Abstract
Posttranslational mechanisms drive fidelity of cellular processes. Phosphorylation and ubiquitination of substrates represent very common, covalent, posttranslational modifications and are often co-regulated. Phosphorylation may play a critical role both by directly regulating E3-ubiquitin ligases and/or by ensuring specificity of the ubiquitination substrate. Importantly, many kinases are not only critical regulatory components of these pathways but also represent themselves the direct ubiquitination substrates. Recent data suggest the role of CUL3-based ligases in both proteolytic and non-proteolytic regulation of protein kinases. Our own recent study identified the mitotic kinase PLK1 as a direct target of the CUL3 E3-ligase complex containing BTB-KELCH adaptor protein KLHL22.1 In this study, we aim at gaining mechanistic insights into CUL3-mediated regulation of the substrates, in particular protein kinases, by analyzing mechanisms of interaction between KLHL22 and PLK1. We find that kinase activity of PLK1 is redundant for its targeting for CUL3-ubiquitination. Moreover, CUL3/KLHL22 may contact 2 distinct motifs within PLK1 protein, consistent with the bivalent mode of substrate targeting found in other CUL3-based complexes. We discuss these findings in the context of the existing knowledge on other protein kinases and substrates targeted by CUL3-based E3-ligases.
Keywords: CUL3, BTB proteins, KLHL22, ubiquitin, kinases, PLK1
Introduction
Posttranslational mechanisms drive fidelity of cellular processes. Phosphorylation and ubiquitination of substrates represent very common, covalent, and reversible, posttranslational modifications. Human genome encodes for about 600 protein kinases that phosphorylate specific substrates and thereby critically regulate various signaling pathways including cell cycle progression.2 Attachment of the small molecule ubiquitin to other proteins is conducted by a coordinated action of different E1, E2, and E3 enzymes.3 There are predicted 600–1000 different E3-ubiquitin ligases in human cells, suggesting huge complexity and specificity of these enzymes. Indeed, unlike most E1 and E2 enzymes, E3 ligases provide substrate specificity, often by association with the adaptor proteins. For different members of the Cullin ring ligases (CRLs), substrate selection is insured by specific protein families, like, for example, F-box protein family members constitute the SCF complexes (Cullin 1 [CUL1]-based), or BTB domain-containing proteins build functional CUL3 E3-ligases.4 Given a high number of the substrate-specific adaptor proteins found in humans (i.e., 200 different BTBs5), numerous ubiquitination targets of these E3-complexes can be predicted. Indeed, recent mass spectrometry approaches suggest existence of about 5000 different ubiquitination substrates in human cells6. On the other hand, another important cell cycle E3-ligase, the anaphase-promoting complex/cyclosome (APC/C) utilizes only 2 distinct adaptor proteins (CDC20 and CDH1) that are each able to bind and target for ubiquitination and subsequent proteolysis many different substrates.7
Interestingly, phosphorylation and ubiquitination pathways are often interconnected. Phosphorylation may play a critical role both by directly regulating E3-ligases and/or by ensuring substrate specificity. For example, the APC/C subunits and the substrate adaptors are differentially phosphorylated during mitosis and mitotic exit, regulating activity of these enzymes.8,9 On the other hand, the substrates of the SCF E3-ligases need to be phosphorylated, creating a so-called “phosphodegron”, in order to be efficiently targeted for ubiquitination10 (Fig. 1). Importantly, many kinases are not only critical regulatory components of these pathways but also represent themselves the direct ubiquitination substrates. In many cases, ubiquitination of the kinases may lead to their subsequent proteasomal degradation, ultimately attenuating their enzymatic activity, as has been demonstrated for some mitotic kinases targeted by the APC/C E3-ligase.11 However, ubiquitination may also serve non-proteolytic functions, leading to regulation of subcellular localization, binding affinities, or even enzymatic activities.12 Recent data suggest the role of CUL3-based ligases in both proteolytic and non-proteolytic regulation of protein kinases (Table 1 and references therein). Our own recent study identified the mitotic kinase PLK1 as a direct target of the CUL3 E3-ligase complex containing BTB-KELCH adaptor protein KLHL22.1 CUL3/KLHL22-mediated ubiquitination serves a non-proteolytic function and regulates the subcellular localization of PLK1 specifically at the kinetochores. Dissociation of the ubiquitinated PLK1 from kinetochores and thereby reduction of its localized kinase activity is an essential event for the anaphase onset in human cells.1
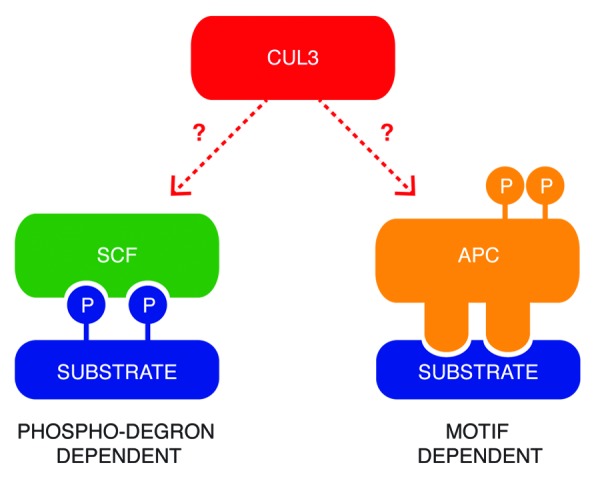
Figure 1. Two common models of substrates recognition by E3-ligase complexes. SCF ligases interact with the phosphorylated residues on the substrates, which create a so-called “phosphodegron”. The APC/C complex recognizes specific short motifs within the amino acid sequence of the targeted proteins, and its activity is regulated by phosphorylation. Little is known about mechanisms of substrate targeting by CUL3-based E3-ligases.
Table 1. The list of protein kinases that are direct substrates of CUL3-based E3-ligases in mammalian cells.
| Substrate specific adaptors | Substrates | Processes | Functions | References |
|---|---|---|---|---|
| KLHL9/13/21 |
Aurora B |
Mitosis and cytokinesis |
Non-proteolytic |
24 and 35 |
| KLHL18 |
Aurora A |
Mitotic entry |
Non-proteolytic |
22 |
| KLHL22 |
PLK1 |
Chromosome segregation |
Non-proteolytic |
1 |
| SPOP |
PIPKIIβ |
Phosphoinositide signaling |
Non-proteolytic |
20 |
| KEAP1/KLHL19 |
IKKβ |
NF-κB signaling |
Proteolytic |
19 and 34 |
| KLHL20 |
DAPK |
IFN-induced cell death |
Proteolytic |
21 |
| KLHL3 | WNK1, WNK4 | Blood pressure | Proteolytic | 33 and 36 |
The table summarizes the current knowledge of the kinases, which were identified as the ubiquitination substrates of CUL3-based E3-ligases in mammalian cells. The identity of the substrate specific adaptor proteins, the roles in the physiological process as well as the function of the ubiquitination events are depicted.
In this study, we aim at gaining mechanistic insights into CUL3-mediated regulation of the substrates, in particular protein kinases (Fig. 1). By analyzing mechanisms of interaction between KLHL22 and PLK1, we conclude that kinase activity of PLK1 is redundant for its targeting for CUL3-ubiquitination. Moreover, we find that CUL3/KLHL22 may contact at least 2 distinct motifs within PLK1 protein. We discuss these findings in the context of the existing knowledge on other protein kinases (Table 1) and substrates targeted by CUL3-based complexes.
Results and Discussion
Postranslational modifications of substrates and recognition by CUL3 E3-ligases
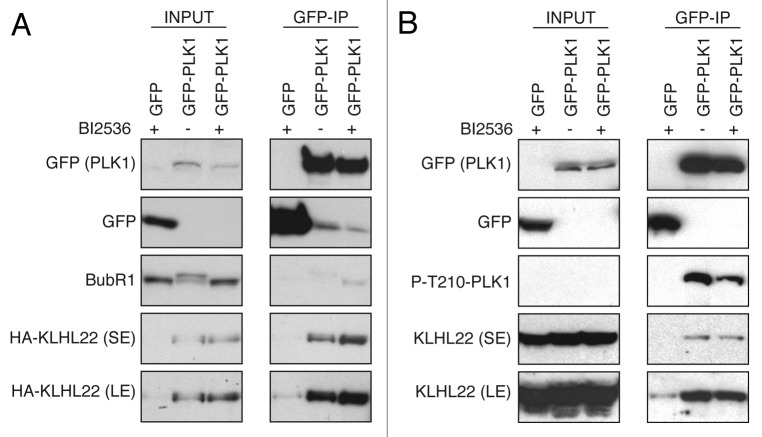
Several substrate-binding mechanisms were identified within the Cullin-RING E3-ligases family. For instance, the SCF E3-ligases require previous phosphorylation of targeted proteins to create a phosphodegron necessary for accurate binding between both components.4,10 In contrast, the APC/C binds to short specific motifs (D-Box, A-box, KEN-box)7 (Fig. 1). The existing data on the regulation of the CUL3 substrate, transcription factor Nrf2, suggest that Nrf2 may not require prior postranslational modification for its binding to CUL3/KEAP113,14 and is constitutively targeteted for ubiquitination under non-stressed cellular conditions. However, little is known about the recognition mechanisms by CUL3-based ligases toward other reported substrates (Fig. 1) in particular protein kinases (Table 1). In order to gain insights into molecular basis of the kinases binding by CUL3, we have tested if the kinase activity of the recently reported substrate PLK11 is required for its binding to CUL3 adaptor KLHL22. For this purpose, GFP tag-protein alone or GFP-tagged PLK1 were expressed in HeLa cells. Subsequently, cells were synchronized in the mitotic stage by addition of Taxol or specific small-molecule inhibitor of PLK1, BI2536,15 and GFP proteins were immunoprecipitated (Fig. 2). As expected, the treatment with BI2536 abolished the phosphorylation-dependent mobility shift of the PLK1 substrate, kinetochore protein BubR116 (Fig. 2A), and reduced amount of the autophosphorylated form of PLK1 (Fig. 2B). In contrast, the HA-tagged KLHL22 (Fig. 2A) and the endogenous KLHL22 (Fig. 2B) efficiently co-immunoprecipitated with PLK1 under these conditions, suggesting that kinase activity of PLK1 is redundant for its recognition by CUL3/KLHL22 E3-ligase. These data are consistent with the fact that strong, salt-resistant interaction was observed in vitro between PLK1 and KLHL22 expressed in bacterial cells, and that downregulation of CUL3/KLHL22 did not modulate kinase activity of PLK1.1 Interestingly, a number of studies suggest that unlike for many SCF E3-ligases,4,10 there is no evidence that posttranslational modifications are required for substrates interactions with CUL3 E3-ligases. Intriguingly in the case of 2 BTB adaptor proteins, KEAP117 and SPOP,18 substrate phosphorylation inhibits rather than promotes their recruitment. Also, regulation of IKKβ by KEAP1 appears to be independent of IKKβ activity,19 and the kinase-dead mutant of PIPKIIβ was more efficiently ubiquitinated, and this CUL3/SPOP-mediated ubiquitination was further enhanced by expression of specific phosphatases.20 A similar situation was observed for DAPK, as the kinase-defective and kinase-active mutants bound KLHL20 as effectively as the wild-type DAPK.21 During mitotic entry, Aurora A kinase is regulated by the CUL3/KLHL18 E3 ligase,22 and this ubiquitination event seems to proceed the activation of Aurora A, suggesting that KLHL18 interacts with an inactive, unmodified kinase. Taken together, all these findings are in a sharp contrast to SCF-mediated mechanisms, in which substrate modification induced by a stimuli switches on the ubiquitination event. One could speculate that for CUL3-mediated ubiquitination events, some specific extra- or intracellular factors generate an “off” signal for substrate targeting. Indeed, Nrf2 is constitutively recognized and ubiquitinated by CUL3/KEAP1, and oxidative stress conditions interfere with E3-ligase activity.13,14 Similarly, DAPK is constitutively targeted to the CUL3-based ligase under basal conditions.21 In both cases, regulation of the substrate ubiquitination occurs at the level of the substrate-specific adaptor rather than substrate itself. While KEAP1 is oxidized on the specific cysteine residues,13,23 KLHL20 is sequestered away from the substrate into PML nuclear bodies under IFN-stress conditions.21 Interestingly, subcellular localization of other mitotic BTB-Kelch proteins involved in CUL3 complexes appears to be regulated in a timely manner during cell cycle progression.1,22,24,25
Figure 2. Catalytic activity of PLK1 is not required for its interaction with KLHL22-adaptor protein. (A) Cells expressing GFP alone and GFP-PLK1 were transfected with HA-KLHL22 and synchronized in mitosis using Taxol or PLK1 inhibitor BI2536. Extracts were immunoprecipitated using GFP-Trap beads. Inputs and immunoprecipitates were analyzed by western blot. (B) Cells expressing GFP alone and GFP-PLK1 only were synchronized and analyzed as in (A). The short (SE) and long (LE) exposures of the representative blots are shown.
Insights into molecular architecture of CUL3 ligases
Unlike SCF ligases that interact with the phosphorylated residues on the substrates, the APC/C complex recognizes specific short motifs within the amino acid sequence of the targeted proteins. Recent studies offer an insight into molecular architecture of the CUL3-based E3-ligase complexes18,26 and suggest a quaternary assembly model, where dimers of BTB-adaptor proteins are engaged in a single complex, supporting a bivalent mode of interaction with the substrate proteins. Indeed, it has been demonstrated that 2 specific acidic motifs within Nrf2 protein are in a direct contact with the β-propeller constituted of the Kelch repeats within KEAP protein,17,27 and the SPOP adaptor is also able to engage multiple-binding sites in one substrate, which could be explained by complex structural flexibility and ability to dimerize.18 However, little is known about the molecular basis of interaction between CUL3-complexes and protein kinases. Interestingly, mutagenesis of the A-box and D-box for APC/C-dependent degradation within Aurora A kinase did not interfere with CUL3-mediated ubiquitnation.22
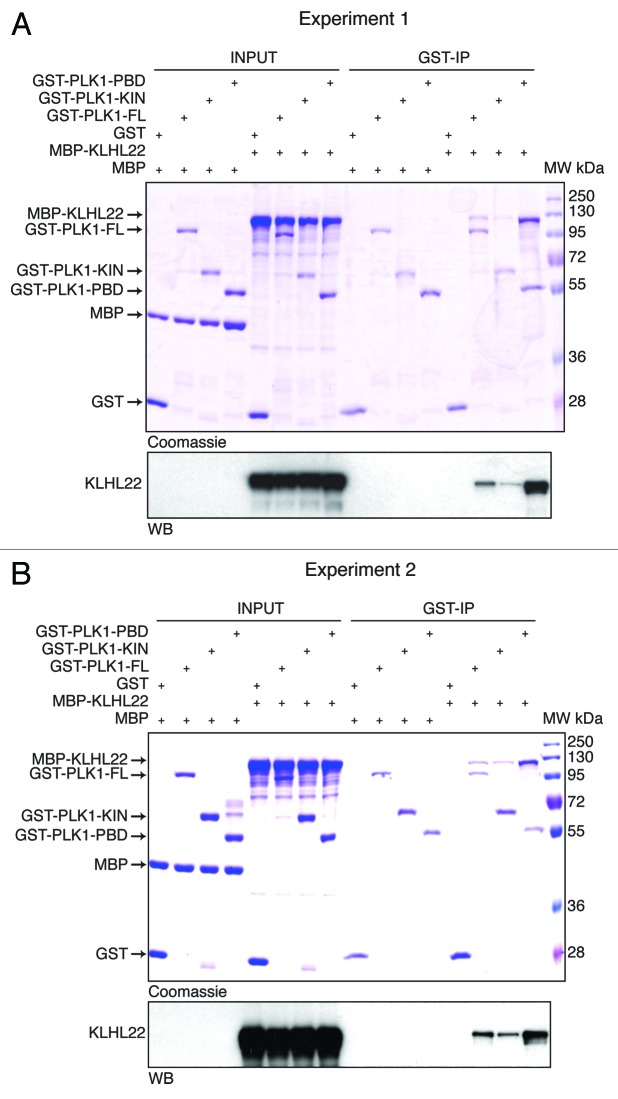
Therefore, we aimed at understanding if a bivalent mode of substrate-CUL3 interaction is also utilized by PLK1. To this end, we expressed and purified the GST-tagged fragments corresponding to entire kinase domain and the regulatory domain, the Polo-box domain (PBD) from bacterial cells. Following incubation with the full-length, bacterially derived MBP-tagged KLHL22 protein, we immunoprecipitated the complexes using GSH-Sepharose. As expected, the full-length PLK1 protein efficiently interacted with the MBP-KLHL22 protein (Fig. 3), consistent with a redundancy for prior posttranslational modifications for this interaction and with the previous findings.1 Interestingly, both PLK1 fragments were able to interact with the full-length KLHL22 protein in vitro (Fig. 3). To corroborate these findings, we expressed the corresponding fragments fused to the GFP-tag in human cells. Following cell synchronization in mitosis by Taxol, we immunoprecipitated the complexes formed. In contrast to the GFP-protein alone, all GFP-fused forms of PLK1 protein were able to efficiently interact with the endogenous KLHL22 protein (Fig. 4). These results suggest that PLK1 utilizes at least 2 distinct binding interfaces located within 2 functional domains of the protein (Fig. 5), consistent with the bivalent model of substrate interaction with the KEAP and SPOP substrate adaptors.17,18,27 While KLHL22 binding to the kinase domain may, in principle, resemble interactions within other BTB-KELCH/kinase complexes, interaction with the PBD domain is likely to be more specific to the family of Polo kinases. The PBD can indeed be only found within PLKs from different species and is critically involved in targeting these kinases to specific subcellular localizations, acting as a phosphoreceptor-biding domain.28-32 It is interesting that the CUL3/KLHL22-mediated ubiquitination takes place at the specific lysine residue (K492) located within the PBD domain and interferes with the phospho-receptor-dependent interactions of PLK1 at the kinetochores.1
Figure 3. KLHL22 interacts with 2 domains of PLK1 in vitro. (A and B) Recombinant GST or GST fused to full-length PLK1 (GST-PLK1), kinase domain fragment (GST-PLK1-KIN), and PBD fragment (GST-PLK1-PBD) were incubated with recombinant MBP or MBP fused to full-length KLHL22 (MBP-KLHL22), and immunoprecipitated using glutathione-sepharose beads. Immunoprecipitates (GST-IP) were analyzed by coomassie blue staining and western blot. (A and B) represent 2 independent experiments.
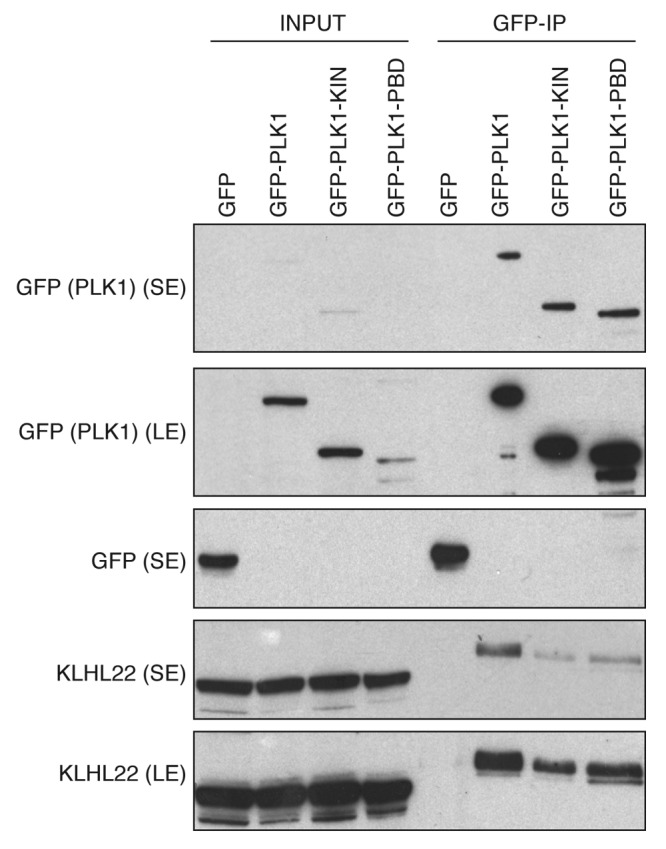
Figure 4. KLHL22 interacts with 2 domains of PLK1 in cells. Cells expressing GFP alone, GFP fused to full-length PLK1 (GFP-PLK1), kinase domain fragment (GFP-PLK1-KIN), and PBD fragment (GFP-PLK1-PBD) were synchronized in mitosis using Taxol. Extracts were immunoprecipitated using GFP-Trap beads. Inputs and immunoprecipitates were analyzed by western blot. The short (SE) and long (LE) exposures of the representative blots are shown.
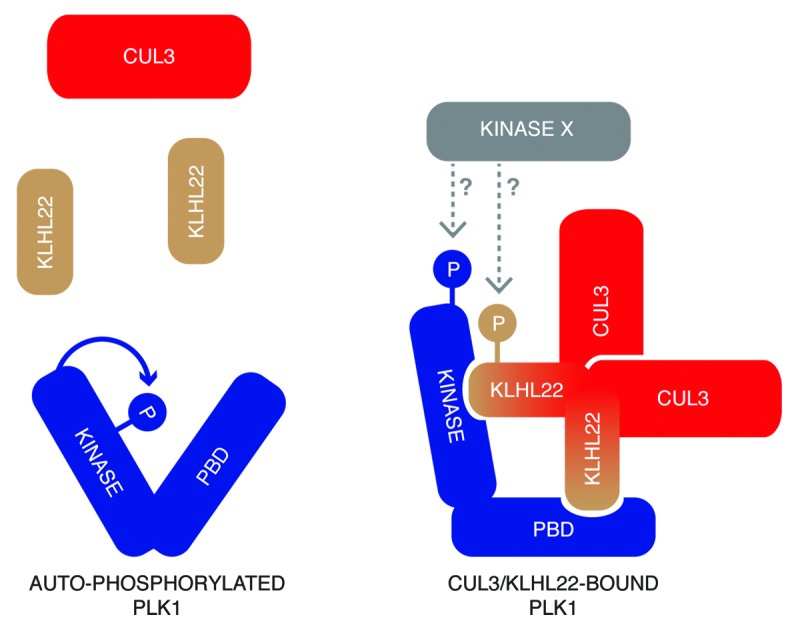
Figure 5. A hypothetical model of the architecture of CUL3/KLHL22 in a complex with PLK1. PLK1 interacts with the substrate specific adaptor protein KLHL22 independent of its kinase activity. It is possible that specific conformational change or modifications by unidentified factors, including upstream kinases (Kinase X), regulate affinity of PLK1 to KLHL22. KLHL22 binds to 2 distinct domains of PLK1 and may utilize 2 similar or different motifs within its primary sequence.
While it cannot be rigorously excluded that postranslational modifications (i.e., phosphorylation), by yet-to-be-identified factors, play a regulatory role for CUL3 complex assembly, our results are consistent with the possibility that specific motifs within substrate proteins exist that mediate their binding to CUL3-complexes (Fig. 5). Indeed, a small acidic region within WNK kinases mediating its interaction with KLHL3 adaptor was recently identified,33 and, similar to Nrf2,17,27 IKKβ possess an acidic (D/N)XE(T/S)GE motif within its kinase domain that is required for KEAP1 binding.19,34 Before any general rules can be formulated, future studies are needed to identify the sequence motifs involved in the substrate binding to CUL3/KLHL22 and other CUL3-based complexes. As many kinases are found to be targeted by these E3-ligases (Table 1), these studies may also lead to better understanding of the general mode of kinase regulation.
Material and Methods
cDNAs
The full-length KLHL22 was cloned into pMal-C2X (New England BioLabs) and pcDNA3.1 (Invitrogen) in fusion with the N-terminal HA-tag as described previously.1 The full-length PLK1 (1–603), the PLK1 kinase domain (PLK1-KIN, 1–320), and the PBD domain (PLK1-PBD, 321–603) were cloned into pGex-6P1 (GE Healthcare) using EcoRI/XhoI restriction sites and into pEGFP-N1 (Clontech) using XhoI/KpnI. The following primers were used for cloning into pGex-6P1: PLK1 N-term (forward): 5′-GCGGAATTCAGTGCTGCAGTGACTGCAGG-3′; PLK1 C-term (reverse) 5′-CGCCTCGAGTTAGGAGGCCTTGAGACGGT-3′; PLK1-KIN (reverse): 5′-CGCCTCGAGAATGGTCAGGCAGGTGAT-3′; PLK1-PBD (forward): 5′-GCGGAATTCCCACCAAGGTTTTCGATTG-3′ and for cloning into pEGFP-N1: PLK1 N-term (forward): 5′-CGCCTCGAGATGAGTGCTGCAGTGACTGC-3′; PLK1 C-term (reverse): 5′-CGGGGTACCCCGGAGGCCTTGAGACGG-3′; PLK1-KIN (reverse): 5′-CGGGGTACCCCGGTCAGGCAGGTGATGG-3′; PLK1-PBD (forward): 5′-CGCCTCGAGATGCCACCAAGGTTTTCG-3′.
Recombinant protein expression
E. coli BL21 (DE3) bacteria were transformed with the different pGex-6P1 and pMal-C2X constructs. Once cultures reached OD600 = 0.4–0.6, they were cooled down at 20°C for 30 min. Expression was subsequently induced overnight at 20°C with 1 mM IPTG. Cells were harvested by centrifugation. GST-fusion proteins were resuspended in lysis buffer (50 mM NaCl, 10 mM TRIS-HCl pH 8, 1 mM DTT, Complete Protease Inhibitor Coktail [Roche]), lysed by sonication, and supernatant was cleared by centrifugation at 40 000 rpm for 1 h using 50.2Ti rotor. The supernatant was incubated for 2 h with 500 µl of Glutathione Sepharose 4B (GE Healthcare) per 1 liter of culture, previously equilibrated in lysis buffer. Beads were washed with 50 ml of lysis buffer, and elution was done with 20 mM glutathione. The elution fraction was dialyzed overnight into lysis buffer to remove glutathione and concentrated using 10 kDa cut-off centrifugal filter units (Amicon). For expression of the recombinant MBP-KLHL22 the following lysis buffer was used (PBS-EDTA (0.5 mM), 1 mM DTT, Complete Protease Inhibitor Coktail [Roche]). The cleared lysate was incubated with 2 ml amylose resin (New England Biolabs) per 1 liter of culture. Elution was performed by supplementing lysis buffer with 15 mM maltose. The eluate was dialysed in PBS and concentrated using 50 kDa cut-off centrifugal filter units (Amicon).
Cell culture, transfections, and synchronization
HeLa Kyoto were cultured as previously described35 and transfected using lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. For mitotic synchronizations, cells were treated with 200 nM Taxol (paclitaxel) (Sigma) or 100 nM PLK1 inhibitor BI2536 (Axon Medchem) for 13 h before harvesting.
Western blotting, immunoprecipitation, and antibodies
Preparation of HeLa cells extracts, immunoprecipitation, and western blotting were described previously.1 The following antibodies were used in this study: rabbit polyclonal KLHL22,1 rabbit polyclonal GFP (abcam ab290, 1:2000), mouse monoclonal BubRI (BD Biosciences 612502, 1:1000), rat monoclonal HA (Roche 11867423001, 1:1000), and rabbit polyclonal pThr210PLK1 (Cell Signaling, 1:1000).
Acknowledgments
The authors wish to thank the members of the entire Sumara group for careful reading of the manuscript and for helpful discussions. The research in the laboratory of Izabela Sumara is sponsored by the IGBMC, ATIP-AVENIR program from CNRS, and INSERM, “Programme de Mécénat” from Sanofi-Aventis, University of Strasbourg and Fondation ARC pour la recherche sur le cancer.
Glossary
Abbreviations:
- PLK1
polo-like kinase 1
- DUBs
deubiquitinating enzymes
- CRLs
cullin ring ligases
- CUL
cullin
- APC/C
anaphase promoting complex/cyclosome
- PBD
polo box domain
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25369
References
- 1.Beck J, Maerki S, Posch M, Metzger T, Persaud A, Scheel H, et al. Ubiquitylation-dependent localization of PLK1 in mitosis. Nat Cell Biol. 2013;15:430–9. doi: 10.1038/ncb2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 3.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 4.Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–70. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 5.Stogios PJ, Privé GG. The BACK domain in BTB-kelch proteins. Trends Biochem Sci. 2004;29:634–7. doi: 10.1016/j.tibs.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–40. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–56. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 8.Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, et al. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sumara I, Maerki S, Peter M. E3 ubiquitin ligases and mitosis: embracing the complexity. Trends Cell Biol. 2008;18:84–94. doi: 10.1016/j.tcb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 11.Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol. 2004;164:233–41. doi: 10.1083/jcb.200309035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–51. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–5. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lénárt P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–15. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 16.Elowe S, Hümmer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21:2205–19. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006;25:3605–17. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell. 2009;36:131–40. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunce MW, Boronenkov IV, Anderson RA. Coordinated activation of the nuclear ubiquitin ligase Cul3-SPOP by the generation of phosphatidylinositol 5-phosphate. J Biol Chem. 2008;283:8678–86. doi: 10.1074/jbc.M710222200. [DOI] [PubMed] [Google Scholar]
- 21.Lee YR, Yuan WC, Ho HC, Chen CH, Shih HM, Chen RH. The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J. 2010;29:1748–61. doi: 10.1038/emboj.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moghe S, Jiang F, Miura Y, Cerny RL, Tsai MY, Furukawa M. The CUL3-KLHL18 ligase regulates mitotic entry and ubiquitylates Aurora-A. Biol Open. 2012;1:82–91. doi: 10.1242/bio.2011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–53. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maerki S, Olma MH, Staubli T, Steigemann P, Gerlich DW, Quadroni M, et al. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J Cell Biol. 2009;187:791–800. doi: 10.1083/jcb.200906117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maerki S, Beck J, Sumara I, Peter M. Finding the midzone: the role of ubiquitination for CPC localization during anaphase. Cell Cycle. 2010;9:2921–2. doi: 10.4161/cc.9.15.12740. [DOI] [PubMed] [Google Scholar]
- 26.Canning P, Cooper CD, Krojer T, Murray JW, Pike AC, Chaikuad A, et al. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J Biol Chem. 2013;288:7803–14. doi: 10.1074/jbc.M112.437996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–9. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/S0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 29.Cheng KY, Lowe ED, Sinclair J, Nigg EA, Johnson LN. The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J. 2003;22:5757–68. doi: 10.1093/emboj/cdg558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanisch A, Wehner A, Nigg EA, Silljé HH. Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting. Mol Biol Cell. 2006;17:448–59. doi: 10.1091/mbc.E05-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee KS, Park JE, Kang YH, Zimmerman W, Soung NK, Seong YS, et al. Mechanisms of mammalian polo-like kinase 1 (Plk1) localization: self- versus non-self-priming. Cell Cycle. 2008;7:141–5. doi: 10.4161/cc.7.2.5272. [DOI] [PubMed] [Google Scholar]
- 32.Park JE, Soung NK, Johmura Y, Kang YH, Liao C, Lee KH, et al. Polo-box domain: a versatile mediator of polo-like kinase function. Cell Mol Life Sci. 2010;67:1957–70. doi: 10.1007/s00018-010-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohta A, Schumacher FR, Mehellou Y, Johnson C, Knebel A, Macartney TJ, et al. The CUL3-KLHL3 E3 ligase complex mutated in Gordon’s hypertension syndrome interacts with and ubiquitylates WNK isoforms: disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem J. 2013;451:111–22. doi: 10.1042/BJ20121903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JE, You DJ, Lee C, Ahn C, Seong JY, Hwang JI. Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell Signal. 2010;22:1645–54. doi: 10.1016/j.cellsig.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Sumara I, Quadroni M, Frei C, Olma MH, Sumara G, Ricci R, et al. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell. 2007;12:887–900. doi: 10.1016/j.devcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 36.Wakabayashi M, Mori T, Isobe K, Sohara E, Susa K, Araki Y, et al. Impaired KLHL3-Mediated Ubiquitination of WNK4 Causes Human Hypertension. Cell Rep 2013. [DOI] [PubMed] [Google Scholar]