Abstract
Proteomic analysis of human body fluids is highly challenging, therefore many researchers are redirecting efforts towards secretome profiling. The goal is to define potential biomarkers and therapeutic targets in the secretome that can be traced back in accessible human body fluids. However, currently there is a lack of secretome profiles of normal human primary cells making it difficult to assess the biological meaning of current findings. In this study we sought to establish secretome profiles of human primary cells obtained from healthy donors with the goal of building a human secretome atlas. Such an atlas can be used as a reference for discovery of potential disease associated biomarkers and eventually novel therapeutic targets. As a preliminary study, secretome profiles were established for six different types of human primary cell cultures and checked for overlaps with the three major human body fluids including plasma, cerebrospinal fluid and urine. About 67% of the 1054 identified proteins in the secretome of these primary cells occurred in at least one body fluid. Furthermore, comparison of the secretome profiles of two human glioblastoma cell lines to this new human secretome atlas enabled unambiguous identification of potential brain tumor biomarkers. These biomarkers can be easily monitored in different body fluids using stable isotope labeled standard proteins. The long term goal of this study is to establish a comprehensive online human secretome atlas for future use as a reference for any disease related secretome study.
Keywords: secretome, stables istopes, primary cells, glioblastoma, biomarkers, body fluids, signal peptide
Introduction
Human cell secreted proteins control and regulate a multitude of biological and physiological processes thus making them a clinically attractive target for biomarkers and therapeutics discovery. Indeed, in almost every disease condition the extracellular environment is somewhat perturbed creating an environment that can act in favor or against disease progression. A simple example is during bacterial or viral infection where the airway epithelium often secretes a variety of proteins to directly combat the foreign pathogen and also to recruit immune cells to further act on the invading pathogens [1]. In more complex pathogeneses such as those occurring in cancer and degenerative diseases, perturbations of the cell secretome often favors disease progression. For example, aggressive cancer cells often secrete a variety of angiogenic factors, growth factors and proteases to promote tumor progression [2]. Similarly, in degenerative diseases such as Duchenne muscular dystrophy, the lack of expression of dystrophin protein, due to mutations in its gene, results in leaky muscle fibers that lead to perturbed extracellular environment, progressive muscle inflammation and degeneration. Thus, in depth analysis of cellular secretomes in health and disease conditions could potentially bring insights into the mechanisms of progression of diseases and eventually define novel biomarkers and novel therapeutic targets.
Typically the majority of secretome studies published to date are performed in vitro by culturing cells of interest first in serum supplemented medium to obtain sufficient number of cells then in serum free medium for a short period of time [3]. This allows collection of genuine cell secreted proteins while minimizing bovine serum interfering proteins. More recently, secretomes have been established for human cell cultures even in the presence of bovine serum by implementing a combination of SILAC and enrichments strategies [4]. These strategies showed that serum starvation might influence cell secretions. However, both serum and serum supplemented in vitro secretome studies might both be still far from the in vivo situation where cells of interest are surrounded by other cell types and a variety of factors that influence their secretome.
One of the main goals of many secretome studies is to define potential biomarkers that can eventually be traced back in body fluids and used as a clinical measure to monitor progression of diseases and response to treatments. However, it is not known how many of the cell secreted proteins actually make it into circulating body fluids and what is the contribution levels of a tissue or a cell type to the body fluid proteome. This knowledge is important especially in the context of identifying which body fluid is most likely to contain the potential biomarker(s). For example, there is a split focus concerning which body fluid, cerebrospinal fluid (CSF) or serum, is suitable for brain tumor biomarker discovery and validation [5,6]. Even though CSF is proximal to the brain, not all brain tumors are actually in contact with CSF, but most tumors have vasculature and therefor it may be more logical to search for a brain tumor biomarker in serum rather than in CSF.
The majority of published secretome studies are focused on established cell lines, and especially cancer cell lines. Only a few studies have investigated the secretome of healthy human primary cells. In this study we sought to build a human secretome atlas by cataloguing secretomes from several human primary cell cultures. This master list can then be compared to body fluid proteomes to determine the contribution of those cell secreted proteins to the body fluid proteome (e.g. serum, CSF and urine). This secretome atlas could eventually serve as a standard reference for comparison to diseased states.
2. Materials and methods
2.1 Cell cultures
Human primary retinal pigment epithelial (RPE) cells used in this study were prepared from human autopsy eyes as described previously [7]. Cells were seeded on a 0.1% gelatin-coated plate, cultured and maintained in DMEM/F12 supplemented with 10% FBS, 100 U/mL penicillin, and 100 g/mL streptomycin.
Human primary astrocytes were purchased from Cell Systems Corporation (Kirkland, WA) and were cultured and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with with 10% FBS, 100 U/mL penicillin, and 100 g/mL streptomycin.
Human primary bronchial epithelial (BE) cells were purchased from Lonza (Walkersville, MD) and cultured as previously described [8,9] using bronchial epithelial basal media (BEBM) and SingleQuots (proliferation media) from Lonza (Walkersville, MD). Culture media was changed every 48 hours until 100% confluence. Then, air-liquid interface was initiated by feeding cultures daily using ALI media only on the basolateral side for 14 days, at which time cells were fully differentiated. ALI media consisted of 1:1 mixture BEBM/DMEM with SingleQuots (minus BPE and RA) and the following: 65mg BPE, 5 × 10-5M RA and 150 μL of 5mg/mL bovine serum albumin).
Nasal epithelial cells were collected from a healthy donor in accordance with an Institutional Review Board approved protocol at Children's National Medical Center (CNMC) and after obtaining patients’ written informed consent. After brief treatment with topical viscous lidocaine 2%, nasal epithelial cells were collected from between the middle and inferior turbinates with a 3-mm cytology brush by turning the brush 180 degrees 3 times. Cells from both nostrils were combined, initially incubated with 1% pronase in a 1:1 mixture of DMEM/Ham's F-12 supplemented with antibiotics for 18–20 h at 4°C then washed and cultured on a Corning plastic cell culture Flasks for 4 hrs at 37°C to remove contaminating cells. The nonadherent cells (i.e. mostly epithelial cells) were collected and cultured in bronchial epithelial basal media (BEBM) as described above for bronchial epithelial cells.
Human primary Schwann cells were purchased from ScienCell Research Laboratories (Carlsbad, CA) and cultured in basal Schwann Cell Medium (SCM) from ScienCell Research Laboratories (Carlsbad, CA) supplemented with 10% FBS, 100 U/ mL penicillin, 100 μg/ mL streptomycin and 1% Schwann cell growth supplement (SCGS) containing BSA 10 μg, insulin 5 μg, PDGF 5 ng, FGF-2 5 ng, progesterone 20 nM, forskolin 200 nM, T3 60 nM and PMA 20 nM. Growing cells were maintained and passaged using the same proprietary media.
Human primary fibroblast 551 (FB 551), human glioblastoma U87 and T98 cells lines were purchased from ATCC and cultured in DMEM basal medium supplemented with 10% FBS, 100 U/mL penicillin, and 100 g/mL streptomycin.
2.2 Collection of cell secreted proteins
All cells were maintained and cultured in their respective media. After obtaining significant amounts of cultured cells, at least three 75 cm2 of each cell type, the cells were rinsed 6 times with serum free media then incubated in serum free media for another 24 hours. Conditioned media (CM) were then collected, centrifuged at 300× g and the supernatant was further filtered through a 0.22 μm nylon filter (Millipore, Bedford, MA) to remove any cell debris or floating cells. Aliquots (2 mL) from each collected CM were further concentrated by using a 3 kDa cutoff filter (Millipore Corp., Billerica, MA) and processed for SDS-PAGE and mass spectrometry analysis. After the gel was stained (Bio-Safe Coomassie; Bio-Rad, Hercules, CA), each lane was sliced into 30 bands, and each band was in-gel digested with trypsin (Promega, Madison, WI) and the resulting peptides analyzed by LC-MS/ MS as described below.
2.4 Stable isotope labeling by amino acid in cell culture and human serum spike-in strategy
To trace back and quantify potential cell secreted proteins in human serum samples, serum from healthy donors was purchased from Sigma Inc (Saint Louis, MI) and used for spike-in with stable isotope labeled secretome prepared from glioblastoma cells lines U8 7 and T98 [10]. U87 and T98 cell lines were cultured for seven cell doublings in custom made DMEM medium where Arg and Lys were replaced by 13C6, 15N2-Labeled Lys and 13C6,-labeled Arg [11]. Conditioned media from U87 and T98 containing fully labeled secreted proteins were mixed 1 to 1 then an aliquot was concentrated as described above and used as a standard for spiking human serum samples. Three aliquots of human serum samples containing 100 μg of total proteins each were spiked with 50 μg of SILAC labeled U87/ T98 secretome and processed for SDS-PAGE, in-gel digestion and LC-MS/ MS as described above.
2.5 Liquid Chromatography-Tandem Mass Spectrometry analysis
Concentrated peptides from each band were injected via an autosampler (6uL) and loaded onto a Symmetry C18 trap column (5μm, 300 μm i.d. × 23 mm, Waters) for 10 min at a flow rate of 10 μL/min, 100% A. The sample was subsequently separated by a C18 reverse-phase column (3 μm, 200A, 100 μm × 15 cm, Magic C18, Michrom Bioresources) at a flow rate of 300 nL/min using an Eksigent nano-hplc system (Dublin, CA). The mobile phases consisted of water with 0.1% formic acid (A) and 90% acetonitrile (B). A 65 minute linear gradient from 5 to 60% B was employed. Eluted peptides were introduced into the mass spectrometer via Michrom Bioresources CaptiveSpray. The spray voltage was set at 1.4 kV and the heated capillary at 200 °C. The LTQ-Orbitrap-XL (ThermoFisherScientific) was operated in data-dependent mode with dynamic exclusion in which one cycle of experiments consisted of a full-MS in the Orbitrap (300-2000 m/z) survey scan in profile mode, resolution 30,000, and five subsequent MS/MS scans in the LTQ of the most intense peaks in centroid mode using collision-induced dissociation with the collision gas (helium) and normalized collision energy value set at 35%.
2.6 Protein identification and quantification
For secretome profiling of the 6 primary cell cultures we used label free proteome profiling. Each raw file was searched for protein identification using the Sequest algorithm in the Bioworks Browser 3.3.1 software (ThermoFisher Scientific) against the Uniprot database indexed for human species (UniProt release 2012_02) and for fully tryptic peptides, 2 missed cleavages and potential modification of oxidized methionine (15.9949 Da). DTA generation parameters were Peptide Tolerance of 50 ppm and Fragment Ion Tolerance of 1 Da. Search result files were loaded into ProteoIQ software (NuSep, Bogart, GA) and filtered based on the following: XCorr > 1.9, 4 spectra per peptide, 2 unique peptides per protein, 0.98 peptide probability and 0.95 protein probability.
For serum spike-in strategy raw files were processed for protein identification and quantification using Integrated Proteomics Pipeline (IP2) version 1.01 software developed by Integrated Proteomics Applications, Inc. (http://www.integratedproteomics.com/). Mass spectral data were uploaded into IP2 software and searched against the forward and reverse Uniprot human database for tryptic peptides allowing two missed cleavages, possible modification of oxidized methionine (15.99492 Da) and heavy arginine (6.0204 Da) and heavy lysine (8.0142 Da). IP2 uses the Sequest 2010 (06_10_13_1836) search engine. Mass tolerance was set at +/- 30 ppm for MS and +/- 1.5 Da for MS/MS. Data were filtered based on a 1% protein false discovery rate. All the bands from each lane were summed in the analysis. Census software version 1.77, built into the IP2 platform, was used to determine the ratios of unlabeled and labeled peptide pairs using an extracted chromatogram approach. Coefficient of variance between the triplicate experiments was determined using the mean ratio and standard deviation obtained for each measurable protein across the three experimental runs.
2.7 Protein classification and mapping
High-confidence peptide identifications from each of the six secretomes were mapped to UniProt human reference proteome sequences (UniProt release 2012_02), including isoforms, to ensure a current and consistent set of protein accessions were associated with each peptide sequence. The peptides and their proteins from all six datasets were then subjected to a global protein parsimony analysis to infer only those proteins with sufficient peptide evidence and eliminate redundant protein accessions. This global parsimony analysis ensures that inferred proteins are consistently identified by the same UniProt accession across all peptide sources. Inferred proteins were required to be supported by at least two distinct peptide sequences not shared with any other inferred protein, while explaining the maximum number of high-confidence peptide identification sequences. Accessions of proteins annotated with various Gene Ontology or UniProt keyword terms including the words secreted, membrane, plasma were derived directly from the current UniProt Knowledgebase. Accessions of proteins from manuscript supplementary material or where peptide sequences were unavailable were mapped using the UniProt ID Mapper service to current UniProt accessions.
2.7 Prediction of classical and non-classical protein secretion
To define all secreted proteins, including those with signal peptides and those secreted via non-classical pathways, we used a combination of prediction servers [12] including the latest version of SignalP4.1[13] (http://www.cbs.dtu.dk/services/SignalP/) and SecretomeP 2.0 [14] (http://www.cbs.dtu.dk/services/SecretomeP/). Fasta format sequences corresponding to UniProt accessions were downloaded from UniProt and evaluated in batch mode using both signalP 4.1 server and SecretomeP 2.0 server to predict proteins that are secreted via classical and non-classical pathways, respectively.
3. Results
3.1 Secretome profiles of cultured human primary cells
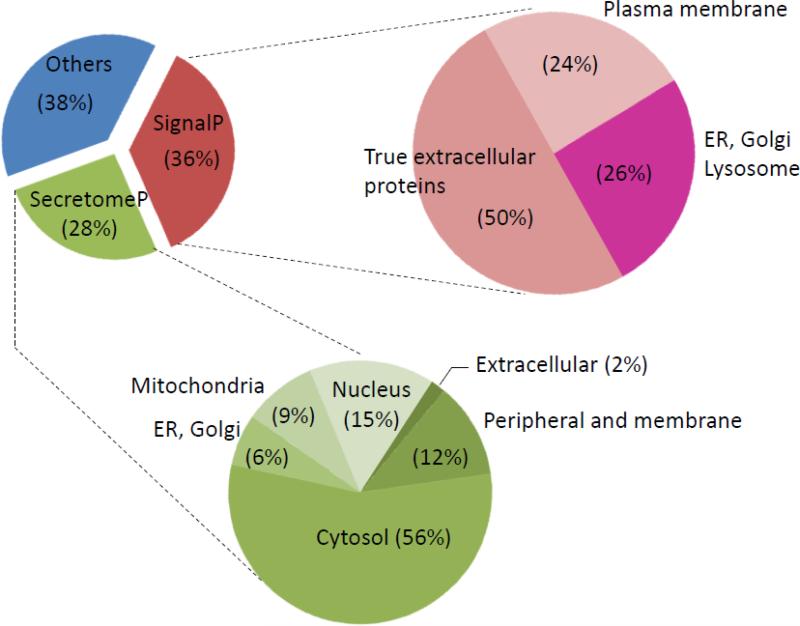
Secretome profiles were established for six human primary cell cultures including astrocytes, fibroblasts, bronchial epithelial cells, nasal epithelial cells, retinal pigment epithelial cells and Schwann cells using high precision LC-MS/MS and spectral counting. There were 1054 non-redundant proteins identified in the conditioned medium of the 6 primary cell cultures combined. Of these, 376 proteins (~36%) had a signal peptide as defined by the SignalP tool and 277 proteins (~26%) were found to be secreted via non-classical pathway as predicted by secretomeP tool. The remaining 400 proteins (~38%) were of intracellular origin from different organelles and compartments, most probably released by cell via vesicular budding or cell leakage. Of the 376 proteins with a signal peptide, 188 are known to be true extracellular proteins, 92 are known to be cell surface or cell membrane proteins while the remaining 96 were of endothelial, lysosome and Golgi origin. The 277 proteins secreted via non-classical pathway were mostly of cytosolic (123 proteins) and peripheral origin (26 proteins) (Figure 1). Interestingly, of the 376 proteins defined as having signal peptide by SignalP, 234 proteins were also predicted as secreted by secretomeP while 142 proteins were not (Supplemental Table S1, sheet “All”).
Figure 1. Pie chart depicting the distribution and the subcellular location of the 1054 proteins identified in the condition media of the 6 human primary cell cultures studied herein.
Fasta sequences were uploaded into SignalP 4.1 server and SecretomeP 2.0 server to predict proteins with a signal peptide and those secreted via non-classical pathway respectively. Accession numbers of all identified proteins in Supplemental Table S1 were uploaded into Uniprot identifier tool and information about their subcellular location was retrieved.
Close examination of the secretome profiles of the 6 primary cell types studied herein revealed specific secretome signatures for specific cell types, thus enabling their characterization and perhaps classification. Of the 1054 identified proteins, 29 proteins were unique to astrocytes with spondin1 and apolipoprotein E (Apo E) being the most abundant (13 and 12 spectral counts, respectively), 33 proteins were unique to fibroblasts with nephronectin, WAP four-disulfide core domain protein and mesothelin being the most abundant (19, 22 and 38 spectral counts, respectively), 10 were unique to bronchial epithelial cells with mucin-4 and lysyl oxidase homolog-4 being the most abundant (7 and 6 spectral counts, respectively), 31 were unique to nasal epithelial cells with laminin subunit beta-3, lysozyme C and urokinase-type plasminogen activator being the most abundant (91, 17 and 10 spectral counts, respectively), 16 were unique to RPE cells with complement factor H, fibromodulin, Nesh-SH3 and complement C4-as being the most abundant (12, 12, 13 and 59 spectral counts, respectively) and 100 were unique to Schwann cells with carboxypeptidase E and collagen ( 26 and 11 spectral counts, respectively). For more details see Supplemental Table S1. Interestingly, only 73 proteins among the overall 1054 identified proteins in the conditioned media were shared by all 6 primary cell cultures. These include mainly proteins of intracellular origin with vimentin, actin, alpha actinin, alpha enolase, prelamin-A/C and moesin as being the most abundant (e.g. 368 to 1531 spectral counts, for more details see Supplemental Table S1)
3.2 Contribution of the Cell secretomes to the human body fluids proteomes
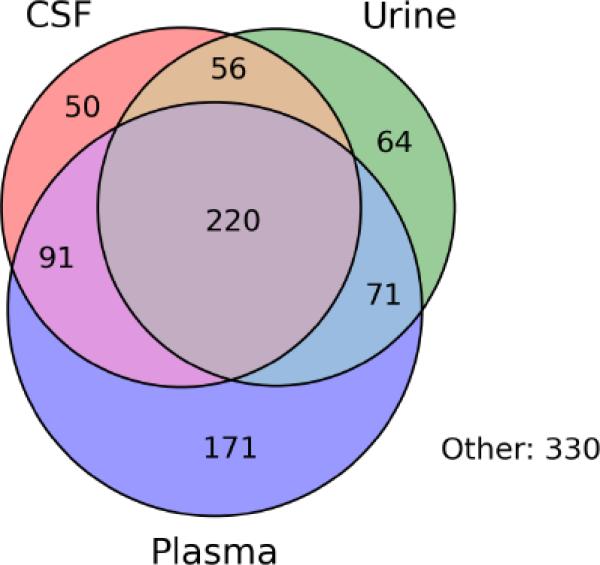
Of the 1054 proteins identified in the condition media of the 6 primary cell cultures studied herein, 723 (69%) were found to occur in the proteomes of at least one of the major human body fluids including plasma proteome [15], CSF proteome [16] and urine proteome [17]. Of these 723 proteins, 220 (30%) occurred in all three body fluids, 91 (13%) occurred in both plasma and CSF proteome but were not detected in urine proteome, 56 (8%) were detected in CSF and urine proteome but not in serum proteome, 71 (10%) were detected in both plasma and urine, 64 (9%) were unique to CSF proteome, 171 (24%) were unique to plasma proteome and 50 (7%) were unique to urine proteome (Figure 2).
Figure 2. Venn diagram showing body fluids distribution of the 1054 proteins identified in the condition media of the 6 human primary cell cultures.
Among the 1054 identified protein, 677 occurred in at least one of the body fluids while the remaining 330 proteins were never reported to exist in any of the body fluids (see Supplemental Table S1, “Bodyfluids proteins” and “interstitial” sheet respectively). Note that among the 677 protein reported in at least one of the body fluids 220 occurred in all three body fluids while 171 were unique to plasma, 50 unique to CSF and 64 unique to urine. The names of these proteins are shown in Supplemental Table S1 sheet “Body fluids proteins”.
Interestingly, among the 723 secretory proteins that were found to occur in human body fluids only 309 (~ 43%) had signal peptides, 175 (~ 24%) were predicted to be secreted via non-classical pathway while the remaining 239 (~ 33%) were of intracellular origin (Supplemental TableS1 “Bodyfluids proteins” sheet). Of the 309 protein with a signal peptide, fibronectin, collagen alpha-1(I) and alpha-2(I) chain, SPARC, complement C3 were abundantly secreted by almost all 6 primary cell cultures and were also reported to be abundant in plasma and other body fluids [15-17]. Of the 175 proteins secreted via non-classical pathway, peripheral proteins such as vimentin, alpha enolase, annexins, myomesin and desomin were abundantly released by all 6 studied primary cell cultures. Of the 239 intracellular proteins circulating in the body fluids, actin, actinins, prelamin A/C, heat shock cognate 71 kDa protein and many glycolitic enzymes were abundantly released by all cultured primary cells (Supplemental TableS1 “Bodyfluids proteins” sheet).
If we omit the keratin proteins, which are probably contaminants, the most abundant cell released proteins that do not have signal peptides but still occurred in body fluids included vimentin, actin, filamin A, actinin, alpha enolase, prelamin-A/C, tropomyosin alpha-4 and moesin with total spectral counts ranging from 370 for moesin to 1531 for vimentin (Supplemental TableS1 “Bodyfluids proteins” sheet). These were followed by moderately abundant proteins such as annexins, galectins and some glycolytic enzymes. These proteins were released by all 6 primary cultures and seem ubiquitous in body fluids.
3.4 Contribution of the cell secretome to interstitial proteome
Of the 1054 proteins identified in the secretome of the 6 human primary cell cultures studied herein, 330 proteins (~32%) were not found in any of the latest comprehensive body fluid proteome lists including plasma proteome [15], CSF proteome [16] or urine proteome [17]. Omitting keratins and intracellular proteins as potential contaminants there were 67 proteins with a signal peptide and 23 proteins were of cell membrane origin but without signal peptides (Supplemental Table S1 sheet “Interstitial proteins”). Some of the most abundant proteins in this category included, fibrillin-2 (108 spectral counts), interstitial collagenases (91 spectral counts), combined laminins (209 spectral counts), stromelysin-2 (77 spectral counts), and pentraxin-related protein (68 spectral counts). The majority of these proteins play a role in cell adhesion and extracellular matrix remodeling and were not observed by others in plasma, CSF nor urine. Except for these truly secreted proteins, most of the remaining 239 proteins were of cytosolic and nuclear origin and were probably shed into the conditioned media due to cell death or via exosomes.
3.3 Contribution of the cell secretome to biomarker discovery
One of the main goals of secretome studies, and especially in the field of cancer research, is to discover biomarkers for diagnosis, prognosis and therapy monitoring. Given the number of differentially secreted proteins usually found between cancerous cells and their non-neoplastic counterpart cells, it is challenging to select the right candidate biomarkers among the hundreds of candidates for further validation in clinical samples [11,18,19]. First it is not known if otherwise normal cells in the body express these same set of candidates as well. Also it is unknown if the selected candidates are detected in body fluids, and which body fluid is ideal to target. As proof of principle we compared the secretome of two glioblastoma cell lines U87 and T98, an aggressive type of brain tumor cells, to the human secretome atlas assembled from the 6 healthy primary cells including primary astrocytes, the non-neoplastic counterpart for GBMs. Comparing GBMs to astrocytes, 440 proteins were detected in the condition media of the U87 and T98 but not in the condition medium of primary astrocyte. However, 281 proteins of these 440 proteins were unique to glioblastoma cell lines when compared to the rest of primary cells (see supplemental Table S2 “Glioma Biomarkers” sheet). Thirteen potential GBM biomarkers were selected form these 281 proteins based on their abundance and occurrence in body fluids. These are listed in Table 1 with their potential role in glioblastoma and location in body fluids. Three out of the 13 potential GBM biomarkers are accessible for measurement in all three body fluids (e.g. CSF, plasma and urine) including chitinase-3-like protein-1 (CHI3L1), semaphorin-7 and macrophage colony-stimulating factor-1 (MCSF). Another 3 potential GMB biomarkers seem accessible for measurement in CSF and plasma but not in urine and included insulin-like growth factor-binding protein-5 (IGFBP5), 72 kDa type IV collagenase (MMP2) and neuropilin-1. Two other potential GBM biomarkers, MHC class I antigen A2 and TGF-beta1-binding protein-1, are accessible for measurement in plasma and urine but not in CSF while peripherin, heat shock-related 70 kDa protein-2 and tumor necrosis factor receptor 11B might be accessible for measurement in urine only. The role of these proteins in glioblastoma is documented in the corresponding references listed in Table 1.
Table 1.
List of potential glioblastoma biomarkers
| Accession n° and Protein Name | Occurrence | Role in glioma | Ref |
|---|---|---|---|
| P24593 Insulin-like growth factor-binding protein 5 | CSF, Plasma | Progression | [39,40] |
| P36222 Chitinase-3-like protein 1 | CSF, Plasma, Urine | Invasion | [11,39-41] |
| O75326 Semaphorin-7A | CSF, Plasma, Urine | Progression | [42] |
| P08253 72 kDa type IV collagenase or MMP2 | CSF, Plasma | Migration | [43] |
| O14786 Neuropilin-1 | CSF, Plasma | Progression/invasion | [44] |
| P41219 Peripherin | Urine | Unknown | |
| P09603 Macrophage colony-stimulating factor-1 | CSF, Plasma, Urine | Invasion | [45] |
| P01892 MHC class I antigen A*2 | Plasma, Urine | Evade immune response | [46] |
| P54652 Heat shock-related 70 kDa protein 2 | Urine | Survival | [47] |
| P08254 Stromelysin-1 or MMP3 | ? | Invasion | [48] |
| Q58FF7 Putative heat shock protein HSP 90 | ? | Survival | [49] |
| Q14766 TGF-beta1-binding protein 1 | Plasma, Urine | Malignancy | [50] |
| O00300 Tumor necrosis factor receptor 11B | Urine | Survival | [51] |
| P33908 MAN1A1 | CSF, Plasma, Urine | Growth | [52] |
| P12111 Collagen alpha-3(VI) chain | CSF, Plasma, Urine | Infiltration | [53] |
| P04406 Glyceraldehyde-3-phosphate dehydrogenase | Plasma, Urine | Growth | [54] |
| P06748 Nucleophosmin | Plasma | Survival | [55] |
| P11413 Glucose-6-phosphate 1-dehydrogenase | Plasma | Tumorigenicity | [56] |
| O60568 PLOD3 | CSF | Unknown | |
| Q8NBJ7 Sulfatase-modifying factor 2 | CSF | Unknown | |
| Q9GZP0 Platelet-derived growth factor D | Urine | Survival/Mitogenic | [57] |
| Q15389 Angiopoietin-1 | Urine | Prognostic Marker | [58] |
3.4 Use of SILAC labeled cell secretome to quantify potential biomarkers in serum samples
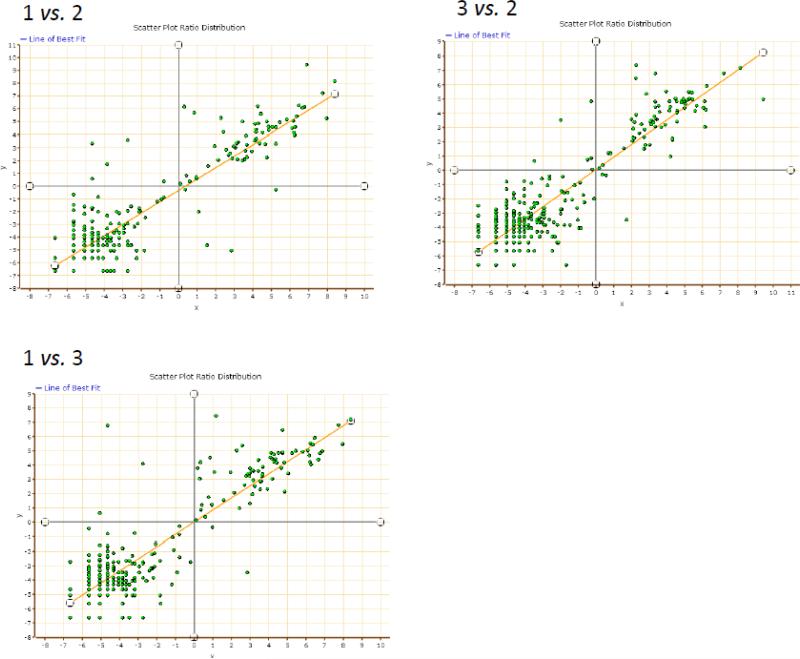
To see if we can target these brain tumor candidate biomarkers in human plasma for quantification we used super SILAC spike-in strategy as described in the method [10]. SILAC labeled secretomes from U87 and T98 cell lines were combined and aliquots were spiked into triplicate serum samples obtained from a healthy donor. As shown in Figure 3, the method was reproducible across the triplicate experiments with coefficient of variance better than 25%. The majority of the potential brain tumor biomarkers in Table 1 were accessible for measurement in human serum except for peripherin and putative heat shock protein HSP 90. The measurable biomarkers are listed in Table 2 with their relative ratios to the spike-in standard. As seen in Table 2, measurements were accurate across the triplicate serum samples with narrow standard error for each protein thus suggesting robustness of the method. All these potential GBM biomarkers occurred in low abundance in serum from a healthy donor and their levels are yet to be verified in serum of individuals with glioblastoma.
Figure 3. Regression plots of ratios measured across triplicate serum samples spiked each with the same amounts of stable isotope labeled U87 + T98 secretome.
Three aliquots from the same serum sample containing 100 μg of total proteins each were spiked with 50 μg SILAC labeled proteins collected from U87 and T98 condition media. Each plot represents a log regression plot of ratios between two separate experiments.
Table 2.
List of potential glioblastoma biomarkers and their amounts in normal serum relative to the spike-in SILAC labeled protein standard from SILAC labeled U87 and T98 secretome.
| Accession n° | Protein Name | Ratio* [serum/(U87+T98)] | peptide count |
|---|---|---|---|
| P24593 | Insulin-like growth factor-binding protein 5 | 0.01 ± 0.00 | 11 |
| P36222 | Chitinase-3-like protein 1 | 0.05 ± 0.04 | 4 |
| O75326 | Semaphorin-7A | 0.03 ± 0.00 | 1 |
| P08253 | 72 kDa type IV collagenase or MMP2 | 0.06 ± 0.01 | 10 |
| O14786 | Neuropilin-1 | 0.05 ± 0.00 | 5 |
| P41219 | Peripherin | nd | |
| P09603 | Macrophage colony-stimulating factor-1 | 0.00 | 2 |
| P01892 | MHC class I antigen A*2 | 0.05 ± 0.01 | 2 |
| P54652 | Heat shock-related 70 kDa protein 2 | 0.07 ± 0.00 | 8 |
| P08254 | Stromelysin-1 or MMP3 | 0.05 ± 0.00 | 1 |
| Q58FF7 | Putative heat shock protein HSP 90 | nd | |
| Q14766 | TGF-beta1-binding protein 1 | 0.16 ± 0.07 | 2 |
| O00300 | Tumor necrosis factor receptor 11B | # NA | 2 |
| P12111 | Collagen alpha-3(VI) chain | 0.01 ± 0.00 | 9 |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | 0.04 ± 0.00 | 10 |
| P06748 | Nucleophosmin | 0.04 ± 0.01 | 3 |
| P11413 | Glucose-6-phosphate 1-dehydrogenase | nd | |
| O60568 | PLOD3 | # NA | 7 |
| Q8NBJ7 | Sulfatase-modifying factor 2 | # NA | 2 |
| Q9GZP0 | Platelet-derived growth factor D | # NA | 3 |
| Q15389 | Angiopoietin-1 | # NA | 3 |
Ratio are determined between unlabeled peptide and its labeled partner originated from serum and form the spiked-in SILAC labeled (U87+T98) secretome respectively. Values are average obtained from triplicate experiments ± standard deviation. nd: not detected, # NA: not applicable to measure ratios because only spiked in labeled peptides are observed and the serum unlabeled peptides were undetectable.
Discussion
Even though body fluids are fairly accessible for biomarker studies, discovering diseases associated biomarkers directly in these body fluids has been challenging owing the wide dynamic range of their proteome and the inherent low abundance of potential biomarkers. Thus, efforts have been turned toward the source from where these biomarkers can originate before they can get diluted in the body fluids circulation. By studying secretome of given cells, investigators hope to define ideal biomarkers that can be traced back in the body fluids using a more sensitive and targeted analyses. Furthermore, seceretome studies of diseased cells versus their healthy counterpart cells can bring insight into the pathogenesis studied.
While efforts are being made to improve secretome studies in vivo [3,20] several in vitro studies have generated valuable data that can be interrogated in vivo using human tissue biopsies and/or human body fluids [3]. However, one of the challenges in in vitro secretome studies is discriminating truly cell released proteins from proteins that are released due to cell death or leakage during culturing. Most importantly, the identified biomarkers in the cell secretome should be measurable in at least one the major circulating body fluids (e.g. serum, CSF and urine). This is especially of interest in the context of biomarker discovery for monitoring disease progression and response to treatment. Therefore it is important to build a human secretome atlas database from as many cell types as possible and compare to the proteome of the major body fluids to determine how many secreted proteins can actually reach circulation. This secretome atlas database will also serve as a reference for past and future secretome studies of different diseases.
As preliminary initiative we sought to define the secretome of 6 healthy human primary cell cultures toward building a human secretome atlas. These primary cell cultures were obtained from healthy donors and included astrocytes, fibroblasts, bronchial epithelial cells, nasal epithelial cells, RPE cells and Schwann cells. These cells were chosen because they were currently studied in our laboratory in the context of different diseases such as brain tumors (astrocytes and GBM cell lines) [11], asthma (bronchial and nasal epithelial cells) [21], age related macular degeneration (RPE cells) [7], and neurofibromatosis type-1 (Schwann cells) [22]. Fibroblasts cells were included as a reference to the primary cell cultures because often primary cell cultures contain fibroblasts cells that might confound the true secretome.
Some of the interesting outcomes from the analysis of the secretome profiles of these 6 human primary cells cultures are cell specific secretome signatures and the distribution of the secreted proteins among the major body fluids (e.g. plasma, CSF and urine).
Cell specific secretome signatures were obtained for specific cell types. For example spondin-1 and ApoE proteins were uniquely secreted by astrocytes but not by any other cell type evaluated. These same proteins were also secreted by cultured mouse astrocytes [23] thus supporting their specificity to the astrocyte secretome. ApoE is ubiquitous in plasma and is secreted by liver as well. In this study we did not analyze the secretome of human primary hepatocytes but ApoE was indeed detected in the conditioned media of cultured human primary hepatocytes [24]. However, ApoE is also expressed in relatively large amounts in the brain and it has long been known to play a major role in neuronal development and repair after brain injury [25-27]. Polymorphisms in ApoE gene and especially in the epsilon 4 allele are strongly associated with risk of developing late onset Alzheimer's disease [28,29]. However, in our secretome profiling we were not able to distinguish between ApoE isoforms due the high sequence homology. Whether the amount of this protein changes with disease condition remains to be carefully examined. Nevertheless, a more recent study has shown an association between the plasma levels of ApoE and other apolipropteins with cognitive status in older individuals [30].
Human bronchial epithelial cells were characterized by specific secretion of mucins, a family of heavily O-glycosylated proteins that are secreted by several epithelial cells including bronchial epithelial cells and form a protective gelatinous film [31]. There are about 18 mucin isoforms secreted by different epithelial cell types. In normal healthy lung, mucin 1 and mucin 4 are expressed and released apically while the mucin 5AC and mucin 2 are expressed by superficial goblet cells [32]. In our secretome profile of human bronchial cells we only detected mucin 4 and 16 probably due the lack of goblet cells in the culture. The other types of mucins are usually expressed under different disease conditions such as in cystic fibrosis [33] and in chronic otitis media fluid [34]. Similarly RPE cells were characterized by specific secretion of complement component factors such complement factor H (CFH) and Complement C4-A that are known to be key regulators of the alternative pathway [35]. Interestingly, mutation in the genes encoding for complement components are strongly associated with age related macular degeneration thus suggesting potential specific role of these factors in the retinal epithelium [36,37]. In this article, we are not going to further describe the specificity of secreted protein in each cell type but the overall conclusion is that specific cell types secrete specific sets of proteins that play a major role in the biology and pathophysiology of the corresponding organ. Most of these proteins are accessible for measurement in body fluids.
Perhaps another interesting observation in this study is the contribution of the cell secretomes to the body fluids proteomes. About 67% of the cell secreted proteins studied herein seem to occur in at least one of the major human body fluids, further supporting the idea of using secretome profiles to define potential biomarkers that can be traced back in body fluids. This was tested using set of potential glioblastoma biomakers that can be targeted in serum samples using a spike-in strategy with a stable isotope labeled reference sample. Here we used normal serum as proof a principle, but this same technique can be used to screen serum samples from patients.
Intriguingly, about 30% of these body fluid circulating proteins were of intracellular origin and did not possess a signal peptide or were predicted to be secreted via non-classical pathway (Supplemental Table S1, sheet “Body fluid proteins”). The roles of these proteins in circulating body fluids are not known. Whether these proteins were released via an active unconventional mechanism [38] or just due to cell death and leakage in the body fluid remain to be carefully examined. Most of the proteins under this category were released by all cultured cells and they are relatively abundant in circulating body fluids [15]. For example vimentin and alpha enolase are one of the most abundantly released proteins by cultured cells. These proteins were also found to be relatively abundant in plasma as well, 28 μg/mL and 55 ng/mL, respectively [11]. These intracellular proteins that are released by all types of cultured cells might not be suitable for biomarker development because of lack of specificity.
Conclusions
Contribution of the cell secretome to biology and biomedical science is becoming apparent as the number of secretome profiles for different cell types and different disease conditions are becoming available. However with the increasing number of secretome catalogs, creation of a public domain for data mining is becoming highly important for meaningful biomarkers and therapeutic target discovery.
Supplementary Material
Highlights.
Cultured human primary cells secrete a variety of proteins
Specific secretome signature was observed for specific cell type
The majority of cell released proteins enter circulating body fluids
Several proteins are released in non-conventional pathway
Cancer cells have perturbed secretome
Acknowledgments
This work was partially supported by NIH grant K12 HL090020-05 and NIH core grants: NCMRR/NINDS 2R24HD050846-06 (National Center for Medical Rehabilitation Research) and IDDRC 5P30HD040677-10 (Intellectual and Developmental Disabilities Research Center) and by NIH NCRR UL1RR031988 (GWU-CNMC CTSI). The authors would like to thank Dr. Jyoti Jaiswal for his helpful discussion and inputs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eisele NA, Anderson DM. Host Defense and the Airway Epithelium: Frontline Responses That Protect against Bacterial Invasion and Pneumonia. J Pathog. 2011;2011:1–15. doi: 10.4061/2011/249802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 3.Brown KJ, Formolo CA, Seol H, et al. Advances in the proteomic investigation of the cell secretome. Expert Rev Proteomics. 2012;9:337–345. doi: 10.1586/epr.12.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eichelbaum K, Winter M, Diaz MB, Herzig S, Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat Biotechnol. 2012;30:984–990. doi: 10.1038/nbt.2356. [DOI] [PubMed] [Google Scholar]
- 5.Jung CS, Unterberg AW, Hartmann C. Diagnostic markers for glioblastoma. Histol Histopathol. 2011;26:1327–1341. doi: 10.14670/HH-26.1327. [DOI] [PubMed] [Google Scholar]
- 6.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 7.An E, Sen S, Park SK, Gordish-Dressman H, Hathout Y. Identification of novel substrates for the serine protease HTRA1 in the human RPE secretome. Invest Ophthalmol Vis Sci. 2010;51:3379–3386. doi: 10.1167/iovs.09-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freishtat RJ, Watson AM, Benton AS, et al. Asthmatic airway epithelium is intrinsically inflammatory and mitotically dyssynchronous. Am J Respir Cell Mol Biol. 2011;44:863–869. doi: 10.1165/rcmb.2010-0029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 10.Boersema PJ, Geiger T, Wisniewski JR, Mann M. Quantification of the N-glycosylated secretome by super-SILAC during breast cancer progression and in human blood samples. Mol Cell Proteomics. 2013;12:158–171. doi: 10.1074/mcp.M112.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Formolo CA, Williams R, Gordish-Dressman H, MacDonald TJ, Lee NH, Hathout Y. Secretome signature of invasive glioblastoma multiforme. J Proteome Res. 2011;10:3149–3159. doi: 10.1021/pr200210w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Min XJ. Evaluation of Computational Methods for Secreted Protein Prediction in Different Eukaryotes. J Proteomics Bioinform. 2010;3:143–147. [Google Scholar]
- 13.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 14.Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 15.Farrah T, Deutsch EW, Omenn GS, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10:M110 006353. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schutzer SE, Liu T, Natelson BH, et al. Establishing the proteome of normal human cerebrospinal fluid. PLoS One. 2010;5:e10980. doi: 10.1371/journal.pone.0010980. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marimuthu A, O'Meally RN, Chaerkady R, et al. A comprehensive map of the human urinary proteome. J Proteome Res. 2011;10:2734–2743. doi: 10.1021/pr2003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marimuthu A, Subbannayya Y, Sahasrabuddhe NA, et al. SILAC-based quantitative proteomic analysis of gastric cancer secretome. Proteomics Clin Appl. 2012 doi: 10.1002/prca.201200069. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu CC, Hsu CW, Chen CD, et al. Candidate serological biomarkers for cancer identified from the secretomes of 23 cancer cell lines and the human protein atlas. Mol Cell Proteomics. 2010;9:1100–1117. doi: 10.1074/mcp.M900398-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korf J, Huinink KD, Posthuma-Trumpie GA. Ultraslow microdialysis and microfiltration for in-line, on-line and off-line monitoring. Trends Biotechnol. 2010;28:150–158. doi: 10.1016/j.tibtech.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Stemmy EJ, Benton AS, Lerner J, Alcala S, Constant SL, Freishtat RJ. Extracellular cyclophilin levels associate with parameters of asthma in phenotypic clusters. J Asthma. 2011;48:986–993. doi: 10.3109/02770903.2011.623334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen HL, Seol H, Brown KJ, et al. Secretome Survey of Human Plexiform Neurofibroma Derived Schwann Cells Reveals a Secreted form of the RARRES1 Protein. Int J Mol Sci. 2012;13:9380–9399. doi: 10.3390/ijms13079380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowell JA, Johnson JA, Li L. Identification of astrocyte secreted proteins with a combination of shotgun proteomics and bioinformatics. J Proteome Res. 2009;8:4135–4143. doi: 10.1021/pr900248y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slany A, Haudek VJ, Zwickl H, et al. Cell characterization by proteome profiling applied to primary hepatocytes and hepatocyte cell lines Hep-G2 and Hep-3B. J Proteome Res. 2010;9:6–21. doi: 10.1021/pr900057t. [DOI] [PubMed] [Google Scholar]
- 25.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. 1988. [DOI] [PubMed] [Google Scholar]
- 26.Poirier J. Apolipoprotein E in the brain and its role in Alzheimer's disease. J Psychiatry Neurosci. 1996;21:128–134. [PMC free article] [PubMed] [Google Scholar]
- 27.Roses AD. Apolipoprotein E in neurology. Curr Opin Neurol. 1996;9:265–270. doi: 10.1097/00019052-199608000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Alonso Vilatela ME, Lopez Lopez M, Yescas Gomez P. Genetics of Alzheimer's Disease. Arch Med Res. 2012;43:622–631. doi: 10.1016/j.arcmed.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Sorbi S. Molecular genetics of Alzheimer's disease. Aging (Milano) 1993;5:417–425. doi: 10.1007/BF03324196. [DOI] [PubMed] [Google Scholar]
- 30.Song F, Poljak A, Crawford J, et al. Plasma apolipoprotein levels are associated with cognitive status and decline in a community cohort of older individuals. PLoS One. 2012;7:e34078. doi: 10.1371/journal.pone.0034078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marin F, Luquet G, Marie B, Medakovic D. Molluscan shell proteins: primary structure, origin, and evolution. Curr Top Dev Biol. 2008;80:209–276. doi: 10.1016/S0070-2153(07)80006-8. [DOI] [PubMed] [Google Scholar]
- 32.Voynow JA, Gendler SJ, Rose MC. Regulation of mucin genes in chronic inflammatory airway diseases. Am J Respir Cell Mol Biol. 2006;34:661–665. doi: 10.1165/rcmb.2006-0035SF. [DOI] [PubMed] [Google Scholar]
- 33.Kreda SM, Davis CW, Rose MC. CFTR, mucins, and mucus obstruction in cystic fibrosis. Cold Spring Harb Perspect Med. 2012;2:a009589. doi: 10.1101/cshperspect.a009589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preciado D, Goyal S, Rahimi M, et al. MUC5B Is the predominant mucin glycoprotein in chronic otitis media fluid. Pediatr Res. 2010;68:231–236. doi: 10.1203/PDR.0b013e3181eb2ecc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.James K. Complement: activation, consequences, and control. Am J Med Technol. 1982;48:735–742. [PubMed] [Google Scholar]
- 36.Bradley DT, Zipfel PF, Hughes AE. Complement in age-related macular degeneration: a focus on function. Eye (Lond) 2011;25:683–693. doi: 10.1038/eye.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gehrs KM, Anderson DH, Johnson LV, Hageman GS. Age-related macular degeneration--emerging pathogenetic and therapeutic concepts. Ann Med. 2006;38:450–471. doi: 10.1080/07853890600946724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nickel W. Unconventional secretory routes: direct protein export across the plasma membrane of mammalian cells. Traffic. 2005;6:607–614. doi: 10.1111/j.1600-0854.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- 39.Santosh V, Arivazhagan A, Sreekanthreddy P, et al. Grade-specific expression of insulin-like growth factor-binding proteins-2, -3, and -5 in astrocytomas: IGFBP-3 emerges as a strong predictor of survival in patients with newly diagnosed glioblastoma. Cancer Epidemiol Biomarkers Prev. 2010;19:1399–1408. doi: 10.1158/1055-9965.EPI-09-1213. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Zhang W, Fuller GN. Overexpression of IGFBP5, but not IGFBP3, correlates with the histologic grade of human diffuse glioma: a tissue microarray and immunohistochemical study. Technol Cancer Res Treat. 2006;5:195–199. doi: 10.1177/153303460600500303. [DOI] [PubMed] [Google Scholar]
- 41.Ku BM, Lee YK, Ryu J, et al. CHI3L1 (YKL-40) is expressed in human gliomas and regulates the invasion, growth and survival of glioma cells. Int J Cancer. 2010;128:1316–1326. doi: 10.1002/ijc.25466. [DOI] [PubMed] [Google Scholar]
- 42.Law JW, Lee AY. The role of semaphorins and their receptors in gliomas. J Signal Transduct. 2012;2012:1–14. doi: 10.1155/2012/902854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mou L, Kang Y, Zhou Y, Zeng Q, Song H, Wang R. Neurokinin-1 receptor directly mediates glioma cell migration by up-regulation of matrix metalloproteinase-2 (MMP-2) and membrane type 1 matrix metalloproteinase (MT1-MMP). J Biol Chem. 2013;288:306–318. doi: 10.1074/jbc.M112.389783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu B, Guo P, Bar-Joseph I, et al. Neuropilin-1 promotes human glioma progression through potentiating the activity of the HGF/SF autocrine pathway. Oncogene. 2007;26:5577–5586. doi: 10.1038/sj.onc.1210348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coniglio SJ, Eugenin E, Dobrenis K, et al. Microglial stimulation of glioblastoma invasion involves epidermal growth factor receptor (EGFR) and colony stimulating factor 1 receptor (CSF-1R) signaling. Mol Med. 2012;18:519–527. doi: 10.2119/molmed.2011.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wischhusen J, Friese MA, Mittelbronn M, Meyermann R, Weller M. HLA-E protects glioma cells from NKG2D-mediated immune responses in vitro: implications for immune escape in vivo. J Neuropathol Exp Neurol. 2005;64:523–528. doi: 10.1093/jnen/64.6.523. [DOI] [PubMed] [Google Scholar]
- 47.Li G, Xu Y, Guan D, Liu Z, Liu DX. HSP70 protein promotes survival of C6 and U87 glioma cells by inhibition of ATF5 degradation. J Biol Chem. 2011;286:20251–20259. doi: 10.1074/jbc.M110.211771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laurent M, Martinerie C, Thibout H, et al. NOVH increases MMP3 expression and cell migration in glioblastoma cells via a PDGFR-alpha-dependent mechanism. FASEB J. 2003;17:1919–1921. doi: 10.1096/fj.02-1023fje. [DOI] [PubMed] [Google Scholar]
- 49.Ohba S, Hirose Y, Yoshida K, Yazaki T, Kawase T. Inhibition of 90-kD heat shock protein potentiates the cytotoxicity of chemotherapeutic agents in human glioma cells. J Neurosurg. 2010;112:33–42. doi: 10.3171/2009.3.JNS081146. [DOI] [PubMed] [Google Scholar]
- 50.Tritschler I, Gramatzki D, Capper D, et al. Modulation of TGF-beta activity by latent TGF-beta-binding protein 1 in human malignant glioma cells. Int J Cancer. 2009;125:530–540. doi: 10.1002/ijc.24443. [DOI] [PubMed] [Google Scholar]
- 51.Naumann U, Wick W, Beschorner R, Meyermann R, Weller M. Expression and functional activity of osteoprotegerin in human malignant gliomas. Acta Neuropathol. 2004;107:17–22. doi: 10.1007/s00401-003-0772-4. [DOI] [PubMed] [Google Scholar]
- 52.Fiaux H, Popowycz F, Favre S, et al. Functionalized pyrrolidines inhibit alpha-mannosidase activity and growth of human glioblastoma and melanoma cells. J Med Chem. 2005;48:4237–4246. doi: 10.1021/jm0409019. [DOI] [PubMed] [Google Scholar]
- 53.Han J, Daniel JC, Pappas GD. Expression of type VI collagen during glioblastoma cell invasion in brain tissue cultures. Cancer Lett. 1995;88:127–132. doi: 10.1016/0304-3835(94)03627-u. [DOI] [PubMed] [Google Scholar]
- 54.Appelskog IB, Ammerpohl O, Svechnikova IG, Lui WO, Almqvist PM, Ekstrom TJ. Histone deacetylase inhibitor 4-phenylbutyrate suppresses GAPDH mRNA expression in glioma cells. Int J Oncol. 2004;24:1419–1425. [PubMed] [Google Scholar]
- 55.Gimenez M, Marie SK, Oba-Shinjo SM, et al. Quantitative proteomic analysis and functional studies reveal that nucleophosmin is involved in cell death in glioblastoma cell line transfected with siRNA. Proteomics. 2012;12:2632–2640. doi: 10.1002/pmic.201200034. [DOI] [PubMed] [Google Scholar]
- 56.Ramao A, Gimenez M, Laure HJ, et al. Changes in the expression of proteins associated with aerobic glycolysis and cell migration are involved in tumorigenic ability of two glioma cell lines. Proteome Sci. 2012;10:1–12. doi: 10.1186/1477-5956-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res. 2002;62:3729–3735. [PubMed] [Google Scholar]
- 58.Sie M, Wagemakers M, Molema G, Mooij JJ, de Bont ES, den Dunnen WF. The angiopoietin 1/angiopoietin 2 balance as a prognostic marker in primary glioblastoma multiforme. J Neurosurg. 2009;110:147–155. doi: 10.3171/2008.6.17612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.