Abstract
During our studies on the diverse endophytic fungi resident on conifer needles, many species of Cladosporium previously unreported in Korea were encountered. In this paper, we report on two species of Cladosporium from the needles of pine trees (Pinus spp.). Based on analyses of internal transcribed spacer gene sequence, and cultural and micromorphological characteristics, they were identified as C. oxysporum and C. sphaerospermum. Both species have not been hitherto reported in Korea.
Keywords: Cladosporium, Endophytic fungi, ITS gene sequence, Morphological characteristics, Pine tree
Cladosporium constitutes one of the largest genera of hypomycetes, comprising more than 772 names (Dugan et al., 2004). These species are among the most common fungi isolated from the environment almost anywhere in the world (Farr et al., 1989; Flannigan, 2001; Mullins, 2001; Schubert, 2005). Many species are plant pathogens (Kwon et al., 2001), while others are regularly encountered as contaminants and spoilage agents in food or industrial products, as well as being frequently associated with asthmatic complaints and endophytic fungi (Riesen and Sieber, 1985; Brown et al., 1998; El-Morsy, 2000; Schubert, 2005). Few investigations to date have determined the incidence of endophytic Cladosporium from pine trees.
Endophytic fungi are taxonomically and biologically diverse but all share the character of colonizing internal plant tissues without causing apparent harm to their host (Wilson, 1995). They have proven to be promising sources of new and biologically active natural products, which are of interest for specific medicinal and agrochemical applications (Strobel, 2002).
During our studies on diversity of endophytic fungi from needles of pine trees in Korea, Cladosporium spp. was encountered frequently. They were identified based on the internal transcribed spacer (ITS) sequences, and cultural and morphological characteristics. In this paper, we report on two species of Cladosporium from the needles of pine trees that have hitherto not been reported in Korea.
Materials and Methods
Sampling
Healthy needles of pine trees (Pinus densiflora, P. rigida) were collected from mountain areas in Daejeon, Korea during July and August, 2006. Pine trees were selected randomly and needles were brought to the laboratory in separate sterile polyethylene bags.
Isolation and culture of endophytic fungi
Samples were cleaned under running tap water to remove debris and then air dried and processed within 5 h of collection. From each sample, 10 segments of 1 cm length were separated and treated as replicates. The segments were surface sterilized by immersion in 95% ethanol for 1 min, sodium hypochlorite (4% available chlorine) for 3 min and 95% ethanol for 30 s. The surface sterilized samples were washed in sterile water three times to remove the surface sterilization agents. Samples were allowed to dry on a paper towel in a laminar air flow chamber. Ten segments per sample were placed horizontally on separate Petri dishes containing potato dextrose agar (PDA) supplemented with the antibiotic streptomycin sulfate 0.4 mg/ml and dichloran rose bengal chloramphenicol agar (DRBC). After incubation at 25℃ for 5, 10 and 25 days, individual hyphal tips of the developing fungal colonies were collected and placed onto PDA, incubated for 8~10 days and checked for culture purity. Eventually, cultures of Cladosporium were separated from other fungi based on their conidia. They were transferred to PDA slant tubes and 20% glycerol stock solution.
DNA extraction and polymerase chain reaction (PCR) amplification
For determination of the ITS region of the rDNA of the isolates, genomic DNA was extracted as previously described (Park et al., 2005). PCR amplification was carried out for ITS1 and ITS4 (White et al., 1990) in an i-cycler (Bio-Rad, Hercules, CA, USA) for 30 cycles of 94℃ for 1 min (denaturing), 55℃ for 1 min (annealing) and 72℃ for 150 s (extension). Initial denaturing at 94℃ was extended to 5 min and the final extension was for 10 min at 72℃. PCR product was purified using a Wizard PCR prep kit (Promega, Madison, WI, USA). Purified double stranded PCR fragments were directly sequenced with BigDye terminator cycle sequencing kits (Applied Bipsystems, Foster City, CA, USA) by following the manufacturer's instructions. The same PCR primer sets as used for PCR amplification were used to sequence both DNA strands. Gel electrophoresis and data collection were performed on an ABI Prism 310 genetic analyzer (Applied Biosystems).
Phylogenetic analysis
The sequences were compared with the ITS sequence of rDNA available in the GenBank database by a BLAST search. Sequences generated from materials in this study and retrieved from GenBank were initially aligned using the CLUSTAL X program (Thompson et al., 1997) and the alignment was refined manually using the PHYDIT program version 3.2 (Chun 1995; available at http://plaza.snu.ac.kr/jchun/phydit). A neighbor-joining tree was reconstructed with Kimura's 2-parameter distance model (Kimura, 1980) using the PHYLIP 3.57c package (Felsenstein, 1985). Bootstrap analysis using 1000 replications was performed to assess the relative stability of the branches.
Morphological observations
Cladosporium isolates were inoculated on PDA and malt extract agar (MEA) media in three regions of 9 cm-diameter plastic Petri dishes incubated for 7 days at 25℃ in the dark. Colony appearance, exudate production, pigmentation and reverse coloration were assessed and colony diameters were recorded after one week at 25℃. These isolates were identified with the help of keys developed by Schubert (2005) and Zalar et al. (2007). The examination and measurements of conidiophores and conidia were made from slide preparations stained with lacto-phenol. Differential interference contrast microscopy was used for the observation and measurement of conidiophores and conidia.
Results and Discussion
Molecular analysis
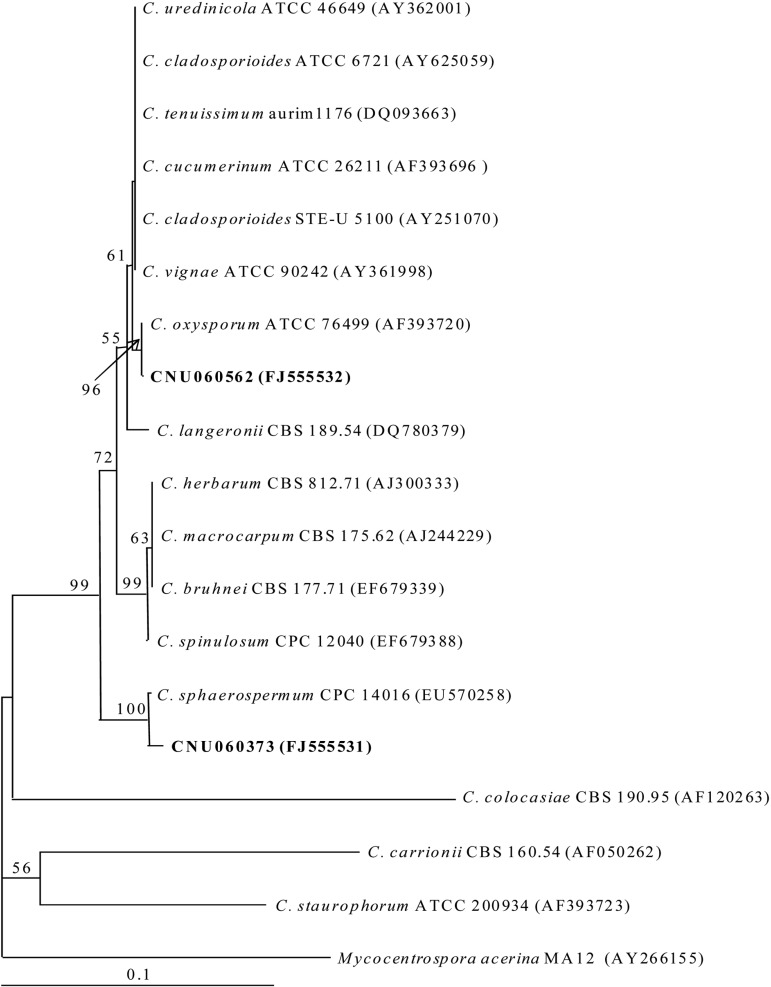
The isolates assumed to be Cladosporium were confirmed on the basis of their ITS gene sequence analysis (Fig. 1). To determine the phylogenetic relationship between the Cladosporium isolates from the needles of pine trees and previously described species, ITS rDNA was sequenced and compared. The CNU060562 isolate and C. oxysporum ATCC 76499 clustered together in a group, which was well supported with a bootstrap value of 89%. The CNU060373 isolate was most closely related to C. sphaerospermum CBS 102045. CNU060373 and C. sphaerospermum CBS 102045 formed a monophyletic group, which was well supported with a bootstrap value of 100%; both were clearly distinguished from other Cladosporium species (Fig. 1).
Fig. 1.
Neighbor-joining tree based on the sequence of the ITS region showing the relationship between endophytic isolates and Cladosporium species. The number above each branch indicates bootstrap values obtained after a bootstrap test with 1000 replications. The bar represents 0.1 substitutions per site. Numbers in parenthesis are GenBank accession numbers.
Morphological observations
Two isolates selected as Cladosporium on the basis of their ITS gene sequence analysis were used for the morphological observations. According to cultural and morphological characteristics, the Cladosporium species were identified as C. oxysporum and C. sphaerospermum (Schubert 2005; Gugnani et al., 2006; Zalar et al., 2007; MycoBank (http://www.mycobank.org/). Taxonomic descriptions, photos of colonies and conidial and conidiophores structures of each species are given below.
Cladosporium oxysporum Berk. & Curt. Figs. 2(A~B), 3(A~D)
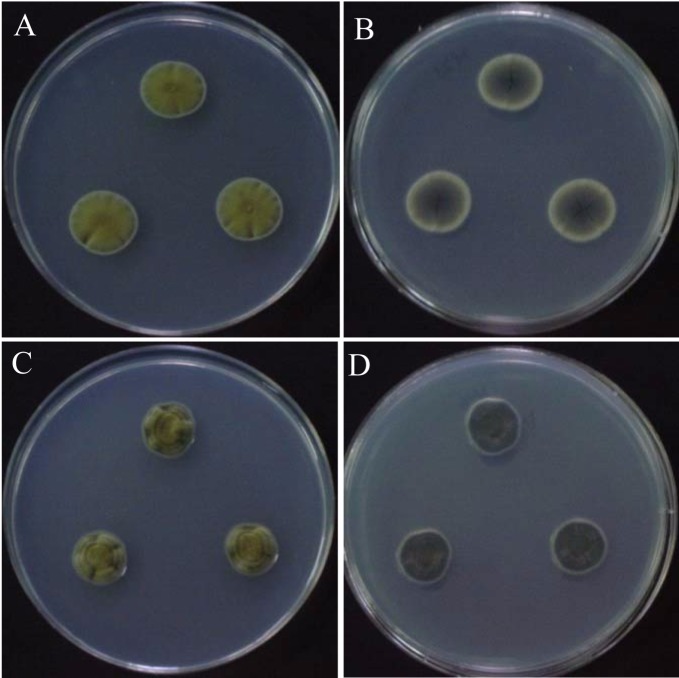
Fig. 2.
Cladosporium colonies on MEA after 7 days of incubation at 25℃ in the dark: C. oxysporum (A, B), C. sphaerospermum (C, D); obverse (A, C), reverse (B, D).
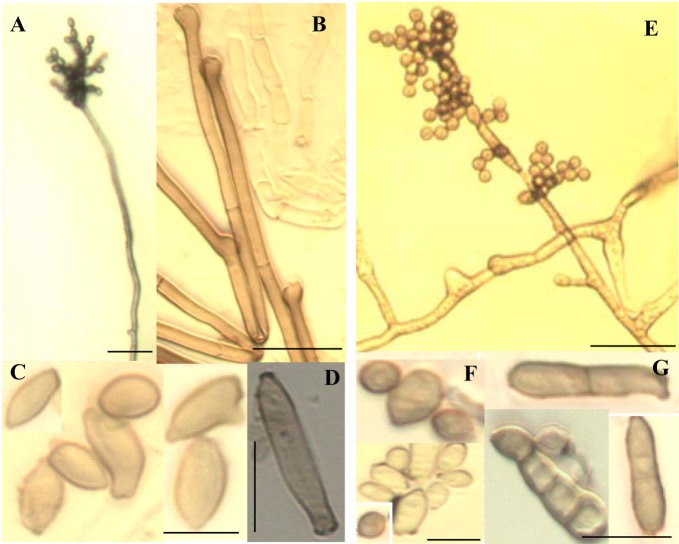
Fig. 3.
Conidiophores (A, B, E), conidia (C, F) and ramoconidia (D, G) of C. oxysporum (A, B, C, D) and C. sphaerospermum (E, F, G). Scale bar = 20 µm (A, B, E, F), 5 µm (C, D, G).
Colonies on MEA were moderately expanding, velvety and often floccose at the center, olivaceous to grey olivaceous; later the colonies turned dark olivaceous with reverse greenish-black. On PDA, the colonies were oliva-reverse dark green. The diameter of colonies on PDA and MEA were 16.5~17.5 mm and 15.0~17.0 mm, respectively. Conidiophores were straight to slightly flexuose, 3.5~5.5 µm wide, distinctly nodose with terminal and intercalary swellings, bearing branched conidial chains. Ramoconidia were present, single cells were common but double celled ramoconidia were rarely evident, with dimensions of 14.5~21.5 × 3.5~4.5 µm. Conidia were spherical to subpherical, smooth-walled, and the length and width of conidia were 2.5~10.0 × 2.0~4.5 µm.
Isolates examined: On needles of P. densiflora; CNU060562.
Notes: Colony characteristics and micromorphology of the fungus agreed well with the description of C. oxysporum (Schubert, 2005; Gugnani et al., 2006). The species is closely related to C. borassi and C. colocasiae. C. borassi produces nodulose conidiophores but swellings are not regular, neither separated nor distinct from each other, and C. calocasiae bears 0~3 septate conidia, which makes them different from C. oxysporum (Schubert and Braun, 2004). The fungus has been reported from Cuba, Mexico, India and Texas (Schubert and Braun, 2004), but not hitherto in Korea.
Cladosporium sphaerospermam Penz. Figs. 1(C~D), 2(E~G)
Colonies on PDA and MEA were 9.5~11.0 mm and 10.0~13.0 mm in diameter, respectively, after 7 days at 25℃. Colonies were olivaceous to grey-olivaceous and powdery on PDA, and velvety to powdery, olive-green to grey-olivaceous on MEA; reverse side of the colonies were greenish-black, with a margin that was either regular or aracnoid, radially furrowed, having a wrinkled colony center and formed a crater-like structure. Conidiophores were variable in length, up to 300 µm long and 3~5 µm wide, smooth-walled or verrucose and not nodose. Secondary ramoconidia were present, possessed 0~4 septa, elongate, smooth-walled or verrucose, and the ramoconidial size were 8.5~20.0 × 3.0~6.0 µm. Conidia were spherical, ellipsoidal to cylindrical with sounded ends, single celled, verrucose with dimensions of 2.0~8.0 × 2.0~4.0 µm.
Isolates examined: On needles of P. densiflora; CNU060373.
Notes: Colony characteristics and micromorphology of the fungus agreed well with the description of C. sphaerospermum (Zalar et al., 2007). The species is similar to C. halotoleraus and C. velox in colonial morphology (Zalar et al., 2007). The fungus, however, has a white margin in colony, is usually irregular shaped and secondary ramoconidia possess 0~4 septa, which distinguishes it from other two fungi. The fungus has been reported from hypersaline water in the Mediterranean and the tropics, soil and plants in temperate climates, indoor wet cells and from humans (Zalar et al., 2007). This is the first record of C. sphaerospermum in Korea.
Table 1.
Comparison of morphological and cultural characteristics of Cladosporium oxysporum and the isolate of endophytic Cladosporium
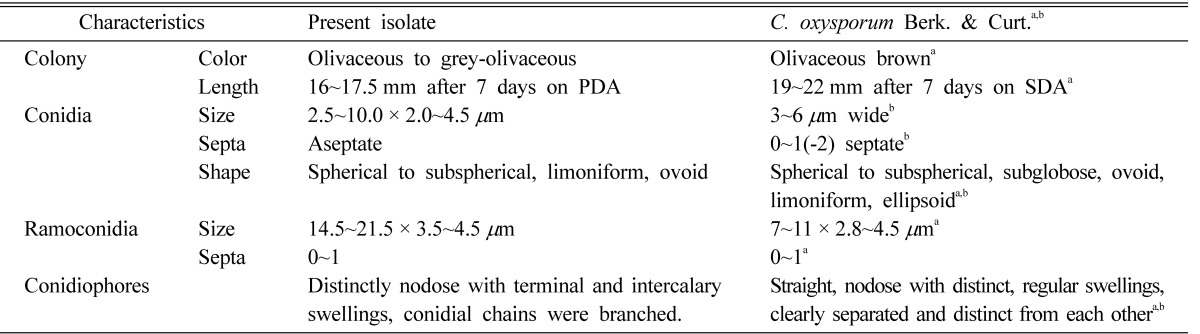
Table 2.
Comparison of morphological and cultural characteristics of Cladosporium sphaerospermum and the isolate of endophytic Cladosporium
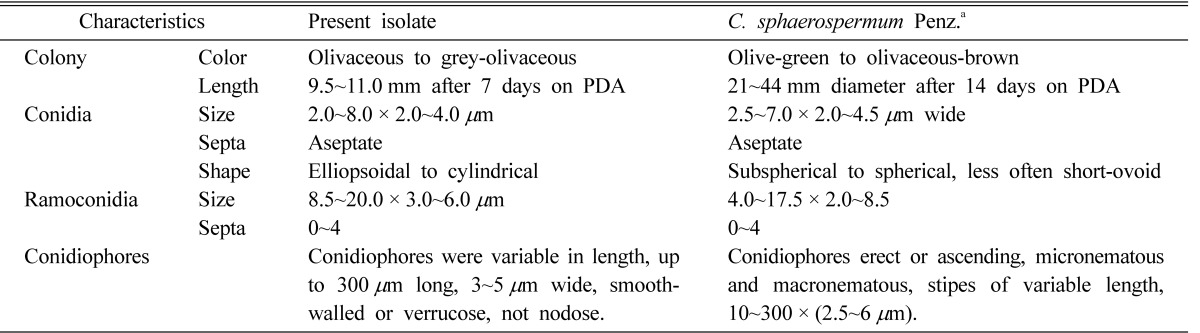
aSource of reference: Zalar et al. (2007).
Acknowledgements
This work was supported by grants from BioGreen 21 Program and National Agrobiodiversity Center of Rural Development Administration (RDA), Korea.
References
- 1.Brown KB, Hyde KD, Guest DI. Preliminary studies on endophytic fungal communities of Musa acuminata species complex in Hong Kong and Australia. Fung Divers. 1998;1:27–51. [Google Scholar]
- 2.Chun J. Computer-assisted classification and identification of actinomycetes. Newcastle Upon Tyne, UK: University of Newcastle; 1995. Ph.D. Thesis. [Google Scholar]
- 3.Dugan FM, Schubert K, Braun U. Check-list of Cladosporium names. Schlechtendalia. 2004;11:1–103. [Google Scholar]
- 4.El-Morsy EM. Fungi isolated from the endorhizosphere of halophytic plants from the red sea coast of Egypt. Fung Divers. 2000;5:43–54. [Google Scholar]
- 5.Farr DF, Rossman AY, Palm ME, McCray EB. Fungi on Plants and Plant Products in the United States. St. Paul, MN: APS press; 1989. [Google Scholar]
- 6.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 7.Flannigan B. Microorganisms in indoor air. In: Flannigan B, Samson RA, Miller JD, editors. Microorganisms in Home and Indoor Work Environments: Diversity, Health Impacts, Investigation and Control. London: Taylor and Francis; 2001. pp. 17–31. [Google Scholar]
- 8.Gugnani HC, Ramesh V, Sood N, Guarro J, Moin-Ul-Haq, Paliwal-Joshi A, Singh B, Makkar R. Cutaneous phaeohyphomycosis caused by Cladosporium oxysporum and its treatment with potassium iodide. Med Mycol. 2006;44:285–288. doi: 10.1080/13693780500294824. [DOI] [PubMed] [Google Scholar]
- 9.Kimura M. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequence. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 10.Kwon JH, Kang SW, Kim JS, Park CS. Scab of tea (Thea sinensis) caused by Cladosporium herbarum in Korea. Plant Pathol J. 2001;17:350–353. [Google Scholar]
- 11.Mullins J. Microorganisms in outdoor air. In: Flannigan B, Samson RA, Miller JD, editors. Microorganisms in Home and Indoor Work Environments: Diversity, Health Impacts, Investigation and Control. London: Taylor and Francis; 2001. pp. 3–16. [Google Scholar]
- 12.Park MS, Seo GS, Bae KS, Yu SH. Characterization of Trichoderma spp. associated with green mold of oyster mushroom by PCR-RFLP and sequence analysis of ITS regions of rDNA. Plant Pathol J. 2005;21:229–236. [Google Scholar]
- 13.Riesen T, Sieber T. Endophytic Fungi in Winter Wheat (Triticum aestivum L.) Zurich: Swiss Federal Institute of Technology; 1985. [Google Scholar]
- 14.Schubert K. Morphotaxonomic revision of foliicolous Cladosporium species (hypomycetes) Germany: Martin Luther University; 2005. Ph.D. Thesis. [Google Scholar]
- 15.Schubert K, Braun U. Taxonomic revision of the genus Cladosporium s. lat. 2. Cladosporium species occurring on hosts of the families Bignoniaceae and Orchidaceae. Sydowia. 2004;56:296–317. [Google Scholar]
- 16.Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Hill CF, Zalar P, Hoog GS, Crous PW. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardization of methods for Cladosporium taxonomy and diagnostics. Stud Mycol. 2007;58:105–156. doi: 10.3114/sim.2007.58.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strobel GA. Rainforest endophytes and bioactive products. Crit Rev Biotechnol. 2002;22:315–333. doi: 10.1080/07388550290789531. [DOI] [PubMed] [Google Scholar]
- 18.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. ClustalX: windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–4878. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal DNA for phylogenetics. In: Innes MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to the methods and applications. New York: Academic Press; 1990. p. 315. [Google Scholar]
- 20.Wilson D. Endophyte-the evolution of a term and clarification of its use and definition. Oikos. 1995;73:274–276. [Google Scholar]
- 21.Zalar P, Hoog GS, Schroers HJ, Crous PW, Groenewald JZ, Gunde-Cimerman N. Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Stud Mycol. 2007;58:157–183. doi: 10.3114/sim.2007.58.06. [DOI] [PMC free article] [PubMed] [Google Scholar]