Snapshots of computer systems that are stored and shared ‘in the cloud’ could make computational analyses more reproducible
Scientific findings must be reproducible for them to be formally accepted by colleagues, practitioners, policy makers and the broader lay public. Although it was once thought that computers would improve reproducibility because they yield repeatable results given the same set of inputs, most software tools do not provide mechanisms to package a computational analysis such that it can be easily shared and reproduced. This inability to easily exchange the computational analyses behind published results is a substantial roadblock to reproducibility across scientific disciplines1.
A number of innovative solutions to this problem have been proposed. Most efforts have focused on software tools that aim to standardize the creation, representation and sharing of computational ‘workflows’2 that tie several software tools together into a single analysis. These workflow tools provide a way to represent discrete computational tasks (e.g. processing an input data file) as computational modules that can be connected into workflows by linking the output of one module with the input of another (e.g. the output from the input data processing module connects to the input of a data normalization module)3. Several of these tools allow researchers to build complex computational workflows through drag-and-drop visual interfaces and to share standardized representations of workflows. Examples include GenePattern4 for analyzing genomic data, the Trident Scientific Workflow Workbench for oceanography (http://www.microsoft.com/mscorp/tc/trident.mspx), Teverna5 for generalized scientific computation, and web-based resources such as myExperiment6 for sharing workflows. Recently, a software solution was described7 that embeds access to computational systems directly into digital representations of scientific papers.
Human nature trumps technology
Existing software applications have not become established solutions to the problem of computational reproducibility. This is not due to any technical shortcomings of the software because in fact, many programs are technically well designed. Rather, the failures are a consequence of human nature and the realities of data-driven science, including (i) efforts are not rewarded by the current academic research and funding environment8, 9; (ii) commercial software vendors tend to protect their markets through proprietary formats and interfaces10; (iii) investigators naturally tend to want to ‘own’ and control their research tools; (iv) even the most generalized software will not be able to meet the needs of every researcher in a field; and finally (v) the need to derive and publish results as quickly as possible precludes the often slower standards-based development path9.
The consequence is that non-standardized research computational pipelines will continue to be developed, especially as new types of molecular measurements generate quantities of data that most standardized software packages cannot handle 11. We conclude that any effort to establish standard protocols that require investigators to adopt a particular software system, creating a real or perceived threat of ‘lock in’ to a single lab’s or commercial vendor’s software, is likely to be met with disregard or even resistance and will fail to drastically change the current behavior of researchers.
Whole system snapshot exchange
Given these realities, we propose capturing and exchanging computational pipelines using complete digital representations of the entire computing environment needed to execute the pipeline. In this approach, which we call whole system snapshot exchange (WSSE), the computer system(s) used by researchers to produce experimental results are copied in their entirety, including the operating system, application software and databases, into a single digital image that can be exchanged with other researchers. Through WSSE, researchers would be able to obtain precise replicas of a computational system used to produce the published results, and have the ability to restore this system to the precise state in which the system existed when the experimental results were generated. Even subtle variances caused by discrepancies between specific versions of software or programming languages are avoided by the WSSE approach. In principle, the input data processed by the pipeline to produce the published results could be exchanged along with the pipeline.
Readers will be quick to realize that the WSSE approach could involve the exchange of data files that might range in size from tens of gigabytes to several terabytes or more. Even with the fastest internet connections, it is not feasible to share such large files easily and efficiently. Thus, it is not our intention that WSSE operate desktop-to-desktop, but rather we propose that data and computational pipelines will be exchanged exclusively using cloud computing, defined here as “computing in which dynamically scalable and virtualized resources are provided as a service over the Internet” 13.
In cloud computing, computation and data are ‘virtualized’, meaning that software and data are not tied to physical computing resources, such as a specific server with hard drives plugged into it. Instead, cloud computing infrastructures are comprised of large and often geographically disparate clusters of computing hardware that are made to appear as a single, homogeneous computational environment14. Since software and computations run across computing resources virtually in the cloud, it is possible for others to clone and reconstitute them in the cloud without any concern for the specifics of the underlying hardware. The virtualization of data in the cloud makes it possible move or copy ‘snapshots’ of large data sets from point-to-point within the cloud at high transfer rates, without the need to associate particular machines or storage drives at either the source or destination of the data transfer. Some cloud providers offer technologies that enable the creation of large databases that can persist as omnipresent data resources that can be accessed by any computation in the cloud.
Concerns have been voiced12 that scientific computing in the cloud could make results less reproducible. Primarily, there are concerns that cloud computing will serves as a large computing ‘black box’, potentially obfuscating important factors or details underlying the production of scientific results by software running on proprietary cloud architectures. Although this is an important concern, we argue that cloud computing infrastructures can serve to dramatically increase the transparency of scientific computing. The virtualization of data and systems in the cloud makes the handover and scrutiny of entire computational infrastructures underlying computational results practical. Historically, software and data used to derive results would need to be modified and repackaged for distribution, and the receiver would be required to possess the infrastructure necessary for peer evaluation of computational methods, which could be substantial. Furthermore, wholesale digital ‘snapshots’ of data and source code are easily produced and distributed within the cloud, which can serve as a basic means to track the provenance of data and source code in scientific computing in lieu of more formalized procedures -- which historically have proven difficult to gain in general acceptance or widespread use.
Cloud-based support for reproducibility
Cloud computing could support reproducibility in several ways, which correspond to different ‘layers’ for accessing computing resources (Fig. 1).
Figure 1.
Layers of reproducible computing in the cloud
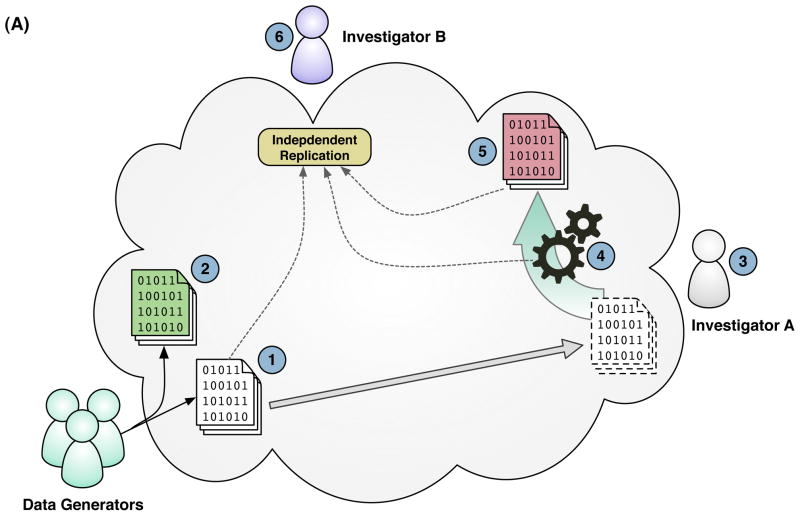
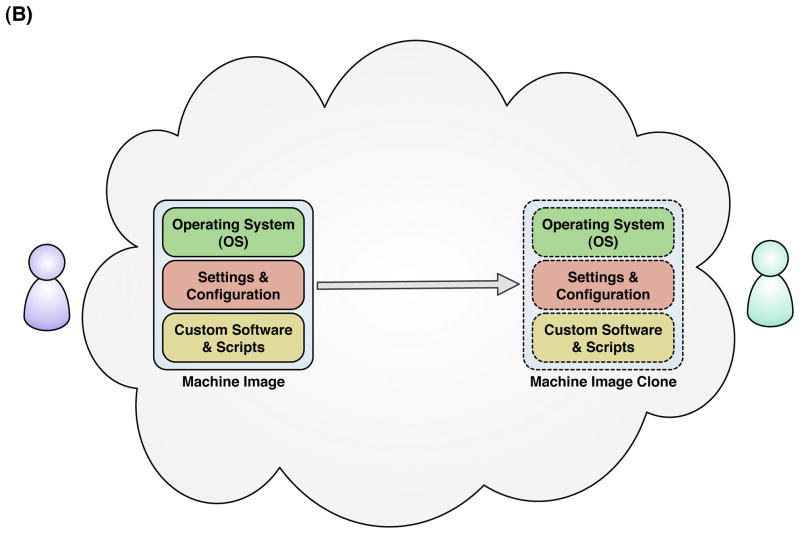
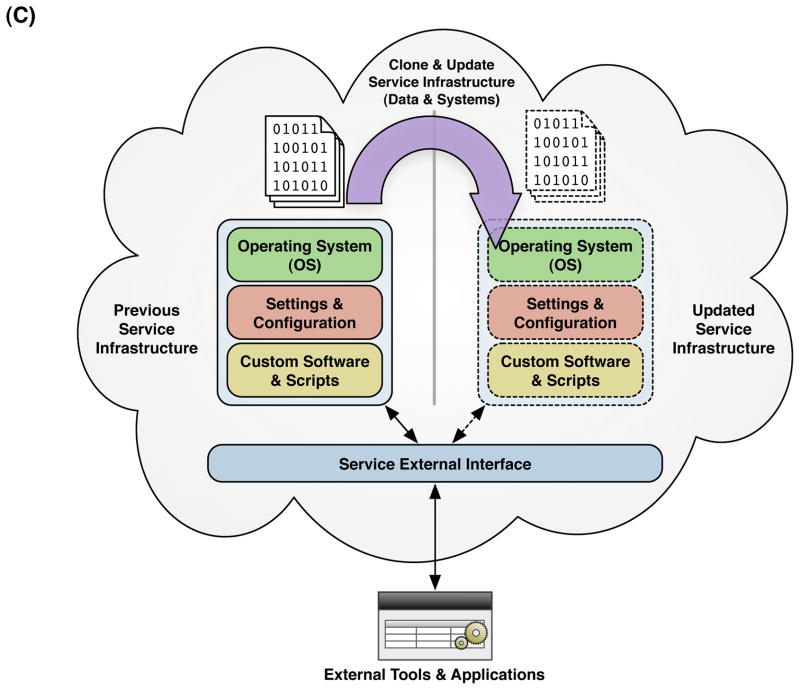
The reproducibility of scientific compuitng in the cloud can be understood at three layers of scientific computation. (A) Data Layer: Generators of large scientific data sets can publish their data to the cloud as large data volumes (1) and substantial updates to these data volumes can exist in parallel without loss or modification of the previous volume (2). Primary investigators can clone entire data volumes within the cloud (3) and apply custom scripts or software computations (4) to derive published results (5). An indepdendent investigator can obtain digital replicates of the original primary data set, software, and published results within the cloud to replicate a published analysis and compare with published results (6). (B) System Layer: Investigators can set up and conduct scientific computations using cloud-based virtual machine images, incorporating all the software, configuration, and scripts necessary to execute the analysis. The customized machine image can be cloned wholesale and shared with other investigators within the cloud for replicate analyses. (C) Service Layer: Instead of making in-place modifications or updates to the systems or data comprising the underlying infrastructure of a scientific computing service, the entire infrastructure can be virrtualized in the cloud and cloned prior to update or modification to retain the state and characteristics of the previous version of the service. Requests made by external tools or applications through the external service interface could incorporate a version parameter into requests to the service, so that published results citing previous versions of the service can be evaluated for reproducibility.
First, at the data layer, cloud computing enables data sets to be easily stored and shared virtually—that is, without necessarily copying it to another computer. Most importantly, when a data set has been used to derive published results, it can be copied and archived in the cloud. This would be facilitated if large public data repositories are regularly versioned and moved to the cloud. One cloud computing vendor, Amazon Web Services (AWS), has already taken the initiative to archive many such public data sets. Their extensive online catalog (viewable at: http://aws.amazon.com/publicdatasets/) contains large public data sets from various domains of science, including astronomy, biology, chemistry and climatology. Second, at the system layer, cloud computing allows snapshots of complete computer systems to be exchanged, as proposed in WSSE. This addresses observations that computer systems can substantially confound the reproducibility of analyses15
A higher-level layer of scientific computation in the cloud is the service layer, in which computational services are exposed to external applications through some form of programming interface16. Examples include the Entrez Utilities from NCBI (http://eutils.ncbi.nlm.nih.gov) that provide access to data and computational resources from NCBI’s repertoire of bioinformatics applications and databases. From the standpoint of reproducibility, there are problems introduced by such a service-oriented approach to scientific computing: the underlying application supporting the computational service may be altered significantly without any apparent change to the public-facing service interface. If the applications and infrastructure supporting such services were migrated to a cloud computing environment, the application service providers could run and maintain replicate instances of their entire application to maintain access to previous versions without the need to duplicate the hardware infrastructure underlying the service. Alternatively, the component data and systems underlying a computational service could be archived as ‘images’ in the cloud in a non-active state, which would provide an economical means of preservation for the service provider while maintaining an efficient route of access to previous versions of a computational service for investigators. This may make it more feasible for authors to cite specific versions of data, systems, and services in publications describing their results, and have a means to easily direct colleagues to these resources for reproducibility and reuse.
Reproducibility through preservation
One often-overlooked barrier to reproducibility is the loss or abandonment of grant-funded databases and other computational resources17. Therefore, one advantage of cloud computing is that virtual machines and cloud-based data storage can offer a means to sustain bioinformatics projects after funding cuts, project termination or abandonment. Although it may be easy to demonstrate the need to make biomedical databases available after their original purpose has been fulfilled, from a practical standpoint, funding the maintenance of these databases has become a contentious issue for funding agencies, who must balance maintenance with declining budgets and the need to fund new investigators and initiatives. For example, the Arabidopsis Information Resource (TAIR), used by thousands of researchers each day, recently fell into disarray after its funding was terminated after 10 years of support from the National Science Foundation18. Instead of turning off these servers and losing access to these resources, virtual machine images of the server could have been created and distributed across the Internet, and executed on- demand through cloud computing, ensuring that investments in bioinformatics resources such as TAIR could be used for years after discontinuation.
The costs associated with preserving data in the a cloud computing environment are relatively minimal, making it feasible for research groups and institutions to preserve data through extended gaps in funding, or to fulfill commitments to collaborators or stakeholders well beyond the defunding of the primary database infrastructure. To illustrate, the current rate for storage space in Amazon’s cloud service is $0.10 USD per gigabyte of storage per month. To maintain even an exceptionally large data set comprising 1 terabyte of data would require nominal costs of approximately $100 USD per month. As the per gigabyte cost of storage is expected to decrease with time, it is likely that a 1 terabyte biomedical database could be preserved in accessible form in the cloud for more than 10 years at a total cost well below $10,000 USD. The cost of this active preservation could be written into the budgets of all proposals for creation and renewal of biomedical databases to address the problem of preservation and access to data beyond proposed funding periods. Moreover, having a working virtual machine server along with open source code could enable distributed teams of investigators to informally support these disbanded projects.
Towards a cloud-based scientific computing commons
Although the cloud computing technology required to facilitate straightforward approaches to reproducible computing, such as WSSE, are now widely available, there are a number measures that could be taken to help and encourage the utilization of cloud-based resources by the broader scientific community. As nearly every scientific discipline is becoming data-driven, one of the most enticing benefits of cloud computing is the means to aggregate scientific data sets efficiently and economically. The aggregation and centralization of public scientific data into the cloud, which offers a unified, location-independent platform for data and computation, might work to establish a shared virtual commons for reproducible scientific computing. For example, it would be possible to develop and implement cloud-based scientific computational analysis that assume and source public data from centralized, cloud-based master catalogs. This computational analysis could then be shared and distributed within the cloud as part of a standardized system image, After reconstitution of the system image, the computational analysis could be reproducibly executed, because the same source code would be executing within the same system environment, drawing the same data from the master data catalog. Journals and funding agencies could support this vision by mandating that more types of measurements be deposited into approved cloud-based data catalogs, and funding could be provided for existing consortia to move their valuable data from isolated data silos into centralized cloud-based catalogs. As cloud computing becomes commonplace, we expect technologies to emerge that allow one to move data between cloud computing providers, to prevent ‘lock in’ 19. For example, the open-source Eucalyptus platform (http://www.eucalyptus.com) is enables one to move cloud-based resources out of the Amazon Web Services commercial platform into a private cloud infrastructure.
Some have already called for the creation of a publicly-funded cloud computing infrastructure14; however, we suggest that it would be most prudent to focus funds and effort into building cloud-based scientific computing software on top of existing cloud infrastructures in such a way that they portable across vendors. Given the economic and competitive pressures facing commercial cloud computing vendors, as well as their substantial technical and capital resources, it is not clear that a publicly funded cloud infrastructure would offer any significant technical or economic advantages over commercial clouds. Furthermore, we suggest that, as has been observed in many aspects of computing, standards enabling cross-cloud interoperability are likely to emerge as cloud computing becomes more prominent and additional vendors increase competition in the market. Groups such as the Open Cloud Consortium (http://opencloudconsortium.org) have already been formed to explore this issue. Notably, many of the popular tools for managing and interacting with cloud-based infrastructures, such as Chef (http://www.opscode.com/chef), are designed in such a way that they are independent from the specifics of any one cloud computing vendor’s infrastructure. Peer-to-peer technologies such as BitTorrent, which is already being leveraged in the bioinformatics community20, can be used as a failover solution to ensure that data sets persist and remain accessible independent of the viability of any single cloud computing provider.
In our domain of biomedicine, a number of efforts towards the development and distribution of cloud-based tools, systems and other reproducible resources for cloud-based biomedical computing have recently emerged21. Several groups have created standardized machine images with software tools and configuration settings optimized for biomedical research. The most ambitious among these so far is the J. Craig Venter Institute Cloud Bio-Linux project (http://www.jcvi.org/cms/research/projects/jcvi-cloud-biolinux/), which aims to provide a comprehensive and coherent system capable of a broad range of bioinformatics functionality. These pre-configured machine images are made publicly available to the research community, providing individual investigators with a standardized, tuned bioinformatics platform for cloud-based computational analyses. Other efforts have gone into the creation of more comprehensive multipart systems for facilitating biocomputing in the cloud22, 23. A significant effort in this area is the Galaxy project24, which provides a platform for large-scale genomic analysis.
Although we suggest that the WSSE approach is a pragmatic and substantial first step towards enabling reproducible scientific computing in the cloud, we acknowledge that it does not address all aspects hindering reproducibility. Foremost, software licensing constraints could prevent the use of WSSE. Many commercial software applications are licensed to specific individuals, and will often constrain the number of software application instances that can run simultaneously per license, or restrict execution to a number of processors per license. These constraints might prevent an investigator from sharing some or all the system or software components of used to produce published results, limiting the effectiveness of WSSE. We also recognize that there is a need for continued research and development into reproducibility enhancements at levels above WSSE. For example, problems in data organization, systematic provenance tracking, standardization and annotation are not solved by WSSE. Nonetheless, we suggest that WSSE can serve to initiate a value-driven movement towards general reproducible scientific data and computing into the cloud, and that solutions to problems of reproducibility not solved by WSSE – many of which already exist in some form – will follow WSSE into the cloud towards the realization of a comprehensive platform for reproducible computing enabled by the technical innovations of cloud computing.
Table 1.
Features of reproducible scientific computing in the cloud
| Traditional challenges | Cloud computing solutions | |
|---|---|---|
| Data Sharing |
|
|
| Software & Applications |
|
|
| System & Technical |
|
|
| Access & Preservation |
|
|
Acknowledgments
J. Dudley is supported by NLM Biomedical Informatics Training Grant (T15 LM007033) and A. Butte is supported by the National Institute for General Medical Sciences (R01 GM079719). We thank Deepak Singh for valuable discussion regarding cloud computing technology.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Gentleman R. Reproducible research: a bioinformatics case study. Stat Appl Genet Mol Biol. 2005;4:Article2. doi: 10.2202/1544-6115.1034. [DOI] [PubMed] [Google Scholar]
- 2.Gil Y, et al. Examining the Challenges of Scientific Workflows. Computer. 2007;40:24–32. [Google Scholar]
- 3.Barker A, van Hemert J. Parallel Processing and Applied Mathematics. 2008:746–753. [Google Scholar]
- 4.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 5.Hull D, et al. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Roure D, Goble C, Stevens R. The design and realisation of the Virtual Research Environment for social sharing of workflows. Future Generation Computer Systems. 2009;25:561–567. [Google Scholar]
- 7.Mesirov JP. Computer science. Accessible reproducible research. Science. 2010;327:415–416. doi: 10.1126/science.1179653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball CA, Sherlock G, Brazma A. Funding high-throughput data sharing. Nat Biotechnol. 2004;22:1179–1183. doi: 10.1038/nbt0904-1179. [DOI] [PubMed] [Google Scholar]
- 9.Brooksbank C, Quackenbush J. Data standards: a call to action. OMICS. 2006;10:94–99. doi: 10.1089/omi.2006.10.94. [DOI] [PubMed] [Google Scholar]
- 10.Wiley HS, Michaels GS. Should software hold data hostage? Nat Biotechnol. 2004;22:1037–1038. doi: 10.1038/nbt0804-1037. [DOI] [PubMed] [Google Scholar]
- 11.Lynch C. Big data: How do your data grow? Nature. 2008;455:28–29. doi: 10.1038/455028a. [DOI] [PubMed] [Google Scholar]
- 12.Osterweil LJ, Clarke LA, Ellison AM. Forecast for Reproducible Data: Partly Cloudy. Science. 2009;325:1622-b. doi: 10.1126/science.325_1622b. [DOI] [PubMed] [Google Scholar]
- 13.Bateman A, Wood M. Cloud computing. Bioinformatics. 2009;25:1475. doi: 10.1093/bioinformatics/btp274. [DOI] [PubMed] [Google Scholar]
- 14.Nelson MR. Building an Open Cloud. Science. 2009;324:1656–1657. doi: 10.1126/science.1174225. [DOI] [PubMed] [Google Scholar]
- 15.Schwab M, Karrenbach M, Claerbout J. Making scientific computations reproducible. Computing in Science and Engg. 2000;2:61–67. [Google Scholar]
- 16.Wagener J, Spjuth O, Willighagen EL, Wikberg JE. XMPP for cloud computing in bioinformatics supporting discovery and invocation of asynchronous web services. BMC Bioinformatics. 2009;10:279. doi: 10.1186/1471-2105-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merali Z, Giles J. Databases in peril. Nature. 2005;435:1010–1011. doi: 10.1038/4351010a. [DOI] [PubMed] [Google Scholar]
- 18.Access denied? Nature. 2009;462:252. doi: 10.1038/462252a. [DOI] [PubMed] [Google Scholar]
- 19.Gu Y, Grossman RL. Sector and Sphere: the design and implementation of a high-performance data cloud. Philos Transact A Math Phys Eng Sci. 2009;367:2429–2445. doi: 10.1098/rsta.2009.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langille MG, Eisen JA. BioTorrents: a file sharing service for scientific data. PLoS One. 2010;5:e10071. doi: 10.1371/journal.pone.0010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein LD. The case for cloud computing in genome informatics. Genome Biol. 2010;11:207. doi: 10.1186/gb-2010-11-5-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL. Searching for SNPs with cloud computing. Genome Biol. 2009;10:R134. doi: 10.1186/gb-2009-10-11-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schatz MC. CloudBurst: highly sensitive read mapping with MapReduce. Bioinformatics. 2009;25:1363–1369. doi: 10.1093/bioinformatics/btp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giardine B, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halligan BD, Geiger JF, Vallejos AK, Greene AS, Twigger SN. Low cost, scalable proteomics data analysis using Amazon’s cloud computing services and open source search algorithms. J Proteome Res. 2009;8:3148–3153. doi: 10.1021/pr800970z. [DOI] [PMC free article] [PubMed] [Google Scholar]