Abstract
Population genetics theory predicts that X (or Z) chromosomes could play disproportionate roles in speciation and evolutionary divergence, and recent genome-wide analyses have identified situations in which X- or Z-linked divergence exceeds that on the autosomes (the “faster-X effect”). Here, we summarize the current state of both the theory and data surrounding the study of faster-X evolution. Our survey indicates that the faster-X effect is pervasive across a taxonomically diverse array of evolutionary lineages. These patterns could be informative of the the dominance/recessivity of beneficial mutations and the nature of genetic variation acted upon by natural selection. We also identify several aspects of disagreement between these empirical results and the population genetic models used to interpret them. However, there are clearly delineated aspects of the problem for which additional modeling and collection of genomic data will address these discrepancies and provide novel insights into the population genetics of adaptation.
Keywords: X chromosome, natural selection, genetics of adaptation, dominance
Motivations for Studying Faster-X Evolution
The widespread availability of population and comparative genomic data has made it possible to estimate rates of molecular evolution and gene expression divergence in entire genomes, across broad swaths of the tree of life. These data, when considered within a statistical population genetic framework, can shed light on the biology of speciation, adaptation, and divergence, by permitting inferences about the processes contributing to evolutionary change [1-5]. The tools of evolutionary genomics produce the most useful insights when they can connect patterns of divergence with causal evolutionary processes, an objective that remains a considerable challenge.
Molecular evolutionary contrasts between the X (or Z) chromosome and autosomes are often motivated by such goals. Classical population genetics theory shows that the evolutionary dynamics of an allele depend, in part, on its mode of inheritance [6, 7]. Under specific parameterizations of allelic dominance, selection in males versus females, mutation, recombination, and effective population size, X-linked genes can be more divergent between species than autosomal genes—a phenomenon known as the “faster-X effect” [7-14]. Analyses of the relative divergence rates between X-linked and autosomal genes may therefore provide insights into the population genetic basis of neutral and adaptive evolution, conditional on our ability to effectively and realistically link pattern and process through evolutionary theory.
The past few years have witnessed considerable growth in theory and data on faster-X evolution, yet the fit between the two has nevertheless become rather complicated. In this review, we emphasize the important assumptions and limitations of current theory, and then we reconsider the diverse array of published data within this theoretical foundation. Along the way, we outline several paths forward.
Theoretical Background
Evolution proceeds by the fixation of neutral, slightly deleterious, and beneficial alleles. Although substitution rates (i.e., total divergence between species) reflect the cumulative fixation process of alleles of all three classes, most X versus autosome theory has emphasized, for two primary reasons, beneficial substitutions and the conditions leading to faster-X adaptive evolution. First, beneficial substitution rates on the X and autosomes are interesting for what they might tell us about the population genetics of adaptation [13]. Second, more elaborate theory is required to characterize the evolutionary dynamics and genetics of adaptation, relative to the comparably simple theory of substitution by genetic drift. While we emphasize the fixation of beneficial alleles in our outline of the theory of faster-X evolution, the predictions with respect to neutral and slightly deleterious substitutions are also discussed.
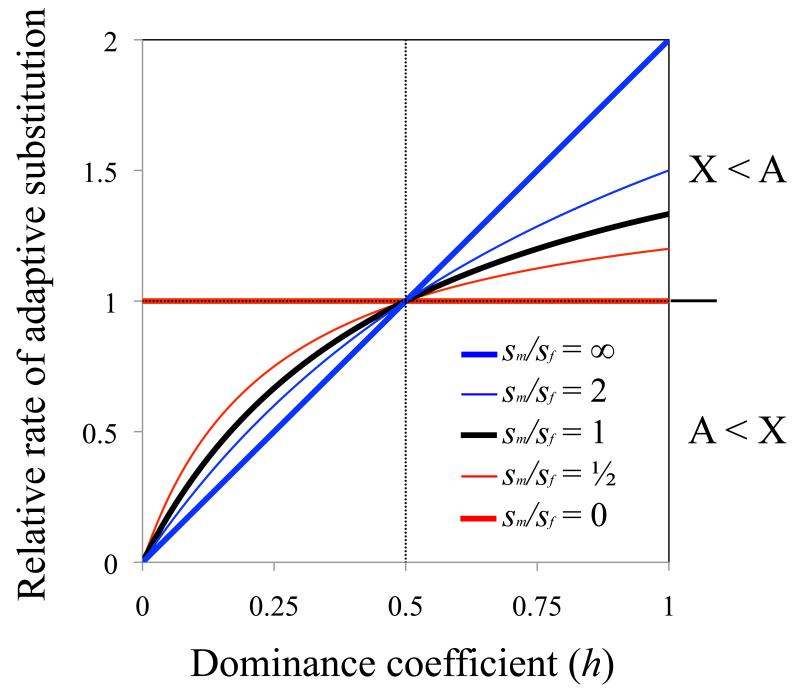
Charlesworth et al. [8] analyzed several models of substitution, including one for beneficial mutations, that is referenced in most molecular evolution studies that contrast the X and autosomes (Boxes 1 and 2). This model builds upon the pioneering work of Kimura and Ohta [15, 16], which characterizes the substitution rate as the product of mutational input per generation and the fixation probability of each mutation. The simplest form of the model is based upon three conditions: (i) unique, beneficial mutations occur at a rate of u per gene copy, per generation; (ii) each beneficial mutation increases fitness by the amount sh in heterozygotes and s in hemi- and homo-zygotes (1 ⪢ s > 0; 1 > h > 0); and (iii) the effective population size (Ne) of the X is three quarters that of the autosomes (NeX/NeA = 3/4). From these conditions, the relative rate of adaptive substitution of an autosomal gene (RA) versus an X-linked gene (RX) will be:
| (1) |
(see eq. (2a) of Charlesworth et al. [8]). Faster-X evolution (RA/RX < 1) occurs when beneficial mutations are partially or completely recessive (0 < h < 1/2); otherwise adaptive evolution is faster on the autosomes (see Figure 1; eq. (1) corresponds to the black curve).
Box 1. X-linked and autosomal fixation probabilities of beneficial mutations.
Theory for the adaptive substitution rate of X-linked versus autosomal genes is heavily influenced by the early work of J.B.S. Haldane [6], who was the first to characterize the fixation probabilities for beneficial mutations (i.e., the probability that individual mutations eventually reach a population frequency of one), and the population genetic dynamics associated with X-linkage, dominance, and sex differences in selection. Haldane showed that the fixation probability of a unique beneficial mutation is Π ≈ 2t, where t represents the average fitness benefit provided to individuals that carry a single copy of the mutation. The model assumes that benefits are small and population size, N = Ne, is large ( ⪡ t ⪡ 1; when N ≠ Ne, Π ≈ 2t(Ne/N) [16]).
To incorporate sex differences in selection on a mutation [7], t can be replaced with the weighted averages of male and female fitness effects of a mutant. The fixation probability of a unique autosomal mutation is
where smAhmA and sfAhfA represent the fitness effect of carrying a single autosomal copy of the mutation in males and females, respectively, and smA and sfA represent the fitness effect of carrying two copies of the mutation (sjA represents the autosomal selection coefficient in homozygotes, and hjA is the dominance coefficient, which determines the fitness of heterozygotes relative to homozygotes; j = {m, f}). The probability of fixation for a unique X-linked mutation is
Contrasts between ΠA and ΠX reveal two basic differences between the X and autosomes. First, because males are haploid, the fixation probability on the X is less sensitive to a mutation’s dominance (hmA, hfA) than is the autosomal fixation probability. Second, the relative importance of selection in males, versus selection in females, differs between chromosomes. For autosomal mutations, selection in males and females carries equal weight because inheritance is symmetric between the sexes (mothers and fathers transmit equally to off spring). X-linked transmission occurs twice as often through females than males, which upwardly biases the importance of female selection on X-linked mutations (though the absence of dominance in males can counteract this asymmetry).
Box 2. Adaptive substitution rates of X-linked and autosomal genes.
Here, we describe how fixation probabilities of Box 1 relate to the adaptive substitution rates of individual genes (i.e., the tempo of nucleotide changes over time), as predicted from the influential theory of X versus autosome adaptive substitution developed by Charlesworth et al. [8]. A gene’s substitution rate is modeled as the product of the population’s beneficial mutation rate and the fixation probability of each mutation, which is assumed to be unique [7, 8, 15, 16]. Autosomal genes mutate to a beneficial allele at rate uA; with N individuals in the population, 2NuA mutations are expected to arise during each generation. The adaptive substitution rate for the autosomal gene is RA ≈ 2NuAΠA, where ΠA ≈ 2tA (Box 1). The adaptive substitution rate for an X-linked gene is RX ≈ NXuXΠX, where NX is the number of X chromosomes in the population (NX/N = 3/2 is assumed [8]), uX is the beneficial mutation rate at the X-linked gene, and ΠX ≈ 2tX (Box 1). Given sex-specific beneficial mutation rates of uf and um, uA = (um + uf)/2 and uX = (2uf + um)/3 [10].
Mutation and selection parameters are likely to be variable across genes, so that average rates of substitution among X-linked and autosomal genes will be 〈RA〉 ≈ 2N〈uAΠA〉 and 〈RX〉 ≈ NX〈uXΠX〉, where 〈〉 denotes the expectation. The relative rate of adaptive substitution will be
where 〈ujΠj〉 = 〈uj〉〈Πj〉+cov(uj, Πj). To express 〈RA〉/〈RX〉 as a simple function of the dominance coefficient, models of X versus autosome substitution must make assumptions about the distribution of beneficial mutation parameters and their covariances. This issue is usually sidestepped by assuming fixed parameter values (i.e., terms of u, s, and h are treated as constants), leading to:
Eq. (1) is obtained when parameters are identical between chromosomes (uA = uX, h = hmA = hfA = hfX, and s = smA = smX = sfA = sfX).
Effects of sex-biased mutation can be controlled by scaling the adaptive substitution rate against the neutral rate [11]. If vm and vf are the male and female mutation rates per silent site, then the neutral substitution rate on the X and autosomes will be vX = (2vf + vm)/3 and vA = (vf + vm)/2, respectively. Assuming that um, uf, vm and vf are constant, and um/uf = vm/vf, then the rescaled ratio of autosome to X-linked adaptive substitution will be:
In practice, this is accomplished by comparing dN/dS between X-linked and autosomal genes (see main text).
Figure 1. Dominance, sex differences in selection, and faster-X adaptive substitution.
Curves show the theoretical predictions for the relative rates of adaptive substitutions at autosomal and X-linked genes, based on the model framework of Charlesworth et al. [8] (Box 2). The y-axis shows the autosome-to-X rate of adaptive evolution, (RA/vA)/(RX/vX), which corrects for sex-biased mutation rates. The dominance coefficient of a beneficial mutation is assumed to be the same for males and females and for the X and autosomes (i.e., h = hmA = hfA = hfX), and beneficial selection coefficients (sm in males, and sf in females) are treated as constants.
Eq (1) relies upon five simplifying assumptions, with violation of each altering the predicted relationship between dominance and RA/RX. These assumptions are as follows:
1. Selection parameters are equal between males and females
The faster-X effect emerges due to selection on recessive beneficial mutations within the heterogametic (i.e., hemizygous) sex, in which there is no masking effect for X-linked alleles. However, the theoretical predictions of faster-X evolution change when mutations have asymmetric fitness effects between the sexes. Faster-X effects are slightly more pronounced when beneficial substitutions have stronger fitness effects in males than females, and there is no predicted faster-X effect when selection only acts in females (Figure 1) [8].
2. Mutation rates are equal between the sexes
The male germline often has more mitoses than the female germline, which can increase the mutation rate in males relative to females [17]. Because the X chromosome spends more time in the female germline, a higher male mutation rate will decrease the relative mutation rate of X-linked genes, which could decrease the rate of evolution of the X chromosome [10, 18]. Effects of sex-biased mutation on faster-X evolution can nevertheless be controlled for by scaling gene substitution rates against the divergence rates at linked neutral sites [11] (as in Figure 1).
3. The effective population size ratio of the X to the autosomes is three-quarters (NeX/NeA = 3/4)
Ne scales positively with the fixation probability of beneficial mutations [15, 16]. Increasing the effective size of X-linked relative to autosomal genes (NeX relative to NeA) enhances opportunities for faster-X evolution [11] (the curves in Figure 1 shift down with NeX/NeA > 3/4); decreasing NeX/NeA reduces the faster-X parameter space. By estimating NeX/NeA from neutral diversity data, the effect of NeX/NeA on faster-X evolution can be disentangled from other contributing factors [12], yet such corrections could prove misleading if selective sweeps are frequent [19].
4. Substitution rates are limited by the fixation probabilities of individual beneficial mutations
Correlations between the adaptive substitution rate and fixation probabilities of unique mutants may be reduced or eliminated if (i) adaptation uses “standing genetic variation” (i.e., it fixes segregating alleles that were neutral or deleterious prior to an environmental change), or (ii) mutations are recurrent [20]. Adaptation using standing genetic variation causes faster-autosome substitution, independent of the dominance of beneficial alleles, provided autosomal loci harbor greater amounts of genetic diversity [8, 9, 14]. Under recurrent mutation, dominance only influences RA/RX if multiple genes, spread across the genome, compete to fix beneficial mutations during individual bouts of adaptation [14]. Experimental evolution experiments and genetic studies of natural populations indicate that individual bouts of adaptation are sometimes highly constrained, with beneficial substitutions recruited from a very small subset of genes [21, 22]. Such scenarios of adaptation will tend to equalize the X and autosomal substitution rates over a wide range of dominance conditions [14].
5. The distribution of mutant fitness effects (DMFE) is the same, on average, for X-linked and autosomal genes
This condition may be violated under plausible biological scenarios, and it is difficult to effectively control for. For example, dosage compensation mechanisms, which vary between species [23], may systematically affect X-linked fitness effects. Opportunities for faster-X evolution are expected to decrease in species without dosage compensation [8] and increase in species with somatic X-inactivation (as in therian mammals), which generates haploid expression within individual female cells [12]. Gene content also differs between the X and autosomes [24], which can bias opportunities for adaptation between chromosomes. Finally, recent theory suggests that haploid versus diploid inheritance can differentially shape the DMFE [25], raising the possibility that X versus autosome differences might be a fundamental property of ploidy differences between chromosomes.
Empirical Tests of the Faster-X Effect
Tests of faster-X evolution typically fall into two categories. First, comparative genomic approaches test whether X-linked loci accumulate more substitutions than autosomal loci. “Faster-X divergence” is said to occur when dN/dS values for X-linked genes are greater than those of autosomal genes, where dN is the rate of non-synonymous (amino acid changing) substitutions and dS is the rate of synonymous (silent or neutral) substitutions in a gene. Although dN/dS is useful for comparing X versus autosome divergence rates, it is important to note that dN captures both nonadaptive (neutral and slightly deleterious) and adaptive substitutions. This approach is therefore ill equipped to differentiate between adaptive and nonadaptive causes of faster-X evolution. The second approach combines within-species polymorphisms and between-species divergence data to estimate adaptive substitution rates (i.e., within the analytical framework of the McDonald-Kreitman or “MK” test [26-28]), which tests for “faster-X adaptation”.
Faster-X divergence
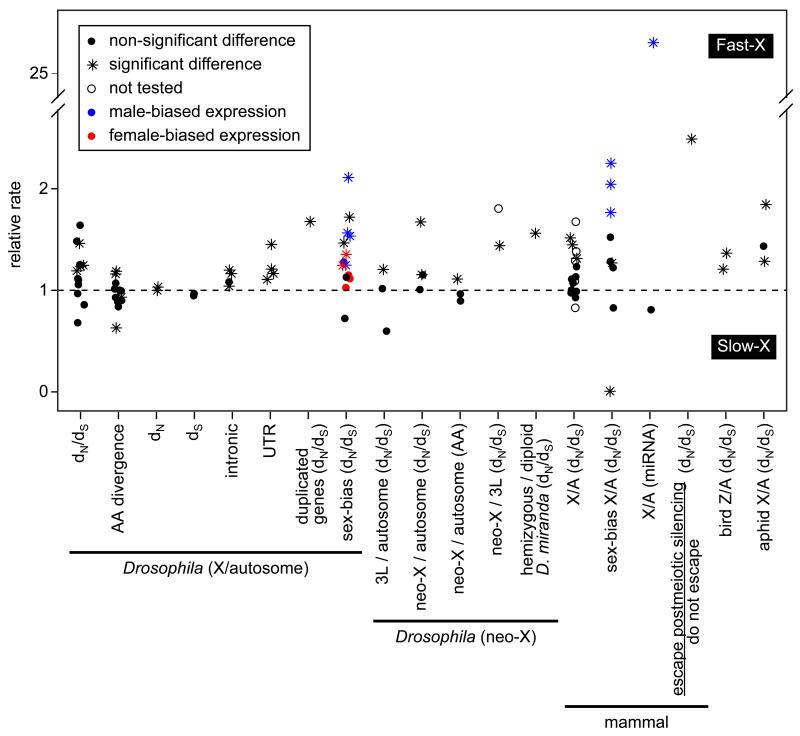
Many of the earliest tests for faster-X divergence were performed in the genus Drosophila (Figure 2), where support for elevated dN/dS in X-linked genes was varied [1, 29-34]. Of particular note were analyses that utilized natural autosome-to-X translocations to control for gene content effects (i.e., the D. pseudoobscura and D. willistoni neo-X chromosomes correspond to autosomes in D. melanogaster), though these studies also failed to reach consensus on the faster-X effect [30-35] (Figure 2). The more recently arisen D. miranda neo-X chromosome allows for an additional test of faster-X evolution by comparing genes that retain a Y-linked homolog (effectively diploid in males) and those that are hemizygous. Consistent with the predictions if beneficial mutations are recessive, hemizygous neo-X-linked protein-coding genes evolve faster than diploid genes on the D. miranda neo-X [36] (Figure 2). Finally, Drosophila X-linked duplicated genes have elevated dN/dS relative to autosomal duplicates [37] (Figure 2), and the amount of chromosomal rearrangement divergence in many taxa, including Drosophila [38], is higher on the X chromosome [8].
Figure 2. Tests for faster-X divergence.
The relative rate of evolution is plotted for different classes of nucleotide sites and chromosomes in Drosophila [29-31, 33, 35-37, 39], mammals [12, 18, 40, 42, 60, 62, 64, 65], birds [48], and aphids [49]. The rate of evolution is measured as either dN/dS, amino acid (AA) divergence, or nucleotide divergence at different classes of cites (indicated on the x-axis). Relative rates are either X/autosome, D. melanogaster chromosome arm 3L (homolog of the D. pseudoobscura neo-X)/autosome, D. pseudoobscura neo-X/autosome, D. pseudoobscura neo-X/D. melanogaster 3L, hemizygous/diploid genes on the D. miranda neo-X, or mammalian genes that escape postmeiotic silencing/those that do not escape. The expectation if X-linked and autosomal genes evolve at equal rates is represented by the dashed line. Significant deviation from unity in the relative rate is indicated by an asterisk, whereas non-significant differences or studies in which significance was not reported are indicated by a circle. In experiments where expression was measured (indicated by “sex-bias” in the x-axis label), the color of the point indicates the expression class of the genes (black=non-sex-biased, blue=male-biased, red=female-biased).
Recent availability of high quality genomes from the closely related species D. melanogaster and D. simulans has allowed for tests of faster-X divergence at many different classes of sites (Figure 2). Intriguingly, this comparison revealed that X-linked protein coding sites and many non-coding sites evolve faster than autosomal sites in the same functional class [1, 39]. However, after using gene ontology classifications to control for gene content, dN at X-linked coding sequences is no longer significantly elevated [39]. A signal of faster-X divergence remains among many classes of non-coding sites [39], which could be driven by a higher mutation rate on the X chromosome or the adaptive fixation of recessive beneficial mutations that affect the transcription of nearby genes. In addition, the faster-X divergence of non-coding sites could be responsible for the faster-X evolution of gene expression (see below).
Comparative genomic studies in other taxa reveal more consistent support for faster-X divergence (Figure 2). Mean dN/dS is higher for X-linked genes in comparisons between human and chimpanzee [40-42] and in rodents [12, 43]. In birds and moths, where females are the heterogametic sex (ZW), Z-linked genes have elevated dN/dS relative to autosomal genes [44-47]. However, faster-Z divergence in birds may not be due to positive selection [48], as described below. Aphids, which have XO males (i.e., no Y chromosome), also show evidence for faster-X divergence in dN/dS estimates [49].
Faster-X adaptation
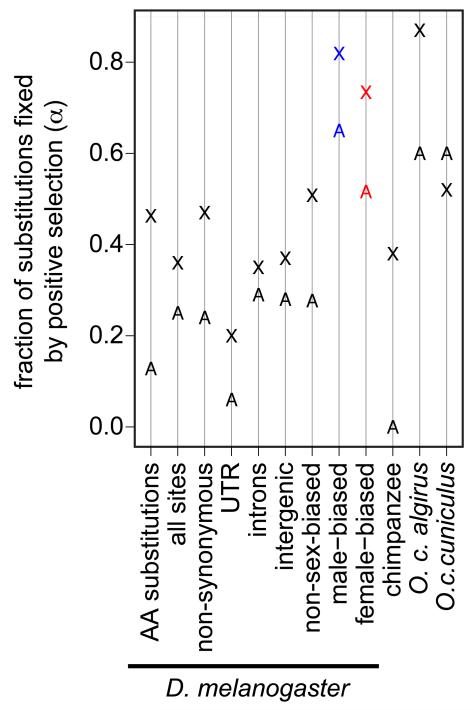
Comparisons of polymorphism and divergence can be used to infer the proportion of substitutions that are fixed by positive selection (α) and the strength of selection [27, 28]. Early implementations of the MK-test, using subsets of the D. melanogaster and D. simulans genomes, provided mixed support for faster-X adaptation [50-53], with the strongest evidence among genes with male-biased expression [52] (see below). More recent, whole-genome analyses reveal robust evidence for elevated frequencies of adaptive substitution among Drosophila X-linked genes [4, 54] (Figure 3). X-linked duplicated genes similarly accumulate more adaptive substitutions than autosomal duplicates [55] (Figure 3). Although demographic events could differentially affect X-linked and autosomal genetic diversity, demographic history alone cannot explain the evidence for faster-X adaptation in D. melanogaster or the elevated divergence relative to polymorphism on the D. melanogaster X chromosome [54].
Figure 3. Tests for faster-X adaptation.
The fraction of substitutions fixed by positive selection (α) is plotted for X-linked (X) and autosomal (A) loci. Estimates of α in the D. melanogaster genome were calculated for amino acid (AA) substitutions [54]; all nucleotide sites, non-synonymous sites, 3′ and 5′ untranslated regions (UTRs), introns, and intergenic regions separately [4]; genes with non-sex-biased, male-biased (blue), or female-biased (red) expression [59]. Estimates of α for the chimpanzee genome [42] and two species of european rabbit [56] reflect the fraction of amino acid substitutions fixed by positive selection.
Support for faster-X adaptation in vertebrate species is less clear than in Drosophila. While X-linked genes in the human-chimpanzee comparison harbor more signatures of positive selection when compared to autosomal genes, based on dN/dS analysis [41], this has not, to our knowledge, been examined in the MK framework (possibly because of the relatively poor quality of DNA sequence polymorphism data for the human X chromosome). However, a recent MK-based, whole-genome analysis found evidence for faster-X adaptation within the chimpanzee lineage, following its split from the human lineage [42] (Figure 3). MK-tests performed on wild mouse populations also yield support for faster-X adaptation [43] (A. Kousathanas et al., unpublished). On the other hand, support for faster-X adaptation in the European rabbit, Oryctolagus cuniculus, is limited to the subspecies with larger Ne, O. c. algirus [56] (Figure 3). In addition, there is evidence for faster-Z adaptation in silkmoth [47], and reduced variation and excess divergence on the Z chromosome in flycatcher birds [44, 57, 58] is consistent with faster-Z adaptation. Although a similar pattern was initially observed for the chicken Z chromosome [45], subsequent work indicates that faster-Z divergence in the chicken lineage may be due to relaxed constraints rather than adaptive evolution [48]. Overall, these results demonstrate that lineages with faster-X divergence do not necessarily exhibit faster-X adaptation, and vice versa. This may reflect differences among taxa in the role of neutral and adaptive causes of faster-X divergence [11, 12], as described below.
Faster-X evolution of male reproductive genes
Several studies emphasize that faster-X effects should be most pronounced in genes with male-biased expression (i.e., primarily expressed in males) or male-limited functions [29, 35, 52, 59], assuming that mutations in these genes have larger fitness effects in males than females (e.g., Figure 1, blue curves). Consistent with this prediction, in both Drosophila and mammals, the strongest evidence for faster-X divergence and adaptation is observed in genes expressed primarily in male reproductive tissues [35, 52, 59-62] (Figure 2, 3). Faster-X effects have also been observed in primate genes expressed in cancer and testis cells [63], microRNAs expressed in mammalian testis [64], and human genes that escape postmeiotic transcriptional silencing [65] (Figure 2).
Although the classical theory predicts a slight elevation in the magnitude of faster-X evolution for male-limited beneficial substitutions, relative to substitutions with similar effects on both sexes, this effect will not be nearly as great as the difference between substitutions beneficial to both sexes and those with female-limited effects (Figure 1). Intriguingly, although we do not expect faster-X divergence or adaptation amongst genes under selection only in females (Figure 1), there is evidence for faster-X evolution amongst Drosophila female-biased genes [35, 59] (Figure 2, 3). Genes with female-biased expression are, however, often expressed in males, and mutations in female-biased genes can have fitness effects in males [66]. Selection on recessive X-linked beneficial mutations in males may therefore drive the faster-X evolution of female-biased genes. Alternatively, there could be something fundamentally different about the genetic basis of adaptation in genes from different functional or expression classes. For example, they might differentially utilize de novo mutations versus standing genetic variation, during adaptation, which can alter the influence of dominance on faster-X adaptation [9].
Effective population size and the faster-X effect
X-to-autosome substitution rates are a function of adaptive, neutral, and slightly deleterious substitutions, which each contribute to total divergence [8, 11, 12]. differential accumulation of each substitution type between the X and autosomes can further complicate the interpretation of faster-X divergence patterns. Lineages with large Ne, such as Drosophila, should experience e cient positive and purifying selection, leading to a high proportion of substitutions driven by positive selection [2]. Conversely, small Ne will limit the accumulation of adaptively fixed mutations [5, 15, 16]. Relative divergence rates of X-linked and autosomal genes could similarly reflect the differential accumulation of adaptive, neutral, and deleterious substitutions among lineages with different Ne [12].
Factors such as mating system variation [67], recombinational differences between the X and autosomes [68, 69], and genetic hitchhiking and background selection [68, 70] can affect the NeX/NeA ratio. This could further affect the X-to-autosome divergence rates or the relative proportions of adaptive versus neutrally fixed substitutions. High variance in male reproductive success decreases NeZ/NeA in ZZ/ZW taxa, permits a higher rate of nearly-neutral evolution of Z-linked genes, and contributes to faster-Z divergence in birds [48]. The NeX/NeA ratio is near one in D. melanogaster, but close to 3/4 in D. pseudoobscura [70]. If NeX/NeA is typically large across the Drosophila phylogeny, it could explain why there is robust evidence of faster-X adaptation in Drosophila [4, 54] (Figure 3). However, the lack of conclusive evidence for faster-X divergence in Drosophila remains perplexing (Figure 2).
On the other hand, larger populations are more polymorphic than small ones. This can increase the probability of adaptation using standing genetic variation [71], which should reduce or eliminate opportunities for faster-X evolution [8, 9, 14]. The interaction between Ne and faster-X evolution is a particularly interesting research area, although it demands additional data. Readers interested in learning more about this topic should consult two recent, comprehensive treatments of the subject [11, 12].
Faster-X evolution of gene expression
Recent work demonstrates that expression level divergence is greater for X-linked than autosomal genes, in both mammals and Drosophila [62, 72-75], leading to a faster-X effect for gene expression. Because the expression of a gene is dependent on DNA sequences both at that locus (acting in cis) and elsewhere in the genome (acting in trans), elevated gene expression divergence in X-linked genes cannot entirely be attributed to rapid sequence evolution on the X chromosome. However, because trans factors should affect both X-linked and autosomal gene expression, whereas cis divergence should specifically affect expression divergence on a single chromosome, the faster-X divergence of gene expression is likely the result of faster-X evolution of cis regulatory sequences. This hypothesis is supported by the faster-X divergence of non-coding sequences [39]. The faster-X evolution of gene expression can inform our general understanding of expression evolution [72, 73, 75] and shed light on the nature of reproductive isolation between species [74].
Applying the MK-test framework to gene expression, if faster-X expression evolution were the result of positive selection, we would not expect to see elevated expression polymorphism amongst X-linked genes [76]. There is no such elevation of gene expression polymorphism on the D. melanogaster X chromosome [73, 75], suggesting that the faster-X divergence of gene expression in Drosophila is driven by faster-X adaptation in cis regulatory sequences that affect X-linked expression levels. This result further suggests that many mutations that affect gene expression have recessive fitness effects, and additional empirical and theoretical work is needed to examine this hypothesis. However, this conclusion comes with the caveat that applying an MK framework to gene expression evolution requires simplifying assumptions about cis and trans variation that may not be biologically realistic.
Lastly, although the faster-X divergence of gene expression in both mammals and Drosophila is detected across multiple tissue types and developmental stages [72, 73, 75], it is especially pronounced amongst genes expressed in male reproductive tissues [62, 74, 75], which is similar to faster-X effects in protein coding genes (Figure 2). As with the protein-coding faster-X effect, it is unclear why genes with male-biased expression should represent the outlier gene category for faster-X divergence (Figure 1).
Future Considerations
As in most areas of molecular evolution and population genetics, theory outpaced data in the early study of faster-X evolution. The genome sequencing projects completed in the past decade have allowed for the first comprehensive tests of faster-X divergence and adaptation, but there still remains disagreement between theoretical predictions and empirical tests of the faster-X effect. For example, much of the theory contrasts rates of adaptive fixation on the X and autosomes, whereas most of the evidence for faster-X divergence combines both adaptive and nonadaptive substitutions. We therefore anticipate additional progress in this area by integrating divergence estimates (dN) with calculations of the frequency of adaptive fixations ( ) so that the rate of adaptive evolution relative to neutral substitutions (!a) [5] can be compared between X-linked and autosomal genes. Comparing rates of adaptive evolution will allow for a more coherent evaluation of empirical results within the framework of faster-X theory.
Although the increasing availability of population genomic data provides a much greater scope for testing hypotheses of faster-X adaptive evolution, it also introduces new challenges that must be overcome before inferences of faster-X adaptation can be accepted (e.g., those from Figure 3). MK-based approaches are useful for estimating the adaptively fixed component of amino acid substitutions, yet these tests yield biased results under several plausible evolutionary scenarios, including (i) population size changes, (ii) non-neutrality of non-synonymous polymorphisms and/or synonymous mutations, and (iii) hitchhiking under recurrent selective sweeps [77-79]. The importance of these biases are likely to differ between the X and autosomes, potentially generating false signatures of faster-X adaptation [78]. Selection on synonymous mutations represents an important and well-studied bias of this sort within Drosophila, where codon usage bias is higher on the X [33]. This limits the utility of dS as a mutation rate index in Drosophila because codon-bias could disproportionately inflate X-linked relative to autosomal dN/dS values [34] and upwardly bias MK-based estimates of X-linked adaptation. Novel statistical approaches that can control for systematic biases between chromosomes should therefore have great value for future faster-X studies.
We also foresee continued e orts to identify the consequences of faster-X molecular evolution on higher level evolutionary process, such as intra-genomic conflict, speciation, and phenotypic evolution [74, 80]. Gene expression represents a particularly promising phenotype for study within the faster-X context, as gene and phenotype (mRNA transcription level) are coupled (provided that transcriptional changes are in cis [73, 75]). On the other hand, faster-X models are largely framed in terms of nucleotide substitution rates rather than tempos of phenotypic change. Adopting different theoretical frameworks for phenotypic evolution, including gene expression, may therefore be warranted. These may include quantitative genetics models [8] or others that link genotype, phenotype, and fitness across distinct genetic systems [25].
Glossary Box.
Heterogametic sex: The sex carrying only one copy of the X or the Z chromosome; the homogametic sex carries two copies of the X (as in Drosophila and mammals) or the Z (as in birds and moths).
Beneficial mutation/allele: Genetic variant that confers increased fitness.
Neutral mutation/allele: Genetic variant that does not affect fitness.
Slightly deleterious mutation/allele: Genetic variant whose deleterious effect is below the level detectable by natural selection (typically s < ).
Genetic drift: The process whereby allele frequencies change as a result of random sampling of genes within a population of finite size. The effect of genetic drift at a gene is inversely proportional to its effective population size (Ne).
Effective population size (Ne): The size of an idealized population (with constant size, random mating, and no natural selection) that experiences a similar amount of genetic drift as a natural population.
Fixation probability: The probability that an allele that is present within a population eventually reaches a frequency of one within that population (i.e., it is eventually carried by every individual in the population/species).
Substitution rate: The rate at which genetic changes accumulate within an evolutionary lineage. Substitutions may become fixed by the action of natural selection or by random genetic drift.
dN: The number of non-synonymous (amino acid changing) substitutions standardized by the number of possible non-synonymous mutations in a gene.
dS: The number of synonymous (non-amino-acid-changing) substitutions standardized by the number of possible synonymous mutations in a gene.
McDonald-Kreitman (MK) test: Statistical test comparing non-synonymous (pN) and synonymous (pS) polymorphism and substitutions within a gene (dN and dS), often for the purpose of detecting a signature of historical adaptive evolution.
α: Proportion of substitutions that are fixed by positive selection, inferred within a MK-test framework, α = 1 − (dSpN)/(dNpS).
Highlights.
Differences between X chromosomes and autosomes illuminate the process of adaptation.
Theory predicts that X-linked genes may evolve faster than autosomal genes.
The evidence for the faster evolution of X-linked genes is mixed.
New theory and data analysis are needed to bridge this divide.
Acknowledgements
We thank Andy Clark, Rhiannon Macrae, and two anonymous reviewers for helpful comments on this manuscript. A. Kousathanas kindly shared unpublished work with us. We apologize to everyone whose work was excluded from this review because of space restrictions. The authors were supported by National Institutes of Health Grant R01 GM064590 to A. G. Clark and A. B. Carvalho during the writing of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- [1].Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sella G, et al. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:0. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mackay TFC, et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gossmann TI, et al. The effect of variation in the effective population size on the rate of adaptive molecular evolution in eukaryotes. Genome Biol. Evol. 2012;4:658–667. doi: 10.1093/gbe/evs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Haldane JBS. A mathematical theory of natural and artificial selection, Part V: selection and mutation. Proc. Camb. Philos. Soc. 1927;28:838–844. [Google Scholar]
- [7].Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat. Rev. Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- [8].Charlesworth B, et al. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 1987;130:113–146. [Google Scholar]
- [9].Orr HA, Betancourt AJ. Haldane’s sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kirkpatrick M, Hall DW. Male-biased mutation, sex linkage, and the rate of adaptive evolution. Evolution. 2004;58:437–440. [PubMed] [Google Scholar]
- [11].Vicoso B, Charlesworth B. effective population size and the faster-X effect: an extended model. Evolution. 2009;63:2413–26. doi: 10.1111/j.1558-5646.2009.00719.x. [DOI] [PubMed] [Google Scholar]
- [12].Mank JE, et al. Effective population size and the faster-X effect: empirical results and their interpretation. Evolution. 2010;64:663–674. doi: 10.1111/j.1558-5646.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- [13].Orr HA. The population genetics of beneficial mutations. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2010;365:1195–1201. doi: 10.1098/rstb.2009.0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Connallon T, et al. Impact of genetic architecture on the relative rates of X versus autosomal adaptive substitution. Mol. Biol. Evol. 2012;29:1933–1942. doi: 10.1093/molbev/mss057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kimura M, Ohta T. On the rate of molecular evolution. J. Mol. Evol. 1971;1:1–17. doi: 10.1007/BF01659390. [DOI] [PubMed] [Google Scholar]
- [16].Kimura M, Ohta T. Theoretical aspects of population genetics. Princeton University Press; 1971. [PubMed] [Google Scholar]
- [17].Ellegren H. Characteristics, causes and evolutionary consequences of male-biased mutation. Proc. R. Soc. Lond., B, Biol. Sci. 2007;274:1–10. doi: 10.1098/rspb.2006.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xu K, et al. Lineage-specific variation in slow- and fast-X evolution in primates. Evolution. 2012;66:1751–1761. doi: 10.1111/j.1558-5646.2011.01556.x. [DOI] [PubMed] [Google Scholar]
- [19].Weissman DB, Barton NH. Limits to the rate of adaptive substitution in sexual populations. PLoS Genet. 2012;8:e1002740. doi: 10.1371/journal.pgen.1002740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Charlesworth B, Charlesworth D. Elements of Evolutionary Genetics. Roberts and Company; 2010. [Google Scholar]
- [21].Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Christin PA, et al. Causes and evolutionary significance of genetic convergence. Trends Genet. 2010;26:400–405. doi: 10.1016/j.tig.2010.06.005. [DOI] [PubMed] [Google Scholar]
- [23].Mank JE, et al. Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution. 2011;65:2133–2144. doi: 10.1111/j.1558-5646.2011.01316.x. [DOI] [PubMed] [Google Scholar]
- [24].Meisel RP, et al. Disentangling the relationship between sex-biased gene expression and X-linkage. Genome Res. 2012;22:1255–1265. doi: 10.1101/gr.132100.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sellis D, et al. Heterozygote advantage as a natural consequence of adaptation in diploids. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20666–20671. doi: 10.1073/pnas.1114573108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- [27].Eyre-Walker A. The genomic rate of adaptive evolution. Trends Ecol. Evol. 2006;21:569–575. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- [28].Fay JC. Weighing the evidence for adaptation at the molecular level. Trends Genet. 2011;27:343–349. doi: 10.1016/j.tig.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Betancourt AJ, et al. A test for faster X evolution in Drosophila. Mol. Biol. Evol. 2002;19:1816–1819. doi: 10.1093/oxfordjournals.molbev.a004006. [DOI] [PubMed] [Google Scholar]
- [30].Counterman BA, et al. Using comparative genomic data to test for fast-X evolution. Evolution. 2004;58:656–660. [PubMed] [Google Scholar]
- [31].Musters H, et al. A genomic comparison of faster-sex, faster-X, and faster-male evolution between Drosophila melanogaster and Drosophila pseudoobscura. J. Mol. Evol. 2006;62:693–700. doi: 10.1007/s00239-005-0165-5. [DOI] [PubMed] [Google Scholar]
- [32].Thornton K, et al. X chromosomes and autosomes evolve at similar rates in Drosophila: no evidence for faster-X protein evolution. Genome Res. 2006;16:498–504. doi: 10.1101/gr.4447906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Singh ND, et al. Contrasting the e cacy of selection on the X and autosomes in Drosophila. Mol. Biol. Evol. 2008;25:454–467. doi: 10.1093/molbev/msm275. [DOI] [PubMed] [Google Scholar]
- [34].Vicoso B, et al. A multispecies approach for comparing sequence evolution of X-linked and autosomal sites in Drosophila. Genet. Res. 2008;90:421–431. doi: 10.1017/S0016672308009804. [DOI] [PubMed] [Google Scholar]
- [35].Grath S, Parsch J. Rate of amino acid substitution is influenced by the degree and conservation of male-biased transcription over 50 million years of Drosophila evolution. Genome Biol. Evol. 2012;4:346–359. doi: 10.1093/gbe/evs012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhou Q, Bachtrog D. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science. 2012;337:341–345. doi: 10.1126/science.1225385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Thornton K, Long M. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol. Biol. Evol. 2002;19:918–925. doi: 10.1093/oxfordjournals.molbev.a004149. [DOI] [PubMed] [Google Scholar]
- [38].Bhutkar A, et al. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics. 2008;179:1657–1680. doi: 10.1534/genetics.107.086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hu TT, et al. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 2013;23:89–98. doi: 10.1101/gr.141689.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lu J, Wu CI. Weak selection revealed by the whole-genome comparison of the X chromosome and autosomes of human and chimpanzee. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4063–4067. doi: 10.1073/pnas.0500436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hvilsom C, et al. Extensive X-linked adaptive evolution in central chimpanzees. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2054–2059. doi: 10.1073/pnas.1106877109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Baines JF, Harr B. Reduced X-linked diversity in derived populations of house mice. Genetics. 2007;175:1911–1921. doi: 10.1534/genetics.106.069419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Borge T, et al. Contrasting patterns of polymorphism and divergence on the Z chromosome and autosomes in two Ficedula flycatcher species. Genetics. 2005;171:1861–1873. doi: 10.1534/genetics.105.045120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mank JE, et al. Fast-X on the Z: rapid evolution of sex-linked genes in birds. Genome Res. 2007;17:618–624. doi: 10.1101/gr.6031907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ellegren H. Genomic evidence for a large-Z effect. Proc. R. Soc. Lond., B, Biol. Sci. 2009;276:361–366. doi: 10.1098/rspb.2008.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sackton TB, et al. Positive selection drives faster-Z evolution in silkmoths. in review. arXiv, 1304.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mank JE, et al. Faster-Z evolution is predominantly due to genetic drift. Mol. Biol. Evol. 2010;27:661–670. doi: 10.1093/molbev/msp282. [DOI] [PubMed] [Google Scholar]
- [49].Jaquiery J, et al. Accelerated evolution of sex chromosomes in aphids, an X0 system. Mol. Biol. Evol. 2012;29:837–847. doi: 10.1093/molbev/msr252. [DOI] [PubMed] [Google Scholar]
- [50].Presgraves DC. Recombination enhances protein adaptation in Drosophila melanogaster. Curr. Biol. 2005;15:1651–1656. doi: 10.1016/j.cub.2005.07.065. [DOI] [PubMed] [Google Scholar]
- [51].Connallon T. Adaptive protein evolution of X-linked and autosomal genes in Drosophila: implications for faster-X hypotheses. Mol. Biol. Evol. 2007;24:2566–2572. doi: 10.1093/molbev/msm199. [DOI] [PubMed] [Google Scholar]
- [52].Baines JF, et al. Effects of X-linkage and sex-biased gene expression on the rate of adaptive protein evolution in Drosophila. Mol. Biol. Evol. 2008;25:1639–1650. doi: 10.1093/molbev/msn111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Langley CH, et al. Genomic variation in natural populations of Drosophila melanogaster. Genetics. 2012;192:533–598. doi: 10.1534/genetics.112.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Thornton K, Long M. Excess of amino acid substitutions relative to polymorphism between X-linked duplications in Drosophila melanogaster. Mol. Biol. Evol. 2005;22:273–284. doi: 10.1093/molbev/msi015. [DOI] [PubMed] [Google Scholar]
- [56].Carneiro M, et al. Evidence for widespread positive and purifying selection across the european rabbit (Oryctolagus cuniculus) genome. Mol. Biol. Evol. 2012;29:1837–1849. doi: 10.1093/molbev/mss025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ellegren H, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- [58].Hogner S, et al. Increased divergence but reduced variation on the Z chromosome relative to autosomes in Ficedula flycatchers: differential introgression or the faster-Z effect? Ecol. Evol. 2012;2:379–396. doi: 10.1002/ece3.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Müller L, et al. Inter- and intraspecific variation in Drosophila genes with sex-biased expression. Int. J. Evol. Biol. 2012;2012:963976. doi: 10.1155/2012/963976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Torgerson DG, Singh RS. Sex-linked mammalian sperm proteins evolve faster than autosomal ones. Mol. Biol. Evol. 2003;20:1705–1709. doi: 10.1093/molbev/msg193. [DOI] [PubMed] [Google Scholar]
- [61].Torgerson DG, Singh RS. Enhanced adaptive evolution of sperm-expressed genes on the mammalian X chromosome. Heredity. 2006;96:39–44. doi: 10.1038/sj.hdy.6800749. [DOI] [PubMed] [Google Scholar]
- [62].Khaitovich P, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- [63].Stevenson B, et al. Rapid evolution of cancer/testis genes on the X chromosome. BMC Genomics. 2007;8:129. doi: 10.1186/1471-2164-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Guo X, et al. Rapid evolution of mammalian X-linked testis microR-NAs. BMC Genomics. 2009;10:97. doi: 10.1186/1471-2164-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sin HS, et al. Human postmeiotic sex chromatin and its impact on sex chromosome evolution. Genome Res. 2012;22:827–836. doi: 10.1101/gr.135046.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Connallon T, Clark AG. Association between sex-biased gene expression and mutations with sex-specific phenotypic consequences in Drosophila. Genome Biol Evol. 2011;3:151–155. doi: 10.1093/gbe/evr004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- [68].Betancourt AJ, et al. A pseudohitchhiking model of X vs. autosomal diversity. Genetics. 2004;168:2261–2269. doi: 10.1534/genetics.104.030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Vicoso B, Charlesworth B. Recombination rates may a ect the ratio of X to autosomal noncoding polymorphism in African populations of Drosophila melanogaster. Genetics. 2009;181:1699–1701. doi: 10.1534/genetics.108.098004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Charlesworth B. The role of background selection in shaping patterns of molecular evolution and variation: evidence from variability on the Drosophila X chromosome. Genetics. 2012;191:233–246. doi: 10.1534/genetics.111.138073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- [73].Kayserili MA, et al. An excess of gene expression divergence on the X chromosome in Drosophila embryos: implications for the faster-X hypothesis. PLoS Genet. 2012;8:e1003200. doi: 10.1371/journal.pgen.1003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Llopart A. The rapid evolution of X-linked male-biased gene expression and the large-X effect in Drosophila yakuba, D. santomea, and their hybirds. Mol. Biol. Evol. 2012;29:3873–3886. doi: 10.1093/molbev/mss190. [DOI] [PubMed] [Google Scholar]
- [75].Meisel RP, et al. Faster-X evolution of gene expression in Drosophila. PLoS Genet. 2012;8:e1003013. doi: 10.1371/journal.pgen.1003013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Meiklejohn CD, et al. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9894–9899. doi: 10.1073/pnas.1630690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Charlesworth J, Eyre-Walker A. The McDonald-Kreitman test and slightly deleterious mutations. Mol. Biol. Evol. 2008;25:1007–1015. doi: 10.1093/molbev/msn005. [DOI] [PubMed] [Google Scholar]
- [78].Campos JL, et al. Codon usage bias and effective population sizes on the X chromosome versus the autosomes in Drosophila melanogaster. Mol. Biol. Evol. 2013;30:811–823. doi: 10.1093/molbev/mss222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Messer PW, Petrov DA. The McDonald-Kreitman test and its extensions under frequent adaptation: problems and solutions. in review. arXiv, 1211.0060. [Google Scholar]
- [80].Meiklejohn CD, Tao Y. Genetic conflict and sex chromosome evolution. Trends Ecol. Evol. 2010;25:215–223. doi: 10.1016/j.tree.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]