9. [DOI] [PubMed] [Google Scholar]
- 9.Gow A, Doctor A, Mannick J, Gaston B. S-Nitrosothiol measurements in biological systems. J Chromatogr B. 2007;851:140–151. doi: 10.1016/j.jchromb.2007.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giustarini D, Milzani A, Dalle-Donne I, Rossi R. Detection of S-nitrosothiols in biological fluids: a comparison among the most widely applied methodologies. J Chromatogr B. 2007;851:124–139. doi: 10.1016/j.jchromb.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 11.MacArthur PH, Shiva S, Galdwin MT. Measurement of circulating nitrite and S-nitrosothiols by reductive chemiluminescense. J Chromatogr B. 2007;851:93–105. doi: 10.1016/j.jchromb.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singel D, Valeri CR, Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc Natl Acad Sci USA. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia L, Bonaventura C, Bonaventura J, Stamler JS. S-Nitrosohemoglobin: a dynamic activity of blood involved in vascular control. Nature. 1996;380:221–226. doi: 10.1038/380221a0. [DOI] [PubMed] [Google Scholar]
- 14.Hausladen A, Privalle CT, Keng T, DeAngelo J, Stamler JS. Nitrosative Stress: activation of the transcription factor OxyR. Cell. 1996;86:719–729. doi: 10.1016/s0092-8674(00)80147-6. [DOI] [PubMed] [Google Scholar]
- 15.Eu JP, Sun J, Xu L, Stamler JS, Meissner G. The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell. 2000;102:499–509. doi: 10.1016/s0092-8674(00)00054-4. [DOI] [PubMed] [Google Scholar]
- 16.Riccio DA, Nutz ST, Schoenfisch MH. Visible photolysis and amperometric detection of S-nitrosothiols. Anal Chem. 2012;84:851–856. doi: 10.1021/ac2031805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirza UA, Chait BT, Lander HM. Monitoring reactions of nitric oxide with peptides and proteins by electrospray ionization-mass spectrometry. J Biol Chem. 1995;270:17185–17188. doi: 10.1074/jbc.270.29.17185. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko R, Wada Y. Mass spectrometric features of S-nitrosylated peptides. J Mass Spectrom Soc Jpn. 2002;50:223–225. [Google Scholar]
- 19.Kaneko R, Wada Y. Decomposition of protein nitrosothiols in matrix-assisted laser desorption/ionization and electrospray ionization mass spectrometry. J Mass Spectrom. 2003;38:526–530. doi: 10.1002/jms.466. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Liu T, Wu C, Li H. A strategy for direct identification of protein S-nitrosylation sites by quadrupole time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1353–1360. doi: 10.1016/j.jasms.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SJ, Lee JR, Kim YH, Park YS, Park SI, Park HS, Kim KP. Investigation of tyrosine nitration and nitrosylation of angiotensin II and bovine serum albumin with electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:2797–2804. doi: 10.1002/rcm.3145. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, Xin C, Eu JP, Stamler JSG. Meissner Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc Natl Acad Sci USA. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsushita K, Morrell CN, Cambien B, Yang SX, Yamakuchi M, Bao C, Hara MR, Quik RA, Cao W, O’Rourke B, Lowenstein JM, Pevsner J, Wagner DD, Lowenstein CJ. Nitric oxide regulates exocytosis by S-nitrosylation of N-ethylmaleimide-sensitive factor. Cell. 2003;115:139–150. doi: 10.1016/s0092-8674(03)00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gow AJ, Chen Q, Hess DT, Day BJ, Ischiropoulos H, Stamler JS. Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J Biol Chem. 2002;277:9637–9640. doi: 10.1074/jbc.C100746200.For a protocol see: Gow AJ, Davis CW, Munson D, Ischiropoulos H. Immnohistochemical detection of S-nitrosylated proteins. Met Mol Biol. 2004;279:167–172. doi: 10.1385/1-59259-807-2:167.
- 25.Munson DA, Grubb PH, Kerecman JD, McCurnin DC, Yoder BA, Hazon SL, Shaul PW, Ischiropoulos H. Pulmonary and systematic nitric oxide metabolites in a baboon model of neonatal chronic lung disease. Am J Respir Cell Mol Biol. 2005;33:582–588. doi: 10.1165/rcmb.2005-0182OC. [DOI] [PubMed] [Google Scholar]
- 26.Greco TM, Hodara R, Parastatidis I, Heijen HF, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci USA. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia HY, Liu Y, Zhang XJ, Han L, Du LB, Tian Q, Xu YC. Potential oxidative stress of gold nanoparticles by induced-NO releasing in serum. J Am Chem Soc. 2009;131:40–41. doi: 10.1021/ja808033w. [DOI] [PubMed] [Google Scholar]
- 28.Faccenda A, Bonham CA, Vacratsis PO, Zhang X, Mutus B. Gold nanoparticle enrichment method for identifying S-nitrosylation and S-glutathionylation sites in proteins. J Am Chem Soc. 2010;132:11392–11394. doi: 10.1021/ja103591v. [DOI] [PubMed] [Google Scholar]
- 29.(a) Saville B. A scheme for the colorimetric determination of microgram amount of thiols. Analyst. 1958;83:670–672. [Google Scholar]; (b) Bramanti E, Jacovozzi K, D’Ulivo L, Vecoli C, Zamboni R, Mester Z, D’Ulivo A. Determination of S-nitrosoglutathione and other nitrosothiols by p-hydroxymercurybenzoate derivatization and reverse phase chromatography coupled with chemical generation atomic fluorescence detection. Talanta. 2008;77:684–694. doi: 10.1016/j.talanta.2009.03.030. [DOI] [PubMed] [Google Scholar]
- 30.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanism for protein S-nitrosylation. Proc Natl Acad Sci USA. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doulias PT, Tenopoulou M, Greene JL, Raju K, Ischiropoulos H. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci Signal. 2013;6:rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Xian M. Fast reductive ligation of S-nitrosothiols. Angew Chemie Intl Ed. 2008;47:6598–6601. doi: 10.1002/anie.200801654. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Wang H, Xian M. An unexpected bis-ligation of S-nitrosothiols. J Am Chem Soc. 2009;131:3854–3855. doi: 10.1021/ja900370y. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Xian M. Facile formation of dehydroalanine from S-nitrosocysteine. J Am Chem Soc. 2009;131:13238–13239. doi: 10.1021/ja905558w. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Li S, Zhang D, Wang H, Whorton AR, Xian M. Reductive ligation mediated one-step disulfide formation of S-nitrosothiols. Org Lett. 2010;12:4208–4211. doi: 10.1021/ol101863s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bechtold E, Reisz JA, Klomsiri C, Tsang AW, Wright MW, Poole LB, Furdui CM, King SB. Water-soluble triarylphosphines as biomarkers for protein S-nitrosation. ACS Chem Biol. 2010;5:405–414. doi: 10.1021/cb900302u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D, Davarie-Baez NO, Pan J, Wang H, Xian M. One-Pot thioether formation from S-nitrosothiols. Org Lett. 2010;12:5674–5677. doi: 10.1021/ol102491n. [DOI] [PMC free article] [PubMed] [Google Scholar]