Abstract
Hepatitis C virus (HCV) remains a major medical problem. In-depth study of HCV pathogenesis and immune responses is hampered by the lack of suitable small animal models. The narrow host range of HCV remains incompletely understood. We demonstrate that the entire HCV life-cycle can be recapitulated in mouse cells. We show that antiviral signaling interferes with HCV RNA replication in mouse cells. We were able to infect mouse cells expressing human CD81 and occludin (OCLN) - the minimal set of entry factor factors required for HCV uptake into mouse cells. Infected mouse cells sustain HCV RNA replication in the presence of miR122 and release infectious particles when mouse apoE is supplied. Our data demonstrate that the barriers of HCV interspecies transmission can be overcome by engineering a suitable cellular environment and provide a blue-print towards constructing a small animal model for HCV infection.
Keywords: hepatitis C virus, innate immunity, viral entry, viral assembly, species tropism, HCV animal model
Introduction
Hepatitis C virus (HCV), the causative agent of classically defined non-A, non-B hepatitis, is highly prevalent, with approximately 3% of the worldwide population infected. Acute HCV infection often evades immune-mediated clearance and results in chronic, life-long persistence. Chronic infections can have severe health consequences, including hepatitis, cirrhosis, liver failure, and hepatocellular carcinoma. Treatment options are limited and are often plagued with serious side effects. A preventative or therapeutic vaccine for HCV does not exist.
HCV has been notoriously difficult to study in cell culture and in vivo systems (reviewed in (Dustin and Rice, 2007)), which has hampered development of more tolerable and effective therapies. Few species are known to be susceptible to HCV infection, including humans, chimpanzees and tree shrews (reviewed in (Bukh, 2012)). The HCV life cycle is blocked or insufficiently supported at multiple steps in murine cells and the barriers for interspecies transmission remain poorly defined (reviewed in (Sandmann and Ploss, 2013)). To enter hepatocytes, HCV utilizes several host proteins including glycosaminoglycans (GAGs) (Barth et al., 2003; Koutsoudakis et al., 2006), the low density lipoprotein receptor (LDLR) (Agnello et al., 1999; Molina et al., 2007; Monazahian et al., 1999; Owen et al., 2009),, the high density lipoprotein receptor scavenger receptor class B type I (SCARB1; (Scarselli et al., 2002)), tetraspanin CD81 (Pileri et al., 1998), and two tight junction (TJ) proteins, claudin-1 (CLDN1; (Evans et al., 2007)) and occludin (OCLN; (Liu et al., 2009; Ploss et al., 2009)). CD81, SCARB1, CLDN1 and OCLN comprise the minimal set of host factors required for HCV uptake into mouse cell lines, where only CD81 and OCLN must be of human origin to overcome the species barrier in mouse cell lines (Ploss et al., 2009) and genetically humanized mice (Dorner et al., 2011). More recently, additional host factors including the cholesterol absorption receptor Niemann-Pick C1-like 1 (NPC1L1; (Sainz et al., 2012)) and two receptor tyrosine kinases, epidermal growth factor receptor (EGFR; (Lupberger et al., 2011)) and EphrinA2 (Lupberger et al., 2011) have been implicated in the viral uptake pathway into human cells. Mouse and human EGFR and EphrinA2 both share approximately 90% amino acid sequence identity, suggesting that functionality within the HCV entry pathway maybe conserved across species. Interestingly, NPC1L1 is not expressed in the mouse liver (Altmann et al., 2004). Nonetheless, it was demonstrated that mice expressing human CD81 and OCLN support HCV uptake into mouse hepatocytes (Dorner et al., 2011) suggesting that lack of NPC1L1 does not limit HCV infection of murine cells.
Following uptake into murine cells, HCV RNA is translated (Dorner et al., 2011; McCaffrey et al., 2002) but does not appear to accumulate, suggesting that viral RNA replication is impaired in mouse cells. HCV RNA replicons, which are selectable HCV RNA genomes, can replicate in murine cell lines (Frentzen et al., 2011; Uprichard et al., 2006; Zhu et al., 2003), demonstrating that interfering dominant negative inhibitors do not appear to exist. These observations also suggest that murine orthologs of host factors critical for HCV replication appear to cooperate sufficiently with the viral replication machinery.
Several reports have hinted that antiviral cellular defenses limit HCV genome propagation. For example, both human hepatoma cells (Blight et al., 2002; Sumpter et al., 2005) as well as human primary hepatocytes impaired in innate immunity are more conducive to viral replication (Andrus et al., 2011; Marukian et al., 2011). It was reported that HCV can counteract innate immune defenses in human cells, e.g. by cleavage of the mitochondrial antiviral signal protein (MAVS) (Meylan et al., 2005) or Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF or TICAM; (Li et al., 2005)). However, these mechanisms may not work with equal efficiency in all cell types and species. In fact, it was recently shown that MAVS from multiple primates is resistant to inhibition by the HCV NS3/4A protease (Patel et al., 2012). This resistance maps to single amino acid changes within the protease cleavage site in MAVS, which protect MAVS proteolytic cleavage by the NS3/4A protease. In murine cells, the NS3/4A protease cleavage motifs of mouse MAVS and TRIF are conserved, but it has not been formally proven that the viral protease actually cleaves the murine ortholog and that this targeted proteolysis translates into increased RNA replication. Thus, it is conceivable that different kinetics and/or a greater magnitude of virally induced innate defenses prevent induction or maintenance of HCV RNA replication in mouse cells. This hypothesis is supported by the previous observation that mouse embryonic fibroblasts (MEFs) with targeted disruptions protein kinase R (PKR; (Chang et al., 2006)) or interferon regulatory factor 3 (IRF3; (Lin et al., 2010b)) are more conducive to HCV RNA replication. In contrast, infectious HCV particles can assemble and be released in mouse cell lines if apolipoprotein E is sufficiently expressed (Long et al., 2011b) suggesting that later stages in the HCV life cycle are not blocked in mouse cells.
In this study we attempted to recapitulate the entire HCV life-cycle in mouse cells. Specifically, we demonstrate that the HCV NS3/4A serine protease is capable of cleaving MAVS and TRIF in mouse cells, thereby creating an environment which more efficiently supports HCV RNA replication. However, this evasion mechanism is not sufficient to readily overcome host defenses interfering with HCV RNA replication in mouse cells. MEFs derived from mouse strains harboring targeted deletions in genes critically involved in type I and III interferon signaling can improve the efficiency of HCV RNA replication, especially in the presence of the liver-specific microRNA 122 (miR122), a host factor that was previously shown to be important for HCV RNA replication (Jopling et al., 2005). In accordance with a previous study (Long et al., 2011a; Scull and Ploss, 2012), expression of human or mouse apoE in mouse fibroblasts infected with a selectable infectious HCV genome with apoE results in production of infectious particles. Taken together, these data show that all steps of the HCV life cycle can be recapitulated in murine cells, providing the framework for an inbred mouse model of HCV infection.
Results
Cleavage of MAVS and TRIF improves HCV RNA replication in mouse cells
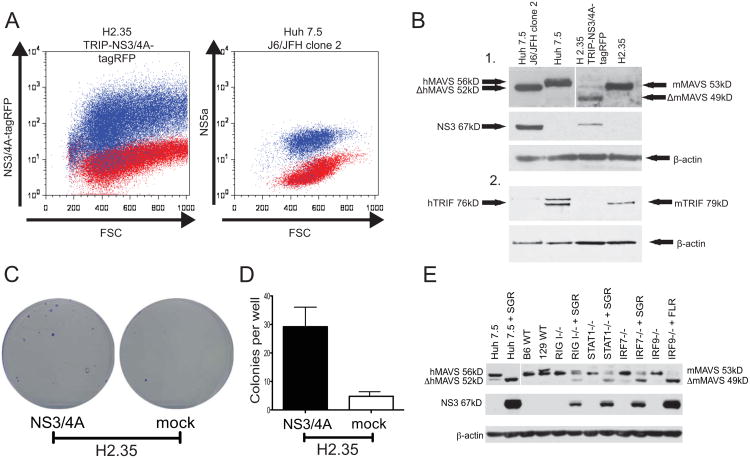
To test whether the NS3/4A serine protease is capable of cleaving mouse MAVS and TRIF, two of the known targets of NS3/4A in human cells, we transduced H2.35 mouse hepatoma cells with a lentivirus TRIP-NS3/4A-TagRFPpuro expressing an enzymatically active HCV NS3/4A (JFH-1) with a TagRFP-puromycin fusion protein expressed in a downstream cistron. This allowed us to sort flowcytometrically a mouse cell population expressing similar levels of NS3/4A to human Huh7.5 cells infected with the robustly replicating J6/JFH1 clone 2 virus (Walters et al., 2009) (Fig. 1A and B). In the parental H2.35 and Huh7.5 cells MAVS and TRIF can be detected using specific antibodies for the respective proteins. In mouse cells overexpressing NS3/4A and Huh7.5 cells infected with HCV, the proteolytic products of mouse and human MAVS and TRIF were readily detectable (Fig. 1B), suggesting that HCV NS3/4A protease mediated immune evasion occurs in mouse cells.
Figure 1. The HCV NS3/4A serine protease efficiently cleaves mouse MAVS and TRIF.
A. (left) Overlay plot of naïve H2.35 cells (red) or H2.35 cells transduced with VSVg-pseudotyped TRIP-NS3/4A (JFH1)-puro2atagRFP and sorted for high expression of the viral protease (blue). (right) Overlay plot of naïve (red) or J6/JFH1 clone 2-infected (blue) Huh7.5 cells stained for HCV NS5A 72 hours following infection. B. Western blot of lysate of naïve and transduced (mouse) and infected (human) cells detecting mouse and human MAVS, TRIF and β-actin. C. Selection of SGR-bsd-JFH1-containing colonies in H2.35/TRIP-NS3/4A-tagRFP and the parental H2.35 cells Representative wells showing colonies of crystal violet stained cells. D. Quantitation of crystal violet-positive colonies per well shown from three independent experiments. E. Western blot detecting mouse and human MAVS in either control or replicon-containing Huh7.5 cells and iMEFs. SGR = SGR-Bsd-JFH1; FLR = FL-Bsd-JFH1.
To assess whether NS3/4A-mediated cleavage of mouse MAVS and TRIF would render mouse cells more permissive to HCV RNA replication, we transfected H2.35 cells with TRIP-NS3/4A-TagRFPpuro and selected for cells highly expressing the HCV protease. We then transfected NS3/4A-expressing cells and H2.35 controls with a selectable subgenomic JFH1 replicon (SGR-bsd-JFH1) encoding the blasticidin resistance gene blasticidin-S-deaminase (bsd) between NS5A/5B. When selected with blasticidin, H2.35 cells expressing NS3/4A formed 8-10 times more colonies (Fig. 1C and D) than NS3/4A-negative controls, suggesting that NS3/4A expression significantly blunted innate antiviral signaling in mouse hepatoma cells, thereby boosting HCV replication.
Blunting antiviral innate defenses improves HCV RNA replication in mouse cells
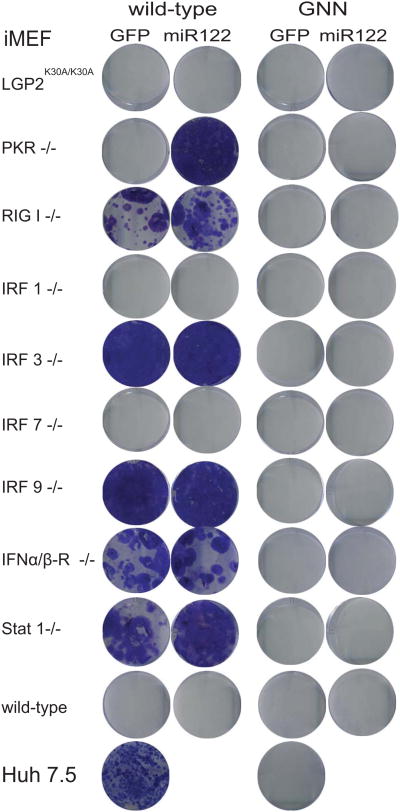
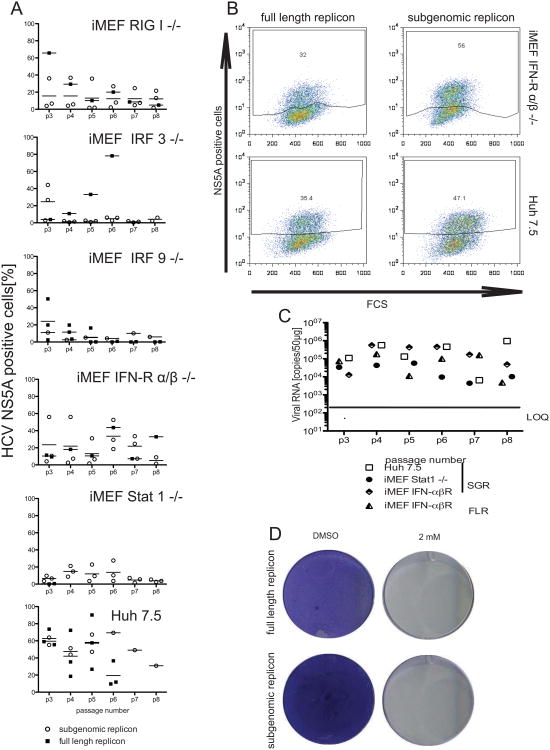
HCV can replicate in mouse cells lines suggesting that murine orthologs of host factors critical for HCV replication cooperate sufficiently with the virally encoded replication machinery. HCV replication is augmented in mouse cells lacking expression of the protein kinase R (Chang et al., 2006) or interferon regulatory factor 3 (IRF3) (Lin et al., 2010b), HCV replication is augmented suggesting that antiviral immunity limits viral replication in mouse cells. Our data above indicate HCVs antiviral evasion mechanisms are functional, but not readily capable of overcoming innate cellular defenses. To identify a murine environment that is more conducive for robust HCV replication we surveyed a larger panel of MEFs deficient in genes critically involved in type I and III interferon responses, including PKR (Balachandran et al., 1998), RIG-I (Kato et al., 2005), IRF-1 (Kimura et al., 1994; Matsuyama et al., 1993), IRF-3 (Sato et al., 2000), IRF-7 (Honda et al., 2005), IRF-9 (Kimura et al., 1996), STAT1 (Durbin et al., 1996), IFNαβ receptor (Muller et al., 1994) or a knock-in strain with a dead mutation in the helicase of LGP2 (Satoh et al., 2010) for their ability to replicate HCV RNA. We generated immortalized MEFs (iMEFs) from these strains as they provide a clean knock-out background for the gene of interest. Liver-specific micro-RNA 122 (miR122) is a critical host factor regulating HCV RNA translation and replication (Henke et al., 2008; Jopling et al., 2005). It has previously been demonstrated that exogenous administration of miR122 can boost HCV RNA replication in non-hepatic human (Da Costa et al., 2012b) and murine cells (Lin et al., 2010a). In order to more closely mimic the murine hepatic environment, we transduced iMEF lines from the various innate immune knockout backgrounds with a lentivirus expressing miR122. This resulted in stable expression of miR122 to similar levels detectable in highly permissive Huh7.5 cells (data not shown). iMEFs expressing miR122 or control cells were transfected with SGR-bsd-JFH1 and drug selection was applied. Transfection of the SGR-bsd-JFH1 replicons into iMEFs derived from wild-type control mice resulted in negligible numbers of blasicidin resistant cells irrespective of the presence of miR122 (Fig. 2). In contrast, in the absence of RIG-I, IRF-3, IRF-9, IFNαβR, STAT1 but not IRF-1 or IRF-7 SGR-bsd-JFH1 conferred resistance to blasticidin selection, indicating that blunting specific antiviral signaling pathways improves HCV RNA replication. iMEF lines stably replicating HCV RNA expressed HCV NS3 which correlated with efficient cleavage of mouse MAVS (Fig. 1E). In order to provide additional evidence for HCV RNA replication we quantified HCV NS5A-positive cells by flow-cytometry over multiple passages in iMEF replicon lines harboring SGR-bsd-JFH1 and maintained under bsd selection (Fig. 3). HCV NS5A protein was detectable in iMEFs lacking RIG-I, IRF-3, IRF-9, IFNαβR and STAT1 harboring either subgenomic JFH1 or full-length (FL) J6-JFH1 replicons (Fig 3A and B; and data not shown). The frequency of NS5A positive cells was expectedly lower in the iMEFs lines harboring SGR-bsd-JFH1 compared to Huh7.5 cells replicating either full-length (FL) or subgenomic replicon-based (SGR) HCV genomes. However, HCV RNA was readily detectable in STAT1-/- and IFNαβR-/- iMEFs (data not shown) harboring SGR or FL replicons at levels comparable to or only slightly lower than replicon-containing Huh7.5 cells (Fig. 3C). When treated with a 2′C methyl adenosine (2′CMA), an inhibitor of the HCV NS5B polymerase, STAT1-/- iMEFs that previously stably replicated SGR or FL genomes did not withstood blasticidin selection, pointing to robust HCV replication on the STAT1-/- background. For all subsequent experiments we primarily used STAT1-/- iMEFs because of their superior stable growth under bsd selection when harboring replicating HCV RNA. Taken together, these data demonstrate that HCV replicates efficiently in non-hepatic mouse cell lines with disrupted type I/III IFN signaling pathways.
Figure 2. Innate immune deficiencies and expression of miR122 facilitate replication of HCV replicons in mouse fibroblasts.
Immortalized MEFs (iMEFs) of the indicated genotypes were lentivirally transduced with GFP or miR122 and then transfected with replication-competent (wild-type) or defective (GNN) bicistronic subgenomic (JFH1) constructs expressing blasticidin S deaminase (bsd) gene that confers resistance blasticidin selection. Cells were selected in blasticidin-containing medium and stained with crystal violet after fixation.
Figure 3. Persistent HCV RNA replication in mouse fibroblasts on immunodeficient backgrounds.
Immortalized MEFs of the indicated genotypes or Huh7.5 cells were transfected with full-length J6/JFH1 (FLR) or subgenomic JFH1 replicons expressing blasticidin S deaminase and cell populations selected in the presence of blasticidin. A. Quantification of HCV NS5A antigen positive cells over six passages following HCV RNA transfection. B. Representative flow cytometry plots of IFNαβR-/- and Huh7.5 cells stably harboring FLR- or SGR-stained for HCV NS5A antigen. C. Quantification of HCV RNA over several passages in Huh7.5 and STAT1-/- iMEFs stably replicating SGRs or FLRs as indicated. D. STAT1-/- iMEFs stably replicating SGRs or FLRs were grown in the presence or absence of 2′CMA (2mM) and stained with crystal violet after fixation. LOQ = limit of quantitation
Initiation of HCV life-cycle following infection of mouse cells expressing human CD81 and OCLN with HCVcc
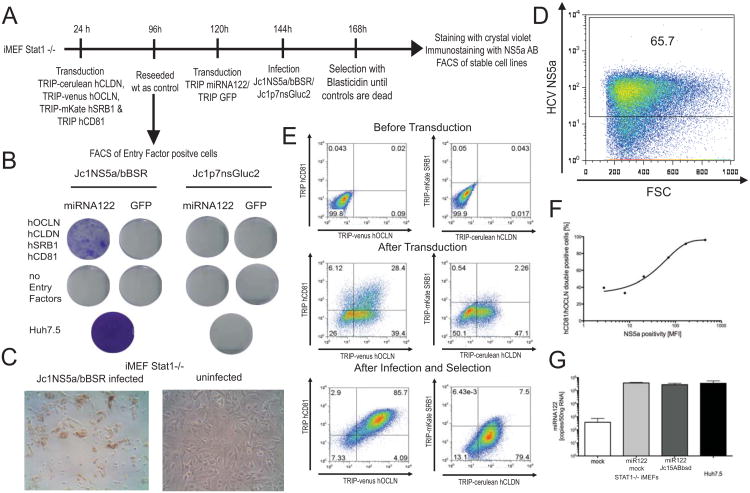
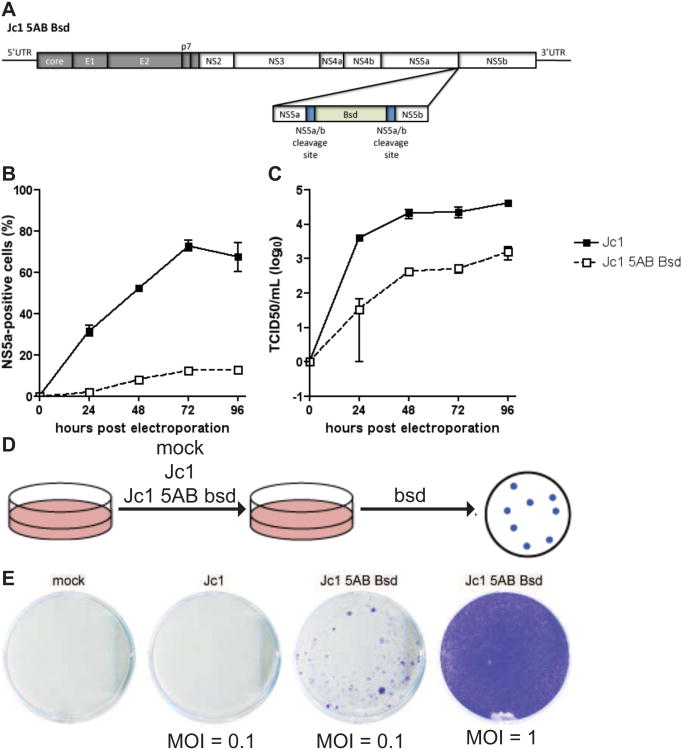
Transfection is likely to introduce considerably more HCV RNA into a cell as compared to a natural infection process. Thus, HCV virion uptake via receptor-mediated endocytosis may prevent putative hyperactivation of antiviral signaling pathways. It was previously demonstrated that CD81, SCARB1, CLDN1 and OCLN are required for HCV entry into rodent cells and CD81 and OCLN constitute the minimal set of human genes required for viral uptake in vitro and in vivo (Dorner et al., 2011; Ploss et al., 2009). To determine whether the HCV life-cycle could be established in iMEFs defective in STAT1, we transduced these cells with human fluorescently-tagged CD81, SCARB1, CLDN1 and OCLN and subsequently with miR122 (Fig. 4A). Expression of human entry factors was confirmed by flow cytometry 3 days following transduction. Approximately 30% of all cells expressed both human CD81 and OCLN and a subset of those in addition human CLDN1 and SCARB1 (Fig 4E middle panels). Similarly, lentiviral delivery of miR122 resulted in robust expression of the microRNA reaching levels equivalent to those in highly permissive Huh7.5 cells (Fig. 4G). To visualize HCV infection more readily we constructed a recombinant HCV genome expressing bsd between NS5A and NS5B, Jc1-5AB bsd (Fig. 5A and (Horwitz et al., 2013)). Jc1-5AB bsd replicates following electroporation in Huh7.5 cells (Fig. 5B), produces infectious particles (Fig. 5C), both albeit lower than the parental Jc1 genome and renders cells replicating the genome resistant to blasticidin selection (Fig. 5D, E).
Figure 4. Stable HCV RNA replication in mouse fibroblasts expressing HCV entry factors and miR122 following infection with Jc1-5AB bsd.
A. Schematic of the experimental set-up. B. Crystal violet stained blasticidin-resistant colonies of the indicated cell populations infected with Jc1-5AB bsd or Jc1p7nsGluc2A. C. Immunohistochemical staining of STAT1-/- iMEFs expressing human HCV entry factors stably infected with Jc1-5AB bsd or non-infected control cells. D. Flow cytometry quantification of HCV NS5A antigen in STAT1-/- iMEFs expressing human HCV entry factors stably infected with Jc1-5AB bsd. E. Flow cytometric analysis of ectopic expression of human CD81, OCLN, CLDN1 and SCARB1 before (upper panels) and after transduction (middle panels) and after infection and selection (lower panels). F. Correlation between human CD81/OCLN and NS5A antigen positivity in Jc1-5AB bsd infected STAT1-/- iMEFs. G. miR122 expression in Jc1-5AB bsd infected or non-infected STAT1-/- iMEFs transduced with miR122 and HCV entry factors, untreated iMEFs and Huh7.5 cells.
Figure 5. Construction of selectable HCVcc by insertion of bsd between NS5A and NS5B.
A. Genome structure of Jc1-5AB bsd illustrating the insertion a blasticidin resistance gene (bsd) by duplicating the NS3/4A protease cleavage site between NS5A and NS5B (NS5A/B CS) flanking the bsd insertion. B. HCV replication following electroporation of in vitro-transcribed RNA of Jc1 or Jc1-5AB bsd into Huh7.5 cells as measured by flow cytometry. C. Longitudinal virus production as measured by end-point limiting dilution following electroporation of Jc1 and Js1-5AB bsd RNA into Huh7.5 cells. D. Schematic depiction of the experimental set-up for the selection of blasticidin resistant colonies. E. Huh7.5 cells infected with Jc1 (MOI = 0.1), Jc1-5AB bsd (MOIs = 0.1 or 1), selected with bsd and stained with crystal violet following fixation.
Following infection of human entry factor- and miR122-transduced STAT1-/- iMEFs with a control virus or Jc1-5AB bsd and application of selection pressure, only cells that expressed HCV entry factors and miR122 and were infected with Jc1-5AB bsd were resistant to the antibiotic (Fig. 4B). Further analysis of the cells that, stably replicated Jc1-5AB bsd revealed that the majority of cells (> 60%) expressed high levels of HCV NS5A antigen -- evidenced by immune-histochemical staining (Fig. 4C) and flow-cytometric quantitation (Fig. 4D). Notably, following bsd selection, expression of HCV entry factors was skewed towards substantially higher expression, in particular human CD81 and human OCLN (> 85% cells) suggesting a beneficial effect presumably due to de novo infection and spread within the culture (Fig. 4E, bottom panels). Indeed, the NS5A staining intensity correlated with the frequency of human CD81/OCLN double positive cells (Fig. 4F). These data demonstrate that HCV replication can be initiated and maintained in murine cells expressing HCV entry factors following uptake of infectious HCV.
Production of infectious virions in mouse cell lines
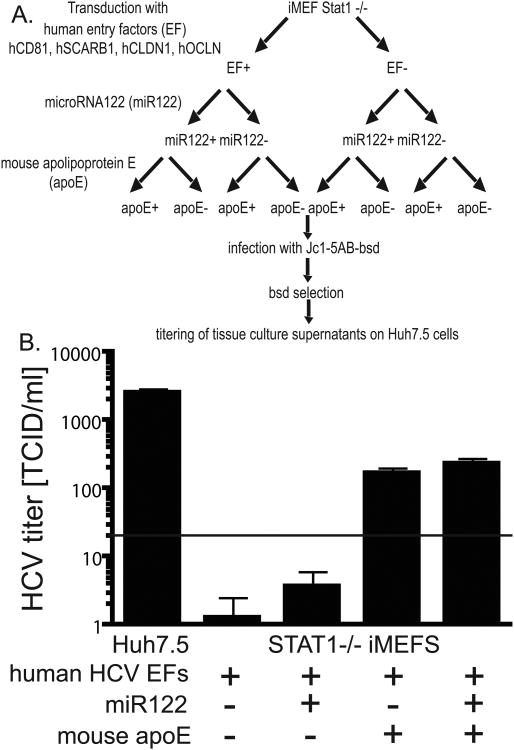
It was recently demonstrated that mouse hepatoma cells stably harboring a subgenomic HCV replicon can produce infectious particles when transcomplemented with the missing HCV structural proteins (Long et al., 2011a). This further affirms that virion production is dependent on an intact VLDL pathway, in particular, expression of apoE, irrespective of human or mouse species origin. We aimed to determine whether mouse cells of non-hepatic origin supplemented with mouse apoE are capable of supporting release of infectious HCV virions. iMEFs lacking STAT1 or Huh7.5 cells were transduced with HCV entry factors, miR122 and/or mouse apoE, infected with Jc1-5AB bsd and subjected to bsd selection (Fig. 6A). To determine whether the STAT1-/- iMEFs supplemented with hCD81, hSCARB1, hCLDN1 and hOCLN, miR122 and/or mouse apoE produced infectious particles, we passed cell culture supernatants on naïve Huh7.5 cells. Only iMEFs expressing apoE released infectious particles, albeit at low levels. These data confirm that mouse cells even of non-hepatic origin can produce infectious HCV simply by overexpressing apoE (Long et al., 2011a). We demonstrate for the first time that this can be achieved with a mono-cistronic HCV genome, which expresses the HCV gene products presumably in stoichiometric ratios as opposed to mouse cells harboring a subgenomic replicon in which structural proteins are overexpressed.
Figure 6. Production of infectious HCV in mouse fibroblasts supplemented with apoE.
A. Schematic of the experimental set-up. B. Infection of naïve Huh7.5 cells with supernatants from cultures of STAT1-/- iMEFs transduced with mouse apoE, eGFP, mock transduced iMEFs, and Huh7.5. NS5A staining was performed 72 h post infection and the number of infectious virions quantified in a limiting dilution assay. LOD = limit of detection
Discussion
HCVs narrow host range, limited to humans and chimpanzees, is incompletely understood. Given the NIH moratorium on ‘non-essential’ chimpanzee research that will severely limit future chimpanzee HCV research, alternative animal models are urgently needed to study HCV associated pathogenesis and to aid HCV vaccine development. Different approaches have been taken to construct small animal models for HCV infection (reviewed in (Ploss and Rice, 2009)): efforts have been made to adapt HCV to infect cells of non-human origin. Alternatively, the murine host can be engineered to provide an environment that is conducive to HCV infection. One approach requires humanization of relevant human tissue compartments, in particular the liver and immune system, via xenotransplantation resulting in so called ‘humanized mice.’ Human liver chimeric mice, in particular, have proven useful to study HCV and to test anti-viral regimens (reviewed in (Meuleman and Leroux-Roels, 2008)). However, generation of xenotransplanted mice requires a high degree of technical skills, is costly and low in throughput, which limits their practical use. In an effort to more systematically define host determinants required to overcome species barriers we have identified the combination of human CD81 and OCLN as the minimal set human cellular factors required to facilitate entry into rodent cells (Ploss et al., 2009). Capitalizing on this discovery we subsequently translated these in vitro observations to a mouse model supporting viral uptake (Dorner et al., 2011; Dorner et al., 2012). While this alternative host adaptation approach demonstrates how barriers of interspecies transmission can be overcome genetically, this particular humanized mouse model can only be used to monitor early steps in the viral life-cycle in vivo as it does not support HCV replication and virion assembly. These data are consistent with previous observations demonstrating that HCV RNA cannot readily replicate in mouse hepatocytes in vivo (McCaffrey et al., 2002). However, dominant-negative restriction factors do not seem to be present in mouse cells as HCV is generally capable, albeit at low levels, to replicate in murine hepatoma cells (Frentzen et al., 2011; Zhu et al., 2003).
Here, we attempted to shed light on restrictions of HCV species tropism in cells derived from non-permissive species. Our data demonstrate that species barriers can be overcome by engineering a suitable cellular environment. It was previously shown that the virally encoded NS3/4A protease is not only critical for processing of the HCV polyprotein but also cleaves numerous cellular targets (reviewed in (Morikawa et al., 2011)). In particular, cleavage of the antiviral signal mediators MAVS and TRIF has been inferred as one way HCV interferes with innate immune responses. Here, we formally demonstrate that the NS3/4A protease does indeed degrade the murine orthologs of both MAVS and TRIF. Consequently, this cleavage moderately increases the ability of HCV to replicate in mouse cell lines. However, this antiviral evasion strategy does not seem to be sufficient for HCV to readily establish persistent RNA replication. In order to interrogate whether more severe interference with antiviral signaling would boost viral replication we surveyed cell lines derived from a number of mouse knock-out lines. In contrast to a knock-down approach which fails to completely abrogate gene expression and can have considerable off-target effects (Buehler et al., 2012; Marine et al., 2012), targeted disruptions of genes provides a clean null background for the allele of interest. We demonstrate that mouse embryonic fibroblast lines derived from mice impaired type I and/or III interferon responses more readily support HCV replication. These data are consistent with previous observations showing that HCV can replicate more efficiently PKR and IRF3 deficient fibroblasts (Chang et al., 2006; Lin et al., 2010b). Besides interfering with innate responses it is conceivable that incompatible host factors modulate HCV RNA replication in mouse cells. It was demonstrated initially in cell culture that the liver specific miRNA122 plays an important role in the HCV life-cycle (Jopling et al., 2005) which was later corroborated in vivo (Lanford et al., 2010). Two sites in the HCV 5′ non-translated region appear to interact with miR122 (Jopling et al., 2008; Jopling et al., 2005). These interactions were shown to be critical for efficient HCV RNA replication, presumably by protecting 5′ terminal viral sequences from nucleolytic degradation or from inducing innate immune responses to the RNA terminus (Machlin et al., 2011), resulting in greater viral RNA abundance in both infected cultured cells and in the liver of infected chimpanzees (Lanford et al., 2010). Not surprisingly, ectopic expression of the liver specific microRNA 122 in MEFs slightly augments HCV RNA replication, which is supported by results from previous studies (Lin et al., 2010b). However, this pronounced effect of miRNA122 overexpression is more likely a reflection of the non-hepatic cellular background used here than a bona-fide species restriction factor. In fact, the exact sequence of miR122 is highly conserved in vertebrate species; comparatively, mouse miR122 is identical in sequence and with 50000 copies per hepatocytes, similar in expressed level to human miR122 (Chang et al., 2004). Consequently, miR122 is not likely to contribute to the restricted host tropism in non-permissive species. Interestingly, HCV RNA replication in PKR deficient iMEFs appeared to be more dependent on the presence of miR122. It is conceivable that in PKR deficient cells, which are presumably less impaired in their ability to mount antiviral innate responses, residual antiviral signaling triggers expression of an ISG profile, which still interferes with HCV RNA replication. In contrast, in cells with more severe innate immune signaling impairments, such as IRF9-/-, IFNabR-/-, or STAT1-/-, early signals upstream innate immune response cascade triggered by HCV RNA terminus are not amplified and thus do not translate into induction of an antiviral program. Likewise, the 5′UTR HCV may also be less susceptible to nucleolytic degradation. Numerous other host factors have been implicated in the HCV replication cycle (reviewed in (Bartenschlager et al., 2010)). Of those, the most convincing experimental evidence has been provided for the critical role of cyclophilin A (CypA) and phosphatidylinositol 4 kinase IIIα (PI4KIIIα) in the HCV life-cycle. However, limited compatibility between the virally encoded replication machinery and these host factors is not likely since the human and murine protein sequences of CypA and PI4KIIIα are highly similar (98.2% and 98.6%, respectively). Nonetheless, it has not been shown whether ectopic expression of human CypA, PI4KIIIα or other host factors may increase HCV RNA replication in mouse cells.
We demonstrate that expression of HCV entry factors in the most permissive immunodeficient cellular backgrounds allows for initiation of replication via infection. Interestingly, we observe a strong skewing towards high expression of human CD81, SCARB1, CLDN1 and OCLN. High-level expression of HCV entry factors may be beneficial for efficient spread in the cell culture. Given the fact that we were only able to measure detectable levels of infectious particles when apoE was expressed infection of neighboring cells may via a direct cell-to-cell transfer in this culture system. Consistent with previous data (Long et al., 2011a) we show that expression of mouse apoE does support assembly of infectious virions. While Long and colleagues used a transcomplementation system (Long et al., 2011a) we show here that mouse cells can assemble and release monocistronic recombinant viruses. It is important to note that apoE, as part of the VLDL pathway, is highly expressed in the mouse liver. Thus, the need for overexpression of apoE to complete the late stages of the HCV life-cycle in cell lines as shown here and in other studies (Da Costa et al., 2012a; Long et al., 2011a) highlights the artificial nature of commonly used cell culture systems.
In summary, we define here the essential elements to recapitulate the entire HCV life-cycle in murine cell lines. Experimental evidence to validate these observations in vivo is underway (Dorner et al. manuscript in revision). Undoubtedly, additional improvements to the genetically humanized mouse model would provide unprecedented opportunities to genetically dissect HCV infection and pathogenesis in vivo and will also serve as tractable, low-cost preclinical platform for testing and prioritizing drug and vaccine candidates.
Materials and Methods
Cells and antiviral drugs
Mouse embryonic fibroblasts (MEFs) were generated from day 12.5 or 13.5 embryos from Irf1tm1Mak (IRF1-/-)(Matsuyama et al., 1993) (obtained from the Jackson Laboratory, Bar Harbor, Maine, USA), Ifnar1tm1Agt (IFNαβR-/-) (Muller et al., 1994) (obtained from B&K Universal Ltd (Hull, UK)) and Stat1tm1Dlv (STAT1-/-) (Durbin et al., 1996) from Taconic (Hudson, NY, USA). Bcl2l12/Irf3tm1Ttg (IRF3-/-) (Sato et al., 2000), Irf7tm1Ttg (IRF7-/-) (Honda et al., 2005) and Irf9tm1Ttg (IRF9-/-) (Kimura et al., 1996) (kindly provided by Tadatsugo Taniguchi, University of Tokyo, Tokyo, Japan), Dhx58tm1(A30K)Aki (LGP2K30A/K30A) (Satoh et al., 2010) (kindly provided by Takashi Satoh and Shizuo Akira, Osaka University, Osaka, Japan), Eif2ak2tm1Cwe (PKR-/-) (Yang et al., 1995) (kindly provided by Adolfo Garcia-Sastre (Mount Sinai School of Medicine, New York, NY, USA) immortalized via transduction with TRIP-SV40 large T antigen. RIG-I MEFs originating from the Akira lab were made available through Alexander Tarakhovsky (The Rockefeller University). Huh 7.5 cells, Huh 7.5.1 cells, immortalized MEFs (iMEFs), 293T cells, and H2.35 cells were cultured in DMEM with 10% fetal bovine serum (FBS) and penicillin/streptomycin, if not noted otherwise. Media were supplemented with blasticidin, puromycin and 2′C methyl adenosine (2′CMA) as indicated. 2′CMA was the gift of. D. Olsen and S. Carroll (Merck Research Laboratories, West Point, PA) and also was obtained from Carbosynth Limited.
Generation of recombinant HCV plasmids
HCV replicons
The full length replicon contains the J6/JFH-1 polyprotein expressed from an encephalomyocarditis virus internal ribosomal entry site (EMCV-IRES). In an upstream cistron, the HCV 5′ untranslated region (UTR) drives expression of the first 19 amino acids of J6 core followed by blasticidin S-deaminase (bsd) containing a C-terminal STOP codon. Transfected into permissive cells, a blasticidin resistant population can be selected and infectious virus produced. The replication-impaired full-length construct contains two mutations in NS5B (GDD → GNN) that render this virus incapable of replication by deactivation of the viral polymerase. Transfected into permissive cells this replicon will be translated but no replication will take place. The other replicon used contains the subgenomic JFH-1 polyprotein including the nonstructural protein set (NS3-NS5B) expressed from an EMCV IRES. In an upstream cistron, the HCV 5′UTR drives expression of the first 19 amino acids of J6 core followed by blasticidin S-deaminase (bsd) containing a C-terminal STOP codon. Transfected into permissive cells, a blasticidin resistant population can be selected, but no infectious virus is released from the cells. Comparable to the full length a replication impaired subgenomic replicon was made. A mutation in NS5B (GDD → GND) renders this construct incapable of replication by deactivation of the viral polymerase. After initial translation no replication of the viral genome occurs.
Infectious viruses
HCVcc containing bsd between NS5A and NS5B
A detailed characterization of the HCV expressing heterologous proteins flanked by NS3/4A cleavage sites within the HCV polyprotein is described elsewhere (Horwitz et al., 2013). Briefly, we generated a Gateway®-compatible destination vector (Invitrogen, Life Technologies, Carlsbad, CA) based upon the fully infectious Jc1 HCV genome, Jc1-5AB-DEST, for insertion of reporter genes between NS5A and NS5B. The 9-amino acid region spanning P7-P2′ of the NS3/4A proteolytic cleavage site between NS5A and NS5B was positioned on both ends of the destination cassette. Jc1-5AB-DEST was generated by PCR amplification of the Gateway® (Invitrogen, Life Technologies, Carlsbad, CA) destination cassette and insertion into the DraIII restriction site at the 3′ end of Jc1(2a) NS5A using standard molecular cloning techniques. Jc1-5AB-BSD was generated by PCR amplification of blasticidin S deaminase (bsd) with primers containing AttB sites for Gateway®-mediated insertion into pDONR™221, and subsequent BP and LR reactions were performed. Jc1FLAG2(p7-nsGluc2A) is a fully-infectious HCVcc virus that has been previously described (Marukian et al., 2008; Pietschmann et al., 2006). J6/JFH Clone2 is a passaged derivative of J6/JFH that contains a number of adaptive mutations that increase infectious titers (Walters et al., 2009).
RNA transcription
In vitro transcripts were generated as previously described (Lindenbach et al., 2005). Briefly, plasmid DNA was linearized by XbaI and purified by using a Minelute column (Qiagen, Valencia, CA). RNA was transcribed from 1 μg of purified template by using the T7 Megascript kit (Ambion, Austin, TX) or the T7 RNA polymerase kit (Promega, Madison, WI). Reaction mixtures were incubated at 37°C for 3 h, followed by a 15-min digestion with 3 U of DNase I (Ambion, Austin, TX). RNA was purified by using a RNeasy Mini kit (Qiagen) with an additional on-column DNase treatment. RNA was quantified by absorbance at 260 nm and diluted to 0.5 μg/μl. Prior to storage at −80°C, RNA integrity was determined by agarose gel electrophoresis and visualization by ethidium bromide staining.
HCVcc generation
Jc1 NS5AB bsd and Jc1FLAG(p7-ns Gluc2aA), which were used in our experiments were generated by electroporating in vitro transcribed RNA into Huh7.5.1 cells. After 72 hours the medium was changed from 5% FBS to 1.5% FBS. The supernatants were collected every 6 hours from 72 to 144 hours after transfection and concentrated through a 100kDa MWCO membrane using a Stirred Ultrafiltration Cell. Supernatant from J6/JFH1 clone 2 electroporated Huh7.5.1 cells was collected daily from 72 to 144 hours but not concentrated. The virus infectivity was determined by limiting dilution as described previously (Lindenbach et al., 2005).
Pseudoparticles
Construction of proviral constructs
A. TRIP-HCV(JFH1)-NS3/4A-TagRFP/puro
This TRIP-based bicistronic lentiviral vector encodes the HCV NS3/4A protease as a single-chain fusion protein driven by a CMV promoter. The downstream cistron is driven by an encephalomyocarditis virus internal ribosomal entry site (EMCV-IRES) and encodes a fusion protein containing a puromycin resistance gene followed by a foot and mouth disease virus autoproteolytic peptide and TagRFP (Puro-2A-TagRFP). Transduction of permissive cell lines with these pseudoparticles results in stable expression of functional NS3/4a and Puro-2A-TagRFP. Transduced cell populations were selected on puromycin, and sorted using fluorescence based life cell sort.
2. TRIP-mApoE-TagRFP
The cDNA clone of murine apolipoprotein E was obtained in pCMV6 (Origene NM_009696). mApoE was PCR amplified from the cDNA vector and flanking attB sites added using primers RU-O-16597 and RU-O-16596, allowing us to insert the resulting product into pDONOR221 (Invitrogen, containing attP sites) using BP clonase II and Gateway cloning technology. mApoE was then shuttled into the destination vector pTRIP.CMV.IVSb.ires.TagRFP-DEST (Schoggins et al., 2011) using LR clonase II (Gateway cloning technology, Life Technologies).
3. TRIP-miRNA122
The proviral plasmid was generated by insertion of the miR122-generating hairpin-loop structure from pCMV-miR122 into the BamHI/XhoI digested pTRIP-GFP.
4. HCV entry factors
Generation of human TRIP-human CD81(Flint et al., 2006), -Venus/YFP-human OCLN and –human OCLN(Ploss et al., 2009), –Cerulean/CFP-human CLDN1 (Ploss et al., 2009), human CLDN1(Evans et al., 2007) and human SCARB1 (Catanese et al., 2010) were described previously. TRIP-mKate-human SCARB1 was generated by overlapping PCR of the coding sequence of mKate, also designated as TagFP635(Shcherbo et al., 2007) (Evrogen) and human SCARB1 resulting into an N-terminal fusion protein which was subsequently cloned into the BamHi/XhoI digested pTRIP-GFP.
Pseudoparticle generation
All pseudoparticles were generated as described previously (Ploss et al., 2009) in 293T cells by FuGENE6 (Roche Applied Science, Indianapolis)-mediated cotransfection of plasmids encoding (1) a minimal HIV provirus (TRIP or CSW) encoding a reporter gene or other transgene, (2) HIV gag-pol and (3) and VSV-G. Pseudoparticle-containing supernatants were harvested at 24, 48 and 72 h were pooled and filtered (0.45 μm mesh). Pseudoparticle infections were performed in the presence of 4 μg ml−1 polybrene. A minimum of 48 h elapsed between transduction and reporter gene quantification or subsequent experiments.
Transfection of HCV RNA into mouse and human cell lines
For all replicon experiments cells were plated in tissue culture treated 6 well (2×10E5 cells) or 10 cm (1×10E6 cells) plates one day or in case of transduction with miRNA122 two days before the transfection. In the latter case, the miR122 or GFP pseudoparticles were applied on the day between seeding and transfection. Mirus TransIT transfection kit was used to transfect 0.5μg (6-well plate) or 3μg (10 cm dish) HCV replicon RNA into the cells. According to manufacture's instructions the transfection medium was changed after six hours. We changed the medium to blasticidin containing selection medium the day after transfection and from there on regularly every 3 days. For all transfection experiments a concentration of 1 μg/ml blasticidin was used for ca 2 weeks before raising the concentration to 2μg/ml blasticidin for an other two weeks. After completing this month the plates were either fixed in PFA and stained using crystal violet or resistant cells were trypsinized and passaged for further experiments. Only the H2.35 cells were immediately exposed to 2μg/ml blasticidin and stained with crystal violet after 2 weeks.
HCV infections
Transduced and control MEFs or Huh7.5 cells were infected with HCVcc (J6/JFH1 clone 2, Jc1 NS-5AB bsd or Jc1FLAG2(p7nsGluc2A as indicated)-containing supernatants one day after cell seeding. For selection experiments, media was replaced the next day and switched to medium containing blasticidin, increasing the bsd concentration every 2 weeks from 1, to 2 to a final concentration of 6μg/ml. Stably infected cells were either fixed in PFA and stained with crystal violet or resistent cells were passaged for further experiments. To quantify HCV infection, cells were fixed in paraformaldehyde, permeabilized with saponin and stained for NS5A with an Alexa488-conjugated 9E10 anti-NS5A antibody (Lindenbach et al., 2005); the signal was quantified by flow-cytometrical analysis.
Flow cytometry
Expression of lentivirally-delivered transgenes was quantified by flow cytometry. CD81 expression was detected by staining with an anti-human CD81-APC (clone JS81, Pharmingen), other proteins were directly fluorescently tagged, i.e. SCARB1 (mKate), CLDN1 (Cerulean) and OCLN (Venus), ApoE, NS3/4A (both TagRFP). All samples were acquired on a LSR2 flow cytometer (Becton Dickinson, Franklin Lakes, NJ).
Western blotting
Cells (Huh7.5, H2.35 and indicated iMEF lines) were trypsinized and the pellet was lysed using the following buffer: TRIS Base 1M, NaCl, Triton-X 20% and Mini EDTA-free protease Inhibitor (Roche). After protein quantification using a Pierce BCA Protein Assay Kit (Thermo Scientific), equal amounts of protein were loaded onto 4–12% or 10% Bis/Tris NuPage polyacrylamide gels (Invitrogen). Proteins were transferred to a nitrocellulose membrane and detected with the respective antibodies: NS3 (Virostat #1878) 1:100, mMAVS (Cell Signaling #4983) 1:1000, hMAVS (Enzo #ALX-804-847) 1:1000, TRIF (Novus #NB120-13810). β-actin was detected as a loading control (Sigma #A3854) 1:200000. After incubation with the respective secondary antibody the proteins were visualized using Super Signal West Pico or Femto (Thermo Scientific).
HCV RNA quantification
After trypsinization, the total RNA from cell pellets was isolated using the RNeasy Mini kit (Qiagen) and the HCV genome copy number was quantified by one-step rtPCR using Multicode-RTx HCV RNA kit (EraGen) and a Roche LC480 light cycler, according to manufacture's instructions.
Highlights.
HCV NS3/4A protease cleaves mouse MAVS and TRIF
Type I and III signaling interferes with HCV RNA replication
Infection of mouse cells expressing human HCV entry factors
Assembly and release of infectious particles in murine cells
Completion of the entire HCV life-cycle in mouse cells
Acknowledgments
The authors thank Adolfo Garcia-Sastre for providing the PKR deficient mice, Tadatsugu Taniguchi for providing the IRF-3, IRF-7 and IRF-9 deficient mice, Takashi Satoh, Taro Kawai and Shizuo Akira for providing the LGP2[K30A/K30A] knock-in mice and RIG-I fibroblasts. The authors also thank Ellen Castillo, Brenna Flatley and Julia Sable and Svetlana Mazel and the staff of the Rockefeller University flow-cytometry core facility for outstanding technical support.
This study was supported in part by award number RC1DK087193 (to C.M.R. and A.P.) from the National Institute of Diabetes and Digestive and Kidney Diseases, R01AI072613 (to C.M.R.) from the National Institute for Allergy and Infectious Disease, The Starr Foundation and the Greenberg Medical Institute. A.V. was supported by a stipend from the International Institute of Life Sciences (IALS), M.A.S by a Ruth L. Kirschstein NRSA postdoctoral fellowship (award number F32AI091207) from the National Institute Of Allergy And Infectious Diseases, M.D. by postdoctoral fellowship from the German Research Foundation (Deutsche Forschungsgesellschaft) and G.G. was supported by the German Academy of Sciences Leopoldina (LPDS2009-9) and by the Human FrontierScience Program (LT000048/2009). A.P. is a recipient of Astella Young Investigator Award from the Infectious Disease Society of America and a Liver Scholar Award from the American Liver Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict-of-interest disclosure: The authors declare the following conflicts of interest, which are managed under University policy: A.P. is a consultant for Apath, LLC and C.M.R. has equity in Apath, LLC, which holds commercial licenses for the Huh-7.5 cell line and the HCV cell culture system.
Authorship: Contribution: A.V., B.M.D., M.D., G.G. performed research; J.A.H. and M.A.S. performed research and edited the manuscript; C.M.R. provided reagents, support and edited the manuscript. A.V. and A.P. designed research, analyzed data and A.P. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann SW, Davis HR, Jr, Zhu LJ, Yao X, Hoos LM, Tetzloff G, Iyer SP, Maguire M, Golovko A, Zeng M, Wang L, Murgolo N, Graziano MP. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- Andrus L, Marukian S, Jones CT, Catanese MT, Sheahan TP, Schoggins JW, Barry WT, Dustin LB, Trehan K, Ploss A, Bhatia SN, Rice CM. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54:1901–1912. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Kim CN, Yeh WC, Mak TW, Bhalla K, Barber GN. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 1998;17:6888–6902. doi: 10.1093/emboj/17.23.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Cosset FL, Lohmann V. Hepatitis C virus replication cycle. Journal of hepatology. 2010;53:583–585. doi: 10.1016/j.jhep.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Von Weizsacker F, Blum HE, Baumert TF. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehler E, Khan AA, Marine S, Rajaram M, Bahl A, Burchard J, Ferrer M. siRNA off-target effects in genome-wide screens identify signaling pathway members. Scientific reports. 2012;2:428. doi: 10.1038/srep00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology. 2012;142:1279–1287 e1273. doi: 10.1053/j.gastro.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Catanese MT, Ansuini H, Graziani R, Huby T, Moreau M, Ball JK, Paonessa G, Rice CM, Cortese R, Vitelli A, Nicosia A. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J Virol. 2010;84:34–43. doi: 10.1128/JVI.02199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA biology. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Chang KS, Cai Z, Zhang C, Sen GC, Williams BR, Luo G. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. J Virol. 2006;80:7364–7374. doi: 10.1128/JVI.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa D, Turek M, Felmlee DJ, Girardi E, Pfeffer S, Long G, Bartenschlager R, Zeisel MB, Baumert TF. Reconstitution of the entire hepatitis C virus life cycle in non-hepatic cells. Journal of virology. 2012a doi: 10.1128/JVI.01066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa D, Turek M, Felmlee DJ, Girardi E, Pfeffer S, Long G, Bartenschlager R, Zeisel MB, Baumert TF. Reconstitution of the entire hepatitis C virus life cycle in nonhepatic cells. Journal of virology. 2012b;86:11919–11925. doi: 10.1128/JVI.01066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, Law M, Rice CM, Ploss A. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Rice CM, Ploss A. Study of hepatitis C virus entry in genetically humanized mice. Methods. 2012 doi: 10.1016/j.ymeth.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- Dustin LB, Rice CM. Flying under the radar: the immunobiology of hepatitis C. Annu Rev Immunol. 2007;25:71–99. doi: 10.1146/annurev.immunol.25.022106.141602. [DOI] [PubMed] [Google Scholar]
- Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- Flint M, von Hahn T, Zhang J, Farquhar M, Jones CT, Balfe P, Rice CM, McKeating JA. Diverse CD81 proteins support hepatitis C virus infection. J Virol. 2006;80:11331–11342. doi: 10.1128/JVI.00104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentzen A, Huging K, Bitzegeio J, Friesland M, Haid S, Gentzsch J, Hoffmann M, Lindemann D, Zimmer G, Zielecki F, Weber F, Steinmann E, Pietschmann T. Completion of Hepatitis C Virus Replication Cycle in Heterokaryons Excludes Dominant Restrictions in Human Non-liver and Mouse Liver Cell Lines. PLoS Pathog. 2011;7:e1002029. doi: 10.1371/journal.ppat.1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Horwitz JA, Dorner M, Friling T, Donovan BM, Vogt A, Loureiro J, Oh T, Rice CM, Ploss A. Expression of heterologous proteins flanked by NS3-4A cleavage sites within the hepatitis C virus polyprotein. Virology. 2013 doi: 10.1016/j.virol.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Tarutani M, Tan RS, Takasugi T, Matsuyama T, Mak TW, Noguchi S, Taniguchi T. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells. 1996;1:115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- Kimura T, Nakayama K, Penninger J, Kitagawa M, Harada H, Matsuyama T, Tanaka N, Kamijo R, Vilcek J, Mak TW, et al. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science. 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80:5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LT, Noyce RS, Pham TN, Wilson JA, Sisson GR, Michalak TI, Mossman KL, Richardson CD. Replication of subgenomic hepatitis C virus replicons in mouse fibroblasts is facilitated by deletion of interferon regulatory factor 3 and expression of liver-specific microRNA 122. J Virol. 2010a;84:9170–9180. doi: 10.1128/JVI.00559-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LT, Noyce RS, Pham TN, Wilson JA, Sisson GR, Michalak TI, Mossman KL, Richardson CD. Replication of subgenomic hepatitis C virus replicons in mouse fibroblasts is facilitated by deletion of interferon regulatory factor 3 and expression of liver-specific microRNA 122. J Virol. 2010b;84:9170–9180. doi: 10.1128/JVI.00559-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete Replication of Hepatitis C Virus in Cell Culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G, Hiet MS, Windisch MP, Lee JY, Lohmann V, Bartenschlager R. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011a doi: 10.1053/j.gastro.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Long G, Hiet MS, Windisch MP, Lee JY, Lohmann V, Bartenschlager R. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011b;141:1057–1066. doi: 10.1053/j.gastro.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset FL, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoel M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin ES, Sarnow P, Sagan SM. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine S, Bahl A, Ferrer M, Buehler E. Common seed analysis to identify off-target effects in siRNA screens. Journal of biomolecular screening. 2012;17:370–378. doi: 10.1177/1087057111427348. [DOI] [PubMed] [Google Scholar]
- Marukian S, Andrus L, Sheahan TP, Jones CT, Charles ED, Ploss A, Rice CM, Dustin LB. Hepatitis C virus induces interferon-lambda and interferon-stimulated genes in primary liver cultures. Hepatology. 2011;54:1913–1923. doi: 10.1002/hep.24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, Dustin LB. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008;48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig TM, Amakawa R, Kishihara K, Wakeham A, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- McCaffrey AP, Ohashi K, Meuse L, Shen S, Lancaster AM, Lukavsky PJ, Sarnow P, Kay MA. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther. 2002;5:676–684. doi: 10.1006/mthe.2002.0600. [DOI] [PubMed] [Google Scholar]
- Meuleman P, Leroux-Roels G. The human liver-uPA-SCID mouse: A model for the evaluation of antiviral compounds against HBV and HCV. Antiviral Res. 2008 doi: 10.1016/j.antiviral.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P, Smolarsky M, Funaro A, Malavasi F, Larrey D, Coste J, Fabre JM, Sa-Cunha A, Maurel P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411–419. doi: 10.1016/j.jhep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Monazahian M, Bohme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. Journal of Medical Virology. 1999;57:223–229. doi: 10.1002/(sici)1096-9071(199903)57:3<223::aid-jmv2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Morikawa K, Lange CM, Gouttenoire J, Meylan E, Brass V, Penin F, Moradpour D. Nonstructural protein 3-4A: the Swiss army knife of hepatitis C virus. Journal of viral hepatitis. 2011;18:305–315. doi: 10.1111/j.1365-2893.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Owen DM, Huang H, Ye J, Gale M., Jr Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99–108. doi: 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MR, Loo YM, Horner SM, Gale M, Jr, Malik HS. Convergent evolution of escape from hepaciviral antagonism in primates. PLoS Biol. 2012;10:e1001282. doi: 10.1371/journal.pbio.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL, Bartenschlager R. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploss A, Rice CM. Towards a small animal model for hepatitis C. EMBO Rep. 2009 doi: 10.1038/embor.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B, Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann L, Ploss A. Barriers of hepatitis C virus interspecies transmission. Virology. 2013;435:70–80. doi: 10.1016/j.virol.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scull MA, Ploss A. Exiting from uncharted territory: hepatitis C virus assembles in mouse cell lines. Hepatology. 2012;55:645–648. doi: 10.1002/hep.24716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nature methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uprichard SL, Chung J, Chisari FV, Wakita T. Replication of a hepatitis C virus replicon clone in mouse cells. Virol J. 2006;3:89. doi: 10.1186/1743-422X-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KA, Syder AJ, Lederer SL, Diamond DL, Paeper B, Rice CM, Katze MG. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS Pathog. 2009;5:e1000269. doi: 10.1371/journal.ppat.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams BR, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Guo JT, Seeger C. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J Virol. 2003;77:9204–9210. doi: 10.1128/JVI.77.17.9204-9210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]