Abstract
Intrinsic affinity tags are useful tools for the study of macromolecular targets. Although polypeptide affinity tags are routinely used in purification and detection of protein complexes, there has been a relative lack of powerful RNA affinity tags that can be embedded within RNA sequences. Here, the preparation and use of two RNA affinity tags against Sephadex or streptavidin are described. The two tags have different strengths that make them appropriate for slightly different uses. One is a high-affinity ligand for streptavidin that can be specifically eluted by competition with biotin under otherwise native binding conditions. The other tag binds selectively to Sephadex beads, and can be eluted by competition with the soluble dextran that composes Sephadex. When properly placed within another RNA molecule, the tags can be used to effect dramatic purification of RNA or ribonucleoprotein complexes from complex mixtures of cellular RNA.
Keywords: Aptamer, SELEX, RNA tag, Streptavidin, RNA purification, Ribonucleoprotein
1. Introduction
The use of protein affinity tags has long been routine for the purposes of either purification or detection of the tagged protein in such applications as Western blots or in situ microscopy. There are a variety of popular protein affinity tags that can be fused to the protein of interest, with different properties depending on the desired application. Some, like glutathione S-transferase (GST) [1] and protein A [2], are full-length proteins; others are short polypeptides, like polyhistidine [3,4] or FLAG [5]. These tags all bind with high affinity to a ligand that either can be immobilized on a chromatography resin for the purposes of purification (see also article by Rodgers et al. [25]) or can be fused to a detection system. In the best cases, the tag can be released from the affinity resin by a small molecule competitor cleaved off of the protein of interest under native (mild) conditions, allowing recovery of the protein of interest with all associated macromolecules.
The study of RNA and RNA–protein complexes (ribonucleoproteins or RNPs) can be greatly facilitated by the ability to attach analogous small RNA motifs to RNA molecules for in vivo and in vitro studies. To some extent RNA affinity tags have not been pursued vigorously because RNA can be tagged through other means. If RNA is synthesized in vitro, it can be tagged by the incorporation of biotin, fluorescent dyes, and other compounds. For detection or purification of in vivo-produced RNAs under denaturing conditions, it is possible to hybridize synthetic, tagged DNA or RNA oligonucleotides. However, for the purification or detection of native in vivo complexes it is useful to have a purely RNA affinity motif that can be incorporated during synthesis of the RNA in vivo. To some extent RNA-based affinity tags have already been identified that either use artificially selected motifs [6] or model naturally occurring RNA–protein interactions [7]. Recently, we have identified two artificially selected RNA motifs that have small, defined structures and particularly useful properties for RNP isolation. The use of both RNA motifs is described here, as they have different strengths and weaknesses. The identification of the RNA motifs by in vitro selection has been described in detail elsewhere [8,9].
Several factors were considered when deciding on target ligands for the RNA affinity tags. Cost and availability of the potential affinity resins were major concerns. The ability to elute bound RNA under native conditions without coeluting nonspecifically bound contaminants was also desired. Lastly, it was important that the affinity matrix not have a high affinity for non-specific RNAs and RNPs, providing a relatively low background. The two ligands that were used to meet these criteria were dextran B512 (in the insoluble form of Sephadex beads) and streptavidin. The relative strengths and weaknesses of these tags are summarized in Table 1, and are described in more detail in the ensuing text.
Table 1.
Advantage–disadvantage comparison of two RNA affinity tags
| D8 Sephadex RNA motif | S1 Streptavidin RNA motif | |
|---|---|---|
| Advantages | Sephadex (G-200 is best choice) is cheap and the concentration of ligand on the beads is nearly infinite; purification from large starting quantities of cell extract is practical; elution can be either with denaturants (such as urea) or by competition with soluble dextran B512 (average molecular weight 10,000) or enzymatically synthesized dextran (Mr ~ 1500). | Affinity is high for streptavidin (Kd ~ 70 nM), but not for egg white avidin (allows blocking of cellular biotin and biotinylated proteins with avidin); avidin and streptavidin reagents for affinity purification and detection are readily available from multiple commercial sources; it elutes cleanly and quickly with biotin under native conditions; binding is stable to high salt (400 mM NaCl). |
| Disadvantages | Affinity of RNA for the antigen is not as high as with the streptavidin tag, so extensive washing of the resin after binding leads to slow loss of bound RNA; native elution by competition with dextran leaves dextran in the eluate, which is harder to remove than biotin. | Resin is more expensive; number of binding sites per bead is much lower than with Sephadex; egg white avidin is usually needed to block biotin in crude cellular lysates. |
2. Description of method
2.1. The motifs that bind Sephadex and streptavidin
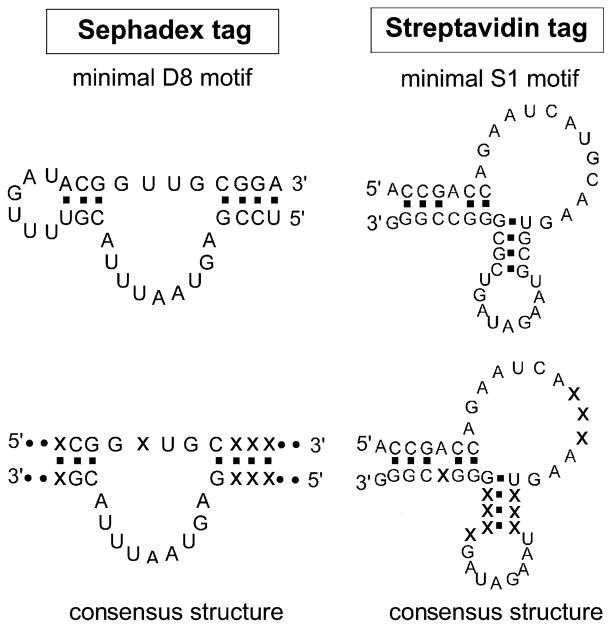
RNA affinity tags capable of binding to Sephadex and streptavidin were developed using in vitro selection or SELEX (systematic evolution of ligands by exponential enrichment) [10,11]. With this powerful technique, DNA or RNA ligands with affinity toward the desired target molecules can be selected out of random-sequence DNA or RNA libraries, and the technique has been used to identify RNA ligands (aptamers) that recognize a wide range of target molecules, from small molecules to more complex macromolecules (reviewed in [12–15]). Two types of aptamers with affinity to Sephadex or streptavidin have been developed, and they are shown to have a potential use as RNA affinity tags [8,9]. D8 and S1 aptamers are the representatives of Sephadex- and streptavidin-binding aptamers, respectively. They are recommended for use as RNA affinity tags because they have high affinities to their targets and have been extensively studied. Predicted secondary structures of D8 and S1 aptamers are shown in Fig. 1. These structures are the minimal motifs that are able to bind to Sephadex or streptavidin as efficiently as the full-length 84-nucleotide aptamers. Fig. 1 also shows the proposed consensus structures of these minimal motifs, deduced from the sequence and secondary structure analyses of the aptamer sequences belonging to multiple aptamer isolates with similar properties.
Fig. 1.
Minimal binding motifs and consensus structures of Sephadex-and streptavidin-binding RNA affinity tags. The actual sequences of the D8 and S1 tags are shown, along with the consensus structures showing conserved stem and loop regions versus conserved nucleotide identities (× indicates nonconserved nucleotides).
The D8 Sephadex-binding RNA minimal motif has 33 nucleotides (5′ UCCGAGUAAUUUACGUUUU GAUACGGUUGCGGA 3′). Since the tag was originally published [8], the indicated minimal structural motif has been discovered. D8 tag was shown to bind specifically to Sephadex G-100 (Pharmacia), which is a commonly used gel filtration matrix, produced by crosslinking of dextran B512 with epichlorohydrin to form beads [16,17]. In contrast, the D8 tag does not bind to other similar matrices such as Sepharose and Seph-acryl. This is an important consideration when combining a Sephadex affinity step with additional steps that might involve other types of chromatography resin or solid supports. The binding of the tag to Sephadex can be efficiently competed with dextran B512, which is a homopolymer of glucose connected mainly via α-1,6 glucosidic linkages (95%) and occasional α-1,3 linkages (5%) [16]. Although the D8 tag was originally selected using Sephadex G-100 as a target, it can bind Sephadex G-200 with equal efficiency. When D8 is used as an affinity tag to purify large RNAs/RNPs, Sephadex G-200 (40–120 μm in bead diameter), which has a fractionation range of 5–600 kDa, is a recommended affinity resin because, for unknown reasons, these beads bind larger complexes more efficiently than Sephadex beads having smaller pore sizes such as Sephadex G-10–G-50. (We suspect that the interior of the beads can be used with the larger pore size. It is possible that when a complex dissociates from an internal bead site, it has an increased tendency to rebind because it is transiently trapped inside the bead.)
The S1 streptavidin-binding RNA motif has 44 nucleotides (5′ ACCGACCAGAAUCAUGCAAGUGCG UAAGAUAGUCGCGGGCCGGG 3′) and was originally selected to bind to streptavidin in either streptavidin–agarose bead assays or polyacrylamide gel electrophoretic mobility shift assays [9]. Streptavidin is a homotetrameric protein from Streptomyces avidinii, that can bind to its natural ligand, d-biotin, with an extraordinarily high affinity [Kd ~ 10−14 M] [18]. S1 tag binds to streptavidin with a Kd of ~70 nM. Binding can be disrupted in the presence of d-biotin, probably because the binding site might be at or near biotin-binding pockets of streptavidin or the conformation of biotin-bound streptavidin might not be recognizable by the RNA tag. Bound RNA tags can thus be released from streptavidin under otherwise native binding conditions by the inclusion of biotin in the binding buffer. Because of an unusually high affinity of biotin to streptavidin, elution of the RNA tag with biotin is very efficient and complete. Once formed, the biotin/streptavidin interaction is essentially irreversible, preventing any rebinding of the RNA back to streptavidin. In addition, because of its small size (244.31 Da), biotin can be easily removed from the eluate fractions by dialysis or ultrafiltration.
A useful feature of the S1 tag is that binding is weak or nonexistent to egg white avidin, a protein that also binds biotin very tightly. This property is particularly helpful when purifying tagged RNPs from crude cellular extracts. Cells tend to contain both free biotin and biotinylated proteins, which would irreversibly bind to streptavidin and block the binding of the RNA tag. Competition from biotin can be reduced by incubating the extracts with avidin shortly prior to the binding step, to allow biotin to be absorbed by avidin instead of streptavidin. Alternatively, it is possible to remove most of the free and protein-associated biotin by using a conventional purification step before the streptavidin–agarose affinity step.
2.2. Tagging the RNA to be purified
There are three main considerations when deciding where to tag the RNA of interest: (i) folding, (ii) steric blockage, and (iii) keeping the tag on the RNA prior to purification. Each of these issues is discussed briefly below, but in short, it is recommended that the RNA affinity tag be inserted either at one end of the RNA or into the terminal loop of a long, nonconserved stem that is known to protrude into solution (see examples in Fig. 2). The folding problem is “simply” a matter of inserting the tag in such a way that both the tag and the RNA of interest remain correctly folded. RNA folding algorithms are usually used to help predict the folding of the RNA tags and recipient RNAs to obtain the tagged RNA with proper folding. However, such predictions are sometimes not reliable and it is often necessary to generate and test several tagged-RNA constructs to ensure that they have normal biological functions and are still able to bind to the affinity matrix. The steric blockage problem arises when the tag is partially or completely covered by either the folded structures of the RNA or its associated protein subunits, thus obstructing access of the tag to the affinity matrix. Therefore, if the information about the structure or accessibility of the RNA of interest is available (e.g., from RNA footprinting study), it will be very helpful in choosing the insertion site that is less likely to have the steric blockage problem. However, if the information is not available, insertion into a protruding stem in predicted structures might avoid the steric hindrance. It is sometimes also useful to place a short spacer between the tag and the main body of the RNA, whether placing it in a stem or at the end of the RNA. A complete folding check is recommended to ensure that the spacer also does not interfere with folding of the tag or the main RNA.
Fig. 2.
Affinity tag insertions into the RNA subunit of ribonuclease P. Both the D8 Sephadex tag and the S1 streptavidin tag have been inserted into the yeast RPR1 gene encoding the RNA subunit of the nuclear tRNA processing enzyme, RNase P. The minimal tag sequences are shown in boxes, although in the case of S1, the larger, original tag was used [9]. Tags were inserted into nonessential loops at the end of stems that were known to protrude into solution. Both insertions gave correct assembly of functional enzyme in vivo, allowing purification of the enzyme from cells containing the modified gene as the only source of RPR1 RNA.
The last problem, keeping the tag on the RNA, refers to placing the tag in such a way that it is not removed by either RNA processing in the cell or nucleases in cell-free extracts. The solution to this is dependent on the particular RNA and expression system, but there are some generic solutions. Degradation of some transcripts by 3′-5′-exonucleases is especially rapid; the tag might be removed when it is placed at the 3′ end of the RNA, making it unusable. This problem can be prevented by placing a strong stem (GC-rich, with a tetraloop) at the 3′ end of the transcript, which makes the end of the transcript more resistant to the nucleases. The tag is inserted internal to this sealing stem. The other way to avoid this problem is to place the tag at the terminal loop of an internal stem that is accessible in solution and does not otherwise serve a function (see tagging of RPR1 RNA below). From our experience, the tags inserted in these regions seem to be quite stable.
2.3. Isolation of tagged complexes
The Sephadex and streptavidin binding motifs were originally identified under roughly physiological solution conditions (50 mM Hepes, pH 7.4, 10 mM MgCl2, 100 mM NaCl), so that the tags could be used for isolation of RNAs or RNP complexes formed in vivo. However, it seems that both tags are able to bind to the corresponding ligands under a variety of solution conditions other than the binding buffer solution used during the aptamer selections. For example, the S1 aptamer can stably bind to streptavidin in a solution containing up to 400 mM NaCl. At this concentration, nonspecific binding of proteins to the affinity matrix can be reduced, resulting in a less contaminated eluate. Therefore, different buffer compositions (e.g., lower or higher salt concentration, addition of glycerol or detergents) should be tested and might be used successfully as binding buffer with the aptamer tags to suit specific requirements of various systems.
Crude lysates containing the tagged complexes can be used directly to bind to the affinity matrix without prior purification steps. The amount of affinity matrix used to isolate the tagged complexes should really be determined experimentally, but as a rough guide to purification of complexes from crude yeast lysates, approximately 10–20 μl Sephadex G-200 (Pharmacia) or streptavidin–agarose (Sigma) is used for each milligram of protein in the lysates. The binding step is usually carried out at 4 °C for 1 h. For isolation of S1-tagged complexes, incubation of the lysates with egg white avidin before the binding step is recommended, especially for yeast lysates. Generally, 5–20 μg of egg white avidin (Sigma) is incubated with each milligram of protein for 10 min at 4 °C before incubating with streptavidin–agarose. However, if the highest yield is desired, it may be helpful to experimentally determine the amount of avidin needed to absorb biotin in the lysates because the level of free biotin and biotinylated proteins may vary widely in different lysates.
Binding with affinity matrices is usually performed at 4 °C for 1 h, followed by washing with binding buffer for 15–30 min. Because the tags, particularly the S1 tag, bind efficiently to the corresponding targets, the duration of wash may be increased up to 2–3 h with only a slight loss of the tagged complexes. Therefore, the duration of the washing step or the amount of buffer used can be varied depending on the desired purity of the eluted product, stability of the target complex, or convenience. For S1-tagged complexes, the elution step is accomplished under either denaturing or native conditions by incubating with 8 M urea or binding buffer containing 5 mM d-biotin (Sigma), respectively, at 4 °C for 30 min. The D8-tagged complexes can also be eluted with urea or competed off Sephadex with 2 resin vol of binding buffer containing 50 mg/ml of dextran B512 for 30 min at 4 °C. Alternatively, binding buffer containing lower-molecular-weight dextran (enzymatically synthesized dextran, Mr ~ 1500, from Fluka) at 100 mg/ml can be used to elute the D8-tagged complexes with efficiency comparable to that of dextran B512. Lower-molecular-weight dextran can be removed more easily from the elution fractions than larger dextran B512.
2.4. Examples of RNP isolations from crude mixtures
We previously reported examples of using tags to recover either the tag (alone) from total cellular RNA (deproteinized) or ribonucleoprotein complexes from crude cellular lysates [8,9]. Here we directly compare the one-step isolation of a single yeast RNP, ribonuclease P (RNase P), after tagging the large RNA subunit with either the D8 Sephadex aptamer or the S1 streptavidin aptamer. RNase P has been purified by conventional means from yeast nuclei [19] and shown to have nine tightly associated protein subunits, in addition to the RNA subunit, RPR1 RNA. Yeast are estimated to have only 200–400 molecules of the unstable enzyme per cell, so isolation is challenging. In addition, the large number of tightly bound subunits suggests that, like the nuclear RNA polymerases, the functional form of RNase P might be more transiently associated with additional “holoenzyme” proteins.
Fig. 2 shows the positions in the RPR1 RNA at which the minimal D8 Sephadex tag or the S1 streptavidin tag was inserted. The approximate structure of the RPR1 RNA had previously been experimentally determined [20,21], and it was determined that the sequences at both insertion sites were nonessential [22,23]. Recombinant genes containing either the S1 streptavidin aptamer plus the D8 Sephadex aptamer or the S1 streptavidin aptamer alone were created using in vitro overlap-extension PCR [9,24; Srisawat and Engelke, manuscript in preparation]. The recombinant genes were inserted into a low-copy plasmid in a haploid Saccharomyces cerevisiae strain (W3031A) in which the wild-type chromosomal copy of the RPR1 gene had been deleted.
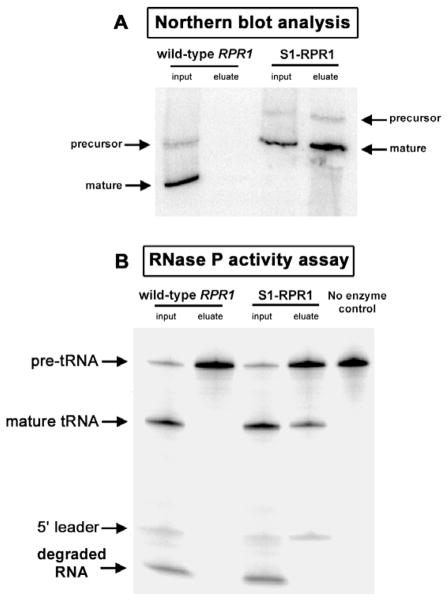
Yeast lysates from either the S1- plus D8-tagged RPR1 or the S1-tagged RPR1 strain were subjected to affinity isolation using Sephadex or streptavidin–agarose followed by gentle elution with biotin or dextran, respectively. Both Sephadex and streptavidin tags enable the specific and rapid isolation of tagged RNase P from crude yeast lysates as shown by Northern blot analyses of RPR1 RNA (Figs. 3A, 4A). In addition, native elution conditions using binding buffer containing dextran or biotin result in a recovery of intact and active enzyme as shown from RNase P activity assays (Figs. 3B, 4B); the eluted enzyme can cleave pre-tRNA substrates into 5′ leader and mature tRNA products. The enzyme in the eluates is less contaminated with nonspecific nucleases compared with that in the crude lysates, which show a nonspecific degradation of radiolabeled substrates into smaller RNA by abundant ribonucleases present in the crude lysates. RNase P-recovered in the eluates using either tag is about 20–30% of the total RNase P in the starting material, and the enrichment of RNase P-specific activity over crude extracts can be as high as 2500-fold, depending on the stringency of purification. This yield is reasonable considering that RNase P is present in very low amounts and the holoenzyme is isolated directly from the crude yeast lysates, that contain an abundance of nonspecific competitors for resin and RNA binding. Near-quantitative yields of tagged RNA can be achieved in purified systems in which there is less nonspecific binding competition.
Fig. 3.
Isolation of RNase P using the D8 Sephadex tag. RNase P in crude lysates from either wild-type or D8-tagged RPR1 strains was isolated using Sephadex as follows: Briefly, 5 mg of crude lysates was incubated with 100 μl of Sephadex G-200 beads at 4 °C for 1 h in binding buffer (50 mM Hepes, pH 7.4, 10 mM MgCl2, 100 mM NaCl, 1 mM DTT, 0.1% Triton X-100, and 10% glycerol). The beads were then washed with 1000 μl of binding buffer for five times, 3 min each time, before being eluted with 200 μl of binding buffer containing 50 mg/ml dextran for 30 min at 4 °C. Northern blot analysis of RPR1 RNA and the RNase P activity assays were performed as described [9]. (A) Northern blot analysis shows the presence of RPR1 RNA, which is present as mature and precursor forms in Saccharomyces cerevisiae, in yeast lysates from both the wild-type and D8-tagged RPR1 strains. Only RNase P RNA containing D8-tagged RPR1 RNA can be specifically recovered. (B) RNase P activity assays show that the D8-tagged RNase P can be gently eluted from Sephadex with dextran, while its enzymatic activity is still preserved. The active enzyme is shown to cleave the pre-tRNA substrates into 5′ leader and mature tRNA products. Note the high level of degradation of radiolabeled substrates and products, particularly the 5′ leader, into smaller RNA or nucleotides in the yeast lysates due to a high abundance of contaminating nucleases, which is largely removed in the eluate fraction. The asterisk shows positions of nonspecific degradation products of pre-tRNA substrates, probably due to the contaminating nucleases in the binding buffer containing dextran used during the elution step.
Fig. 4.
Isolation of RNase P using the S1 streptavidin tag. Soluble extracts from either the wild-type or S1-tagged RPR1 strains were used for RNase P isolation using streptavidin–agarose, followed by elution using binding buffer containing biotin as previously described in [9]. (A) The presence of RPR1 RNA in the input and the eluate fractions was analyzed by Northern blot analysis. Only RNase P containing the S1-tagged RPR1 RNA can be isolated using streptavidin–agarose. (B) RNase P activity assays show that the S1-tagged RNase P, which is gently eluted with biotin, is also enzymatically active. Reproduced, with permission, from Srisawat and Engelke [9].
Final purification of the RNase P from crude extracts has not yet been achieved using only one of the affinity tags, possibly because complete purification of the enzyme is expected to require at least 100,000-fold enrichment over crude cell lysate [19]. It should be possible to double-tag an RNA with the D8 and S1 aptamers and perform two sequential purification steps to attain high levels of purity in a matter of a few hours.
3. Concluding remarks
Here we have shown examples of using Sephadex and streptavidin RNA affinity tags to provide rapid and substantial enrichment of RNP complexes from cellular lysates under mild conditions. Because the isolation of a tagged RNP is done entirely under native elution conditions over short periods, the complexes might be recovered complete with loosely associated proteins. This could enable the identification of the interacting proteins that might otherwise be lost during a lengthy conventional purification. In addition, the RNA tags can be used to rapidly and specifically isolate a particular precursor or product form of RNA of interest. For example, to study the precursor of the RNase P holoenzyme, an RNA tag has been placed at the 5′ leader of RPR1 RNA, enabling separation of the precursor from the mature form of the holoenzyme (Srisawat and Engelke, in preparation). Another potential use for the RNA affinity tags is for specific isolation and characterization of RNAs or RNPs containing lethal mutations. Cells harboring such mutants are unable to grow unless the wild-type gene is present. To overcome this problem, tags can be added exclusively to the mutant RNA in cells carrying a wild-type gene. After affinity purification with Sephadex or streptavidin–agarose, only the tagged, mutant RNAs are specifically isolated from the corresponding wild-type RNA or RNP, making it possible to study the subunit composition and function of those lethal mutants.
Although affinity isolation of RNPs is useful, it should also be possible to use tags for a variety of additional purposes. Examples include isolation of deproteinized RNA species from crude cellular RNA, tagging positions on macromolecular complexes, and detecting either at a molecular level with colloidal gold–streptavidin or at a subcellular level with fluorescent protein–streptavidin fusions.
References
- 1.Simons PC, Vander Jagt DL. Methods Enzymol. 1981;77:235–237. doi: 10.1016/s0076-6879(81)77031-9. [DOI] [PubMed] [Google Scholar]
- 2.Stahl S, Nygren PA. Pathol Biol. 1997;45:66–76. [PubMed] [Google Scholar]
- 3.Porath J, Carlsson J, Olsson I, Belfrage G. Nature. 1975;258:598–599. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- 4.Porath J. Protein Expression Purif. 1992;3:263–281. doi: 10.1016/1046-5928(92)90001-d. [DOI] [PubMed] [Google Scholar]
- 5.Prickett KS, Amberg DC, Hopp TP. BioTechniques. 1989;7:580–589. [PubMed] [Google Scholar]
- 6.Bachler M, Schroeder R, von Ahsen U. RNA. 1999;5:1509–1516. doi: 10.1017/s1355838299991574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das R, Zhou Z, Reed R. Mol Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- 8.Srisawat C, Goldstein IJ, Engelke DR. Nucleic Acids Res. 2001;29:e4. doi: 10.1093/nar/29.2.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srisawat C, Engelke DR. RNA. 2001;7:632–641. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 11.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 12.Jayasena SD. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 13.Wilson DS, Szostak JW. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 14.Hermann T, Patel DJ. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 15.Patel DJ, Suri AK, Jiang F, Jiang L, Fan P, Kumar RA, Nonin S. J Mol Biol. 1997;272:645–664. doi: 10.1006/jmbi.1997.1281. [DOI] [PubMed] [Google Scholar]
- 16.Robyt JF. Essentials of Carbohydrate Chemistry. Springer-Verlag; New York: 1998. [Google Scholar]
- 17.Misaki A, Torii M, Sawai T, Goldstein IJ. Carbohydr Res. 1980;84:273–285. [Google Scholar]
- 18.Green NM. Methods Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 19.Chamberlain JR, Lee Y, Lane WS, Engelke DR. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tranguch AJ, Engelke DR. J Biol Chem. 1993;268:14045–14055. [PubMed] [Google Scholar]
- 21.Tranguch AJ, Kindelberger DW, Rohlman CE, Lee JY, Engelke DR. Biochemistry. 1994;33:1778–1787. doi: 10.1021/bi00173a022. [DOI] [PubMed] [Google Scholar]
- 22.Pagan-Ramos E, Tranguch AJ, Kindelberger DW, Engelke DR. Nucleic Acids Res. 1994;22:200–207. doi: 10.1093/nar/22.2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindahl L, Fretz S, Epps N, Zengel JM. RNA. 2000;6:653–658. doi: 10.1017/s1355838200992574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling MM, Robinson BH. Anal Biochem. 1997;254:157–178. doi: 10.1006/abio.1997.2428. [DOI] [PubMed] [Google Scholar]
- 25.Rodgers ND, Jiao X, Kiledjian M. Methods. 2002;26 doi: 10.1016/S1046-2023(02)00014-2. [DOI] [PubMed] [Google Scholar]