Abstract
n-3 Polyunsaturated fatty acids (n-3 PUFAs) have anti-obesity effects that may modulate risk of obesity, in part, through interactions with genetic factors. Genome-wide association studies (GWAS) have identified genetic variants associated with body mass index (BMI); however, the extent to which these variants influence adiposity through interactions with n-3 PUFAs remains unknown. We evaluated 10 highly replicated obesity GWAS single nucleotide polymorphisms (SNPs) for individual and cumulative associations with adiposity phenotypes in a cross-sectional sample of Yup’ik people (n = 1,073) and evaluated whether genetic associations with obesity were modulated by n-3 PUFA intake. A genetic risk score (GRS) was calculated by adding the BMI-increasing alleles across all 10 SNPs. Dietary intake of n-3 PUFAs was estimated using nitrogen stable isotope ratio (δ15N) of red blood cells, and genotype–phenotype analyses were tested in linear models accounting for familial correlations. GRS was positively associated with BMI (p = 0.012), PBF (p = 0.022), ThC (p = 0.025), and waist circumference (p = 0.038). The variance in adiposity phenotypes explained by the GRS included BMI (0.7 %), PBF (0.3 %), ThC (0.7 %), and WC (0.5 %). GRS interactions with n-3 PUFAs modified the association with adiposity and accounted for more than twice the phenotypic variation (~1–2 %), relative to GRS associations alone. Obesity GWAS SNPs contribute to adiposity in this study population of Yup’ik people and interactions with n-3 PUFA intake potentiated the risk of fat accumulation among individuals with high obesity GRS. These data suggest the anti-obesity effects of n-3 PUFAs among Yup’ik people may, in part, be dependent upon an individual’s genetic predisposition to obesity.
Electronic supplementary material
The online version of this article (doi:10.1007/s12263-013-0340-z) contains supplementary material, which is available to authorized users.
Keywords: BMI, Adiposity, Alaska Native, SNP, δ15N, rs9939609, rs7647305, FTO, ETV5, Genetic risk score, CANHR, Gene-by-environment interactions
Introduction
Obesity contributes to a series of metabolic abnormalities including elevated triglycerides and inflammatory cytokines, leading to insulin resistance and the development of type 2 diabetes (T2D) and other diseases (Ogden et al. 2007). Given the increasing public health burden associated with obesity and the difficulty maintaining long-term weight loss (Curioni and Lourenço 2005; Dansinger et al. 2007; Tsai and Wadden 2005), it is important to elucidate mechanisms that influence body weight.
The heritability of obesity has been estimated to between 40 and 70 % (Maes et al. 1997; Stunkard et al. 1990). A recent meta-analysis of obesity genome-wide association studies (GWAS) conducted primarily in European samples has identified thirty-two single nucleotide polymorphisms (SNPs) associated with variation in BMI (Speliotes et al. 2010). Although a large number of SNPs are associated with excess body weight in European samples, the extent to which these loci are associated with adiposity phenotypes in other ethnic groups is unclear (Hester et al. 2012; Li et al. 2008). Furthermore, the heritability of obesity explained by the aggregate of these loci remains low (<2 %) (Speliotes et al. 2010).
Increasingly, gene-by-environment interactions have been suggested to account for the “missing heritability” in complex traits (Van IJzendoorn et al. 2011). Genetic association studies that recruit isolated populations, characterized by an environment that is less heterogeneous and reduced genetic admixture, may have a unique opportunity to delineate genetic and environmental factors that impact the heritable component of obesity (Heutink and Oostra 2002). Empirical evidence that demonstrates modifiable environmental factors interact with SNPs associated with obesity and partially account for the heritability of obesity may elucidate mechanisms that modulate the risk of obesity-related comorbidities through lifestyle interventions (Marti et al. 2008).
Dietary fat is a modifiable environmental factor that has been implicated in the development of obesity (Bray et al. 2004); however, a growing body of evidence indicates that not all dietary fatty acids are equally obesigenic (Storlien et al. 2001). Regular consumption of n-3 polyunsaturated fatty acids (n-3 PUFAs) may reduce adiposity in humans (Buckley and Howe 2010), in part, by inhibiting adipogenesis (Kopecký et al. 2009) and stimulating fat oxidation (Couet et al. 1997). Animal models have shown significant reductions in fat mass when dietary n-3 PUFAs, namely eicosapentaenoic (EPA) and docosahexaenoic acids (DHA), are substituted for saturated fats (Hainault et al. 1993), monosaturated fats (Su and Jones 1993), and n-6 polyunsaturated fats (Jones 1989), after controlling for caloric intake. These findings have largely been confirmed by human studies (Thorsdottir et al. 2007); however, information regarding whether the anti-obesity effects attributed to n-3 PUFA is dependent upon a specific genotype remains limited (Jourdan et al. 2011). Knowledge about the interplay between genetic factors and consumption of n-3 PUFA in isolated populations with widely varying n-3 PUFA intake may facilitate the choice of more effective and specific measures of obesity prevention based upon individualized genetic makeup.
Given the anti-obesigenic effects of n-3 PUFAs, the widely varying intake of n-3 PUFA in this isolated study population of Yup’ik people (nearly 50-fold range of EPA), and the availability of a precise biomarker for n-3 PUFA intake (δ15N; (O’Brien et al. 2009)), we evaluated whether n-3 PUFA intake modifies the association between GWAS SNPs and adiposity phenotypes. The individual and cumulative effects of 10 SNPs reproducibly associated with BMI (Frayling et al. 2007; Loos et al. 2008; Scuteri et al. 2007; Thorleifsson et al. 2009; Willer et al. 2009) at genome-wide significance (p < 5 × 10−8) were tested in a sample of Yup’ik people from the Center for Alaska Native Health Research (CANHR) study. We evaluated the cumulative effects of these SNPs using a genetic risk score (GRS) that estimates an individual’s genetic predisposition to obesity by adding the BMI-increasing alleles across all 10 SNPs.
Methods
Participants and study design
The CANHR studies genetic, behavioral, and dietary risk factors underlying obesity and their relationship to diabetes and cardiovascular disease among Yup’ik people (Mohatt et al. 2007). Recruitment of Yup’ik participants was initiated in 2003 and continues in 11 Southwest Alaskan communities. All residents ≥14 years old were invited to participate, and the resulting distribution of age in our study sample reflects the age distribution among eligible participants according to 2000 US census data. Participants signed informed-consent documents before entering the study using protocols that were approved by the University of Alaska Institutional Review Board, the National and Alaska Area Indian Health Service Institutional Review Boards, and the Yukon Kuskokwim Human Studies Committee. Summary statistics regarding family data were calculated using PEDINFO in the Statistical Analysis for Genetic Epidemiology (S.A.G.E., 2009) software. The analyses in this report included 1,073 non-pregnant Yup’ik participants (41 founders, 920 non-founders, and 112 singletons) with ages ranging between 14 and 94 years at enrollment. There were 195 pedigrees in this data set with a mean size 5.52 individuals (range, 1–849) and 696 sibships with a mean size of 1.32 (range, 1–9).
Anthropometric and biochemical measurement
Anthropometric measurements were obtained by trained staff using protocols from the NHANES III Anthropometric Procedures Manual (Lohman and Roche 1988) as previously described (Boyer et al. 2007). These measurements included height, weight, and 4 circumferences (waist, hip, triceps, and thigh). Percent body fat (PBF) was measured by electrical bioimpedance using a Tanita TBF-300A bioimpedance analyzer (Tanita Corp, Arlington Heights, IL, USA).
Biomarker of n-3 PUFA intake
n-3 PUFA intake was assessed using the nitrogen stable isotope ratio (δ15N) of red blood cells (RBC), which has been validated as a biomarker for EPA and DHA intake as previously described (O’Brien et al. 2009). The time to 50 % turnover of RBC is approximately 45 days therefore, the mean RBC δ15N values reflect a mean n-3 PUFA intake over 1.5 months. A volume of 1.8 μl aliquots of RBC were pipetted into 3.5 × 3.75 mm tin capsules, autoclaved for 20 min at 121 °C to destroy blood-borne pathogens, and dried to a constant mass of 0.2–0.4 mg. Capsules were crushed into a ball for loading into an autosampler. Samples were analyzed at the Alaska Stable Isotope Facility by continuous-flow isotope ratio mass spectrometry, using a Costech ECS4010 Elemental Analyzer (Costech Analytical Technologies, Valencia, CA, USA) interfaced to a Finnigan Delta Plus XP isotope ratio mass spectrometer via the Conflo III interface (Thermo-Finnigan Inc., Breman, Germany). Isotope ratios are analyzed relative to IAEA-certified reference materials calibrated to atmospheric nitrogen, for which 15N/14N = 0.0036765. By convention and for ease of interpretation, isotope ratios are presented as delta values in “permil” relative to atmospheric nitrogen: δ15N = [(15N/14Nsample − 15N/14Nstandard)/(15N/14Nstandard)] × 1,000 ‰. We concurrently prepared and ran multiple laboratory standards (peptone, δ15N = 7.00) to assess analytical accuracy and precision; these were analyzed after every eighth sample and gave values of δ15N = 7.01 ± 0.24 ‰ (mean ± SD). The range of isotopic variation in our dataset (9 ‰) was very large relative to analytical precision (0.2 ‰). We modeled the effects of n-3 PUFA intake as a categorical variable with four groups by quartile, which is hereafter referred to as δ15N.
SNP selection and genotyping
Due to limited resources available at the time of genotyping, we selected SNPs most reproducibly associated with BMI at genome-wide significance (p < 5 × 10−8) which included the 10 obesity loci in or near FTO, MC4R, TMEM18, GNPDA2, BDNF, NEGR1, SH2B1, ETV5, MTCH2, and KCTD15 (Frayling et al. 2007; Loos et al. 2008; Scuteri et al. 2007; Thorleifsson et al. 2009; Willer et al. 2009) and that were found to be significantly associated with BMI in an obesity GWAS meta-analysis with 249,796 participants (Speliotes et al. 2010). In situations where more than a single SNP has been identified for a given gene, we genotyped the SNP most frequently reported in these studies. Genotyping was carried out at the Broad Institute (Cambridge, MA) by allele-specific primer extension of multiplex amplified products and detection using matrix-assisted laser desorption ionization time-of-flight spectrometry on a Sequenom iPLEX platform (Tang et al. 1999). We compared the frequency of the GWAS BMI-increasing “risk alleles” in Yup’ik people to five HapMap populations (The International HapMap Consortium 2003).
Quality control of phenotypic and genotypic data
Simple linear models were fit to each phenotype using all covariates (age, sex, community group) included in the association models, and the distributions of the residuals for each phenotype were examined for normality with the R statistical programming language (v2.10.1, R Development Core, 2009). We considered a series of transformations (square root, log, inverse, etc.) to improve normality, and the Box-Cox transformation (Box and Cox 1964) was identified as the best procedure for phenotypes whose residuals did not follow a normal distribution. Information regarding the power transformation for each adiposity phenotypes is presented in Supplementary Table I. Familial data were stored in the Progeny database (Progeny Software LLC, South Bend, IN, USA) and merged into a single extended pedigree comprised of multiple independent families using PedMerge (Plaetke and Balbi 2010). Genotypic data were tested for Mendelian inconsistencies using PEDCHECK (O’Connell and Weeks 1998). In this sample, Illumina IV linkage panel (Illumina, Inc., San Diego, CA, USA) genotypes were available from an ongoing linkage study and were used to construct principal components using the EIGENSTRAT analysis package (Price et al. 2006). The first PC did not have an obvious interpretation to the data; however, the second PC discriminated individuals into two groups that corresponded to either coastal or inland communities. Based on this, we defined a dichotomous community group variable that indicates either coastal or inland. We assessed Hardy–Weinberg equilibrium (HWE) in the founders using PLINK (v1.07) (Purcell et al. 2007) and determined minor allele frequency (MAF) for each SNP using the FREQ module in the Statistical Analysis for Genetic Epidemiology (S.A.G.E 2009) program. The present study restricted association and interaction analysis to SNPs with MAF ≥5 % that did not deviate from HWE (p ≤ 0.005). HWE multiple test correction was determined using a Bonferroni correction for 10 tests (α ≤ 0.005).
Association analysis
Each SNP was tested using as additive model for association with obesity-related phenotypes using the ASSOC module in the S.A.G.E. software package (S.A.G.E. 6.3 [2012] http://darwin.cwru.edu/sage/; George and Elston 1987). We included both demographic (age, community group, and sex) and environmental covariates (δ15N) in the association analysis and calculated likelihood ratio statistics to compare 4 nested models. Model 1 included demographic covariates (age, sex and community group); Model 2 included baseline covariates and SNP to test for an additive genetic effect of SNP (defined as the number of minor alleles); Model 3 included demographic covariates, the additive genetic effect of SNP, and δ15N (defined by quartiles of δ15N) as an estimate of n-3 PUFA intake; Model 4 included all covariates from Model 3 and an interactions between the additive genetic effect and δ15N categories. Note that the Model 4 is the only model to test gene–diet interactions. Correction for multiple testing in genetic analyses (Models 1–4) for SNPs with MAF ≥0.05 employed a Bonferroni correction for 10 tests (α ≤ 0.005). Effect sizes (β) were presented for association analyses as the change in transformed phenotypes according to BMI-increasing “risk allele” adjusted for demographic and environmental covariates. Statistical power that accounted for familial correlations (Nyholt 2004) was assessed using SAS version 9.1 (SAS Institute, Cary, NC). The general estimates of power in our sample using an additive genetic model at α = 0.005 for detecting the effect sizes (β) in transformed phenotypes (i.e., BMI) between 0.1 and 1.5 were >90 % when the minor allele frequency was at least 5 %.
The obesity genetic risk score (GRS) was calculated by summing the number of risk alleles that each individual possessed. The obesity risk alleles were defined as those that were associated with higher BMI (defined as BMI ≥25 kg/m2) in previous GWAS studies (Speliotes et al. 2010; Thorleifsson et al. 2009; Willer et al. 2009). To account for missing genotypes, we divided the sum of risk alleles by the number of loci included in the score. The GRS was split into three groups by tertiles (Qi et al. 2009), and a categorical variable was created to allow for nonlinear effects without being unduly influenced by extreme values. We tested the GRS for association with obesity traits using two models (Models 5 and 6) run with the ASSOC module in S.A.G.E. Model 5 included the GRS as a main effect and controlled for demographic (sex, age, and community group) covariates. Model 6 tested the GRS for interaction with δ15N and controlled for demographic covariates and δ15N. Model 7 tested for interactions (δ15N*GRS and δ15N*community group) in a model that included demographic and environmental covariates. Note that Model 7 is the only model to test gene–diet interaction. In a subset of participants (n = 488), we evaluated 24 Hour Recalls (24HR) to estimate total energy (kcal) and total fat intake (% calories) as potential confounding dietary variables in the δ15N*GRS interaction model (Model 7). The variance explained by the fixed effects in the GRS association models were calculated as the difference between the total variance of the trait and the sum of the random effects in the model (i.e., the residual variance and the family effect). The proportion of variance explained by random effects was calculated, whereby the ratio of the difference noted above was included in the numerator and the total variance of the trait was included in the denominator. We defined the proportion of the variance explained by each term in the GRS association model as the difference in proportion of variance explained by models with and without the terms of interest. Results were considered significant in the GRS analyses (Models 5–7) if the p value was ≤0.05 (2-tailed).
Results
Descriptive statistics
Descriptive statistics on Yup’ik men (n = 510) and women (n = 563) enrolled in this study are presented in Table 1. Yup’ik women in this study had a mean age of 38.5 (±1.6) years and had more adiposity than men that reported a mean age of 36.6 (±0.6) years. Specifically, women had more adiposity that included greater BMI (p < 0.0001), percentage body fat (p < 0.0001), and hip circumference (p < 0.0001) compared to men. Interestingly, we did not detect significant differences between women and men with respect to waist circumference.
Table 1.
Descriptive statistics of adiposity phenotypes
| Variables | Women | Men | p values |
|---|---|---|---|
| No. of participants | 563 | 510 | |
| Age (years) | 38.5 ± 1.6 | 36.6 ± 0.6 | 0.0740 |
| BMI (kg/m2) | 28.7 ± 5.6 | 26.3 ± 5.6 | <0.0001 |
| Percentage body fat (%) | 42.8 ± 1.4 | 27.8 ± 1.1 | <0.0001 |
| Waist circumference (cm) | 88.1 ± 21.6 | 87.5 ± 21.5 | 0.5039 |
| Hip circumference (cm) | 105.4 ± 43.0 | 98.7 ± 43.0 | <0.0001 |
| Thigh circumference (cm) | 51.1 ± 21.4 | 50.3 ± 21.3 | 0.0110 |
Values are reported as mean (±SE) predicted from linear model accounting for familial correlations. p values for differences by gender are derived using student t test
Distribution of δ15N in the study population
We assessed n-3 PUFA intake in 1,073 Yup’ik participants using RBC δ15N as a biomarker of EPA and DHA intake. Summary statistics are grouped by gender and δ15N quartiles and reported in Table 2. The mean δ15N value in this study was 9.0 ‰ and ranged from 6.4 to 15.2 ‰. According to the linear relationship between RBC δ15N and RBC EPA described elsewhere for this study population (O’Brien et al. 2009), the corresponding mean EPA (%RBC fatty acids) was 2.66 %. Measurement of δ15N by gender yielded means of 9.1 ‰ for females and 8.7 ‰ for males. The mean RBC δ15N values according to the four groups by quartile were 7.3, 8.2, 9.1, 11.0 ‰ in groups 1–4, respectively. These values correspond to EPA (% RBC fatty acids) group means of: 0.9, 1.8, 2.8, and 4.7 % (O’Brien et al. 2009). The standard deviation of δ15N in this sample did not differ according to gender (1.5 ‰ for both females and males).
Table 2.
Distribution of the RBC nitrogen stable isotope ratio (δ15N)
| Sex | Quartiles of δ15N** | ||||||
|---|---|---|---|---|---|---|---|
| Total | Women | Men | Q1 | Q2 | Q3 | Q4 | |
| No. of participants | 1,073 | 565 | 510 | 262 | 261 | 269 | 281 |
| Mean ± SD (‰) | 9.0 ± 1.5 | 9.1 ± 1.5 | 8.7 ± 1.5 | 7.3 ± 0.3 | 8.2 ± 0.2 | 9.1 ± 0.3 | 11.0 ± 1.1 |
| Maximum | 15.2 | 15.2 | 13.5 | 7.8 | 8.6 | 9.8 | 15.2 |
| Minimum | 6.4 | 6.4 | 6.4 | 6.4 | 7.8 | 8.6 | 9.8 |
| Range (‰) | 8.8 | 8.8 | 7.1 | 1.4 | 0.8 | 1.2 | 5.4 |
Isotope ratios are presented as delta values in “permil” relative to atmospheric nitrogen: δ15N = [(15N/14Nsample − 15N/14Nstandard)/(15N/14Nstandard)] × 1,000 ‰. The relationship between δ15N and EPA follows the linear model: EPA (%RBC fatty acid) = 1.04 × δ15N − 6.7 ‰, as previously described for this population (O’Brien et al. 2009)
Genetic variation among obesity SNPs
Allele frequencies for 10 SNPs that showed significant association with obesity and/or BMI in large-scale GWAS are presented in Table 3. The frequencies for risk alleles observed in this sample of Yup’ik people were comparable to frequencies reported in other populations (Table 3). Notable exceptions include the rs10838738 (MTCH2) and the rs7498665 (SH2B1) risk alleles which are at higher frequencies among this sample of Yup’ik people. All genotyped SNPs were consistent with expected HWE (p > 0.002) and had MAF ≥ 0.05 with the exception of rs6265 (BDNF) which had MAF = 0.04.
Table 3.
Obesity GWAS SNP polymorphisms
| Gene | SNPa | Chr | Risk allele | Frequency of risk allele | Frequency of risk allele in HAPMAP populations | HWE (p value) | References | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| YRI | CEU | HCB | JPT | MEX | |||||||
| NEGR1 | rs2815752 | 1 | A | 0.81 | 0.53 | 0.64 | 0.88 | 0.92 | 0.73 | 0.2007 | Willer et al. (2009) |
| TMEM18 | rs7561317 | 2 | G | 0.85 | 0.79 | 0.85 | 0.91 | 0.88 | 0.88 | 0.1768 | Thorleifsson et al. (2009) |
| ETV5 | rs7647305 | 3 | C | 0.94 | 0.61 | 0.79 | 0.94 | 0.94 | 0.81 | 0.478 | Thorleifsson et al. (2009) |
| GNPDA2 | rs10938397 | 4 | G | 0.15 | 0.21 | 0.45 | 0.25 | 0.37 | NA | 0.2097 | Willer et al. (2009) |
| BDNF | rs6265 | 11 | G | 0.96 | 0.99 | 0.81 | 0.38 | 0.63 | 0.79 | 1 | Willer et al. (2009) |
| MTCH2 | rs10838738 | 11 | G | 0.66 | 0.04 | 0.36 | 0.35 | 0.35 | 0.4 | 0.0037 | Willer et al. (2009) |
| SH2B1 | rs7498665 | 16 | G | 0.66 | 0.21 | 0.38 | 0.15 | 0.13 | 0.38 | 0.462 | Thorleifsson et al. (2009), Willer et al. (2009) |
| FTO | rs9939609 | 16 | A | 0.19 | 0.51 | 0.46 | 0.12 | 0.19 | NA | 0.3223 | Frayling et al. (2007), Scuteri et al. (2007), Willer et al. (2009) |
| MC4R | rs17782313 | 18 | C | 0.08 | 0.31 | 0.27 | 0.14 | 0.24 | 0.14 | 1 | Loos et al. (2008), Willer et al. (2009) |
| KCTD15 | rs29941 | 19 | C | 0.31 | 0.86 | 0.68 | 0.22 | 0.26 | 0.65 | 0.4179 | Thorleifsson et al. (2009) |
Frequency of BMI-increasing allele computed using FREQ module in S.A.G.E. Frequency of BMI-increasing allele among five HapMap populations: Yoruba in Ibadan, Nigeria (YRI), Utah residents with ancestry from northern and western Europe (CEU), Han Chinese in Beijing, China (CHB), Japanese in Tokyo, Japan (JPT), Mexican ancestry in Los Angeles, California. HWE p value computed using founders in PLINK (Purcell et al. 2007)
aSeattle SNPs Genome Variation Server on March 2008 (dbSNP build 126) Version 5.01
Individual SNPs and adiposity phenotypes
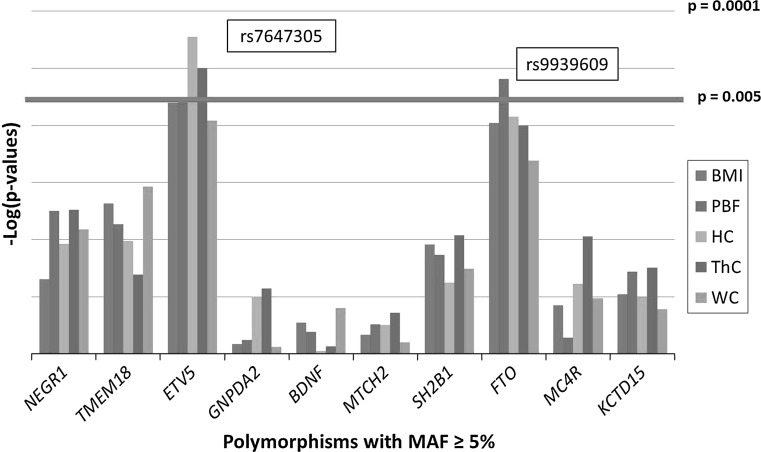
Without adjustment for n-3 PUFA intake (δ15N), no SNPs were significantly associated with adiposity phenotypes after Bonferroni multiple test correction (Supplementary Table II). Although not significant, 8 of the 10 SNPs tested in this study had direction-consistent association trends with BMI that were in agreement with reports from GWAS. After adjusting our analyses for n-3 PUFA intake (δ15N), we found that rs9939609 (FTO) and rs7647305 (ETV5) risk alleles were associated with adiposity phenotypes (Fig. 1; Table 4). Specifically, rs9939609 (FTO) was positively associated with percent body fat (PBF, p = 0.004), and rs7647305 (ETV5) was negatively associated with hip and thigh circumference (HC, p = 0.002 and ThC, p = 0.003, respectively). We did not detect significant SNP interactions with n-3 PUFA intake that modified any associations with adiposity phenotypes (Supplementary Table III).
Fig. 1.
Obesity GWAS polymorphisms associated with adiposity phenotypes. Association of SNPs in a linear regression model adjusted for age, sex, community membership, and n-3 PUFA intake. The red line represents multiple test correction that was estimated using the spectral decomposition of LD matrix (Nyholt 2004). Body mass index (BMI), percent body fat (PBF), hip circumference (HC), thigh circumference (ThC), and waist circumference (WC)
Table 4.
Obesity polymorphisms associated with adiposity after δ15N adjustment
| Gene | SNP | BMI | PBF | HC | ThC | WC |
|---|---|---|---|---|---|---|
| NEGR1 | rs2815752 | 0.222 | 0.056 | 0.109 | 0.055 | 0.081 |
| (β = −0.3, SE = 0.2) | (β = −0.4, SE = 0.2) | (β = −0.4, SE = 0.2) | (β = −0.6, SE = 0.3) | (β = −0.5, SE = 0.3) | ||
| TMEM18 | rs7561317 | 0.048 | 0.073 | 0.103 | 0.202 | 0.034 |
| (β = 0.5, SE = 0.2) | (β = 0.4, SE = 0.2) | (β = 0.4, SE = 0.2) | (β = 0.4, SE = 0.3) | (β = 0.6, SE = 0.3) | ||
| ETV5 | rs7647305 | 0.006 | 0.006 | 0.002 | 0.003 | 0.009 |
| (β = −1.5, SE = 0.6) | (β = −1.4, SE = 0.5) | (β = −1.8, SE = 0.6) | (β = −2.0, SE = 0.7) | (β = −1.9, SE = 0.7) | ||
| GNPDA2 | rs10938397 | 0.821 | 0.761 | 0.315 | 0.267 | 0.867 |
| (β = 0.1, SE = 0.2) | (β = 0.1, SE = 0.2) | (β = 0.2, SE = 0.2) | (β = 0.3, SE = 0.3) | (β = 0.1, SE = 0.3) | ||
| BDNF | rs6265 | 0.531 | 0.646 | 0.943 | 0.859 | 0.396 |
| (β = 0.4, SE = 0.6) | (β = 0.2, SE = 0.5) | (β = 0.0, SE = 0.6) | (β = 0.1, SE = 0.7) | (β = 0.6, SE = 0.7) | ||
| MTCH2 | rs10838738 | 0.682 | 0.550 | 0.556 | 0.440 | 0.797 |
| (β = 0.1 SE = 0.2) | (β = 0.1, SE = 0.2) | (β = 0.1, SE = 0.2) | (β = 0.2, SE = 0.2) | (β = 0.1, SE = 0.2) | ||
| SH2B1 | rs7498665 | 0.111 | 0.136 | 0.239 | 0.092 | 0.179 |
| (β = 0.3, SE = 0.2) | (β = 0.2, SE = 0.2) | (β = 0.2, SE = 0.2) | (β = 0.4, SE = 0.2) | (β = 0.3, SE = 0.2) | ||
| FTO | rs9939609 | 0.010 | 0.004 | 0.008 | 0.010 | 0.020 |
| (β = 0.6, SE = 0.2) | (β = 0.6, SE = 0.2) | (β = 0.6, SE = 0.2) | (β = 0.7, SE = 0.3) | (β = 0.7, SE = 0.3) | ||
| MC4R | rs17782313 | 0.377 | 0.726 | 0.243 | 0.094 | 0.328 |
| (β = 0.3, SE = 0.3) | (β = 0.1, SE = 0.3) | (β = 0.4, SE = 0.3) | (β = 0.6, SE = 0.4) | (β = 0.4, SE = 0.4) | ||
| KCTD15 | rs29941 | 0.301 | 0.192 | 0.319 | 0.177 | 0.405 |
| (β = 0.2, SE = 0.2) | (β = 0.2, SE = 0.2) | (β = 0.2, SE = 0.2) | (β = 0.3, SE = 0.2) | (β = 0.2, SE = 0.3) |
p values for association of obesity SNPs adjusted for age, sex, community group, and n-3 PUFA intake. Estimates of effect size (β) are reported for additive model and BMI-increasing alleles using transformed phenotypes. Results significant at p ≤ 0.005 are highlighted in bold. Body mass index (BMI), percent body fat (PBF), hip circumference (HC), thigh circumference (ThC), and waist circumference (WC)
Genetic risk score and adiposity phenotypes
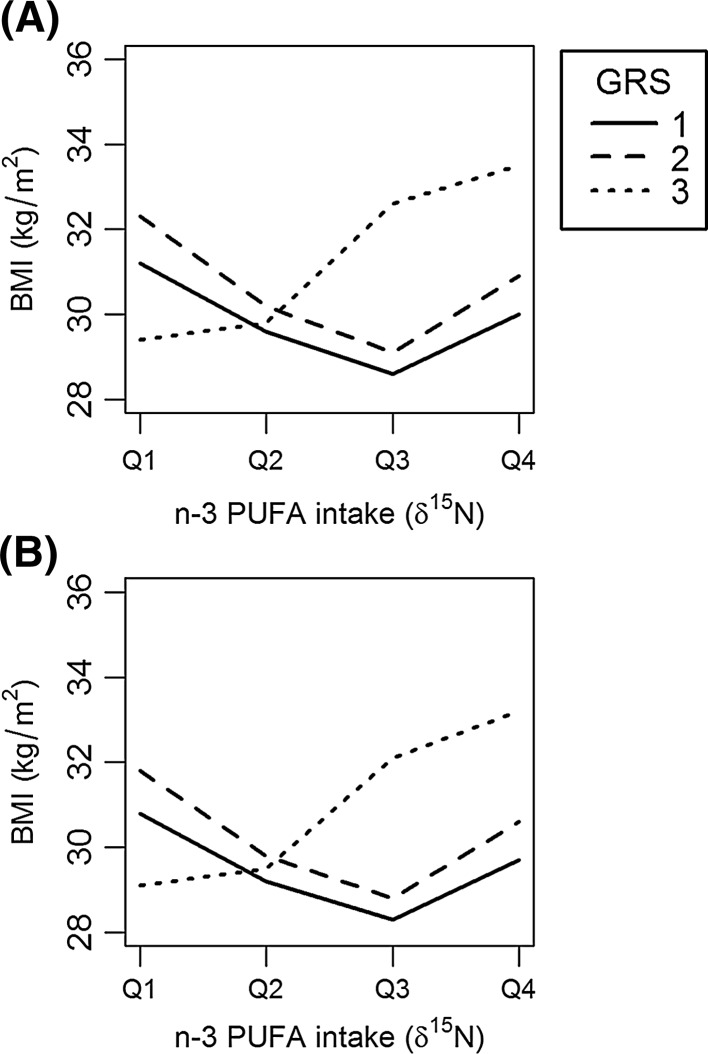
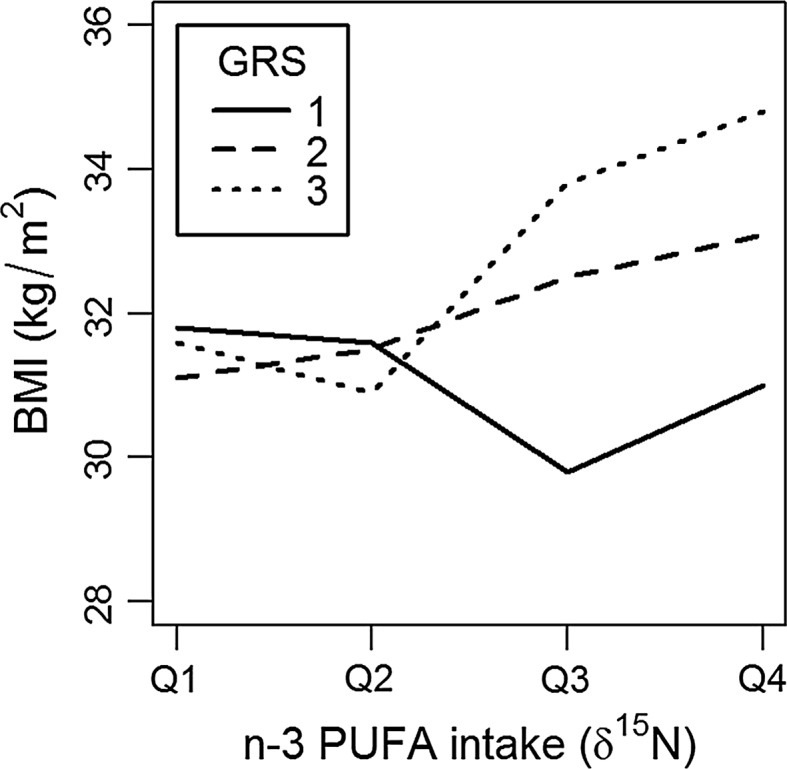
The GRS was positively associated with adiposity as measured by BMI (p = 0.012), PBF (p = 0.022), ThC (p = 0.025), and WC (p = 0.038) after adjusting for n-3 PUFA intake (δ15N) (Table 5). We did not observe significant interactions between the GRS and community location that modified adiposity phenotypes. The GRS interactions with n-3 PUFA intake were significant for BMI (p = 0.011), PBF (p = 0.025), and WC (p = 0.018). Figure 2 show that n-3 PUFA intake significantly modifies the GRS association with BMI, such that people with the highest BMI also had high levels n-3 PUFA intake (Q4) and a high GRS (T3). In contrast, our results show that people with a low GRS (T1) and high levels of n-3 PUFA intake (Q4) had a lower BMI, relative to individuals with the same GRS (T1) and low n-3 PUFA (Q1) intake (Fig. 2). The GRS interaction with n-3 PUFA explained approximately twice the phenotypic variation in adiposity phenotypes relative to GRS associations with adiposity alone (Table 6). Figure 3 demonstrates the δ15N*GRS interaction corrected for either (A) total energy (kcal) or (B) total fat intake (% calories) in a subset of participants (n = 488) did not significantly change the trend or strength of the δ15N*GRS interaction (Supplementary Table IV).
Table 5.
Interaction with n-3 PUFA intake modifies the GRS association with adiposity phenotypes
| Outcome | Main effecta | GRS*n-3 PUFA intakeb | GRS*community groupc |
|---|---|---|---|
| BMI (kg/m2) | 0.012 | 0.011 | 0.480 |
| Percentage body fat (%) | 0.022 | 0.025 | 0.469 |
| Hip circumference (cm) | 0.08 | 0.068 | 0.916 |
| Thigh circumference (cm) | 0.025 | 0.084 | 0.555 |
| Waist circumference (cm) | 0.038 | 0.018 | 0.517 |
Results significant at p ≤ 0.05 are highlighted in bold
ap values for GRS analysis adjusted for sex, age, community group, and n-3 PUFA intake
bp values for interaction analysis (GRS*n-3 PUFA) adjusted for sex, age, community group, and n-3 PUFA intake
cp values for interaction analysis (GRS*community group) adjusted for sex, age, community group, and n-3 PUFA intake
Fig. 2.
n-3 PUFA intake modifies the GRS association with BMI (p = 0.011). n-3 PUFA intake (δ15N) is represented as a categorical variable with four groups by quartile (Q1–Q4) where Q1 is lowest level of n-3 PUFA intake and Q4 is the highest level of n-3 PUFA intake. The GRS is represented as a categorical variable with three groups by tertiles (T1–T3) where T1 is lowest obesity GRS and T3 is highest obesity GRS
Table 6.
Variation (%) in adiposity phenotypes explained by GRS association and GRS interactions with n-3 PUFA intake
| Variable | BMI | PBF | HC | ThC | WC |
|---|---|---|---|---|---|
| GRS | 0.7 | 0.3 | 0.5 | 0.7 | 0.5 |
| GRS*δ15N | 1.7 | 0.8 | 1.4 | 1.5 | 1.4 |
Results significant at p ≤ 0.05 are highlighted in bold. Variation attributed to GRS after adjustment for age, sex, community group, and n-3 PUFA intake. Variation attributed to GRS*n-3 PUFA interaction after adjustment for age, sex, community group, and n-3 PUFA intake
Fig. 3.
n-3 PUFA intake modifies the GRS association with BMI after correction for either a total fat intake (p = 0.034) or b total energy intake (p = 0.033). In a subset of participants (n = 488), we evaluated whether total fat intake (kcal) or total energy intake (% calories) confounded the GRS*n-3 PUFA intake interaction with BMI by including these variables, derived from 24 Hour Recalls (24HR), as an additional covariate in the δ15N*GRS interaction model. n-3 PUFA intake (δ15N) is represented as a categorical variable with four groups by quartile (Q1–Q4) where Q1 is lowest level of n-3 PUFA intake and Q4 is the highest level of n-3 PUFA intake. The GRS is represented as a categorical variable with three groups by tertiles (T1–T3) where T1 is lowest obesity GRS and T3 is highest obesity GRS
Discussion
In this study, we evaluated 10 of the 32 loci identified in a previous meta-analysis of obesity GWAS (Speliotes et al. 2010) for individual and cumulative associations with adiposity phenotypes in a cohort of Yup’ik people and assessed whether exposure to n-3 PUFAs modified these associations. After corrections for n-3 PUFA consumption, we found rs9939609 (FTO) was positively associated PBF and rs7647305 (ETV5) was negatively associated with ThC and HC. Interestingly, controlled feeding studies in rodents have shown that n-3 PUFAs decrease adiposity by reducing central fat mass (Tiryaki-Sönmez et al. 2011); however, epidemiological evidence in humans supporting the anti-obesity effects of n-3 PUFAs remains controversial and is often limited to n-3 PUFA supplementation studies (Buckley and Howe 2010). Our analyses indicate that genetic predisposition to obesity, as measured by the GRS, was positively associated with adiposity and interactions with n-3 PUFA intake modified these associations whereby among individuals with a low GRS, n-3 PUFA intake have either no effect on BMI or they are anti-obesigenic. However, individuals with moderate (2) or high (3) GRS, n-3 PUFAs intake is obesigenic. Taken together, these results suggest the anti-obesity effects of n-3 PUFA in humans may be dependent upon an individual’s genetic predisposition to obesity.
GWAS have consistently identified SNPs associated with BMI in European populations that are located within the first intron of FTO, such as rs9939609 (Frayling et al. 2007; Speliotes et al. 2010; Willer et al. 2009). To date, most FTO candidate gene studies have been conducted on the rs9939609 SNP mainly because this variant has the strongest known association of any SNP with BMI (Tung and Yeo 2011). Subsequent to the discovery of the rs9939609 SNP, studies on the association of FTO with BMI in European (Cauchi et al. 2009), African (Adeyemo et al. 2010), and Asian (Cha et al. 2008) study populations have largely confirmed the importance of FTO as a candidate gene for obesity, although this is not without exception (Karns et al. 2011; Li et al. 2008). Our data indicate the rs9939609 (FTO) risk allele was positively associated with PBF in Yup’ik people and supports other studies indicating that variation in FTO contributes to fat accumulation (Moore et al. 2011).
In contrast to rs9939609 (FTO), candidate gene studies that have replicated the rs7647305 (ETV5) association with obesity remain limited (Elks et al. 2010; Li et al. 2011; Sandholt et al. 2011). A prospective sample of 7,146 European children showed the ETV5 (rs7647305) SNP was associated with increased BMI and body weight (Elks et al. 2010), and a cohort of 18,014 middle-aged Danish adults demonstrated the ETV5 (rs7647305) SNP was associated with increased odds of being overweight and/or obese (Sandholt et al. 2011). Finally, a prospective study among 20,428 European adults with an average follow-up of 12.9 years demonstrated the ETV5 (rs7647305) SNP was associated with protection from developing T2D, and this association was stronger after correcting for BMI (Li et al. 2011).
Our study was not able to evaluate whether the ETV5 (rs7647305) SNP was associated with protection from developing T2D in Yup’ik people due to our cross-sectional study design. Interestingly, we found the ETV5 (rs7647305) associations with adiposity phenotypes in Yup’ik people were “opposite” to those reported in participants of European ancestry (Elks et al. 2010; Sandholt et al. 2011). Although this lack of consistency can be attributed to type I error, theoretical modeling has argued that “flip-flop” association can be attributed to population differences that occur when the SNP of interest is correlated with the causal variant through linkage disequilibrium (Lin et al. 2007). Specifically, our results demonstrate the ETV5 (rs7647305) C allele was associated with reduced HC and ThC and near associations for reduced BMI, PBF, and WC. Moreover, despite the fact that overweight and obesity prevalence among Yup’ik people resembles the prevalence of overweight and obesity in the general US population, T2D prevalence remains low (~3 %) (Mohatt et al. 2007). These results are largely consistent with the finding of Li et al. (2011) and together suggest the ETV5 (rs7647305) loci may protect against the development of T2D by reducing obesity.
The GRS, which evaluates the cumulative association of 10 obesity GWAS loci, was positively associated with several measures of obesity (BMI, PBF, ThC, and WC) in our study population. Consistent with previous studies, our analysis demonstrated the GRS predicts <1 % of the BMI variation in this sample of Yup’ik people (0.7 % for BMI). Li et al. (2010) included 12 SNPs that explained 0.9 % of BMI variation in Europeans, Takeuchi et al. (2011) calculated a GRS with 14 SNPs that explained 0.65 % among Japanese, and Peterson et al. (2011) included 56 SNPs explained 0.66 % of the variation in BMI among a mixed sample of European- and African-Americans. We calculated a GRS in this study using 5 obesity GWAS SNPs that overlap with Li et al. (2010), 7 obesity GWAS SNPs that overlap with Takeuchi et al. (2011), and 9 obesity GWAS SNPs that overlap with Peterson et al. (2011). Taken together, these results suggest that while considerable progress has been made in gene discovery, obesity SNPs highly replicated in GWAS do not yet predict a significant proportion of the heritability attributed to adiposity phenotypes.
Increasingly, experimental designs are considering gene–environment interactions to account for the “missing heritability” associated with complex phenotypes like obesity (Van IJzendoorn et al. 2011). Given the established contribution of n-3 PUFAs to changes in adiposity phenotypes and the availability of a precise biomarker for n-3 PUFA intake (δ15N) in Yup’ik people (O’Brien et al. 2009), we tested whether individuals SNPs and the GRS associations with adiposity phenotypes were modified by consumption of n-3 PUFA. Our analysis demonstrated that elevated n-3 PUFA intake with an increased GRS strengthened the association with adiposity phenotypes. Although n-3 PUFA intake (δ15N) differs according to community group (Supplementary Table V), we did not detect significant GRS*community group interactions that modified adiposity phenotypes. Interestingly, the interaction of GRS with n-3 PUFA intake accounted for approximately twice the phenotypic variation as the genetic risk score alone. These results are consistent with gene-by-environment analysis conducted by Li et al. (2010) that demonstrated physical activity interactions with a GRS, calculated from 12 obesity risk alleles, accounted for twice as much phenotypic variation in obesity-related traits relative to the genetic risk score alone.
The strengths of this study include a sample size with adequate statistical power (>90 %) to detect SNPS associated with adiposity phenotypes, and a statistical approach that accounts for relationships among participants while also allowing for covariates. Our isolated study population of Yup’ik people were ideally suited to test the contribution of interactions between n-3 PUFA intake (Makhoul et al. 2010, 2011) and genetic factors (Lemas et al. 2012) to changes in adiposity phenotypes. Given the 50-fold range of n-3 PUFA consumption from naturally harvested foods in the region that this study population resides in (Bersamin et al. 2008), an additional strength of this study includes a biomarker of EPA and DHA intake (δ15N) that can be precisely estimated in large epidemiological studies (O’Brien et al. 2009). A potential limitation of our study was that n-3 PUFA intake may be confounded by total fat and total energy intake due to a traditional diet pattern that is high in fat. Given this limitation, it is possible that the δ15N*GRS interaction with adiposity phenotypes may have been influenced by total fat or total energy intake. We evaluated this limitation in a subset of participants (n = 488) by including total energy (kcal) and total fat intake (% calories), derived from 24 Hour Recalls (24HR), as an additional covariate in the δ15N*GRS interaction model (Model 7). Our follow-up analysis did not find that including total energy and total fat or intake in the δ15N*GRS interaction model significantly changed the results. These data indicate the GRS*n-3 PUFA interactions were not appreciably confounded by total fat and total energy intake and suggest that n-3 PUFA intake may modulate the risk associated with obesity susceptibility genes.
In conclusion, cross-sectional studies of this nature have potential to elucidate novel gene–diet interactions that may ultimately help account for some of the “missing heritability” associated with obesity (Manolio et al. 2009). To the extent that genetic predisposition to obesity is estimated by 10 SNPs reproducibly identified through GWAS, our results suggest that including n-3 PUFA interactions increased the ability to detect the contribution of these SNPs to changes in BMI. In contrast, individuals with no genetic predisposition to obesity, as measured by GRS, had less adiposity in the presence of high n-3 PUFA intake. Although animal and human studies have reported that consumption of n-3 PUFAs may reduce adiposity (Buckley and Howe 2010), our results indicate the anti-obesity effects of n-3 PUFAs may, in part, be dependent upon an individual’s genetic predisposition to obesity. Characterizing gene–diet interactions in with high intakes of n-3 PUFAs may help identify individuals that are likely to benefit from specific dietary interventions. Interestingly, these analyses do suggest anti-obesity effects of n-3 PUFA intake are, in part, dependent upon an individual’s genotype and may play an essential role in determining the risk associated with obesity susceptibility genes among Yup’ik people. Additional genomic studies will be required to replicate these results in large circumpolar populations with widely varying intake of n-3 PUFA in order to determine the validity and public health implication.
Electronic supplementary material
Acknowledgments
This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Award R01-DK074842 (PI: B.B.B) and R01-DKO74842-02S1 (PI: B.B.B); and by National Center for Research Resources (NCRR) and the National Institute of General Medical Sciences Awards P20RR016430 (PI: B.B.B) and P30DK056336 (PI: D.B.A). Some of the results presented in this paper were obtained by using the program package S.A.G.E., which is supported by a US Public Health Service Resource Grant RR-03655 (PI: R.C.E) from the National Center for Research Resources (NCRR). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. The authors also thank the community field research assistants who helped with the study recruitment and data collection as well as Dr. Andrea Bersamin that generously provided assistance regarding dietary analysis. Finally, the CANHR team would like to express our sincere appreciation to all of our study participants and their communities for welcoming us and teaching us so much about the Yup’ik way of life. Quyana!
Conflict of interest
Dr. Allison has received, anticipates, or has had financial interests with the Frontiers Foundation; Vivus, Inc; Kraft Foods; University of Wisconsin; University of Arizona; Paul Weiss, Wharton & Garrison LLP; and Sage Publications. No other co-authors on this manuscript declare conflict of interest.
Contributor Information
Dominick J. Lemas, Email: dominck.lemas@ucdenver.edu
Yann C. Klimentidis, yann@email.arizona.edu
Howard H. Wiener, Email: hwiener@uab.edu
Diane M. O’Brien, Email: dmobrien@alaska.edu
Scarlett E. Hopkins, Email: sehopkins2@alaska.edu
David B. Allison, Email: dallison@uab.edu
Jose R. Fernandez, Email: jose@uab.edu
Hemant K. Tiwari, Email: htiwari@uab.edu
Bert B. Boyer, Phone: +907-474-7733, FAX: +907-474-5712, Email: bboyer@alaska.edu
References
- Adeyemo AA, Chen G, Zhou J, et al. FTO genetic variation and association with obesity in West Africans and African Americans. Diabetes. 2010;59:1549–1554. doi: 10.2337/db09-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersamin A, Luick BR, King IB, et al. Westernizing diets influence fat intake, red blood cell fatty acid composition, and health in remote Alaskan Native communities in the center for Alaska Native health study. J Am Diet Assoc. 2008;108:266–273. doi: 10.1016/j.jada.2007.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box GEP, Cox DR. An analysis of transformations. J Roy Stat Soc Ser B (Methodol) 1964;26:211–252. [Google Scholar]
- Boyer BB, Mohatt GV, Plaetke R, et al. Metabolic syndrome in Yup’ik Eskimos: the Center for Alaska Native Health Research (CANHR) Study. Obesity. 2007;15:2535–2540. doi: 10.1038/oby.2007.302. [DOI] [PubMed] [Google Scholar]
- Bray GA, Paeratakul S, Popkin BM. Dietary fat and obesity: a review of animal, clinical and epidemiological studies. Physiol Behav. 2004;83:549–555. doi: 10.1016/j.physbeh.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Buckley JD, Howe PRC. Long-chain omega-3 polyunsaturated fatty acids may be beneficial for reducing obesity—a review. Nutrients. 2010;2:1212–1230. doi: 10.3390/nu2121212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchi S, Stutzmann F, Cavalcanti-Proenca C, et al. Combined effects of MC4R and FTO common genetic variants on obesity in European general populations. J Mol Med (Berl) 2009;87:537–546. doi: 10.1007/s00109-009-0451-6. [DOI] [PubMed] [Google Scholar]
- Cha SW, Choi SM, Kim KS, et al. Replication of genetic effects of FTO polymorphisms on BMI in a Korean population. Obesity. 2008;16:2187–2189. doi: 10.1038/oby.2008.314. [DOI] [PubMed] [Google Scholar]
- Couet C, Delarue J, Ritz P, et al. Effect of dietary fish oil on body fat mass and basal fat oxidation in healthy adults. Int J Obes Relat Metab Disord. 1997;21:637–643. doi: 10.1038/sj.ijo.0800451. [DOI] [PubMed] [Google Scholar]
- Curioni CC, Lourenço PM. Long-term weight loss after diet and exercise: a systematic review. Int J Obes. 2005;29:1168–1174. doi: 10.1038/sj.ijo.0803015. [DOI] [PubMed] [Google Scholar]
- Dansinger ML, Tatsioni A, Wong JB, et al. Meta-analysis: the effect of dietary counseling for weight loss. Ann Intern Med. 2007;147:41–50. doi: 10.7326/0003-4819-147-1-200707030-00007. [DOI] [PubMed] [Google Scholar]
- Elks CE, Loos RJF, Sharp SJ, et al. Genetic markers of adult obesity risk are associated with greater early infancy weight gain and growth. PLoS Med. 2010;7:e1000284. doi: 10.1371/journal.pmed.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George VT, Elston RC. Testing the association between polymorphic markers and quantitative traits in pedigrees. Genet Epidemiol. 1987;4:193–201. doi: 10.1002/gepi.1370040304. [DOI] [PubMed] [Google Scholar]
- Hainault I, Carolotti M, Hajduch E, et al. Fish oil in a high lard diet prevents obesity, hyperlipidemia, and adipocyte insulin resistance in rats. Ann N Y Acad Sci. 1993;683:98–101. doi: 10.1111/j.1749-6632.1993.tb35696.x. [DOI] [PubMed] [Google Scholar]
- Hester JM, Wing MR, Li J, et al. Implication of European-derived adiposity loci in African Americans. Int J Obes. 2012;36:465–473. doi: 10.1038/ijo.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heutink P, Oostra BA. Gene finding in genetically isolated populations. Hum Mol Genet. 2002;11:2507–2515. doi: 10.1093/hmg/11.20.2507. [DOI] [PubMed] [Google Scholar]
- Jones PJ. Effect of fatty acid composition of dietary fat on energy balance and expenditure in hamsters. Can J Physiol Pharmacol. 1989;67:994–998. doi: 10.1139/y89-156. [DOI] [PubMed] [Google Scholar]
- Jourdan C, Kloiber S, Nieters A, et al. Gene-PUFA interactions and obesity risk. Br J Nutr. 2011;106:1263–1272. doi: 10.1017/S0007114511001541. [DOI] [PubMed] [Google Scholar]
- Karns R, Viali S, Tuitele J, et al. Common variants in FTO are not significantly associated with obesity-related phenotypes among Samoans of Polynesia. Ann Hum Genet. 2011;76:17–24. doi: 10.1111/j.1469-1809.2011.00686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecký J, Rossmeisl M, Flachs P, et al. n-3 PUFA: bioavailability and modulation of adipose tissue function. Proc Nutr Soc. 2009;68:361–369. doi: 10.1017/S0029665109990231. [DOI] [PubMed] [Google Scholar]
- Lemas DJ, Wiener HW, O’Brien DM, et al. Genetic polymorphisms in carnitine palmitoyltransferase 1A gene are associated with variation in body composition and fasting lipid traits in Yup’ik Eskimos. J Lipid Res. 2012;53:175–184. doi: 10.1194/jlr.P018952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wu Y, Loos RJF, et al. Variants in the fat mass- and obesity-associated (FTO) gene are not associated with obesity in a Chinese Han population. Diabetes. 2008;57:264–268. doi: 10.2337/db07-1130. [DOI] [PubMed] [Google Scholar]
- Li S, Zhao JH, Luan J, et al. Cumulative effects and predictive value of common obesity-susceptibility variants identified by genome-wide association studies. Am J Clin Nutr. 2010;91:184–190. doi: 10.3945/ajcn.2009.28403. [DOI] [PubMed] [Google Scholar]
- Li S, Zhao JH, Luan J, et al. Genetic predisposition to obesity leads to increased risk of type 2 diabetes. Diabetologia. 2011;54:776–782. doi: 10.1007/s00125-011-2044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P-I, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TG, Roche AF, Martorell R (1988) Anthropometric standardization reference manual. Human kinetics books, Illinois, pp 39–70
- Loos RJF, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27:325–351. doi: 10.1023/A:1025635913927. [DOI] [PubMed] [Google Scholar]
- Makhoul Z, Kristal AR, Gulati R, et al. Associations of very high intakes of eicosapentaenoic and docosahexaenoic acids with biomarkers of chronic disease risk among Yup’ik Eskimos. Am J Clin Nutr. 2010;91:777–785. doi: 10.3945/ajcn.2009.28820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhoul Z, Kristal AR, Gulati R, et al. Associations of obesity with triglycerides and C-reactive protein are attenuated in adults with high red blood cell eicosapentaenoic and docosahexaenoic acids. Eur J Clin Nutr. 2011;65:808–817. doi: 10.1038/ejcn.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti A, Martinez-González AM, Martinez JA. Interaction between genes and lifestyle factors on obesity. Proc Nutr Soc. 2008;67:1–8. doi: 10.1017/S002966510800596X. [DOI] [PubMed] [Google Scholar]
- Mohatt GV, Plaetke R, Klejka J, et al. The Center for Alaska Native Health Research Study: a community-based participatory research study of obesity and chronic disease-related protective and risk factors. Int J Circumpolar Health. 2007;66:8–18. doi: 10.3402/ijch.v66i1.18219. [DOI] [PubMed] [Google Scholar]
- Moore SC, Gunter MJ, Daniel CR, et al. Common genetic variants and central adiposity among Asian-Indians. Obesity. 2011;20:1902–1908. doi: 10.1038/oby.2011.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien DM, Kristal AR, Jeannet MA, et al. Red blood cell delta15N: a novel biomarker of dietary eicosapentaenoic acid and docosahexaenoic acid intake. Am J Clin Nutr. 2009;89:913–919. doi: 10.3945/ajcn.2008.27054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CL, Yanovski SZ, Carroll MD, Flegal KM. The epidemiology of obesity. Gastroenterology. 2007;132:2087–2102. doi: 10.1053/j.gastro.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Peterson RE, Maes HH, Holmans P, et al. Genetic risk sum score comprised of common polygenic variation is associated with body mass index. Hum Genet. 2011;129:221–230. doi: 10.1007/s00439-010-0917-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaetke R, Balbi F. PedMerge: merging pedigrees to facilitate family-based genetic statistical analyses. Bioinformatics. 2010;26:2790–2791. doi: 10.1093/bioinformatics/btq523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale BM, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Cornelis MC, Zhang C, et al. Genetic predisposition, Western dietary pattern, and the risk of type 2 diabetes in men. Am J Clin Nutr. 2009;89:1453–1458. doi: 10.3945/ajcn.2008.27249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandholt CH, Vestmar MA, Bille DS, et al. Studies of metabolic phenotypic correlates of 15 obesity associated gene variants. PLoS One. 2011;6:e23531. doi: 10.1371/journal.pone.0023531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen W-M, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlien LH, Huang XF, Lin S, et al. Dietary fat subtypes and obesity. World Rev Nutr Diet. 2001;88:148–154. doi: 10.1159/000059778. [DOI] [PubMed] [Google Scholar]
- Stunkard AJ, Harris JRR, Pedersen NLL, McClearn GEE. The body-mass index of twins who have been reared apart. N Engl J Med. 1990;322:1483–1487. doi: 10.1056/NEJM199005243222102. [DOI] [PubMed] [Google Scholar]
- Su W, Jones PJH. Dietary fatty acid composition influences energy accretion in rats. J Nutr. 1993;123:2109–2114. doi: 10.1093/jn/123.12.2109. [DOI] [PubMed] [Google Scholar]
- Takeuchi F, Yamamoto K, Katsuya T, et al. Association of genetic variants for susceptibility to obesity with type 2 diabetes in Japanese individuals. Diabetologia. 2011;54:1350–1359. doi: 10.1007/s00125-011-2086-8. [DOI] [PubMed] [Google Scholar]
- Tang K, Fu D-J, Julien D, et al. Chip-based genotyping by mass spectrometry. Proc Natl Acad Sci USA. 1999;96:10016–10020. doi: 10.1073/pnas.96.18.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Thorsdottir I, Tomasson H, Gunnarsdottir I, et al. Randomized trial of weight-loss-diets for young adults varying in fish and fish oil content. Int J Obes. 2007;31:1560–1566. doi: 10.1038/sj.ijo.0803643. [DOI] [PubMed] [Google Scholar]
- Tiryaki-Sönmez G, Schoenfeld B, Vatansever-Ozen S. Omega-3 fatty acids and exercise: a review of their combined effects on body composition and physical performance. Biomed Hum Kinet. 2011;3:23–29. [Google Scholar]
- Tsai AG, Wadden TA. Systematic review: an evaluation of major commercial weight loss programs in the United States. Ann Intern Med. 2005;142:56–66. doi: 10.7326/0003-4819-142-1-200501040-00012. [DOI] [PubMed] [Google Scholar]
- Tung Y-CL, Yeo GSH. From GWAS to biology: lessons from FTO. Ann N Y Acad Sci. 2011;1220:162–171. doi: 10.1111/j.1749-6632.2010.05903.x. [DOI] [PubMed] [Google Scholar]
- Van IJzendoorn MH, Bakermans-Kranenburg MJ, Belsky J et al (2011) Gene-by-environment experiments: a new approach to finding the missing heritability. Nat Rev Genet 12:881 [DOI] [PubMed]
- Willer CJ, Speliotes EK, Loos RJF, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.