Abstract
It is now well established that tumor cell invasion through tissue is strongly regulated by the microstructural and mechanical properties of the extracellular matrix (ECM). However, it remains unclear how these physical microenvironmental inputs are jointly processed with oncogenic lesions to drive invasion. In this study, we address this open question by combining a microfabricated polyacrylamide channel (μPAC) platform that enables independent control of ECM stiffness and confinement with an isogenically-matched breast tumor progression series in which the oncogenes ErbB2 and 14-3-3ζ are overexpressed independently or in tandem. We find that increasing channel confinement and overexpressing ErbB2 both promote cell migration to a similar degree when other parameters are kept constant. In contrast, 14-3-3ζ overexpression slows migration speed, and does so in a fashion that dwarfs effects of ECM confinement and stiffness. We also find that ECM stiffness dramatically enhances cell motility when combined with ErbB2 overexpression, demonstrating that biophysical cues and cell-intrinsic parameters promote cell invasion in an integrative manner. Morphometric analysis of cells inside the μPAC platform reveals that the rapid cell migration induced by narrow channels and ErbB2 overexpression both are accompanied by increased cell polarization. Disruption of this polarization by pharmacological inhibition of Rac GTPase phenocopies 14-3-3ζ overexpression by reducing cell polarization and slowing migration. By systematically measuring migration speed as a function of matrix stiffness and confinement, we also quantify for the first time the sensitivity of migration speed to microchannel properties and transforming potential. These results demonstrate that oncogenic lesions and ECM biophysical properties can synergistically interact to drive invasive migration, and that both inputs may act through common molecular mechanisms to enhance migration speed.
Introduction
The malignant progression of breast tumors is a multi-step process triggered in part by specific oncogenic mutations.1 Among the key regulators of malignancy in breast tumors are the oncogenes ErbB2 and 14-3-3ζ, mutations of which are correlated with poor patient survival.2,3 These two oncogenes contribute to the clinical progression of breast tumors in a somewhat synergistic manner; whereas 14-3-3ζ is markedly overexpressed during initial stages of malignant transformation,2 ErbB2 is overexpressed in later stages, where it promotes tumor invasion.4 ErbB2 is a transmembrane receptor kinase of the epidermal growth factor receptor (EGFR) family of proteins,4–6 members of which directly and indirectly associate with integrins and have been closely associated with tumor cell migration and chemotaxis.4–7 Thus, microenvironmental factors that regulate tumor cell migration, such as extracellular matrix (ECM) stiffness and microstructure, might also be expected to influence how ErbB2 lesions control tumorigenesis. For example, Weaver and colleagues found that while concomitant ECM stiffening and ErbB2 overexpression can induce an invasive phenotype in mammary epithelial acini, neither manipulation is capable of doing so on its own.7 Similarly, 14-3-3ζ is a member of the 14-3-3 protein family implicated in survival and apoptosis resistance, both of which are now understood to be strongly regulated by ECM-derived and integrin-mediated biophysical cues.2,3,8 These regulatory relationships are particularly important in the context of tumor metastasis to distant sites, which involves superposition of these and other cell-intrinsic oncogenic lesions upon the ECM, which tumor cells must successfully traverse in order to successfully invade and metastasize.9–12 During this process, cells encounter ECM environments of varying stiffness, degree of confinement, ligand density, and other microstructural parameters that critically regulate cell migration.10–12 While the effects of oncogenic lesions and ECM properties on tumor cell motility have been studied separately, the field’s understanding of how oncogenic lesions interact with ECM microstructural parameters to promote tumor cell invasion remains unclear.
An important challenge in mapping tumor-relevant phenotypic behaviors to specific oncogenic lesions is the need to isolate these effects by placing the lesions on a common genetic background. To this end, Yu and colleagues recently developed an isogenically-matched progression series of human MCF10A mammary epithelial cells (MECs) in which the oncogenes ErbB2 and 14-3-3ζ are overexpressed either independently or in tandem.2,3,13–15 A variety of preclinical and clinical evidence shows that the overexpression of ErbB2 and 14-3-3ζ can suppress p53 expression and contribute to the transformation of ductal carcinoma in situ into invasive breast cancer, and that overexpression of both proteins together has a greater impact than over expression with either one alone.2,3,16 In a soft agar assay, cells co-overexpressing both ErbB2 or 14-3-3ζ formed agar colonies, a characteristic behavior of MCF10A cells exhibiting adhesion and motility, while the cells overexpressing only ErbB2 or 14-3-3ζ remained relatively immobile within soft agar suspension.3 When these four cell lines were grown in 3D Matrigel, cells overexpressing both ErbB2 and 14-3-3ζ showed distinctly increased invasive capacity and disrupted acinar structures compared to the cells overexpressing either ErbB2 or 14-3-3ζ alone.3 Thus, overexpression of each protein can be regarded as lying along a continuum of “transforming potentials” that describes the capacity of each genetic lesion to alter cellular invasive properties.
More recently, the stiffness-dependent motility of this progression series has been characterized using both traditional 2D protein-coated glass ECMs and 3D collagen ECMs in which stiffness was controlled by varying collagen density.14 Zaman and colleagues found that ErbB2 overexpression produced more rapid cell motility while 14-3-3ζ overexpression (both in isolation and in tandem with ErbB2 overexpression) slowed motility on collagen-coated 2D glass surfaces. However, inside 3D collagen gels, this trend was partially reversed, with 14-3-3ζ overexpression modestly promoting motility.14 When a stiffer 3D collagen gel was used, the reversal from 2D became even more complex, with overexpression of either ErbB2 or 14-3-3ζ alone suppressing motility but overexpression of both ErbB2 and 14-3-3ζ modestly increasing it.14 This intriguing result suggests that specific features of the 3D ECM might play an important role in governing phenotypic responses to ErbB2 and 14-3-3ζ expression; however, identifying which features are especially important to this regulation is rather complicated given that it is difficult to vary ECM stiffness without influencing other properties relevant to cell migration. For example, changing collagen stiffness by varying protein concentration also alters integrin ligand density, pore size, and fiber architecture. This exemplifies an important unmet need in the field to understand how individual biophysical properties of 3D ECMs contribute to oncogenic regulation of tumor invasion.
To this end, we recently developed a new paradigm for studying microenvironmental regulation of tumor invasion based on microfabrication of polyacrylamide channels (μPACs),17 which complements a growing set of microchannel-based material systems.18–20 In this system, we fabricate microchannels of defined size from polyacrylamide (PA) gels of defined stiffness, thus enabling us to control stiffness and channel size independently of one another. We then functionalize these channels with full-length ECM proteins to confer adhesive functionality, introduce cells, and track their motility as they migrate through the channels. By decoupling matrix stiffness and matrix pore size, this system offers a unique opportunity to investigate how each of these parameters control tumor cell invasion in response to specific cell-intrinsic or extrinsic factors. We now apply this ECM platform to the MCF10A ErbB2/14-3-3ζ progression series with the goal of addressing the following questions: (1) Does ECM stiffness regulate the motility-regulatory effects of ErbB2 and/or 14-3-3ζ expression in the absence of geometric confinement? (2) Do these regulatory relationships change when confinement is progressively introduced? And finally: (3) Can ECM stiffness alone affect the dependence of cell motility on transforming potential and matrix confinement? To address these questions, we measured cell migration speeds across matrix stiffness and confinement for varying transforming potentials and examined the sensitivity of cell motility to each of three parameters. We find that overexpression of ErbB2 and increased confinement each promote directional cell migration to a much greater degree than increases in matrix stiffness. Furthermore, we show that enhanced migration speed is closely correlated with the morphological polarization of cells inside the channels, both of which vary with transforming potential and may be reduced by inhibition of Rac GTPase.
Results
To independently investigate effects of ECM stiffness and confinement on migration speed, we employed a microfabricated polyacrylamide channel (μPAC) platform we recently introduced (Figure 1).17 In this system, we use standard photolithography methods to fabricate silicon masters with a negative relief of the microchannels and then assemble a polyacrylamide (PA) hydrogel of defined stiffness against these masters to create an array of microchannels with defined stiffness and geometry. We then functionalize these PA surfaces with full-length ECM proteins (in this study, collagen I) to enable cell adhesion. To study how targeted overexpression of the oncogenes ErbB2 and 14-3-3ζ might interact with matrix stiffness and confinement to regulate tumor cell motility, we introduced to this system a previously-characterized isogenically-matched breast tumor progression series of MCF10A mammary epithelial cells in which cells overexpress either ErbB2, 14-3-3ζ, both oncogenes simultaneously, or no protein (empty vector control).14,15
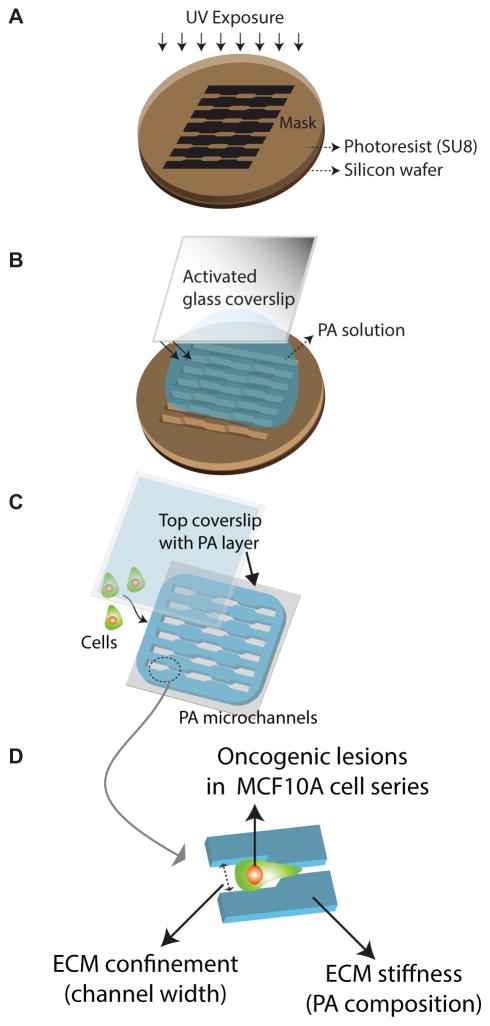
Figure 1. Schematic of cell-ECM model system.
Fabrication procedure of polyacrylamide (PA) microchannels that includes (A) soft photolithography to fabricate silicon master with topographic patterns matching microchannels of desired dimensions, (B) polymerization of PA between an activated glass coverslip and the silicon master that results in PA molding around the microchannels, and (C) seeding cell in prescribed culture conditions on the PA microchannels of desired stiffness and topography. (D) A schematic description of the variables that define the cell-ECM model system used in this study – namely, oncogenic lesions of varying transforming potentials in MCF10A cell lines, ECM confinement enabled by the channel width, and ECM stiffness manipulated by PA composition.
Transforming potential and channel confinement independently regulate cell motility
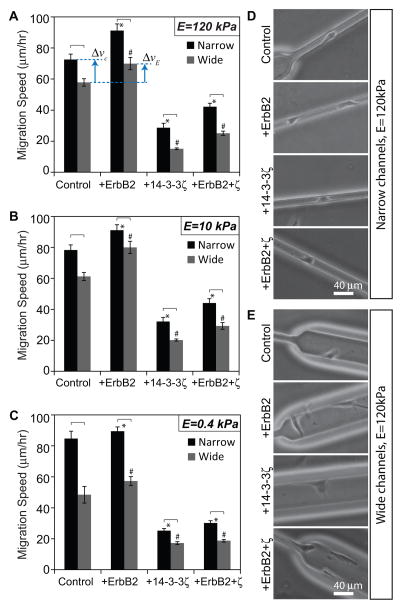
To study the role of ECM confinement on the migration speed of cells of varying transforming potential, we first fabricated μPACs of channel widths 10 and 40 um in a single device with a PA stiffness of 120 kPa. We introduced each of the four MCF10A-derived cell lines (control, +ErbB2, +14-3-3ζ, and +ErbB2+ζ) into the μPAC platform and measured random migration speeds by phase-contrast time-lapse imaging (Figure 2A). These studies revealed that the migration speed of cells in narrow channels was higher than that in wide channels regardless of oncogenic status. While we expected increasing confinement to increase migration speed based on our previous findings,17 we were surprised about the consistency of this trend across the various cell lines which were already genetically “hardwired” to produce varying degrees of invasiveness. Within a given channel width, overexpression of ErbB2 enhanced migration while overexpression of 14-3-3ζ, either alone or in tandem with ErbB2, slowed cell migration, consistent with past observations on unconfined collagen-coated glass surfaces (Fig. 2A).14 We gained additional insight into how ErbB2/14-3-3ζ overexpression and matrix confinement by quantifying percentage changes in migration speed due to channel narrowing and ErbB2 overexpression. Overexpression of ErbB2 alone increased the average migration speed in stiff, wide channels by approximately 21% (illustrated as ΔvE in Fig. 2A). This was qualitatively similar to the increase in migration speed of empty vector cells (without any oncogenic lesions) due solely to channel narrowing (Δvc ≈ 25%; see Fig. 2A). Thus, both transforming potential and matrix confinement, two unrelated parameters, independently affected cell migration to similar degrees. This observation reinforced the idea that physical cues present in the microenvironment can dramatically influence tumor cell invasiveness, a property traditionally often ascribed to specific genetic lesions or soluble factors. The quantitatively comparable influence of a physical parameter and a biological parameter on tumor cell invasiveness is a striking evidence of the importance of microenvironmental factors in the study of tumor invasion and metastasis. To study the effect of ECM stiffness on cell migration speed as a function of transforming potential and channel confinement, we repeated these experiments on μPACs fabricated from PA gels of stiffness 10 kPa and 0.4 kPa (Figures 2B–C). We found that the overall trends in the data were similar across all stiffnesses; specifically, ErbB2 overexpression and channel narrowing increased migration speed, and 14-3-3ζ reduced migration speed.
Figure 2. Migration speed across oncogenic lesions, channel confinements and ECM stiffness.
Mean migration speed of MCF10A cells of varying transforming potentials (control, +ErbB2, +14-3-3ζ, and +ErbB2+ζ) inside narrow and wide channels made of PA of stiffness (A) 120 kPa, (B) 10 kPa, and (C) 0.4 kPa. Phase contrast images of cells of varying transforming potentials inside (D) narrow and (E) wide channels made of stiff (120 kPa) PA. *p<0.05 with respect to control in narrow channels. #p<0.05 with respect to control in wide channels. Statistically different pairs (p<0.05) are indicated by horizontal square brackets. n>30 cells per condition. Scale bar = 40 μm.
Morphological polarization of cells correlates with migration speed
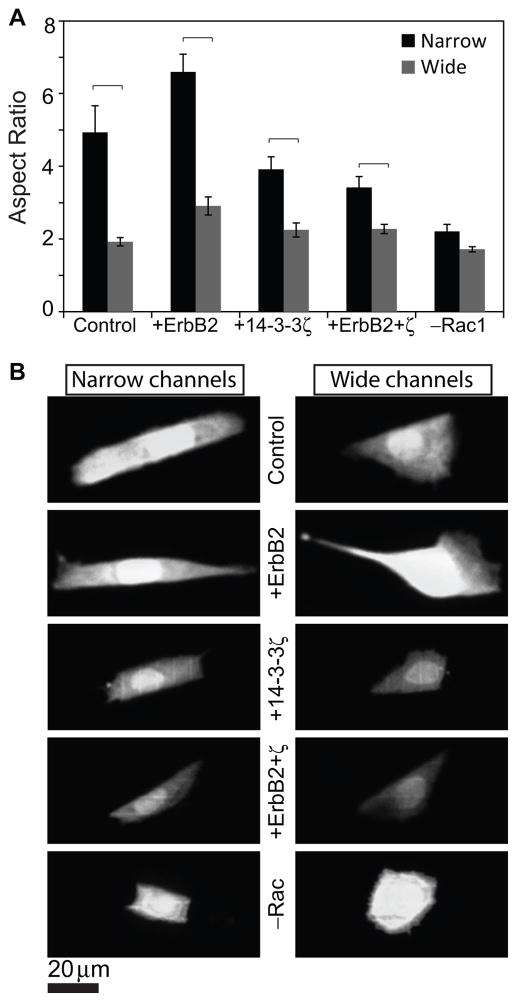
The above studies show that channel width strongly affects migration speed. Previously, we had shown that confinement-induced increases in migration speed are due to polarization of traction forces within the actomyosin cytoskeleton, with these tractions most efficiently directed to the axis of migration when the matrix is highly confined.17 To determine if differences in migration speed in this progression series is similarly associated with morphological differences, we imaged and quantified cell morphology under each condition and asked whether enhancements in migration speed were associated with increases in cell polarity (Fig. 3, Videos 1–4). Migration through narrow channels physically deformed the nucleus into an oblong shape and produced significantly more polarized morphologies (as measured by aspect ratio) than wide channels for all cell lines (transforming potentials). This morphometric analysis revealed that the dependence of migration speed on cell elongation closely mirrored the dependence of migration speed on transforming potential and confinement (compare trends in Figs. 3A and 2A, Fig. S1). While this progression series exhibits strong correlation between morphological polarity and migration speed, this relationship is not universally observed in other biological systems and thus not trivially predictable. For example, cells migrating in an amoeboid mode have rounded morphology yet are able to migrate faster than the polarized cells migrating in a mesenchymal mode.21
Figure 3. Cell morphology versus matrix confinement and transforming potential.
(A) Aspect ratio, a measure of cell polarization, of cells of varying transforming potentials and of Rac inhibited control cells, all inside narrow as well as wide channels made of stiff PA. (B) Confocal images of cell body stains across channel width and transforming potentials. Scale bar = 20 μm.
Rac inhibition slows cell migration
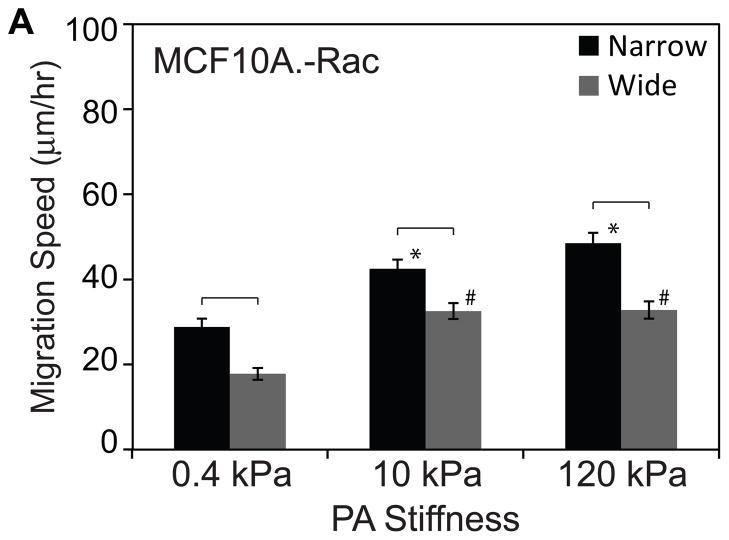
The preceding results suggest that enhanced migration speed is strongly correlated with cell elongation, regardless of whether that enhancement is driven by oncogene overexpression (transforming potential) or matrix confinement. This suggests that polarization of traction forces may serve as a proximal, common regulator through which these cell-intrinsic and cell-extrinsic factors act. Activation of Rac GTPase has been previously implicated in cell elongation, with Rac inhibition reducing polarization and motility by compromising protrusive activity and Rac overexpression slowing motility by giving rise to multiple competing and nonproductive lamellipodia.22–24 While Rac inhibition, cell polarization, and migration speed track together in these previous studies, such a trend cannot be assumed a priori, because both Rac inhibition and cell polarization can influence cell migration in many different ways. To explore a potential mechanistic role for Rac in this system, we pharmacologically inhibited Rac activation using the drug NSC23766. As expected, Rac inhibition reduced cell polarization in both narrow and wide channels (“–Rac1” bar in Fig. 3A). Time-lapse imaging revealed that these Rac-inhibited cells migrated much more slowly than their untreated counterparts across all stiffnesses (Figure 4; compare with Figure 2). The slowed migration combined with reduced cell polarization associated with Rac inhibition mimicked the effects of 14-3-3ζ overexpression and provided another confirmation of the direct correlation between cell shape and migration speed hypothesized earlier. Thus, beyond demonstrating a strong correlation between cell polarization and migration speed, we find that Rac inhibition phenocopies 14-3-3ζ-overexpression, which suggests that the two molecules may lie within a common signaling network.
Figure 4. Effect of Rac inhibition on cell motility.
(A) Mean migration speed versus ECM stiffness and channel width of cells treated with 20 μM NSC23766 inhibiting Rac GTPase activity. *p<0.05 with respect to control in narrow channels. #p<0.05 with respect to control in wide channels. Statistically different pairs (p<0.05) are indicated by horizontal square brackets. n>30 cells per condition.
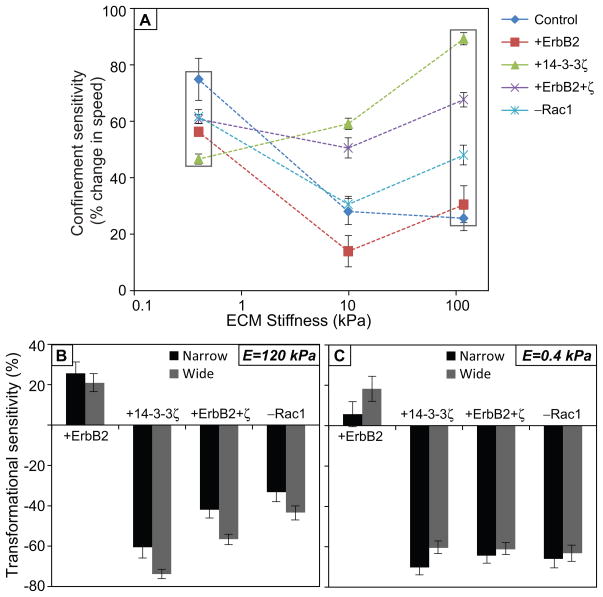
Confinement sensitivity is co-regulated by transforming potential and ECM stiffness
These studies reveal that transformational potential, matrix confinement, and Rac activation all contribute to migration speed. To compare the relative contribution of each parameter, we calculated a “confinement sensitivity” value, which we define as the percentage change in average migration speed from wide to narrow channels for a given transformational potential and ECM stiffness. First, we observed that in stiff microchannels, the confinement sensitivity of intrinsically slow cells (overexpressing 14-3-3ζ, co-overexpressing 14-3-3ζ and ErbB2, or Rac1 inhibited) was higher than those of intrinsically faster cells (control, or overexpressing ErbB2) (Fig. 5A). However, in soft microchannels, these differences in confinement sensitivity values were much less significant (Fig. 5A, squares). In other words, the differences in confinement sensitivity caused by varying transforming potential become more pronounced on stiff ECMs, while those differences collapse on soft ECMs. In an analogous fashion, we also calculated “transformational sensitivity” as the percentage change in migration speed for a given cell line relative to the empty-vector control at a given ECM stiffness and channel width. Overexpression of ErbB2 yielded a positive transformational sensitivity, while all other transformations led to negative sensitivity values independent of stiffness or confinement (Fig. 5B,C). Interestingly, the range of confinement sensitivity values of control cells (~25–75%; see Fig. 5A) strongly overlaps the range of transformational sensitivity values for various oncogenic lesions in stiff channels (~25–60% for narrow channels, ~21–74% for wide channels; Fig. 5B). These observations suggest that matrix confinement and expression of specific oncogenes can enhance tumor cell migration to comparable degrees.
Figure 5. Sensitivity of cell motility to confinement and transforming potentials.
(A) Confinement sensitivity, calculated as percentage change in the mean migration speed between wide and narrow channels, versus ECM stiffness for all transforming potentials. (B, C) Transformational sensitivity, calculated as the percentage change in migration speed for a given cell line relative to the empty-vector control at a given ECM stiffness and channel width.
Discussion
It is now clear that the metastatic invasion of tumor cells in breast cancer and other neoplasms is regulated both by the biophysical properties of the ECM in addition to the oncogenic lesions classically associated with malignant transformation. Several recent studies have explored how breast tumor cells of varying transforming potential interact with their extracellular environment in a variety of settings, including 2D surfaces, 3D collagen gels,14 and in vivo models.3 While these studies have nicely established the general importance of the biophysical microenvironment in regulating tumor invasion, they have largely left open the comparative importance of specific microstructural parameters in to this process. For example, Zaman and colleagues discovered that increasing 3D collagen matrix density reduced migration speed in partially transformed cells but did so to a much lesser extent in more fully transformed cells. Because increasing collagen density concomitantly increases stiffness and reduces pore size, it is challenging to identify the mechanistic origin of the effect. Here we sought to achieve this mechanistic insight by orthogonally varying two cell-extrinsic parameters (matrix stiffness, confinement), and one cell-intrinsic parameter (transforming potential) and comparing their relative contribution to migration speed.
Revisiting the motivation for these studies, we had proposed to investigate three specific regulatory relationships: (1) The effect of ECM stiffness on ErbB2- and 14-3-3ζ-based regulation of motility in the complete absence of confinement; (2) The effect of confinement on ErbB2- and 14-3-3ζ-based regulation of motility; and (3) The role of ECM stiffness alone in dictating how confinement and transforming potential regulate motility. Our findings now allow us to answer these questions in a specific manner. First, we find that in the absence of confinement, ECM stiffness does not change the relative effect of specific oncogenic lesions on migration speed; i.e., lesions that produce rapid migration on stiff matrices also do so on soft matrices. Second, increasing channel confinement increases migration speed across all cell lines and enhances motility to a degree comparable to overexpression of a pro-invasive oncogene (ErbB2). Finally, while these relationships are not grossly altered by changes in ECM stiffness, ECM stiffening does enhance the sensitivity of motility to confinement and transforming potential. Our ability to draw these conclusions was made possible by the use of an isogenically-matched progression series, and this study represents an example of how such series can be combined with microengineered ECMs to dissect genetic and microenvironmental control of motility.
Considering these results in greater depth, we find that migration speed is much more sensitive to matrix confinement and transforming potential than to ECM stiffness, at least in the range of 0.4 – 120 kPa (Figs. 2A–C). Moreover, matrix stiffness exerted the greatest effect on migration speed in the setting of ErbB2 overexpression, which is consistent with the observation of Weaver and colleagues that matrix stiffening and ErbB2 overexpression are both needed to induce an invasive phenotype in MEC acini.7 To demonstrate the cooperative influence of stiffness and ErbB2 on cell invasiveness in our framework, we consider the following three results from wide channels: (1) On soft ECMs, ErbB2 overexpression produced an 18% increase in migration speed over control cells; (2) Control cells produced 26% faster migration on stiff ECMs than soft ECMs; and (3) Cells overexpressing ErbB2 on stiff ECMs migrated 65% faster than control cells on soft ECMs. Thus, a combination of ErbB2 overexpression ECM stiffening produced much greater increases in migration speed than either perturbation alone. This exemplifies how this experimental paradigm can be used to dissect roles of specific matrix properties on cell migration, something that could not be easily achieved in matrix systems in which stiffness and pore size are intertwined.
Our previous study17 demonstrated that gliomblastoma cells migrate faster with increasing channel confinement, and that this effect is most pronounced with increasing matrix stiffness. The fact that we observe similar phenomenology in this study is notable given that the biological context (ECM protein, tissue of origin) is completely different. For example, the different cell types could have been expected to respond to disparate ECM contexts with different degrees of adhesiveness and motility. For instance, if the MECs were disproportionately more adhesive to collagen-coated gels than brain tumors cells were to fibronectin, it is quite likely that narrow channels would have constricted migration. In fact, inside 3D gels, the migration of MCF10A cells has been shown to be slower in collagen gels of narrower pores.14 Along the same lines, one might have expected the highly invasive cell series to exhibit fast migration regardless of the degree of confinement, or the slowest cells not to be rescued by channel narrowing. Our results suggest that the oncogenically-driven differences in motility observed in this progression series do not completely free cells from extrinsic regulation of cell motility. This may have important implications for pathophysiology and therapy and is consistent with recent work showing that suppression of myosin can override the effect of pro-motility growth factors and suppress tumor cell motility.25
Furthermore, we find that overexpression of ErbB2 and channel confinement enhance cell migration independently of one another, with confinement proportionally increasing cell migration speed to the same extent as ErbB2 overexpression. Our morphometric analysis implicates cell polarization as a common predictor of increased migration speed, suggesting that all of these disparate inputs may enhance migration through polarization of traction force. Pharmacologic inhibition of Rac GTPase abolished both cell polarization and confinement-induced enhancement of migration, implying that Rac activation strongly contributes to these effects. While the precise mechanisms through which ErbB2 overexpression and confinement might stimulate Rac activation remain unclear, there is certainly ample evidence for a close relationship between the three parameters. Several lines of evidence illustrate that ErbB2-driven transformation is closely related to Rac-dependent signaling.26 For example, ErbB2 expression correlates with the activation of the Rac effector Pak1 in human breast tumor samples, and overexpression of a dominant negative Rac or Pak1 mutant can effectively rescue ErbB2-mediated transformation of MCF10A-based acini.27 Additionally, the Rac guanine exchange factor (GEF) P-Rex1, which is strongly overexpressed in breast tumors, plays an essential role in ErbB2- and Rac-mediated induction of cell proliferation and motility.28 Similarly, there is much evidence tying Rac to matrix confinement and cell polarity, although the relationships here appear to be quite complex. For example, Rac suppression reduces lamellipodium formation, induces an elongated phenotype, and promotes persistent motility.24 Despite inducing cell elongation, however, Rac suppression does not increase cell motility to the same extent as culturing cells on 1D patterned ECMs, suggesting that elongation may be necessary but not sufficient to increase cell migration.29 Clearly, future studies are needed in which migration speed is systematically studied as a function of Rac activation and matrix confinement. Moreover, it will be important to explore the role of other critical mechanotransductive signals in mediating coupling between biophysical inputs, oncogenic lesions, and mechanotransductive signaling. For example, Rho GTPase is known to directly influence actin polymerization-dependent protrusion and actomyosin contractility30 and thus regulate ECM-dependent motility.23 Combining this platform with genetic and small molecule screening approaches is likely to help identify additional candidates.
In summary, we have used a microfabricated ECM platform to dissect relative contributions of ECM biophysical parameters and ErbB2 and 14-3-3ζ expression (transforming potential) to the invasive migration of breast tumor cells. We find that migration speed is most sensitive to matrix confinement and ErbB2 overexpression, and that the two parameters can act synergistically to enhance motility. While the intrinsic effect of ECM stiffness is modest compared to these other two parameters, matrix stiffening in the presence of ErbB2 overexpression can significantly increase motility, highlighting the notion that ECM biophysical cues ultimately interact with a variety of cell-intrinsic properties to promote invasion. While the molecular connections between matrix confinement, ErbB2 expression, and migration speed remain to be fully elucidated, we find that matrix confinement acts through Rac activation to promote polarization of traction forces. Our study hints at the complex relationship between oncogene expression, matrix parameters, and invasive behavior and points towards the need to understand the mechanisms and implications of these relationships in much greater detail.
Materials and Methods
Fabrication of polyacrylamide microchannels
Standard photolithographic techniques were used to fabricate silicon masters of defined geometry (Fig. 1), where a 4-inch silicon wafer was spin-coated with a 25-μm-thick layer of photoresist (SU-8 2015, Microchem) and exposed with UV light through a transparency mask printed with defined topographical features matching a desired pattern of microchannels. To facilitate easy separation of the silicon master from the polymerized polyacrylamide gel in the subsequent step, silicon master was silanized beforehand (N-octadecyltriethoxysilane, UCT) under vacuum. Next, a volume of PA precursor solution sufficient to achieve a gel of approximately 100 μm thickness was placed between a reactive glass surface and the hydrophobic silicon mold and allowed to polymerize. After approximately 30 min, the coverslip containing the polymerized PA molded to the shape of channels on the silicon master was separated. Thus, the microchannel patterns were transferred from the silicon-SU8 mold onto the PA gel surface. The monomer/crosslinker ratios for the PA solution precursor solutions were chosen based on previous characterization – acrylamide/bisacrylamide (A/B) percentages of 4% A/0.2% B, 10% A/0.3% B, and 15% A/1.2% B corresponding to PA gels of elastic moduli of 0.4, 10, and 120 kPa.31,32 The substrates containing polyacrylamide microchannels were functionalized with 0.1 mg/ml type 1 collagen (Bovine Collagen Solution Type I, Advanced BioMatrix). The same polyacrylamide synthesis was used to prepare a 250-μm-thick layer of soft (0.4 kPa) polyacrylamide gel attached to a coverslip. This coverslip-PA assembly was placed on top of the substrate of channels after the cells had been seeded in the PA-channels (Fig. 1).
Cell culture
All studies in this article were performed using stable lines of human-derived MCF10A mammary epithelial cells (kindly provided by Prof. Muhammad H. Zaman). Cells were cultured in DMEM/F12 growth media as previously described.15 Cell suspensions were introduced in the device of PA-channels at volumetric cell densities sufficient to achieve a subconfluent density of 3000 cells per cm2, and then cells were allowed to adhere to the PA surfaces. At least 8 h after cell seeding, prior to the start of imaging and data acquisition, the top coverslip layered with soft PA was lowered into the device and oriented in a way that the two PA surfaces were in contact (Fig. 1). The Rac GTPase inhibitor NSC23766 (Santa Cruz Biotechnology) was added to the cell culture medium at 20 μM final concentration in relevant experiments at least 14 h after initial cell seeding and the cells were tracked between 2 to 8 hours after the drug addition.
Phase contrast microscopy and data analysis
Live timelapse imaging of the cells was performed using a Nikon TE2000E2 microscope equipped with an incubator chamber for controlled temperature, humidity, and CO2, and a motorized, programmable stage (Prior Scientific, Inc.). Images were recorded with a CCD camera (Photometrics Coolsnap HQ2) interfaced to image acquisition software (SimplePCI, Hamamatsu Corporation). In each experiment, 10x phase contrast images were acquired every 15 min for 10–16 h. These timelapse images were analyzed using manual tracking in ImageJ (National Institutes of Health) to quantify migration speeds across conditions. The migration speed was calculated as total distance divided by total time for any given cell. The data were further processed to obtain a mean speed for a given condition.
Confocal imaging and morphometric analysis
Cells were fixed with 4% paraformaldehyde (Fisher Scientific) in PBS, followed by permeabilization of cell membrane with 0.1% Triton- X 100 (EMD Biosciences) and blocking with 5% goat serum in PBS. To visualize entire cell body of these fixed cells adhered inside channels, a cytoplasmic dye called Cell Tracker Green CMFDA (Molecular Probes, Eugene, OR) was used to perform staining. Confocal images (40X) were captured as z-stacks at an interval of 1 μm using an Olympus BX51WI microscope (Olympus Corporation) equipped with Swept Field Confocal technology (Prairie Technologies, Inc.). The cell shapes were visualized by combining the z-stacks for each cell in ImageJ (NIH) using the Z-Project tool. The resulting z-projected images of the cell shape were analyzed in ImageJ to measure aspect ratio for each cell.
Statistical analysis
Unless specified otherwise, data are reported as mean ± standard error, and statistical comparisons were performed with a one-way ANOVA followed by a Tukey-Kramer HSD (honestly significant difference) test for pairwise comparisons.
Supplementary Material
Cellular aspect ratio (a measure of polarization) vs. mean migration speed for MCF10A cells across all conditions studies: transforming potential, Rac status, channel width, channel stiffness.
Volumetric 3D reconstruction of the body of a ErbB2-overexpressing cell confined in a narrow, 120 kPa channel shows the cell adhered along the walls of the channel. The coordinate system places the X-axis along the length of the channel, the Y-axis along the width of the channel, and the Z-axis towards the bottom surface of the channel.
Volumetric Volumetric 3D reconstruction of the body of a ErbB2-overexpressing cell confined in a wide, 120 kPa channel shows the cell adhered along the walls of the channel. The coordinate system places the X-axis along the length of the channel, the Y-axis along the width of the channel, and the Z-axis towards the bottom surface of the channel.
Volumetric 3D reconstruction of the body of a Rac-inhibited cell confined in a narrow, 120 kPa channel shows the cell adhered along the walls of the channel. The coordinate system places the X-axis along the length of the channel, the Y-axis along the width of the channel, and the Z-axis towards the bottom surface of the channel.
Volumetric 3D reconstruction of the body of a Rac-inhibited cell confined in a wide, 120 kPa channel shows the cell adhered along the walls of the channel. The coordinate system places the X-axis along the length of the channel, the Y-axis along the width of the channel, and the Z-axis towards the bottom surface of the channel.
Acknowledgments
SK gratefully acknowledges the support of the National Institutes of Health (Director’s New Innovator Award 1DP2OD004213; Physical Sciences-Oncology Center Award 1U54CA143836) and the National Science Foundation (CMMI 1105539). Confocal microscopy images were obtained at the CIRM/QB3 Stem Cell Shared Facility.
References
- 1.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Danes CG, Wyszomierski SL, Lu J, Neal CL, Yang W, Yu D. Cancer Res. 2008;68:1760–1767. doi: 10.1158/0008-5472.CAN-07-3177. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, Guo H, Treekitkarnmongkol W, Li P, Zhang J, Shi B, Ling C, Zhou X, Chen T, Chiao PJ, Feng X, Seewaldt VL, Muller WJ, Sahin A, Hung MC, Yu D. Cancer Cell. 2009;16:195–207. doi: 10.1016/j.ccr.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu D, Hung MC. Oncogene. 2000;19:6115. doi: 10.1038/sj.onc.1203972. [DOI] [PubMed] [Google Scholar]
- 5.Hackel PO, Zwick E, Prenzel N, Ullrich A. Curr Opin Cell Biol. 1999;11:184–189. doi: 10.1016/s0955-0674(99)80024-6. [DOI] [PubMed] [Google Scholar]
- 6.Sun R, Gao P, Chen L, Ma D, Wang J, Oppenheim JJ, Zhang N. Cancer Res. 2005;65:1433–1441. doi: 10.1158/0008-5472.CAN-04-1163. [DOI] [PubMed] [Google Scholar]
- 7.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Ling C, Zuo D, Xue B, Muthuswamy S, Muller WJ. Genes Dev. 2010;24:947–956. doi: 10.1101/gad.1896810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guarino M. Int J Biochem Cell Biol. 2007;39:2153–2160. doi: 10.1016/j.biocel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Guiot C, Pugno N, Delsanto PP, Deisboeck TS. Physical Biology. 2007;4:P1–P6. doi: 10.1088/1478-3975/4/4/P01. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S, Weaver V. Cancer Metast Rev. 2009;28:113–127. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pathak A, Kumar S. Integr Biol. 2011;3:267–278. doi: 10.1039/c0ib00095g. [DOI] [PubMed] [Google Scholar]
- 13.Baker EL, Lu J, Yu D, Bonnecaze RT, Zaman MH. Biophys J. 2010;99:2048–2057. doi: 10.1016/j.bpj.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker EL, Srivastava J, Yu D, Bonnecaze RT, Zaman MH. PLoS ONE. 2011;6:e20355. doi: 10.1371/journal.pone.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debnath J, Muthuswamy SK, Brugge JS. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 16.Nofech-Mozes S, Spayne J, Rakovitch E, Hanna W. Adv Anat Pathol. 2005;12:256–264. doi: 10.1097/01.pap.0000184177.65919.5e. [DOI] [PubMed] [Google Scholar]
- 17.Pathak A, Kumar S. Proc Natl Acad Sci U S A. 2012;109:10334–10339. doi: 10.1073/pnas.1118073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, Boggs AE, Martin SS, Konstantopoulos K. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraning-Rush CM, Carey SP, Lampi MC, Reinhart-King CA. Integr Biol. 2013;5:606–616. doi: 10.1039/c3ib20196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson ME, Kota N, Kim Y, Wang Y, Stolz DB, LeDuc PR, Ozdoganlar OB. Lab Chip. 2011;11:1550–1555. doi: 10.1039/c0lc00561d. [DOI] [PubMed] [Google Scholar]
- 21.Lammermann T, Sixt M. Curr Opin Cell Biol. 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Katz BZ, Zohar M, Teramoto H, Matsumoto K, Gutkind JS, Lin DC, Lin S, Yamada KM. Biochem Biophys Res Commun. 2000;272:717–720. doi: 10.1006/bbrc.2000.2853. [DOI] [PubMed] [Google Scholar]
- 23.MacKay JL, Keung AJ, Kumar S. Biophys J. 2012;102:434–442. doi: 10.1016/j.bpj.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, Matsumoto K, Yamada KM. J Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivkovic S, Beadle C, Noticewala S, Massey SC, Swanson KR, Toro LN, Bresnick AR, Canoll P, Rosenfeld SS. Mol Biol Cell. 2012;23:533–542. doi: 10.1091/mbc.E11-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wertheimer E, Gutierrez-Uzquiza A, Rosemblit C, Lopez-Haber C, Sosa MS, Kazanietz MG. Cell Signal. 2012;24:353–362. doi: 10.1016/j.cellsig.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arias-Romero LE, Villamar-Cruz O, Pacheco A, Kosoff R, Huang M, Muthuswamy SK, Chernoff J. Oncogene. 2010;29:5839–5849. doi: 10.1038/onc.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sosa MS, Lopez-Haber C, Yang C, Wang H, Lemmon MA, Busillo JM, Luo J, Benovic JL, Klein-Szanto A, Yagi H, Gutkind JS, Parsons RE, Kazanietz MG. Mol Cell. 2010;40:877–892. doi: 10.1016/j.molcel.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doyle AD, Wang FW, Matsumoto K, Yamada KM. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridley AJ. Int J Biochem Cell Biol. 1997;29:1225–1229. doi: 10.1016/s1357-2725(97)00052-6. [DOI] [PubMed] [Google Scholar]
- 31.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Biophys J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulrich TA, de Juan Pardo EM, Kumar S. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cellular aspect ratio (a measure of polarization) vs. mean migration speed for MCF10A cells across all conditions studies: transforming potential, Rac status, channel width, channel stiffness.
Volumetric 3D reconstruction of the body of a ErbB2-overexpressing cell confined in a narrow, 120 kPa channel shows the cell adhered along the walls of the channel. The coordinate system places the X-axis along the length of the channel, the Y-axis along the width of the channel, and the Z-axis towards the bottom surface of the channel.
Volumetric Volumetric 3D reconstruction of the body of a ErbB2-overexpressing cell confined in a wide, 120 kPa channel shows the cell adhered along the walls of the channel. The coordinate system places the X-axis along the length of the channel, the Y-axis along the width of the channel, and the Z-axis towards the bottom surface of the channel.
Volumetric 3D reconstruction of the body of a Rac-inhibited cell confined in a narrow, 120 kPa channel shows the cell adhered along the walls of the channel. The coordinate system places the X-axis along the length of the channel, the Y-axis along the width of the channel, and the Z-axis towards the bottom surface of the channel.
Volumetric 3D reconstruction of the body of a Rac-inhibited cell confined in a wide, 120 kPa channel shows the cell adhered along the walls of the channel. The coordinate system places the X-axis along the length of the channel, the Y-axis along the width of the channel, and the Z-axis towards the bottom surface of the channel.