Abstract
Amylin is a pancreatic hormone that plays important roles in overall metabolism and in glucose homeostasis. The therapeutic restoration of postprandial and basal amylin levels is highly desirable for patients with diabetes who need to avoid glucose excursions. Protein conjugation with polyethylene glycol (PEG) has long been known to be a convenient approach for extending the biological effects of biopharmaceuticals. We have investigated the reactivity of amylin with methoxy polyethylene glycol succinimidyl carbonate and methoxy polyethylene glycol succinimidyl propionate, which have an average molecular weight of 5 kDa. The reaction, which was conducted in both aqueous and organic (dimethyl sulfoxide) solvents, occurred within a few minutes and resulted in at least four detectable products with distinct kinetic phases. These results suggest a kinetic selectivity for PEGylation by succinimidyl derivatives; these derivatives exhibit enhanced reactivity with primary amine groups, as indicated by an evaluation of the remaining amino groups using fluorescamine. The analysis of tryptic fragments from mono- and diPEGylated amylin revealed that conjugation occurred within the 1-11 amino acid region, most likely at the two amine groups of Lys1. The reaction products were efficiently separated by C-18 reversed phase chromatography. Binding assays confirmed the ability of mono- and diPEGylated amylin to interact with the amylin co-receptor receptor activity-modifying protein 2. Subcutaneous administration in mice revealed the effectiveness of monoPEG-amylin and diPEG-amylin in reducing glycemia; both compounds exhibited prolonged action compared to unmodified amylin. These features suggest the potential use of PEGylated amylin to restore basal amylin levels.
Key words: amylin, diabetes, islet-associated polypeptide, PEGylation, receptor activity-modifying protein
INTRODUCTION
Amylin is a 37 amino acid pancreatic protein hormone that is co-secreted with insulin (1) and typically displays low basal serum levels and higher postprandial concentrations (2). Amylin plays an important role in the control of various physiological and metabolic functions such as glucose and lactate homeostasis and the regulation of insulin and glucagon secretion (2,3).
The use of amylin restoration to the treatment of diabetes was originally suggested (4), shortly after its discovery by two independent groups (5–8). However, human amylin displays limited solubility in an aqueous medium (9), which prompted a proposal to use the triple proline amylinomimetic compound (Pro25, 28, 29) pramlintide (10) after the discovery of more stable proline-rich amylin variants compounds (11). Pramlintide is administered by subcutaneous (s.c.) injection shortly before mealtime, which mimics postprandial amylin levels. However, to date, there is no product available to restore basal amylin levels in the fasting state; the only currently available product is a polymeric system doped with amylin for sustained and controlled hormone release (12).
Bioconjugation with polyethylene glycol (PEG) has long been used as a strategy for modifying the pharmacokinetics of therapeutic proteins (13,14). The addition of one or more PEG chains to a macromolecule can enhance its lifetime circulation and solubility and can also modify its biological activity (13–17). Amylin decays rapidly in plasma, with a half-life (t0.5) of approximately 13 min (10); therefore, increasing the amylin t0.5 would be a desirable strategy to achieve effective basal hormone restoration.
In an attempt to develop a conjugated amylin product with improved control over glycemia, we evaluated the PEGylation strategy using two distinct methoxy polyethylene glycol (mPEG) succinimidyl coupling agents, namely, mPEG succinimidyl carbonate (mPEG-SC; Fig. 1b) and mPEG succinimidyl propionate (mPEG-SPA; Fig. 1c), both in aqueous (phosphate buffered saline (PBS), pH 7.4) and nonaqueous (dimethyl sulfoxide (DMSO)) solvents. Our results show that both coupling agents exhibit similar reaction kinetics and that purified mono- and diPEGylated amylin exert more sustained effects on in vivo glucose homeostasis relative to unmodified amylin. We discuss the therapeutic potential of PEGylated amylin in restoring basal amylin levels and its applicability for the modification of other amylin and amylinomimetic compounds.
Fig. 1.

Chemical structures of methoxy polyethylene glycol succinimidyl agents and amylin. a The amino acids-containing amine groups are in bold. The numbers correspond to the sequence. b A chemical representation of methoxy polyethylene glycol succinimidyl carbonate. c A chemical representation of methoxy polyethylene glycol succinimidyl propionate. d A ribbon representation of amylin (PDB entry 2KJ7). The amino acid side chains are represented by lines. At the top, in sticks, is a detailed view of the lysine amine groups
MATERIAL AND METHODS
Reagents
Carboxy-amidated murine (mouse/rat) amylin (CAS 122384-88-7) with a disulfide bridge between C2 and C7 was obtained from GenScript (USA; cat. no. RP11280, lot nos. 55613-1, 40214-1, and 128567001040611LQ). Methoxyl polyethylene glycol succinimidyl carbonate and propionate (both with average molecular numbers = 5 kDa, polydispersity = 1.08 according to the manufacturer) were both acquired from Nanocs (USA). Fluorescamine (CAS no. 38183-12-9) and fluorescein isothiocyanate (FITC) were purchased from Sigma-Aldrich Brazil. Distilled water was deionized to less than 1.0 μs and filtered prior to use through a 0.22-μm pore membrane in a water purification system. All other reagents were of analytical grade. All buffers and solutions were prepared immediately prior to use.
Amylin PEGylation
Amylin (5 mg/mL) was PEGylated by mixing it with the desired amount of mPEG (as indicated in the “Results” section) in either aqueous solution (PBS buffer, 8.1 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, pH 7.4) or DMSO at 25°C. Aliquots were collected at the indicated times and quenched by the addition of excess l-lysine or by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (18–21).
Electrophoretic Analysis of PEGylation Reaction
The quenched reaction was analyzed on a 22.5% SDS-PAGE gel (18–21), with bovine serum albumin (66 kDa), lysozyme (14 kDa), and insulin B-chain (3.4 kDa) as molecular weight markers. A molecular weight estimation could not be performed by comparison to the ladder due to the anomalous (slower) migration of PEGylated products on gels (18,20). The gels were stained with the Coomassie Brilliant Blue R-250 followed by barium iodine staining for PEG detection (18). The stained gels were digitized for further data analysis by integration with ImageJ (22). Peak analysis was performed through a curve-fitting process using Fityk (23), assuming Gaussian functions for each peak.
Kinetic Analysis of the Amylin PEGylation Reaction
The PEGylation reaction was followed by a derivatization of the remaining primary amines in amylin with fluorescamine, as described in detail elsewhere (12,24). In brief, the PEGylation reaction was aliquoted at varying time intervals as indicated in the figures and was added to freshly prepared fluorescamine solution (0.05% in DMSO), and the resulting mixture was added to PBS. Fluorescence readings were performed using the Jasco FP-6300 spectrofluorometer (Jasco, Inc., USA) with excitation at 390 nm and emission at 475 nm. The readings were corrected for their signal contributions with a blank sample.
PEGylation Product Purification
Preparative bioconjugation and further amylin purification were carried out as follows. Amylin was dissolved in DMSO to yield 10 mg/mL final concentration and then added to an equal volume of mPEG succinimidyl–DMSO solution for a molar excess of 10:1 mPEG/amylin. Reactions conducted with a molar excess of 5:1 or lower resulted in very poor conjugation efficiency, yielding a large amount of free, nonconjugated amylin, even after a 4-h reaction. At given time intervals, 10-μL aliquots were collected for SDS-PAGE analysis, and 2-μL aliquots were collected for fluorescamine analysis (see below). The PEGylation reaction was stopped with l-lysine in water (2 mol l-Lys added for each 1 mol mPEG) and then diluted 1:1 (v/v) with mobile phase A (0.1% trifluoroacetic acid (TFA) in water). The reaction was then chromatographed using the Jasco LC-2000 HPLC System (Jasco, Inc., USA) and the Kromasil C18 column (250 × 10 mm, particle size 5 μm) with a flow rate of 4 mL/min. The chromatography procedure consisted of a 15-min linear gradient of acetonitrile in water containing 0.1% (v/v) TFA, progressing from 40% (after 5 min at 0% B) to 70%. The purification was monitored by following the absorbance at 220 nm. The samples were pooled as indicated, lyophilized, and then dissolved in water for immediate use. The protein content was evaluated by the Bradford assay (25), which did not show any interference from conjugated free mPEG-SC or mPEG-SPA moieties as judged from control analytical curves with free mPEG (not shown).
In-Gel Trypsin Digestion of mPEG-Amylin Products
A total of 10 μg of conjugated amylin from each reaction was separated on a nonreducing 22.5% SDS-PAGE gel. The bands were removed from the gel and subjected to in-gel trypsin proteolysis. The gel pieces were destained with 25 mM ammonium bicarbonate in 50% acetonitrile for 20 h at room temperature. The protein disulfide bonds were reduced with 10 mM dithiothreitol in 25 mM ammonium bicarbonate for 1 h at 56°C and then treated with 55 mM iodoacetamide in 25 mM ammonium bicarbonate for 45 min at room temperature in the dark. The solution was removed, and the gel slices were washed with 25 mM ammonium bicarbonate in 50% acetonitrile for 10 min and then dehydrated with 100% acetonitrile for 5 min. The acetonitrile was removed, and the gel slices were rehydrated in 25 mM ammonium bicarbonate with 100 ng of trypsin (Promega, sequencing grade). The digestion proceeded for 18 h at 37°C, and tryptic peptides were then extracted from the gel pieces twice by sonication for 15 min in a solution containing 5% trifluoroacetic acid and 50% acetonitrile. The solution was pooled together with the first extraction and then concentrated with the SpeedVac System for further matrix-assisted laser desorption and ionization–time-of-flight mass spectrometry (MALDI-ToF-MS) analysis.
Mass Spectrometry
MALDI-ToF-MS was performed as follows. The samples were mixed with equal volumes of 50% acetonitrile in water containing 0.1% TFA and 10 mg/mL sinapinic acid. The samples were then subjected to MALDI-ToF-MS in the Aulex Speed spectrometer (Bruker, USA). The MS spectra were obtained in positive linear mode and further analyzed using mMass (26). The number average molecular weight (Mn) and the weight average molecular weights (Mw) of both free mPEG and conjugated amylin were calculated from the polymer unit distribution from their respective MALDI-ToF-MS as follows:
where Ni is the intensity of the peak, and m/z is equal to Mi. The polydispersity (Pd) index was calculated as follows:
Human Receptor Activity-Modifying Protein 2 (Extracellular Topological Domain)
The DNA sequence encoding the extracellular topological domain (ETD) of the human receptor activity-modifying protein 2 (RAMP2; residues 43–144, UNIPROT ID no. O60895) was synthesized in vitro and cloned into bacterial protein expression vector pET28b (Novagen/EMD Millipore) by GenScript Corporation (Piscataway, NJ, USA) using NdeI and EcoRI. The sequence and identity of the gene in the pET28b-RAMP2 construct was confirmed with DNA sequencing by GenScript using the T7 promoter/terminator primers.
The RAMP2 was produced as a fusion protein containing an N-terminal histidine tag and a thrombin protease cleavage site. The protein was expressed in Escherichia coli BL21DE3pLys (1 mM IPTG, 37°C, 3 h) and purified as described elsewhere (27). In brief, RAMP2-ETD was produced in inclusion bodies. The inclusion bodies were denatured with 7 M guanidinium chloride and then purified using the IMAC Sepharose FastFlow (GE Healthcare, Brazil) and refolded by rapid dilution into 50 mM Tris–HCl buffer (pH 8.3) containing 1 M arginine hydrochloride, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, and 1 mM EDTA and left in place for 24 h at 4°C. After refolding, RAMP2-ETD was dialyzed, concentrated, and stored in 20 mM sodium phosphate buffer and 300 mM NaCl, pH 7.0, in liquid nitrogen until use.
RAMP2/Amylin Isothermal Binding Assay
RAMP2 (20 μM) was labeled with FITC (Sigma-Aldrich; 0.2 mM) in 20 mM sodium phosphate and 300 mM NaCl, pH 7.0, for 1 h at room temperature, and purified from the free FITC using a desalting column (Sephadex G25; GE Healthcare, Brazil) equilibrated with the labeling buffer. Labeling was confirmed by evaluating both contributions from protein and FITC in the absorbance spectra. The labeling efficiency was estimated at 0.1 mol FITC/1 mol RAMP2 as described elsewhere (28).
Amylin and PEGylated amylin binding to RAMP2 was assayed by monitoring the changes in the fluorescence quantum yield of fluorescein conjugated to RAMP2 as a function of amylin. RAMP2 (50 nM) was incubated with varying concentrations of free amylin, PEGylated amylin, or free mPEG-SC (which was previously inactivated by incubating mPEG-SC with 10× molar excess l-Lys for 2 h at 37°C) for 15 min at 25°C, and the fluorescence was measured by setting the excitation at 485 nm and emission at 520 nm in the Spectramax M5 (molecular devices). All assays were performed in 20 mM sodium phosphate with 300 mM NaCl, pH 7.0. The results represent the mean and standard deviations of five measurements. Binding data analysis was performed with the SigmaPlot 11 (Jandel Sci).
Pharmacological Evaluation of PEGylated Amylin
The pharmacological evaluation of amylin products was performed as described elsewhere (12,29). In brief, 5-week-old Swiss male mice (25 ± 1 g) were divided into three groups as follows: control (n = 5), amylin (n = 5), and PEGylated amylin (n = 5). The animals were housed in a temperature-controlled room with a light–dark cycle of 12 h. Water and food were available ad libitum and suspended 4 h before the experiments. All mice were fasted throughout the experiments and kept at 22 ± 2°C. The groups received 100 μL of the formulations (50 nmol/kg), equivalent to 200 μg free amylin/kg by subcutaneous injection using a standard 29-gauge needle (BD™). This dose was chosen on the basis of amylin dose dependence during fasting mice glycemia as described elsewhere (30). The glucose concentrations were monitored following the recommendations of the Mouse Metabolic Phenotyping Center Consortium from the National Institutes of Health (31). Whole blood samples were obtained from the tail tips of conscious, unrestrained mice. The measurements were taken both before injection and at repeated intervals after administration using pre-calibrated point-of-care glucometers (Accu-Chek Active, Roche Diagnostics, Germany; serial no. GN08146937), as described elsewhere (12). This research was in compliance with the Ethics in Animal and Clinical Investigations of the AAPS Journals Ethics Policy. This protocol was approved by the Institutional Bioethics Committee on Animal Care and Experimentation at UFRJ (no. FARMACIA05/2012).
Statistical Analysis
Statistical analysis was performed with a t test using SigmaStat, as implemented in SigmaPlot 11 (Systat Software, Inc.). The significance of the differences in the pharmacological evaluation was calculated, and values of p < 0.05 were considered to be significant.
RESULTS
mPEG Conjugation with Amylin
Amylin has two amine groups, both of which are located in the first amino acid residue, namely, l-lysine (Lys1); one amine group is located at the amino terminus, and the other is adjacent to the ε-carbon in the side chain (Fig. 1a). To investigate amylin modification by mPEG, we made use of two mPEG derivatives with an average molecular weight of 5 kDa: mPEG-SC (Fig. 1b) and mPEG-SPA (Fig. 1c). Both derivatives have been shown to display preferential reactivity with primary amine groups, although succinimidyl derivatives have also been shown to conjugate to hydroxyl groups such as those found in serine, tyrosine, and threonine (32–35).
We first attempted to conjugate amylin with mPEG-SC by using a molar ratio of 2 mPEG-SC to 1 amylin in DMSO. The conjugation reactions were resolved on 22.5% SDS-PAGE gels (18), revealing two major products in addition to free amylin (Fig. 2b). These two conjugate products were formed within 10 min, with no further apparent increment after up to 2 h of reaction. The amylin did not react completely, as observed by the remaining band of free amylin in the SDS-PAGE (Fig. 2b). A densitometric analysis of the SDS-PAGE suggests that approximately one third of the amylin remained after 2 h of reaction, approximately one third of the amylin was converted to the monoPEGylated, and another one third was converted to the diPEGylated amylin products (Fig. 2c). These data demonstrate the highly reactive nature of the amine groups against a succinimidyl mPEG agent as indicated by a fast reaction, although the incomplete reaction leading to the remaining free amylin indicates the need for higher concentration of mPEG in the reaction milieu to obtain better efficacy in amylin conjugation.
Fig. 2.

Amylin conjugation with mPEG-SC 5 kDa. Amylin PEGylation was conducted in DMSO, as described in the “MATERIAL AND METHODS” section, in the presence of a 2:1 M ratio of mPEG-SC-amylin. The reaction was run on a 22.5% SDS-PAGE gel after interruption at varying reaction time as indicated in the top of the gel (0, 1, 10, 30, 60, and 120 min). a The gels were stained with Coomassie Brilliant Blue. The ladder (L) is composed of BSA (66 kDa), lysozyme (14 kDa), and insulin B-chain (3.4 kDa). b Densitometry analysis of the digitalized gel. c The kinetics of the reaction, as followed by the integrated band area. The peaks were integrated, and the relative area was plotted as a function of time. The details are described in the “MATERIAL AND METHODS” section
We then conducted amylin conjugation with both mPEG-SC and mPEG-SPA by using a 10:1 M ratio and by performing the PEGylation reaction in two distinct liquid phases as follows: aqueous buffer (phosphate-buffered saline, pH 7.4) and organic solvent (DMSO). We followed the reaction between mPEG and amylin by evaluating the remaining amine groups (Fig. 1d) through derivatization with fluorescamine. We observed a steep decrease in fluorescamine fluorescence intensity as a function of time within the first 2 to 4 h of reaction in PBS, pH 7.4 (Fig. 3a), and in DMSO (Fig. 3b). This decrease was followed by a slow reaction phase for the remaining groups. Both mPEG-SC and mPEG-SPA showed similar reaction rates in both PBS (Fig. 3a) and DMSO (Fig. 3b), with mPEG-SC reacting faster than mPEG-SPA. To date, we have no explanation for this behavior.
Fig. 3.
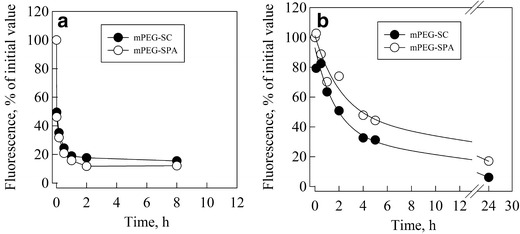
Amylin PEGylation reaction kinetics. Amylin PEGylation was performed at room temperature in the presence of a 10:1 M ratio of mPEG/amylin. The reaction was followed by monitoring the decay of fluorescamine fluorescence through the derivatization of the remaining amine groups, the concentration of which is proportional to the amount of remaining reactive amine groups in amylin (12). Data were normalized to the percentage of initial fluorescence, after proper blank correction, as follows: % fluorescence = 100 × F obs/F initial. Each line represents the best fit for the data with a monoexponential decay combined with a linear function. a The reaction in PBS, pH 7.4. b The reaction in DMSO, mPEG-SC (filled circle) and mPEG-SPA (empty circle). The details are described in the “MATERIAL AND METHODS” section
The conjugation reactions with both mPEG agents were resolved on 22.5% SDS-PAGE gels (18), revealing the appearance of at least four distinct bands for PEGylated amylin products, both in aqueous medium (Fig. 4a, b) and for the reaction taking place in DMSO (Fig. 5a, b). Within 10 min of the reaction, two major products were observed. Within 1 h of the reaction, most of the amylin had been conjugated. Further incubation for longer periods of time (up to 24 h) resulted in the appearance of products with higher molecular weights, both for reactions taking place in PBS (Fig. 4a, b) and for those in DMSO (Fig. 5a, b).
Fig. 4.

Analysis of amylin PEGylation reaction in aqueous solution. Amylin PEGylation was conducted in PBS, pH 7.4, as described in the “MATERIAL AND METHODS” section, in the presence of a 10:1 M ratio of mPEG/amylin, with mPEG-SC (a, c, e) or mPEG-SPA (b, d, f). The reaction was interrupted at the times indicated at the top of the gels and run on a 22.5% SDS-PAGE gel. a, b The gels were stained with Coomassie Brilliant Blue. The ladder (L) is composed of BSA (66 kDa), lysozyme (14 kDa), and insulin B-chain (3.4 kDa). c, d Densitometry analysis of the digitalized gel, as conducted with ImageJ. e, f The kinetics of the reaction, as followed by the integrated band area. The peaks were integrated with Fityk, and the relative area was plotted as a function of time. The details are described in the “MATERIAL AND METHODS” section
Fig. 5.

Analysis of the amylin PEGylation reaction in organic solvent. Amylin PEGylation was conducted in DMSO, as described in the “MATERIAL AND METHODS” section, in the presence of a 10:1 M ratio of mPEG/amylin, with mPEG-SC (a, c, e) or mPEG-SPA (b, d, f). The reaction was interrupted at the times indicated at the top of the gels and run on a 22.5% SDS-PAGE gel. a, b The gels were stained with Coomassie Brilliant Blue. The ladder (L) is composed of BSA (66 kDa), lysozyme (14 kDa), and insulin B-chain (3.4 kDa). c, d Densitometry analysis of the digitalized gel, as conducted with ImageJ. e, f The kinetics of the reaction, as followed by the integrated band area. The peaks were integrated with Fityk, and the relative area was plotted as a function of time. The details are described in the “MATERIAL AND METHODS” section
A detailed evaluation of the PEGylation reaction was obtained from densitometry analysis of the SDS-PAGE results, which revealed a band corresponding to the free amylin, as well as four additional bands. This banding pattern was observed for the conjugation with mPEG-SC in both aqueous (Fig. 4c) and organic media (Fig. 5c) and also for the conjugation with mPEG-SPA in aqueous solution (Fig. 4d) and in DMSO (Fig. 5d). An evaluation of the reaction kinetics through densitometric analysis of the SDS-PAGE results revealed that most of the changes occurred within 2 h of the reaction time in both solution conditions (aqueous and organic) and with both mPEG agents (Fig. 4e, f), with a slow secondary reaction taking place to a small extent during longer incubations. A faint band for the fifth reaction product became evident after 24 h of reaction in both solvents and with both mPEG agents. These data suggest that mPEG succinimidyl agents might be reacting with the two amine groups, as indicated by the fast decay in mPEG availability (Fig. 3) and the slower kinetic rate at which the mPEG agents would be reacting with other nucleophilic groups, such as the hydroxyl moieties of the Ser, Thr, and Tyr residues present in the amylin peptide.
Purification and Characterization of PEGylated Amylin Products
We conducted an amylin PEGylation reaction with mPEG-SC in DMSO for 10 min and subjected it to purification by using the C18 RP-HPLC method (Fig. 6a). Samples were collected and subjected to both MALDI-ToF-MS (Fig. 6b, c) and SDS-PAGE analysis (Fig. 6d–g). We observed a product with a molecular weight of approximately 8∼9 kDa (Fig. 6b, c) in the first fractions (elution time about 9.6 min, Fig. 6a), indicating a monoPEGylated amylin. The subsequent fractions (up to 11 min of elution time) also indicated a major contribution by the monoPEGylated amylin, although they had increasing average molecular weights (Fig. 6b, c), indicating that the purification method was able to separate the mPEG isomers of increasing average molecular weights. Fractions collected between elution times of about 11 and 12.5 min were also identified by MALDI-ToF-MS analysis as a diPEGylated amylin (Fig. 6b, c). Additional samples collected at increasing elution times revealed an amylin conjugated with three or more mPEG moieties (Fig. 6b, c).
Fig. 6.

Purification and characterization of PEGylated amylin products. Amylin (5 mg/mL) was reacted in DMSO with ten times mPEG-SC molar excess for 10 min at 25°C, and the quenched reaction (with excess l-Lys) was purified by C18 RP-HPLC with a water–acetonitrile linear gradient. Collected fractions were subjected to MALDI-ToF-MS and SDS-PAGE analysis. a Chromatogram of the C18 RP-HPLC purification; b, c MALDI-ToF-MS analysis of each 1 mL samples collected from the C18 RP-HPLC purification (Fig. 6a). Each intensity vs m/z mass spectrum in b is identified by the colors as shown in the top part of panel (C), which is the plot of the integrated intensity at each m/z (a gel-like representation). The number of mPEG moieties conjugated to amylin is indicated at the left side of panel (c). D–G SDS-PAGE analysis of each 1-mL sample collected from the C18 RP-HPLC purification (Fig. 6a). The elution times are indicated by the numbers below panels d and e and above panels f and g, and L and mA are the ladder and free amylin, respectively. d, f Images of the SDS-PAGE fractions collected up to 11 min; e, g images of the SDS-PAGE fractions collected after 11 min. Both SDS-PAGE gels were run in parallel in the same electrophoresis system, and the top and bottom parts of the gel in these images are made visible for the sake of clarity. d, e Images from the gel stained with Coomassie Brilliant Blue for polypeptide moiety identification; f, g images from the same gel that were subsequently stained with barium/iodine for PEG moiety identification
The purified mPEG-amylin products were also subjected to SDS-PAGE analysis and stained with both Coomassie Brilliant Blue (for the identification of the polypeptide moiety, Fig. 6d, e) and iodine (for the identification of the PEG moiety, Fig. 6f, g) showing that both products had equivalent distribution patterns in the MALDI-ToF-MS analysis and in terms of electrophoretic mobility, thereby intervalidating the SDS-PAGE as a tool for monitoring PEGylation reaction progress.
Identification of PEGylation Sites
An mPEG-SC amylin conjugation reaction was subjected to SDS-PAGE, and the separated band products were excised from the gel for further in-gel trypsinization. The resulting peptide products were analyzed by MALDI-ToF-MS. The PEGylated amylin resulted in trypsin digestion products with a typical spectrum of a PEGylated product, with a distribution of m/z peaks differing in 44 Da, the molecular weight of an ethylene glycol monomer unit (Fig. 7).
Fig. 7.

Identification of the PEGylation sites in amylin. a MALDI-ToF-MS analysis of tryptic digests from PEGylated amylin. mPEG-SC was allowed to react with amylin (10:1) for 4 h, and the reaction was resolved by SDS-PAGE. Bands were removed from the gel and digested with trypsin for further MALDI-ToF-MS analysis of the resulting peptides. Inset, SDS-PAGE analysis of the reaction products and an indication of the excised bands (labeled #1 and #2) used for tryptic digestion. b MALDI-ToF-MS analysis of free mPEG-SC-NHS. c MALDI-ToF-MS analysis of free mPEG-SPA-NHS. The calculation of the average molecular number (Mn) and the average molecular weight from the MALDI-ToF-MS analysis is shown in Table II. The details are described in the “MATERIAL AND METHODS” section
Amylin has only two trypsin digestion sites, namely, Arg11 and Arg18 (Table I). Cleavage at these sites would result in peptide products with masses of 832 Da (amylin12–18), 1,196 Da (amylin1–11), 1,931 Da (amylin19–37), 2,010 Da (amylin1–18), and 2,745 Da (amylin12–37). As a consequence of the inherent Pd of mPEG derivatives, it is not possible to obtain a single, discrete molecular weight for the PEGylated peptide. From the MS spectra of the PEGylated tryptic digestion products (Fig. 7, inset), we have calculated the m/z for these distributions, with both the Mn and the Mw (Table II) yielding a Pd of 1.002 for both digestion products. The difference between the Mn of the digest products was 11,613 − 6,435 = 5,178, corresponding to the average Mn of the conjugated mPEG. The difference between the Mw of the digest products was 11,643 − 6,448 = 5,195, which is the average Mw of the mPEG conjugated to the peptide.
Table I.
Trypsin Digestion Fragments of Amylin
| Sequence | Molecular weight, Da (sequence numbering) |
|---|---|
| KCNTATCATQRLANFLVRSSNNLGPVLPPTNVGSNTY | 832 (12–18) |
| KCNTATCATQRLANFLVRSSNNLGPVLPPTNVGSNTY | 1,196 (1–11) |
| KCNTATCATQRLANFLVRSSNNLGPVLPPTNVGSNTY | 1,931 (19–37) |
| KCNTATCATQRLANFLVRSSNNLGPVLPPTNVGSNTY | 2,010 (1–18) |
| KCNTATCATQRLANFLVRSSNNLGPVLPPTNVGSNTY | 2,745 (12–37) |
Table II.
Data Analysis of Tryptic Digests from MonoPEGylated and DiPEGylated Amylin
| Tryptic digest | Mna | Mwb | Pdc |
|---|---|---|---|
| Free mPEG-SC-NHS | 5,435 | 5,481 | 1.008 |
| Free mPEG-SPA-NHS | 5,443 | 5,482 | 1.007 |
| monoPEGylated (m/z 5,776 to 7,291) | 6,435 | 6,448 | 1.002 |
| diPEGylated (m/z 10,549 to 12,759) | 11,613 | 11,643 | 1.002 |
| Estimated mass of the conjugated mPEG | 11,613 − 6,435 = 5,178 | 11,643 − 6,448 = 5,195 | |
| Estimated mass of the PEGylated peptide | 6,435 − 5,178 = 1,257 | 6,448 − 5,195 = 1,253 |
Data obtained from the mass spectra as shown in Fig. 7
Mn = Σ(Ni * Mi)/ΣNi
Mw = Σ(Ni * Mi2)/Σ(Ni * Mi)
Pd = Mw/Mn
N i = intensity at given m/z; M i = major m/z peaks
To better estimate the molecular weight of the mPEG agents used in this work, we subjected them to MALDI-ToF-MS analysis, resulting in Mn and Mw of 5,435 and 5,481, respectively, for mPEG-SC (Fig. 7b; Table II), and Mn and Mw of 5,443 and 5,482 for mPEG-SPA, respectively (Fig. 7c; Table II). Taking into account the Mw of the N-hydroxysuccinimide (NHS) (115 Da), the Mn and Mw become 5,320 and 5,366 for mPEG-SC, respectively, and the Mn and Mw are 5,328 and 5,367 for mPEG-SPA, respectively. These values are in close agreement with the estimated value for mPEG conjugated to the peptide of Mn of 5,178 and the Mw of 5,195.
By using 5,178 as the Mw of mPEG conjugated to amylin, we estimate the PEGylated amylin digestion product size as follows:
Alternatively, by using 5,440 − 115 = 5,325 as the Mw of the mPEG (without the NHS), we can also estimate the PEGylated amylin peptide size as released from trypsin digestion as follows:
Both 1,257 and 1,110 Da are thus the approximate average Mw of the amylin fragment where conjugation occurred, which corresponded to the amylin tryptic digestion product composed of amino acids 1∼11 (Mw = 1,196 Da; Table I), which includes the Lys1 and its two primary amines, with the groups expected to be conjugated by mPEG using succinimidyl as a reactive agent. These data suggest that the conjugation site for both monoPEGylated amylin and diPEGylated amylin lies within the amino acids region 1–11. The fast formation of these two PEGylated products (Figs. 4a, b and 5a, b) occurs within a time scale that is compatible with a reduction in the amount of amine-reactive groups (Fig. 1). Collectively, these data indicate that the major PEGylation site in amylin with succinimidyl mPEG derivatives is in the Lys1 residue. We believe that secondary PEGylation reactions taking place at a slower rate might involve hydroxyl groups coupling of the Ser, Thr, and Tyr side chains, but a determination of the exact positional isomers requires further investigation. However, while residue Lys1 has been shown to play no fundamental role in receptor interactions, the remaining portion of amino acids 5 to 37 is involved in self-assembly (36) and in interacting with other molecular partners (2,36,37). Consequently, Lys1 PEGylation is a preferable strategy for amylin conjugation and prevents a loss of function.
Pharmacological Evaluation of PEGylated Amylin Products
To conduct a pharmacological evaluation of the PEGylated amylin products, we first carried out a preparative reaction of both mPEG-SC and mPEG-SPA in DMSO followed by C18 reversed phase HPLC purification. We obtained well-separated peaks for PEGylation reactions conducted with both mPEG-SC (Fig. 8a) and mPEG-SPA (Fig. 8b), with both reactions exhibiting similar elution times for peaks I (∼10.5 min), II (∼11.7 min), and III (13 min) and a fraction (“tail”) collected between peaks I and II. The pooled peaks were analyzed by SDS-PAGE (Fig. 8a, b; inset), which showed high purity for pooled fractions I and II, while the small tail between peaks I and II appeared to be a combination of peaks I and II. To confirm the identity of the PEGylation products, we conducted MALDI-ToF-MS analyses of these peaks (Fig. 8c–f).
Fig. 8.

Purification of mono- and diPEGylated amylin with mPEG-SC and mPEG-SPA in organic solvent. Amylin (5 mg/mL) was reacted in DMSO with ten times mPEG-SC (a, c, e) or mPEG-SPA (b, d, f) molar excess for 4 h at 25°C. The quenched reactions were purified by C18 RP-HPLC, which was followed by MALDI-ToF-MS analysis. a, b Chromatograms from the C18 RP-HPLC purifications. The peaks were collected and freeze-dried for further use. Free indicates the retention time region for unmodified amylin (the chromatogram of free amylin alone is not shown), peak I indicates the monoPEGylated amylin, peak II indicates the diPEGylated amylin, and peak III indicates the pooled remaining fractions containing other PEGylation products. Tail is the fraction collected between peaks “I” and “II,” and R is the quenched reaction before RP-HPLC purification. Inset, SDS-PAGE characterization of the RP-HPLC fractions. c, d MALDI-ToF-MS analysis of monoPEGylated amylin (peak I) e, f MALDI-ToF-MS analysis of diPEGylated amylin (peak II). The details are described in the “MATERIAL AND METHODS” section
Amylin exerts its function by interacting with the RAMP, a protein family composed of three single-transmembrane proteins involved in the intermediation of G protein-coupled receptors (2,38–42). A RAMP2 extracellular topological domain interaction with amylin and PEGylated amylin was performed by an isothermal binding assay with fluorescein-labeled RAMP2 (RAMP2-FITC). RAMP2-FITC (50 nM) was incubated with varying concentrations of murine amylin and with the purified mono and diPEGylated amylin. We first conducted the RAMP2-amylin binding assay by performing fluorescein anisotropy on fluorescein-labeled RAMP2. However, a large change in the fluorescein quantum yield as a function of ligand binding took place, limiting the use of fluorescence anisotropy data as a result of the need for extensive mathematical correction to changes in quantum yield and fluorescence lifetimes (43). Instead, fluorescein change is a direct consequence of the interaction, and thus the isothermal binding assay results could be directly inferred from the dependence of fluorescence changes on the ligand concentrations (43).
We then followed the fluorescence increase of the fluorescein moiety as a function of the ligand concentration (Fig. 9). Changes occurred in the high nanomolar concentration range, reaching a saturation at approximately 3 μM for free amylin and 20 μM for mono- and diPEGylated amylin. To rule out a direct PEG moiety effect on RAMP2, we performed a control assay using mPEG-SC conjugated to l-Lys, resulting in no significant interactions in the same concentration range. Data analysis allowed us to estimate a C50%, i.e., a concentration resulting in a 50% transition, of 0.27 ± 0.07 μM for free amylin and 11.21 ± 1.8 and 12.49 ± 4.9 μM for mono- and diPEGylated amylin, respectively. Collectively, these data demonstrate that both monoPEGylated amylin and diPEGylated amylin are effective in performing direct interaction with the amylin co-receptor RAMP2.
Fig. 9.

PEGylated amylin interaction with RAMP2. Free amylin and mono- and diPEGylated amylin were assayed for the ability to bind to RAMP2 using fluorescein-labeled RAMP2 (RAMP2-FITC). Empty circle, free amylin; inverted triangle, monoPEGylated amylin; empty square, diPEGylated amylin; diamond, free mPEG-SC-l-Lys. Binding was performed in 20 mM sodium phosphate and 300 mM NaCl, pH 7.0, at 25°C. Fluorescence was measured with emission at 520 nm and excitation at 485 nm. Symbols represent the mean and standard deviation of five measurements. Lines are the best adjustments to sigmoid four parameters logistic function. Details are available in the “MATERIAL AND METHODS” section
The PEGylated amylin products were then evaluated for their in vivo biological activity by monitoring the product’s potential to reduce glycemia when administered subcutaneously to mice (Fig. 10). Unmodified amylin (50 nmol/kg) treatment resulted in a marked glycemia reduction, reaching its maximum effect at 2 h after s.c. administration and with complete recovery to control glycemia values at 6 to 8 h after s.c. administration (Fig. 10a). These results are in agreement with previously published work (12). Both the monoPEGylated and diPEGylated amylin products (50 nmol/kg) that were obtained with the mPEG-SC reagent resulted in a sustained glycemia reduction for 8 to 12 h following s.c. administration (Fig. 10b). The PEGylated amylin products that were obtained from mPEG-SPA showed an enhanced effect on glycemia reduction for up to 8 h, while the diPEGylated amylin isomer that was obtained with mPEG-SPA showed no significant difference from the unmodified amylin test group (Fig. 10c).
Fig. 10.

In vivo pharmacological evaluation of mono- and diPEGylated amylin with mPEG-SC and mPEG-SPA. These experiments were performed with a dose of 50 nmol amylin product/kg body weight, with conscious, unrestrained fasting mice. The points and bars represent the means ± standard deviations, respectively (n = 5). *p < 0.05 (unpaired t test). a Free amylin, b amylin conjugated with mPEG-SC, c amylin conjugated with mPEG-SPA. Control (saline/filled circle), free (non-modified) amylin (empty circle), monoPEGylated amylin (filled square), diPEGylated amylin (empty square). The details are described in the “MATERIAL AND METHODS” section
DISCUSSION
Nearly 90 years have passed since the discovery of insulin and the subsequent development of slow/basal insulin products targeting basal levels. Since then, no further significant advances have been observed in the improvement of the available protein therapeutics for the treatment of diabetes (44), and so this goal has yet to be actualized. Islet transplantation (45) and beta-cell regeneration (46) are considered to be optimum target approaches, but they are still under development.
Amylin displays remarkable effects on diabetes physiology, including significant improvements in the tight control of glycemia (2,47,48). It would be highly desirable to perform amylin restoration in conjunction with insulin treatment, preferably with a highly humanized therapy that minimizes injections. Amylin interacts with insulin (36,49,50), and combining insulin and the amylinomimetic compound pramlintide in the same vial is not recommended (51). As a consequence, two injections—including insulin—are needed at mealtime for patients engaged in amylinomimetic compounds restoration. Even in this scenario, a basal amylin level cannot be achieved using the currently available therapeutic products.
In the field of novel biotherapeutic protein design, increasing the in vivo biological activity of compounds has typically been achieved using bioconjugation strategies with large, biocompatible molecular partners, such as fatty acids, albumin, and polyethylene glycol, which directly or indirectly inhibit renal excretion (52–58). With this strategy in mind, we have designed and obtained PEGylated amylin conjugates with high reaction efficiency through the reaction of mPEG-SC and mPEG-SPA to the two amine groups of the Lys1 residue, which has been confirmed by trypsin digestion followed by MALDI-ToF-MS analysis. The major positional isomer of the Lys1-monoPEGylated amylin has yet to be characterized (in terms of whether it was coupled to its α- or ε-amino group), and this feat would most likely be accomplished using NMR spectroscopy of 15N-isotopically labeled amylin (59). The reaction occurred within a few minutes and can be purified by conventional reversed phase chromatography C18 RP-HPLC, resulting in PEGylated amylin products with biological activity. The mPEG-SC mono- and diPEGylated amylin products were effective in depressing glycemia in vivo, an effect lasting approximately 6 h longer than the effect of regular, unmodified amylin. These data indicate a PEGylation enhancement of the amylin circulation half-life, resulting in extended regulation over glucose homeostasis. Further improvements could be achieved using PEG molecules with longer or branched chains. The PEGylation of amylin with derivatives of higher molecular weight (13,14) could also be a strategy for enhancing the persistence of amylin biological activity, providing a more extended effect on glycemia beyond the one observed here with mPEG 5 k. A more in-depth investigation of the pharmacokinetics will be necessary to fully understand this process,
PEGylation reactions with a lower molar ratio were not satisfactory because about half of the amylin did not react. Using a molar ratio of 10 mol PEG/1 mol amylin led to complete amylin conversion within a few minutes, both in aqueous and organic solutions with the conjugated products. We believe that an organic solvent would be a more appropriate milieu for amylin conjugation (60) because of solubility issues in aqueous solution (61), in particular for human amylin (9) and amylin from other organisms, along with amylinomimetic compounds (11,62,63).
CONCLUSIONS
We envision that our current approach could be used for the development of PEGylated human amylin or PEGylated amylinomimetic compounds such as pramlintide. Human amylin displays limited, nanomolar water solubility (9) and quickly aggregates into an inactive form (64). There is an increasing evidence that human amylin aggregation can be hampered by molecular confinement and conjugation (12,65–72). The PEGylation of human amylin could serve as a strategy to improve both its half-life and its water solubility and to avoid protein interactions and aggregation, as previously suggested by the inhibition of amylin by alkylation (73). In fact, previous evidence suggests that peptide PEGylation from the highly amyloidogenic region of human amylin blocks fibrillogenesis (74). We believe that our present method might inspire the development of further amylin bioconjugates.
Acknowledgments
We would like to thank Mr. Eduardo R. dos Santos (CEMBIO-IBCCF-UFRJ) for his technical assistance with the MALDI-ToF-MS measurements. This research was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Nanobiotec-CAPES 04/08, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), IMBEBB, Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro Carlos Chagas Filho (FAPERJ), and PRONEX. The funding agencies had no role in the study design, data collection and analysis, or decision to publish or prepare the manuscript.
Abbreviations
- mPEG
methoxy polyethylene glycol
- mPEG-SC
methoxy polyethylene glycol succinimidyl carbonate
- mPEG-SPA
methoxy polyethylene glycol succinimidyl propionate
- MALDI-ToF-MS
matrix-assisted laser desorption and ionization–time-of-flight mass spectrometry
- RAMP
receptor activity-modifying protein
Footnotes
Luiz Henrique Guerreiro and Mariana F. A. N. Guterres contributed equally to this work.
References
- 1.Hartter E, Svoboda T, Ludvik B, Schuller M, Lell B, Kuenburg E, et al. Basal and stimulated plasma levels of pancreatic amylin indicate its co-secretion with insulin in humans. Diabetologia. 1991;34(1):52–54. doi: 10.1007/BF00404025. [DOI] [PubMed] [Google Scholar]
- 2.Young A. Amylin: physiology and pharmacology. 1. San Diego: Elsevier Academic; 2005. [Google Scholar]
- 3.Cooper GJS. Amylin and related proteins: physiology and pathophysiology. In: Jefferson LS, Cherring AD, editors. Handbook of physiology. New York: Oxford University Press; 2001. [Google Scholar]
- 4.Cooper GJ. Amylin and insulin co-replacement therapy for insulin-dependent (type I) diabetes mellitus. Med Hypotheses. 1991;36(3):284–288. doi: 10.1016/0306-9877(91)90150-W. [DOI] [PubMed] [Google Scholar]
- 5.Cooper GJ, Willis AC, Clark A, Turner RC, Sim RB, Reid KB. Purification and characterization of a peptide from amyloid-rich pancreases of type 2 diabetic patients. Proc Natl Acad Sci USA. 1987;84(23):8628–8632. doi: 10.1073/pnas.84.23.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper GJ, Willis AC, Reid KB, Clark A, Baker CA, Turner RC, et al. Diabetes-associated peptide. Lancet. 1987;2(8565):966. doi: 10.1016/S0140-6736(87)91444-9. [DOI] [PubMed] [Google Scholar]
- 7.Westermark P, Wernstedt C, O’Brien TD, Hayden DW, Johnson KH. Islet amyloid in type 2 human diabetes mellitus and adult diabetic cats contains a novel putative polypeptide hormone. Am J Pathol. 1987;127(3):414–417. [PMC free article] [PubMed] [Google Scholar]
- 8.Westermark P, Wernstedt C, Wilander E, Hayden DW, O’Brien TD, Johnson KH. Amyloid fibrils in human insulinoma and islets of Langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. Proc Natl Acad Sci USA. 1987;84(11):3881–3885. doi: 10.1073/pnas.84.11.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper GJ, Leighton B, Dimitriadis GD, Parry-Billings M, Kowalchuk JM, Howland K, et al. Amylin found in amyloid deposits in human type 2 diabetes mellitus may be a hormone that regulates glycogen metabolism in skeletal muscle. Proc Natl Acad Sci USA. 1988;85(20):7763–7766. doi: 10.1073/pnas.85.20.7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young AA, Vine W, Gedulin BR, Pittner R, Janes S, Gaeta LS, et al. Preclinical pharmacology of pramlintide in the rat: comparisons with human and rat amylin. Drug Development Research. 1996;37:231–248. doi: 10.1002/(SICI)1098-2299(199604)37:4<231::AID-DDR5>3.0.CO;2-M. [DOI] [Google Scholar]
- 11.Westermark P, Engstrom U, Johnson KH, Westermark GT, Betsholtz C. Islet amyloid polypeptide: pinpointing amino acid residues linked to amyloid fibril formation. Proc Natl Acad Sci USA. 1990;87(13):5036–5040. doi: 10.1073/pnas.87.13.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerreiro LH, Da SD, Ricci-Junior E, Girard-Dias W, Mascarenhas CM, Sola-Penna M, et al. Polymeric particles for the controlled release of human amylin. Colloids Surf B Biointerfaces. 2012;94:101–106. doi: 10.1016/j.colsurfb.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Abuchowski A, Van ET, Palczuk NC, Davis FF. Alteration of immunological properties of bovine serum albumin by covalent attachment of polyethylene glycol. J Biol Chem. 1977;252(11):3578–3581. [PubMed] [Google Scholar]
- 14.Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov. 2003;2(3):214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 15.Larson N, Ghandehari H. Polymeric conjugates for drug delivery. Chem Mater. 2012;24(5):840–853. doi: 10.1021/cm2031569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Youn YS, Na DH, Lee KC. High-yield production of biologically active mono-PEGylated salmon calcitonin by site-specific PEGylation. J Control Release. 2007;117(3):371–379. doi: 10.1016/j.jconrel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 17.Veronese FM, Pasut G. PEGylation, successful approach to drug delivery. Drug Discov Today. 2005;10(21):1451–1458. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- 18.Fee CJ. Protein conjugates purification and characterization. In: Veronese FM, editor. PEGylated protein drugs: basic science and clinical applications. 1. Basel: Birkhäuser; 2009. pp. 113–125. [Google Scholar]
- 19.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Freitas DS, Abrahao NJ. Biochemical and biophysical characterization of lysozyme modified by PEGylation. International Journal of Pharmaceutics. 2010;392:111–117. doi: 10.1016/j.ijpharm.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 21.Scaramuzza S, Tonon G, Olianas A, Messana I, Schrepfer R, Orsini G, et al. A new site-specific monoPEGylated filgrastim derivative prepared by enzymatic conjugation: production and physicochemical characterization. J Control Release. 2012;164(3):355–363. doi: 10.1016/j.jconrel.2012.06.026. [DOI] [PubMed] [Google Scholar]
- 22.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wojdyr M. Fityk: a general-purpose peak fitting program. Journal of Applied Crystallography. 2010;43:1126–1128. doi: 10.1107/S0021889810030499. [DOI] [Google Scholar]
- 24.Udenfriend S, Stein S, Bohlen P, Dairman W, Leimgruber W, Weigele M. Fluorescamine: a reagent for assay of amino acids, peptides, proteins, and primary amines in the picomole range. Science. 1972;178(63):871–872. doi: 10.1126/science.178.4063.871. [DOI] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analyt Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Strohalm M, Hassman M, Kosata B, Kodicek M. mMass data miner: an open source alternative for mass spectrometric data analysis. Rapid Communications in Mass Spectrometry. 2008;22(6):905–908. doi: 10.1002/rcm.3444. [DOI] [PubMed] [Google Scholar]
- 27.Kusano S, Kukimoto-Niino M, Hino N, Ohsawa N, Okuda K, Sakamoto K, et al. Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding. Protein Sci. 2012;21(2):199–210. doi: 10.1002/pro.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matozo HC, Santos MAM, Neto MD, Bleicher L, Lima LMTR, Iuliano R, et al. Low-resolution structure and fluorescence anisotropy analysis of protein tyrosine phosphatase eta catalytic domain. Biophys J. 2007;92(12):4424–4432. doi: 10.1529/biophysj.106.094961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panagiotidis G, Salehi AA, Westermark P, Lundquist I. Homologous islet amyloid polypeptide: effects on plasma levels of glucagon, insulin and glucose in the mouse. Diabetes Res Clin Pract. 1992;18(3):167–171. doi: 10.1016/0168-8227(92)90142-E. [DOI] [PubMed] [Google Scholar]
- 30.Guerreiro L, Da Silva D, Mizurini DM, Sola-Penna M, Lima LMTR. Amylin induces hypoglycemia in mice. An Acad Bras Cienc. 2013;85(1):143–148. doi: 10.1590/S0001-37652013005000011. [DOI] [PubMed] [Google Scholar]
- 31.Ayala JE, Samuel VT, Morton GJ, Obici S, Croniger CM, Shulman GI, et al. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis Model Mech. 2010;3(9–10):525–534. doi: 10.1242/dmm.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller BT, Rogers ME, Smith JS, Kurosky A. Identification and characterization of O-biotinylated hydroxy amino acid residues in peptides. Analyt Biochem. 1994;219(2):240–248. doi: 10.1006/abio.1994.1263. [DOI] [PubMed] [Google Scholar]
- 33.Miller BT, Kurosky A. Elevated intrinsic reactivity of seryl hydroxyl groups within the linear peptide triads His-Xaa-Ser or Ser-Xaa-His. Biochem Biophys Res Commun. 1993;196(1):461–467. doi: 10.1006/bbrc.1993.2272. [DOI] [PubMed] [Google Scholar]
- 34.Banks PR, Paquette DM. Comparison of three common amine reactive fluorescent probes used for conjugation to biomolecules by capillary zone electrophoresis. Bioconjug Chem. 1995;6(4):447–458. doi: 10.1021/bc00034a015. [DOI] [PubMed] [Google Scholar]
- 35.Roberts MJ, Bentley MD, Harris JM. Chemistry for peptide and protein PEGylation. Adv Drug Deliv Rev. 2002;54(4):459–476. doi: 10.1016/S0169-409X(02)00022-4. [DOI] [PubMed] [Google Scholar]
- 36.Gilead S, Wolfenson H, Gazit E. Molecular mapping of the recognition interface between the islet amyloid polypeptide and insulin. Angew Chem Int Ed Engl. 2006;45(39):6476–6480. doi: 10.1002/anie.200602034. [DOI] [PubMed] [Google Scholar]
- 37.Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006;443(7113):870–874. doi: 10.1038/nature05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leuthauser K, Gujer R, Aldecoa A, McKinney RA, Muff R, Fischer JA, et al. Receptor-activity-modifying protein 1 forms heterodimers with two G-protein-coupled receptors to define ligand recognition. Biochem J. 2000;351(Pt 2):347–351. doi: 10.1042/0264-6021:3510347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tilakaratne N, Christopoulos G, Zumpe ET, Foord SM, Sexton PM. Amylin receptor phenotypes derived from human calcitonin receptor/RAMP coexpression exhibit pharmacological differences dependent on receptor isoform and host cell environment. J Pharmacol Exp Ther. 2000;294(1):61–72. [PubMed] [Google Scholar]
- 40.Hay DL, Christopoulos G, Christopoulos A, Poyner DR, Sexton PM. Pharmacological discrimination of calcitonin receptor: receptor activity-modifying protein complexes. Mol Pharmacol. 2005;67(5):1655–1665. doi: 10.1124/mol.104.008615. [DOI] [PubMed] [Google Scholar]
- 41.Sexton PM, Morfis M, Tilakaratne N, Hay DL, Udawela M, Christopoulos G, et al. Complexing receptor pharmacology: modulation of family B G protein-coupled receptor function by RAMPs. Ann N Y Acad Sci. 2006;1070:90–104. doi: 10.1196/annals.1317.076. [DOI] [PubMed] [Google Scholar]
- 42.Christopoulos G, Perry KJ, Morfis M, Tilakaratne N, Gao Y, Fraser NJ, et al. Multiple amylin receptors arise from receptor activity-modifying protein interaction with the calcitonin receptor gene product. Mol Pharmacol. 1999;56(1):235–242. doi: 10.1124/mol.56.1.235. [DOI] [PubMed] [Google Scholar]
- 43.Lakowicz JR. Principles of fluorescence spectroscopy. 2. New York: Plenum; 1999. [Google Scholar]
- 44.Eisenstein M. More than insulin. Nat Biotechnol. 2011;29(9):782–785. doi: 10.1038/nbt.1967. [DOI] [PubMed] [Google Scholar]
- 45.Eliaschewitz FG, Aita CA, Genzini T, Noronha IL, Lojudice FH, Labriola L, et al. First Brazilian pancreatic islet transplantation in a patient with type 1 diabetes mellitus. Transplant Proc. 2004;36(4):1117–1118. doi: 10.1016/j.transproceed.2004.04.065. [DOI] [PubMed] [Google Scholar]
- 46.Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, et al. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464(7292):1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lutz TA. The role of amylin in the control of energy homeostasis. Am J Physiol Regul Integr Comp Physiol. 2010;298(6):R1475–R1484. doi: 10.1152/ajpregu.00703.2009. [DOI] [PubMed] [Google Scholar]
- 48.Young DA, Deems RO, Deacon RW, McIntosh RH, Foley JE. Effects of amylin on glucose metabolism and glycogenolysis in vivo and in vitro. Am J Physiol. 1990;259(3 Pt 1):E457–E461. doi: 10.1152/ajpendo.1990.259.3.E457. [DOI] [PubMed] [Google Scholar]
- 49.Wei L, Jiang P, Yau YH, Summer H, Shochat SG, Mu Y, et al. Residual structure in islet amyloid polypeptide mediates its interactions with soluble insulin. Biochemistry. 2009;48(11):2368–2376. doi: 10.1021/bi802097b. [DOI] [PubMed] [Google Scholar]
- 50.Jiang P, Wei L, Pervushin K, Mu Y. pH-Dependent interactions of human islet amyloid polypeptide segments with insulin studied by replica exchange molecular dynamics simulations. J Phys Chem B. 2010;114(31):10176–10183. doi: 10.1021/jp101811u. [DOI] [PubMed] [Google Scholar]
- 51.Pharmaceuticals A. NDA 21-332—Division of Pharmaceutical Evaluation-II. Office of Clinical Pharmacology and Biopharmaceutics. Symlin. Silver Spring: Food and Drug Administration; 2000. [Google Scholar]
- 52.Hashimoto M, Takada K, Kiso Y, Muranishi S. Synthesis of palmitoyl derivatives of insulin and their biological activities. Pharm Res. 1989;6(2):171–176. doi: 10.1023/A:1015992828666. [DOI] [PubMed] [Google Scholar]
- 53.Markussen J, inventor. Novo Industri A/S, assignee. Process for preparing esters of human insulin. Denmark patent US4343898; 1982.
- 54.Markussen J, Havelund S, Kurtzhals P, Andersen AS, Halstrom J, Hasselager E, et al. Soluble, fatty acid acylated insulins bind to albumin and show protracted action in pigs. Diabetologia. 1996;39(3):281–288. doi: 10.1007/BF00418343. [DOI] [PubMed] [Google Scholar]
- 55.Thibaudeau K, Leger R, Huang XC, Robitaille M, Quraishi O, Soucy C, et al. Synthesis and evaluation of insulin-human serum albumin conjugates. Bioconjugate Chemistry. 2005;16(4):1000–1008. doi: 10.1021/bc050102k. [DOI] [PubMed] [Google Scholar]
- 56.Shechter Y, Mironchik M, Rubinraut S, Tsubery H, Sasson K, Marcus Y, et al. Reversible pegylation of insulin facilitates its prolonged action in vivo. European Journal of Pharmaceutics and Biopharmaceutics. 2008;70(1):19–28. doi: 10.1016/j.ejpb.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 57.Drucker DJ, Dritselis A, Kirkpatrick P. Liraglutide. Nature Reviews Drug Discovery. 2010;9:267–268. doi: 10.1038/nrd3148. [DOI] [PubMed] [Google Scholar]
- 58.Hompesch M, Troupin B, Heise T, Elbroend B, Endahl L, Haahr H, et al. Time-action profile of insulin detemir and NPH insulin in patients with type 2 diabetes from different ethnic groups. Diabetes Obes Metab. 2006;8(5):568–573. doi: 10.1111/j.1463-1326.2006.00643.x. [DOI] [PubMed] [Google Scholar]
- 59.Wang YS, Youngster S, Bausch J, Zhang R, McNemar C, Wyss DF. Identification of the major positional isomer of pegylated interferon alpha-2b. Biochemistry. 2000;39(35):10634–10640. doi: 10.1021/bi000617t. [DOI] [PubMed] [Google Scholar]
- 60.Abedini A, Singh G, Raleigh DP. Recovery and purification of highly aggregation-prone disulfide-containing peptides: application to islet amyloid polypeptide. Analyt Biochem. 2006;351(2):181–186. doi: 10.1016/j.ab.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 61.Milton NG, Harris JR. Fibril formation and toxicity of the non-amyloidogenic rat amylin peptide. Micron. 2013;44:246–253. doi: 10.1016/j.micron.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 62.Nilsson MR, Raleigh DP. Analysis of amylin cleavage products provides new insights into the amyloidogenic region of human amylin. J Mol Biol. 1999;294(5):1375–1385. doi: 10.1006/jmbi.1999.3286. [DOI] [PubMed] [Google Scholar]
- 63.Nilsson MR, Nguyen LL, Raleigh DP. Synthesis and purification of amyloidogenic peptides. Analyt Biochem. 2001;288(1):76–82. doi: 10.1006/abio.2000.4887. [DOI] [PubMed] [Google Scholar]
- 64.Young A. Clinical studies. Adv Pharmacol. 2005;52:289–320. doi: 10.1016/S1054-3589(05)52018-0. [DOI] [PubMed] [Google Scholar]
- 65.Patil SM, Xu S, Sheftic SR, Alexandrescu AT. Dynamic alpha-helix structure of micelle-bound human amylin. J Biol Chem. 2009;284(18):11982–11991. doi: 10.1074/jbc.M809085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nanga RP, Brender JR, Xu J, Hartman K, Subramanian V, Ramamoorthy A. Three-dimensional structure and orientation of rat islet amyloid polypeptide protein in a membrane environment by solution NMR spectroscopy. J Am Chem Soc. 2009;131(23):8252–8261. doi: 10.1021/ja9010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh G, Brovchenko I, Oleinikova A, Winter R. Peptide aggregation in finite systems. Biophys J. 2008;95(7):3208–3221. doi: 10.1529/biophysj.108.136226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larson JL, Miranker AD. The mechanism of insulin action on islet amyloid polypeptide fiber formation. J Mol Biol. 2004;335(1):221–231. doi: 10.1016/j.jmb.2003.10.045. [DOI] [PubMed] [Google Scholar]
- 69.Knight JD, Williamson JA, Miranker AD. Interaction of membrane-bound islet amyloid polypeptide with soluble and crystalline insulin. Protein Sci. 2008;17(10):1850–1856. doi: 10.1110/ps.036350.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiltzius JJ, Sievers SA, Sawaya MR, Eisenberg D. Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process. Protein Sci. 2009;18(7):1521–1530. doi: 10.1002/pro.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yonemoto IT, Kroon GJ, Dyson HJ, Balch WE, Kelly JW. Amylin proprotein processing generates progressively more amyloidogenic peptides that initially sample the helical state. Biochemistry. 2008;47(37):9900–9910. doi: 10.1021/bi800828u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cabaleiro-Lago C, Lynch I, Dawson KA, Linse S. Inhibition of IAPP and IAPP(20–29) fibrillation by polymeric nanoparticles. Langmuir. 2010;26(5):3453–3461. doi: 10.1021/la902980d. [DOI] [PubMed] [Google Scholar]
- 73.Rijkers DT, Hoppener JW, Posthuma G, Lips CJ, Liskamp RM. Inhibition of amyloid fibril formation of human amylin by N-alkylated amino acid and alpha-hydroxy acid residue containing peptides. Chemistry. 2002;8(18):4285–4291. doi: 10.1002/1521-3765(20020916)8:18<4285::AID-CHEM4285>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 74.Mazzaglia A, Micali N, Scolaro LM, Attanasio F, Magri A, Pappalardo G, et al. Aggregation properties of the peptide fragments derived from the 17–29 region of the human and rat IAPP: a comparative study with two PEG-conjugated variants of the human sequence. J Phys Chem B. 2010;114(2):705–713. doi: 10.1021/jp908436s. [DOI] [PubMed] [Google Scholar]


