Abstract
The toxin colicin E3 targets the 30S subunit of bacterial ribosomes and cleaves a phosphodiester bond in the decoding center. We present the crystal structure of the 70S ribosome in complex with the cytotoxic domain of colicin E3 (E3-rRNase). The structure reveals how the rRNase domain of colicin binds to the A site of the decoding center in the 70S ribosome and cleaves 16S rRNA between A1493 and G1494. The cleavage mechanism involves the concerted action of conserved residues Glu62 and His58 of the cytotoxic domain of colicin E3 that activate the 16S rRNA for 2′ OH induced hydrolysis. Conformational changes observed for E3-rRNase, 16S rRNA and Helix 69 of 23S rRNA suggest that a dynamic binding platform is required for colicin E3 binding and function.
INTRODUCTION
The vital role of the ribosome in translating genetic information into proteins makes it an obvious target for various natural antibiotics and bacteriocins, which are protein toxins produced by bacteria to inhibit the growth of similar or closely related bacterial strains. Recent progress in ribosome crystallography has provided enormous structural details in understanding how antibiotics interfere with protein translation1-4. However, questions on the specific action of bacteriocins await a high resolution structure of their complexes with ribosomes.
Bacteriocins that target the ribosome include colicins E3, E4, E6 and cloacin DF13, which all show specific 16S rRNase activity5,6. Colicin E3 is a plasmid-encoded 60kDa protein that is produced during times of stress by some strains of Escherichia coli. It contains three domains that are required for the toxin to enter and kill the cell: the N-terminal translocation domain (T), the central receptor-binding domain (R) and the C-terminal cytotoxic domain7. Together with E3, the cell co-expresses and releases an immunity protein (Im3) that binds with high affinity (Kd~pM) to the E3 cytotoxic domain and prevents the producing cell from committing suicide by specifically inactivating the RNase activity8,9,10. The E3-Im3 complex is expressed and released from cells through the SOS response, penetrating a target cell via interactions of the R and T domains with specific receptors and translocators in the outer membrane of E. coli. During the translocation process the Im3 protein dissociates from the complex leaving the E3-rRNase to exert its cytotoxic activity11. E3-rRNase cleaves a single phosphodiester bond between A1493 and G1494 (E. coli numbering, used throughout the manuscript) in the decoding site (A site) in helix 44 of ribosomal 16S rRNA12,13, consequently impairing protein translation and eventually causing cell death.
Biochemical and structural studies on colicin E3 have identified many residues implicated in the catalytic activity of the rRNase, including residues Arg40, Arg42, Asp55, His58, Glu60, Glu62 and Arg90 (refs. 14-17). Furthermore, in vitro assays have shown that ribosomes with E3-cleaved 16S rRNA in the A site show impaired decoding defects resulting in the production of short peptides18. However, despite this plethora of biochemical and structural data, the nature of the high specificity of E3 16S rRNase, its cleavage mechanism, as well as the structural and biological consequences of 16S RNA cleavage, remain speculative and unresolved.
To address these questions at the atomic level, we solved the crystal structure of E3-rRNase bound to Thermus Thermophilus 70S ribosomes in complex with mRNA and tRNAs in the post-cleavage state at 3.2 Å resolution. The structure provides new insights into bacteriocin action on ribosomes.
RESULTS
Overall structure of the 70S – colicin E3-rRNase complex
Intact Thermus Thermophilus 70S ribosomes were purified as previously described19 and incubated with mRNA, tRNAfMet and a mutant of the C-terminal domain of colicin E3 (E3H58A-rRNase, 96 residues with a histidine to alanine mutation at position 58) prior to crystallization. Residue numbering is based on that of the isolated domain, as previously reported15 (Supplementary Fig. 1). E3H58A-rRNase is inactive in in vivo cytotoxicity tests and in vitro transcription-translation assays17 and so was used here to solve the structure of E3-rRNase bound to the ribosome in its pre-cleavage state. However, under our crystallization conditions E3H58A-rRNase (referred to as E3-rRNase in the following) cleaves the 16S rRNA between A1493 and G1494 (Supplementary Fig. 2), so the structure represents the post-cleavage state (Table 1).
Table 1.
Data collection and refinement statistics
| 70S – E3-rRNase(H58A) complex | |
|---|---|
| Data collection | |
| Space Group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | a= 212.04 b= 453.51, c= 616.10 |
| Resolution (Å) | 45-3.2 (3.3-3.2)* |
| Rsym, % | 11.2 (98.8) |
| I / σI | 10.95 (1.5) |
| Completeness, % | 99.4 (98.8) |
| Redundancy | 4.18 (4.17) |
| Refinement | |
| Resolution (Å) | 45.0-3.2 |
| No. of unique reflections | 960332 |
| Rwork / Rfree (%) | 22.8/27.0 |
| No. atoms | |
| RNA | 196158 |
| Protein | 95597 |
| Ions | 2350 |
| B-factors (Å2) | |
| RNA | 107.7 |
| Proteins | 122.8 |
| Proteins including E3-rRNase | 123.1 |
| Ions | 74.5 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.14 |
Highest resolution shell is shown in parenthesis
The overall structure of the ribosome in the 70S-E3-rRNase complex is similar to previously published 70S structures19,20, suggesting that E3-rRNase binding and cleavage do not induce major global changes in the ribosome.
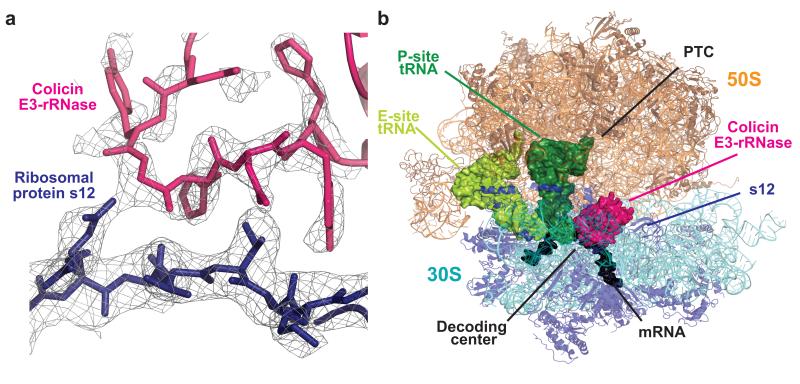
E3-rRNase binds to the A site in the ribosomal decoding center of the 30S subunit and prevents access of both A-site tRNA21 and ribosomal factors such as release and elongation factors. The toxin spans the head and body of the 30S, its overall topology resembling that of the anticodon stem loop (ASL) of an A-site-bound tRNA (Figure 1A and 1B). Its binding site within the post-cleavage state ribosome is delineated by ribosomal components surrounding the decoding center, including 16S rRNA (helices 44, 18, and 30-31), helix 69 of 23S rRNA, mRNA, P-site tRNA as well as the 30S protein S12, all of which are contacted by the E3-rRNase.
Figure 1. Structure of colicin E3-rRNase bound to the 70S ribosome.
(a) Representative electron density from a 3mFo-2dFc map displayed at 0.9 σ, with the refined model of E3-rRNase (pink) and ribosomal protein S12 (blue). (b) Overall view of the complex, with colicin E3-rRNase and P-site and E-site tRNA depicted as surfaces, and rRNA and proteins as cartoons.
Previously reported structures of the E3-rRNase in complex with immunity protein Im3 (refs. 14,15) revealed a compact fold for the E3-rRNase domain, consisting of a six-stranded, highly twisted, antiparallel β-sheet (β1 to β6). When binding the ribosome, E3-rRNase maintains its compact conformation and does not undergo major conformational changes, but slight arrangements are visible where interactions are made to rRNA and to ribosomal protein S12. Complex formation with Im3 does not directly obscure the active site of E3-rRNase, but hinders access of the enzyme to the ribosome, consistent with previous suggestions that Im3 is an exosite inhibitor9,15. Superimposing the E3-rRNase–Im3 complex to the E3-rRNase domain bound to the ribosome reveals that Im3 would severely clash with large parts of the 16S rRNA (helices 18, 34) and ribosomal protein S12. Furthermore, electrostatic repulsion between the negatively charged Im3 and the negatively charged 16S rRNA represents another barrier to the approach of colicin E3 to its substrate. On binding to the ribosome about one third of the solvent-accessible area of E3-rRNase gets buried (2128 Å2), which is about twice the loss as when E3-rRNase binds to Im3 (1257 Å2).
Deformation of the decoding site as a result of E3-rRNase binding
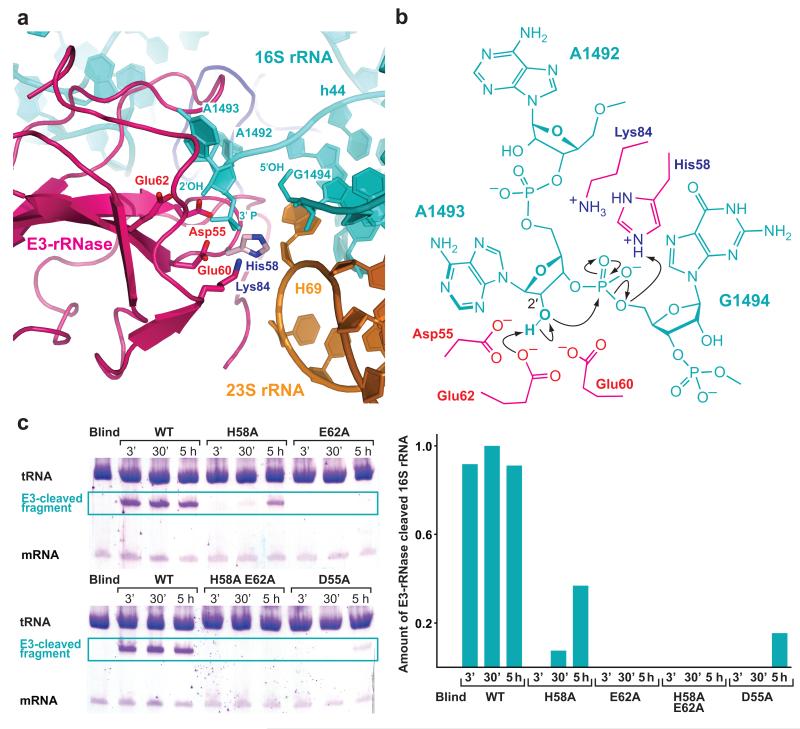
The structure reported here represents the post-cleavage state where the phosphodiester bond between A1493 and G1494 has been hydrolyzed by attack of E3-rRNase. The formal outcome of this reaction is a 2′, 3′-cyclic phosphate at the 3′-terminal end (A1493) and a free 5′-hydroxyl at the 5′-terminal end (G1494). The ~50-nucleotide long 3′-terminal fragment of 16S rRNA does not show deviations from its position in 70S structures with unperturbed 16S rRNA: nucleobase G1494 remains Watson-Crick base-paired to C1407 without major conformational changes (Supplementary Fig. 3a). The end of the 5′-fragment, however, is pushed outwards relative to its path in the uncleaved state and is sandwiched between loop regions β1-β2 (residues 32-39) and β3-β4 (residues 55-62) of E3-rRNase, which constitute the active site of the enzyme and is gripped by conserved residues thereof (Figure 2A). Residue Asp8 contacts the 2′-hydroxyl of G1491 with its carboxyl group, while residue Tyr9 stabilizes the position of nucleotide A1492 by stacking with its nucleobase (Supplementary Fig. 3a). Gln57 and Ser56 are positioned to make interactions with the 2′- and 3′- ribose oxygens of A1492 via their backbone and side chain, respectively. In the structure reported here, the conformation of key nucleotides A1492 and A1493 is somewhat different from that observed in empty ribosomes or when cognate tRNA is bound in the A site19,22,23. Presumably due to shape complementarity, A1492 is pulled fully out of 16S helix 44, and its conformation is reminiscent of the 1492 conformation found in structures with nuclease RelE24 or release factor RF225,26 bound to the A site (Supplementary Fig. 3b). A1493 is also partly extruded and held in place by extensive hydrogen-bonding interactions with E3-rRNase residues. Arg40, conserved in all colicin E3-rRNases, contacts both the nucleobase of A1493 (N7) as well as one of the non-bridging phosphate-oxygens via its guanidinium group (Supplementary Fig. 3a). Gln34 stabilizes the phosphate of 1493 by forming a hydrogen-bond through its backbone amide NH.
Figure 2. A mechanism for 16S rRNA cleavage by colicin E3-rRNase.
(a) The active site of E3-rRNase (pink cartoon) bound to the 70S ribosome with critical amino acids shown as labeled sticks. The structure represents the post-cleavage state, where the phosphodiester bond between A1493 and G1494 has been hydrolyzed, leading to a 3′-phosphate moiety at A1493 and a free 5′ OH at G1494. The E3-rRNase construct used in this study displayed a H58A mutation, residue His58, shown in light pink, has been modeled in this figure. (b) Schematic presentation of the proposed cleavage mechanism with residues from E3-rRNAse shown in pink. Glu62 deprotonates the 2′OH of A1493 activating it for a nucleophilic attack on the adjacent 3′-phosphate. The negatively charged trigonal bipyramidal transition state is likely to be stabilized by His58, which is also thought to finally protonate the leaving 5′-OH RNA fragment. (c) In vitro cleavage assay. E3-rRNase mutants (wt, H58A, E62A, H58A E62A, D55A) were incubated with a 10-fold excess of 70S Th. Thermophilus ribosomes, tRNAfMet and mRNA at 37°C. At the indicated time points the reactions were stopped by addition of phenol/chlorofom, the RNAs concentrated by ethanol precipitation and separated by 16% urea denaturating PAGE. The amounts of E3-cleaved 16S rRNA were determined by scanning the toluidine stained bands with a Typhoon imager and quantified with Image Quant TL (both GE Healthcare). While the wildtype E3-rRNase shows full cleavage activity already after 3 minutes, the mutants where either the proposed general base Glu62 or both the general acid and base (His58/Glu62) have been mutated to alanines decrease the E3-rRNase cleavage activity below the detection limit.
Outside the decoding center, the side chain hydroxyl of universally conserved Ser67 of loop β4-β5 of E3-rRNase contacts one of the non-bridging phosphate-oxygens of nucleotide 954 in helix 30. Furthermore residues Arg65 and Arg49 come very close to nucleotides of helix 30 (954-955) and helix 31 (956-957 and 960) and presumably form hydrogen-bonds to stabilize the E3-rRNase-ribosome interaction in the pre-cleavage state.
Nucleotide G530 (helix 18), which plays a key role in the decoding process by monitoring the correct codon-anticodon interaction in the A site23 was found in a syn conformation rather than in the anti conformation reported in previous 70S structures with A-site substrates. This is the conformation of the base in the open form of the empty 30S subunit, and is consistent with the global conformation of the 30S subunit we observe here. The G530 base is kept in this conformation by making contact to the backbone amide-oxygen of Lys39 of E3-rRNase via a water molecule. In fact, rotation around the N-glycosidic bond to adopt an anti conformation, would lead to a steric clash of base G530 with residues 38-40 of the β1-β2 loop of E3-rRNase. In addition, nucleotide C518 of helix 18 stays in close vicinity to residue Gly38 and the side chain of Lys41 and might form hydrogen-bonds in the pre-cleavage state.
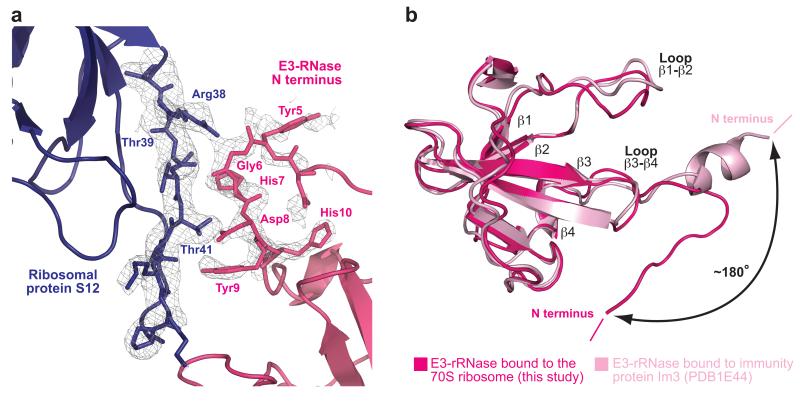
The N terminus of E3-rRNase contacts ribosomal protein S12
The only ribosomal protein that makes interactions with E3-rRNase is protein S12, which constitutes an important part of the shoulder domain of the 30S subunit and is functionally associated with the fidelity rate of translation27. The N-terminal residues 5-9 of E3-rRNase make extensive hydrogen-bonding interactions with residues 38 to 41 of S12, forming a pseudo β-sheet (Figure 3A). The following interactions were observed: Tyr5 and Gly6 of E3-rRNase contact via their backbone amide-oxygens the side chain of Arg38 and the backbone of Thr41 of S12, respectively. The His7 side chain (E3-rRNase) is within hydrogen-bonding distance to the backbone amide-oxygen of Thr39 (S12). In addition the side chains of Asp8 and Tyr9 of E3-rRNase form interactions with the side chain-hydroxyl of Thr41 and the backbone amide oxygen of Ala48 of protein S12, respectively.
Figure 3. The N terminus of E3-rRNase contacts ribosomal protein S12.
(a) The N-terminal residues 5-9 of E3-RNase make hydrogen-bonding interactions with residues 38-41 of S12, forming a pseudo β-sheet. The 3mFo-2dFc electron density map is displayed at 0.9 σ. (b) Comparison of colicin E3-rRNAse bound to the 70S ribosome (this structure) and E3-rRNase bound to its immunity protein Im3 (pdb: 1e44)15 highlights the different conformation and orientation that the N terminus of E3-rRNase adopts upon binding to the ribosome.
In known colicin E3 structures these N-terminal residues (1-10) form an α-helix that interacts extensively with a wide hydrophobic groove at the top of the immunity protein Im314,15. However, in complex with the ribosome, these residues do not form a helix, but are extended (Figure 3B) and act as an interaction platform to ribosomal protein S12. The involved residues of both E3-rRNase and protein S12 show a high degree of conservation, among rRNase homologs E4, E6, Cloacin DF13 and among Th. Thermophilus and E. coli, respectively (Supplementary Fig. 1). Therefore it is possible that the change in the conformation of the E3-rRNase N-terminal residues that allows their interaction with S12 represents part of the mechanism of the specific recognition of the 30S subunit by E3.
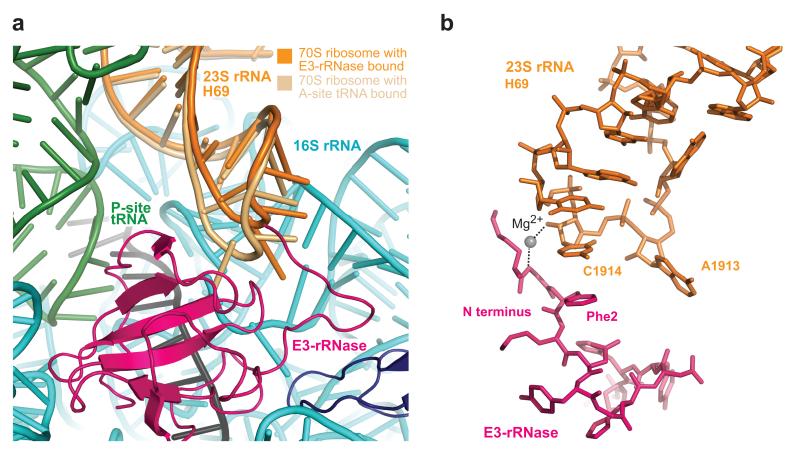
Conformational changes at Helix 69 of 23S rRNA induced by E3-rRNase binding
The only region of 23S rRNA in close proximity to the decoding center and E3-rRNase is Helix 69. Compared to its position in 70S ribosome structures with a bound A-site tRNA19,20, the tip of H69 (nucleotides 1912-1918) is pushed further away from helix 44 in 16S rRNA and the decoding center (Figure 4A). A1913 at the tip of H69, that in previous structures was found inserted into a tight pocket formed by the backbone of h44 and A-site tRNA and thereby involved in formation of the intersubunit salt-bridge B2a, is now flipped down between C1409 and G1491 of h44 and comes close to Gln57 of E3. This position is stabilized by formation of a hydrogen-bond between N6 of A1913 and N1 of G1491, and between N7 of A1913 and the amide amino group of Gln57. Furthermore, we observed a stacking interaction between H69 nucleotide C1914 and Phe2 of E3-rRNase (Figure 4B) as well as a Mg2+-mediated hydrogen-bond of its 2′ OH to the backbone amide of Gly1.
Figure 4. Conformational changes at Helix 69 of 23S rRNA induced by E3-rRNase binding and cleavage.
(a) Comparison of H69 in the 70S ribosome with cognate A-tRNA bound and H69 in this structure highlights the conformational changes at nucleoside A1913. (b) Interactions between N-terminal residues Gly1 and Phe2 of E3-rRNAse with H69 of 23S rRNA.
Interactions of E3-rRNase with mRNA
The binding of E3-rRNase pushes the mRNA A-site codon towards helix 44 (region 1397–1402) and the mRNA thereby shows a more pronounced kink between P- and A-site codons than in previously reported structures19,20. Sequence independent interactions of E3-rRNase with the sugar-phosphate backbone of the mRNA A-site codon involve two conserved E3 residues, Gln34 and Arg90, whose side chains make hydrogen bonds with non-bridging phosphate- or ribose-oxygens of the first two nucleotides in the A-site U19 and U20 (Supplementary Fig. 4).
Interactions of downstream mRNA with small subunit proteins
Apart from the codons at the tRNA binding sites, we saw four additional nucleotides at the 3′ end (A22 to A25), which provide insights into the pathway of the mRNA in this region. Nucleotides A24 and A25 corresponding to mRNA positions +9 and +10 hydrogen-bond via their sugar-phosphate backbone with basic side chains of conserved residues Arg164 and Arg15 of 30S proteins S3 and S5, respectively. Our observations provide evidence at an atomic level that S3 and S5 form a gate for the incoming mRNA guiding it through the downstream tunnel of the small subunit formed by proteins S3, S4 and S5 as suggested from previous structures at lower resolution 28,29.
DISCUSSION
The mechanism of 16S rRNA cleavage by colicin E3-rRNase
On the basis of the post-cleavage structure reported here and supported by in vitro cleavage data, we propose a mechanism of 16S rRNA cleavage by colicin E3-rRNase (Figure 2B and 2C), which is in accordance with what has been proposed based by docking E3 to the ribosome16 and by superposition and alignment of E3-rRNase with the active site of the nonspecific ribonuclease barnase15. In this mechanism, the side chain of Glu62 acts as a general base, deprotonating the 2′ OH of A1493 and thereby activating it for an in-line nucleophilic attack on the adjacent 3′-phosphate. The negatively charged penta-coordinated transition state is likely to be stabilized by the positively charged side chain of His58. Finally, the protonated imidazole moiety of His58 donates a proton to the leaving 5′ OH RNA fragment upon cleavage thereby acting as general acid. Conserved residues Glu60 and Asp55 are within hydrogen-bonding distance of His58 and could both help to orient the histidine side chain and increase its pKa so as to stabilize its positive charge during its role as proton donor in the transphosphorylation reaction. The 2′,3′-cyclic phosphate at A1493 might be further hydrolyzed to a 3′-phosphate by an exogenous water molecule stabilized by His58. However, from the density in our structure at the cleaved 3′-end we cannot unambiguously distinguish between a 2′,3′-cyclic phosphate or a 3′-phosphate. The structure cannot explain previous suggestions of the involvement of Arg90 in biological activity15-17. In our structure Arg90 points towards the A-site codon mRNA (see above). A structure of the pre-cleavage or transition state would help resolve this issue.
Beside the two main catalytic residues His58 and Glu62, which are proposed to act as a general acid-base pair, there are a few other residues in the vicinity of the scissile phosphodiester bond that can be involved in catalysis. Among these, two conserved, acidic residues, Asp55 and Glu60 are in hydrogen-bonding distance to the 2′ OH of A1493 and might function as general bases in the absence of Glu62. The H58A mutant used in this study retained some catalytic activity (especially under our crystallization conditions, Supplementary Fig. 2), indicating that other nearby positively charged residues such as Lys84 might be able to fulfill its functions. To further analyze the importance of these individual residues in catalysis, we constructed D55A, H58A, E62A single mutants as well as E62A H58A double mutants and measured their cleavage efficiencies on programmed 70S Th. thermophilus ribosomes in vitro (Figure 2C). All the mutants tested, compromised cleavage activity by several orders of magnitude, the single mutant E62A and the double mutant H58A E62A having the strongest effect and depriving colicin E3 of its RNase activity. Under our crystallization conditions, however, we were not able to get a complex with uncleaved 16S rRNA even with mutants that compromise cleavage activity (Supplementary Fig. 2, Figure 2C). In order to do so it might be necessary to modify the 16S rRNA nucleotide A1493 by 2′-O-methylation.
Colicin E3 shows a highly specific RNase-activity. Among the more than 1500 phosphodiester bonds in 16S rRNA only the one between A1493 and G1494 is cleaved by E3-rRNase. E3-rRNase can bind to the A site, because this is a region of exposed 16S rRNA devoid of any protein cover and accessible to external factors. The structure reported here shows that binding of colicin E3-rRNAse causes the universally conserved nucleotides A1492 and A1493 to flip out of helix 44. This alters the sugar-phosphate backbone immediately following the flipped-out bases, thereby positioning the 2′ OH of A1493 in a way that it can easily be deprotonated by nearby acidic residues (Asp55, Glu60 and Glu62). Furthermore base flipping of A1492 and A1493 aligns the A1493 2′ OH nucleophile inline with the scissile bond so that it is poised for cleavage (Figure 2B). It has been shown for other endoribonucleases and ribozymes that they use base flipping to switch the substrate from an inactive to an active form30,31.
Implications of E3-mediated 16S rRNA cleavage for decoding and translation
The bacteriocin colicin E3 inhibits bacterial protein synthesis by causing a specific cleavage in the 16S rRNA of the 30S subunit both in vivo12,13 and in vitro32,33. Cleavage of 16S rRNA by colicin E3 has been reported to occur only in the context of the intact 70S ribosome34,35, although other studies indicate that at high concentrations of the toxin isolated small subunits are also cleaved36. Our results do not show any evidence that the reaction in vitro requires the presence of the 50S subunit; incubation of 30S subunits with E3-RNase resulted in specifically cleaved rRNA (data not shown). Nevertheless, attempts to crystallize only the 30S subunit together with E3-rRNase failed.
Recent biochemical data have revealed that the main defect of colicin E3-mediated 16S rRNA cleavage is observed during decoding, whereas peptide bond formation and translocation are largely unaffected18. The cleavage of 16S rRNA seems not to impair monitoring of codon-anticodon complexes or GTPase activity during EF-Tu-dependent binding of aa-tRNA, but decreases the stability of codon-recognition complexes, which leads to a higher rejection rate during the aa-tRNA accommodation step.
The fact that E3-cleaved ribosomes retain their ability to select cognate codon-anticodon complexes implies that on binding of aa-tRNA, nucleosides A1492, A1493 and G530 are still able to make interactions with the minor groove of the codon-anticodon helix. This induces the global ‘domain closure’ of the 30S subunit around the A site, which finally triggers GTP hydrolysis of the EF-Tu-aa-tRNA ternary complex. However, due to the E3-induced 16S rRNA cleavage between A1493 and G1494 the flexibility of the decoding site might be increased, so that even cognate aa-tRNA will not be held as tightly. It is tempting to speculate that the conformational change observed for A1913 at the tip of H69 in 23S rRNA (Figure 4A) which alters the structure of the intersubunit bridge B2a19 between 16S rRNA and 23S rRNA might also be persistent after release of E3-rRNase and thereby disrupt the hydrogen-bonding interaction between A1913 and the 2′ OH of nucleotide 37 in the aa-tRNA, which could explain the lower stability of the codon recognition complex.
By providing a more flexible binding platform in the A site, E3-mediated cleavage may lower both the stabilization of near-cognate and cognate aa-tRNA in the codon recognition complex. This leads to a relatively high proportion of cognate aa-tRNA that is lost during proofreading and explains why ribosomes with cleaved 16S rRNA were found to be hyper-accurate and are able to offset the effect of the error-inducing antibiotic streptomycin37,38. These in turn could lead to a slowly translating ribosome that would also arrest the following ribosomes in a polysome array. The effect of E3-mediated 16S rRNA cleavage on even a small fraction of ribosomes could thus abolish protein synthesis and lead to cell death.
Conclusions
The structure of the 70S – E3-RNase complex at the post-cleavage state reported here provides atomic details on how this bacteriocin site-specifically hydrolyzes a phosphodiester bond in 16S rRNA. The cleavage of 16S rRNA in the A-site renders the decoding center around A1492 and A1493 of 16S rRNA more flexible and leads to defects in acceptance of cognate aa-tRNA, thereby leading to a slow down of protein synthesis that finally leads to cell death.
METHODS
Purification of 70S ribosomes, tRNA, mRNA and E3-RNase
Thermus thermophilus 70S ribosomes and deacylated E. coli tRNAfMet were purified as described19. The mRNA used was chemically synthesized (Dharmacon) and had the sequence 5′-GGC AAG GAG GUA AAA AUG UUC AAA A-3′ with an AUG fMet codon at the P site and an UUC codon in the A-site. The E3-RNase was expressed and purified as previously described10,17.
Complex formation and crystallization
Complexes of E3-RNAse with the 70S ribosome, tRNAfMet and mRNA were prepared as described previously in buffer G (5 mM HEPES pH 7.5, 50 mM KCl, 10 mM NH4Cl, 10 mM magnesium acetate, 6 mM 2-mercaptoethanol)19. To form an E3-RNAse complex, 70S ribosomes at a final concentration of 4.4 μM were incubated with a 2-fold excess of mRNA at 55 °C for 6 min. A 2.5-fold excess of tRNAfMet was added and the complex was incubated at 55 °C for 30 min. The mixture was cooled down to 37°C and incubated with a 5-fold excess of E3-RNase for 10 min at 37°C and 20 min at room temperature. Crystals were grown in sitting drop vapor diffusion experiments in which 2.4 μl of ribosomal complex including 2.8 mM Deoxy Big Chap (Hampton Research) were mixed with 2 μl reservoir solution containing 0.2 M KSCN, 0.1 M Tris-HAc pH 7.2, 3.5-4.3% w/v PEG 20K, and 3.5-4.3% w/v PEG 550 monomethylether (MME) and left to equilibrate at 20°C. The crystals were cryo-protected in three steps (with 8%, 18% and 28% (w/v) PEG 550 MME, respectively) and transferred into the final cryoprotecting solution (0.2 M KSCN, 0.1 M Tris-HAc pH 7.2, 4.0% (w/v) PEG 20K and 28.0% (w/v) PEG 550 MME. The final solution contained 22 μM E3-RNase, and crystals were incubated for 24 h before being frozen by plunging into liquid nitrogen. All data collection was carried out at 100 K.
Data collection, refinement and model building
The final data set was collected from two regions of a single crystal at beamline ID14-4 at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France. Data were integrated and scaled using XDS39. A starting model consisting of the empty 70S ribosome20 (without tRNAs, mRNA and ions) was used for initial refinement and phase calculation using CNS40. The refinement process involved one cycle of rigid body (60 steps) refinement for each of the two 70S molecules in the asymmetric unit, one cycle of minimization (200 steps) and individual and grouped B-factor refinement. Unbiased mFo-DFc difference electron density maps calculated after initial refinement using a complete model of the ribosome, but without mRNA, tRNas or factor allowed modeling of mRNA, P-site tRNAfMet, non-cognate E-site tRNA and E3-rRNAse (E3-rRNase model, PDB code: 1E44). The final model included the entire P-site tRNAfMet, non-cognate E-site tRNA, mRNA (nucleosides 10 to 25) and the E3-RNase residues 0-96 (equal to registry 455-551 of full length colicin E3). The E3-RNase construct used in this study contained seven more residues at the N-terminal end (MEKNKPR). However no density could be observed for these residues indicating that this N-terminus makes no contacts with the ribosome and is rather flexible. Comparison of the 70S structure reported here with previously reported structures19,20 showed r.m.s.d values of ~1.2 Å and ~0.4 Å for 16S and 23S rRNA (derived by least square fit of the phosphate backbones), respectively. The overall low r.m.s.d. values suggest that E3-rRNase binding and cleavage do not induce major global changes in the ribosome. Nevertheless, the higher r.m.s.d. value for the small subunit compared to the large subunit indicates that the 30S undergoes more pronounced conformational changes upon binding and cleavage by E3-RNase. The small subunit of the 70S–E3-rRNase structure reported here adopts an open form similar to what has been observed for native 30S subunits, but distinct to 30S or 70S structures with cognate tRNAs bound to the A-site19,20,41,42. It should be noted, that only one of the E3-RNase molecules in the asymmetric unit could be fully modeled, although with elevated B-factor (average B-factor ~146) compared to the surrounding environment, while the other E3-rRNase molecule could be build (with occupancy = 1) only from residue 1 to 14 due to lack of clear electron density, showing that the small subunit in the open form shows a significant level of conformational variability (especially in the shoulder region). The 16S rRNA in both ribosome molecules, however was cleaved. The N-terminal end of S13 (residues 119-125) was removed due to a lack in density. L31 that previously contained model and registry ambiguity19 was corrected referring to the Genbank registry and was built to contain residue 1 to residue 57 according to the unbiased map. The final refinement in CNS had Rwork/Rfree of 22.8%/27.0%. Data and refinement statistics are reported in Table 1.
In vitro cleavage assays
70S Th. Thermophilus ribosomes were complexed with tRNAfMet, mRNA and E3-rRNase mutants (wt, H58A, E62A, H58A E62A, D55A) in polymix buffer (5mM magnesium acetate, 0.5 mM CaCl2, 5mM NH4Cl, 95 mmM KCl, 8 mM putrescine pH 7.5, 1 mM spermidine, 5 mM potassium phosphate pH 7.3, 1mM dithioerythriol), essentially as described above but only with 0.44 μM of E3-rRNase mutants, so that the ribosomes were in a 10-fold excess over E3-rRNase mutants. Colicin E3-rRNase mutants were added at 37°C and reactions incubated for different time points (3′, 30′ and 5 h). At the indicated time points the reactions were stopped by addition of phenol/chlorofom, concentrated by ethanol precipitation and separated by 16% urea denaturating PAGE. The amounts of E3-cleaved 16S rRNA were determined by scanning the toluidine stained bands with a Typhoon imager and quantified with Image Quant TL (both GE Healthcare).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Cajetan Neubauer, Israel S. Fernandez, Yonggui Gao, Martin Schmeing, Jade Li and Miguel Ortiz-Lombardia for helpful discussions and Nadine Kirkpatrick for technical assistance. VR was supported by the Medical Research Council (UK), the Wellcome Trust, the Louis-Jeantet Foundation and the Agouron Institute. CK acknowledges BBSRC (UK) for funding.
Footnotes
Accession codes The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank with accession IDs 2XFZ, 2XG0, 2XG1, 2XG2.
References
- 1.Brodersen DE, et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 2.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 3.Hansen JL, et al. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 4.Auerbach T, et al. Antibiotics targeting ribosomes: crystallographic studies. Curr Drug Targets Infect Disord. 2002;2:169–186. doi: 10.2174/1568005023342506. [DOI] [PubMed] [Google Scholar]
- 5.Cascales E, et al. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James R, Penfold CN, Moore GR, Kleanthous C. Killing of E coli cells by E group nuclease colicins. Biochimie. 2002;84:381–389. doi: 10.1016/s0300-9084(02)01450-5. [DOI] [PubMed] [Google Scholar]
- 7.Bouveret E, Rigal A, Lazdunski C, Benedetti H. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol Microbiol. 1998;27:143–157. doi: 10.1046/j.1365-2958.1998.00667.x. [DOI] [PubMed] [Google Scholar]
- 8.Jakes KS, Zinder ND. Highly purified colicin E3 contains immunity protein. Proc Natl Acad Sci U S A. 1974;71:3380–3384. doi: 10.1073/pnas.71.9.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleanthous C, Walker D. Immunity proteins: enzyme inhibitors that avoid the active site. Trends Biochem Sci. 2001;26:624–631. doi: 10.1016/s0968-0004(01)01941-7. [DOI] [PubMed] [Google Scholar]
- 10.Walker D, Moore GR, James R, Kleanthous C. Thermodynamic consequences of bipartite immunity protein binding to the ribosomal ribonuclease colicin E3. Biochemistry. 2003;42:4161–4171. doi: 10.1021/bi0273720. [DOI] [PubMed] [Google Scholar]
- 11.Duche D, Frenkian A, Prima V, Lloubes R. Release of immunity protein requires functional endonuclease colicin import machinery. J Bacteriol. 2006;188:8593–8600. doi: 10.1128/JB.00941-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowman CM, Dahlberg JE, Ikemura T, Konisky J, Nomura M. Specific inactivation of 16S ribosomal RNA induced by colicin E3 in vivo. Proc Natl Acad Sci. 1971;68:964–968. doi: 10.1073/pnas.68.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senior BW, Holland IB. Effect of colicin E3 upon the 30S ribosomal subunit of Escherichia coli. Proc Natl Acad Sci. 1971;68:959–963. doi: 10.1073/pnas.68.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soelaiman S, Jakes K, Wu N, Li C, Shoham M. Crystal structure of colicin E3: implications for cell entry and ribosome inactivation. Mol Cell. 2001;8:1053–1062. doi: 10.1016/s1097-2765(01)00396-3. [DOI] [PubMed] [Google Scholar]
- 15.Carr S, Walker D, James R, Kleanthous C, Hemmings AM. Inhibition of a ribosome-inactivating ribonuclease: the crystal structure of the cytotoxic domain of colicin E3 in complex with its immunity protein. Structure Fold Des. 2000;8:949–960. doi: 10.1016/s0969-2126(00)00186-6. [DOI] [PubMed] [Google Scholar]
- 16.Zarivach R, et al. On the interaction of colicin E3 with the ribosome. Biochimie. 2002;84:447–454. doi: 10.1016/s0300-9084(02)01449-9. [DOI] [PubMed] [Google Scholar]
- 17.Walker D, Lancaster L, James R, Kleanthous C. Identification of the catalytic motif of the microbial ribosome inactivating cytotoxin colicin E3. Protein Sci. 2004;13:1603–1611. doi: 10.1110/ps.04658504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lancaster LE, Savelsbergh A, Kleanthous C, Wintermeyer W, Rodnina MV. Colicin E3 cleavage of 16S rRNA impairs decoding and accelerates tRNA translocation on Escherichia coli ribosomes. Mol Microbiol. 2008;69:390–401. doi: 10.1111/j.1365-2958.2008.06283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 20.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufmann Y, Zamir A. Protection of E. coli ribosomes against colicin E3-induced inactivation by bound aminoacyl-tRNA. FEBS Lett. 1973;36:277–280. doi: 10.1016/0014-5793(73)80390-4. [DOI] [PubMed] [Google Scholar]
- 22.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 23.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 24.Neubauer C, et al. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weixlbaumer A, et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korostelev A, et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci U S A. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma D, Cukras AR, Rogers EJ, Southworth DR, Green R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J Mol Biol. 2007;374:1065–1076. doi: 10.1016/j.jmb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 29.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 30.Yang X, Gerczei T, Glover LT, Correll CC. Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping. Nat Struct Biol. 2001;8:968–973. doi: 10.1038/nsb1101-968. [DOI] [PubMed] [Google Scholar]
- 31.Rupert PB, Ferre-D’Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 32.Boon T. Inactivation of ribosomes in vitro by colicin E 3. Proc Natl Acad Sci U S A. 1971;68:2421–2425. doi: 10.1073/pnas.68.10.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowman CM, Sidikaro J, Nomura M. Specific inactivation of ribosomes by colicin E3 in vitro and mechanism of immunity in colicinogenic cells. Nat New Biol. 1971;234:133–137. doi: 10.1038/newbio234133a0. [DOI] [PubMed] [Google Scholar]
- 34.Boon T. Inactivation of ribosomes in vitro by colicin E 3 and its mechanism of action. Proc Natl Acad Sci U S A. 1972;69:549–552. doi: 10.1073/pnas.69.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowman CM. Inactivation of ribosomes by colicin E3 in vitro: Requirement for 50 S ribosomal subunits. FEBS Lett. 1972;22:73–75. doi: 10.1016/0014-5793(72)80222-9. [DOI] [PubMed] [Google Scholar]
- 36.Ohno-Iwashita Y, Imahori K. Colicin E3 induced cleavage of 16s rRNA of isolated 30S ribosomal subunits. J Biochem (Tokyo) 1977;82:919–922. doi: 10.1093/oxfordjournals.jbchem.a131770. [DOI] [PubMed] [Google Scholar]
- 37.Twilt JC, Overbeek GP, van Duin J. Translational fidelity and specificity of ribosomes cleaved by cloacin DF13. Eur J Biochem. 1979;94:477–484. doi: 10.1111/j.1432-1033.1979.tb12915.x. [DOI] [PubMed] [Google Scholar]
- 38.Sander G. Colicin E3 treatment renders ribosomes more resistant to streptomycin and reduces miscoding. FEBS Lett. 1979;97:217–220. doi: 10.1016/0014-5793(79)80087-3. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–200. [Google Scholar]
- 40.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 42.Ogle JM, Ramakrishnan V. Structural Insights into Translational Fidelity. Ann Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.