Summary
Background
Mutations in the pyrazinamidase (PZAse) coding gene, pncA, have been considered as the main cause of pyrazinamide (PZA) resistance in Mycobacterium tuberculosis. However, recent studies suggest there is no single mechanism of resistance to PZA. The pyrazinoic acid (POA) efflux rate is the basis of the PZA susceptibility Wayne test, and its quantitative measurement has been found to be a highly sensitive and specific predictor of PZA resistance. Based on biological considerations, the POA efflux rate is directly determined by the PZAse activity, the level of pncA expression, and the efficiency of the POA efflux pump system.
Objective
This study analyzes the individual and the adjusted contribution of PZAse activity, pncA expression and POA efflux rate on PZA resistance.
Methods
Thirty M. tuberculosis strains with known microbiological PZA susceptibility or resistance were analyzed. For each strain, PZAse was recombinantly produced and its enzymatic activity measured. The level of pncA mRNA was estimated by quantitative RT-PCR, and the POA efflux rate was determined. Mutations in the pncA promoter were detected by DNA sequencing. All factors were evaluated by multiple regression analysis to determine their adjusted effects on the level of PZA resistance.
Findings
Low level of pncA expression associated to mutations in the pncA promoter region was observed in pncA wild type resistant strains. POA efflux rate was the best predictor after adjusting for the other factors, followed by PZAse activity. These results suggest that tests which rely on pncA mutations or PZAse activity are likely to be less predictive of real PZA resistance than tests which measure the rate of POA efflux. This should be further analyzed in light of the development of alternate assays to determine PZA resistance.
Keywords: Mycobacterium tuberculosis, POA efflux rate, PZAse activity, pncA expression level, PZA resistance
1. Introduction
Tuberculosis (TB) remains one of the major causes of disease and death worldwide, exacerbated by HIV-TB co-infection 1 and the emergence of multidrug-resistant TB (MDR-TB) and extensively drug-resistant TB strains in industrialized and developing countries.2 The World Health Organization (WHO) defines MDR-TB as tuberculosis that is resistant to both isoniazid and rifampicin, two of the four first line anti-tuberculous drugs. Pyrazinamide (PZA) is one of the most important drugs in both first- and second-line treatment of TB.3 It is particularly effective against latent TB infection and allows anti-tuberculous treatment to be shortened from 9 to 6 months.4–6 Therefore the emergence of PZA resistant strains represents an important public health problem. In Peru, about 50% of MDR-TB clinical strains are resistant to PZA.7
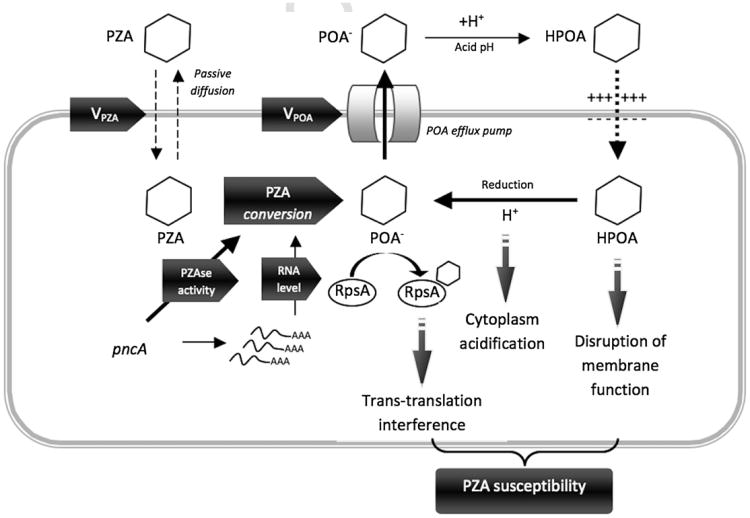
The mechanisms of action and resistance to PZA in Mycobacterium tuberculosis are not entirely understood.8 PZA is a pro-drug, which enters the mycobacteria by passive diffusion and is transformed in the cytoplasm to pyrazinoic acid (POA) by a nicotinamidase that also has PZAse activity.8,9 Pyrazinoic acid, the active drug, accumulates in the cytoplasm and is expelled from the bacilli by an efflux pump that has yet to be identified. In the acidic environment outside the bacilli, POA is protonated and then re-enters the mycobacteria. Once inside the bacilli again, the protons are released, acidifying the cytoplasm10 and allowing POA to accumulate. As a result, mycobacterial membrane permeability and transport are disrupted, causing cellular damage8,11,12 (Figure 1). A recent study identified the ribosomal protein S1 (RpsA), particularly important in latent stage, as a POA target. RpsA is involved in the ribosome-sparing process of trans-translation. POA binds to RpsA resulting in trans-translation inhibition, while it is likely that mutations in RpsA can cause PZA resistance.13
Figure 1.
Schematic representation of the pyrazinamide mechanism of action in Mycobacterium tuberculosis. Pyrazinamide (PZA) is a pro-drug that enters M. tuberculosis through passive diffusion. In the cytoplasm, pyrazinamidase (PZAse) converts PZA to its active form pyrazinoic acid (POA). This conversion is affected by the PZAse activity and the level of pncA expression. POA is pumped out of the mycobacteria through a potential POA efflux pump. Under extracellular acidic conditions, POA is protonated to POAH, which re-enters the bacteria along a potential gradient. Inside the cell, POAH releases its proton. This cycle is repeated resulting in POA accumulation and intracellular acidification. The outcome is a lethal disruption of membrane permeability. A recent study identified that POA interferes with the ribosome-sparing process of trans-translation. POA binds to the Ribosomal protein S1 (RpsA), competing with the tmRNA and inhibiting trans-translation. Vpza, PZA flux rate; Vpoa, POA efflux rate; H+, released proton.
The primary mechanism of PZA resistance according to several studies is the loss of PZAse activity due to mutations in pncA, the PZAse coding gene.9,11,12,14–19 It has been reported that PZA resistance is also associated with an elevated POA efflux, which despite cytoplasmic acidification prevents POA accumulation as occurs in naturally PZA resistant Mycobacterium smegmatis.10,20 Recently, other studies have reported that the POA efflux rate was more predictive of PZA resistance (61%) than the PZAse activity with a high sensitivity and specificity, making it an important predictor of PZA resistance in M. tuberculosis.19–21 This evidence suggests that other factors should be present to account for the remaining unexplained variability of PZA resistance. Although it has not been proved, it is evident and likely that the POA efflux rate at least depends on the PZAse activity, the intracellular PZAse concentration, the efficiency of the efflux pump system, and the concentration of efflux pumps. Whilst the importance of the POA efflux rate to the susceptibility to PZA is recognized and its quantitative effect has been shown to be significant,19–21 the identity of the POA efflux pump system is still unknown.
PZAse/nicotinamidase is a ubiquitous enzyme present in prokaryotes and is expressed constitutively in Mycobacterium tuberculosis,11,12,22,23 Eschericia coli,24–26 Salmonella typhimurium,25 Torula cremoris26 and Borrelia burgdorferi.27 The physiological role of nicotinamidase is to convert nicotinamide to nicotinic acid mononucleotide. Adenylation of this mononucleotide followed by amide formation completes the biosynthesis of Nicotinamide (NAD). NAD and NAD-phosphate (NADP) are essential compounds in over 300 biochemical redox reactions.25 Although PZAse is constitutively expressed in M. tuberculosis, alterations of the pncA expression could impair intracellular PZAse activity and as a consequence the POA efflux rate, resulting in PZA resistance. Mutations in pncA promoter or silent mutations that switch codons and are associated with low levels of tRNA may impair pncA expression and reduce the PZAse intracellular concentration.
In this study we measured concurrently the level of pncA expression, POA efflux rate, and the PZAse activity, from multiple strains and evaluated their adjusted effects on PZA susceptibility. Combining these molecular and biochemical factors in the context of a multivariate analysis allows us to better understand the genetic basis of PZA susceptibility in M. tuberculosis.
2. Materials and methods
2.1. M. tuberculosis isolates
Thirty M. tuberculosis clinical isolates including H37Rv, a PZA susceptible reference strain, were analyzed in this study. Ten strains were pncA wild type, and 20 were pncA mutated (relative to the H37Rv pncA sequence). All isolates had the pncA gene and its promoter region (up to position −100 bp) previously sequenced.19
From the 20 pncA mutated strains, 14 isolates were selected from our strains collection and previously described 19–21 and the remaining 6 were new isolates. From the 10 pncA wild type, 7 isolates were selected from our strain collection because they were resistant to PZA. The other 3 isolates were sensitive to PZA and have been previously described20,21 (Table 1).
Table 1.
Pyrazinamide susceptibility and PZAse-related biochemical parameters of Mycobacterium tuberculosis strains.
| PZAse mutation | Promoter mutation | PZA-MIC 7H9 | Wayne activity | Bactec-growth index% | PZAse activity | pncA expression level | VPOA | POA efflux pump efficiency |
|---|---|---|---|---|---|---|---|---|
| D12A | No | 400(R) | Neg(R) | 77%(R) | 9.24 | 0.67 ±0.11 | 3.61 | 0.58 |
| D12G | No | 400(R) | Neg(R) | 70%(R) | 14 | 1.66 ±0.38 | 1.53 | 0.07 |
| D136G | No | 50(S) | Pos(S) | 3%(S) | 12.34 | 0.32 ±0.03 | 7.30 | 1.85 |
| D49N | No | 400(R) | Neg(R) | 74%(R) | 0.05 | 0.46 ±0.07 | 0.65 | 28.26 |
| F94L | No | 400(R) | Neg(R) | 70%(R) | 21.19 | 0.47 ± 0.07 | 4.18 | 0.42 |
| G24D | No | 200(R) | Neg(R) | 68%(R) | 4.28 | 0.63 ± 0.06 | 0.72 | 0.27 |
| G78C | No | 100(R) | Neg(R) | 27%(R) | 6.96 | 0.47 ± 0.01 | 6.39 | 1.95 |
| H51R | No | 800(R) | Neg(R) | 90%(R) | 0.01 | 8.07 ±2.87 | 0.45 | 5.58 |
| K48T | No | 50(S) | Pos(S) | 20%(R) | 10.45 | 0.41 ±0.05 | 1.33 | 0.31 |
| L4S€ | No | 400(R) | Neg(R) | 99%(R) | 0.02 | 4.21 ± 1.18 | 1.07 | 12.71 |
| P54L€ | No | 400(R) | Neg(R) | 86%(R) | 5.53 | 3.47 ± 1.51 | 0.69 | 0.04 |
| P62L€ | No | 400(R) | Neg(R) | 100%(R) | 10.13 | 1.66 ± 1.01 | 0.67 | 0.04 |
| Q10P€ | No | 900(R) | Neg(R) | 91%(R) | 1.98 | 0.35 ± 0.05 | 0.74 | 1.07 |
| R29P€ | No | ND | Pos(S) | 0%(S) | 2.76 | 0.34 ± 0.06 | 3.54 | 3.77 |
| T135P | No | 800(R) | Neg(R) | 98%(R) | 0.02 | 0.50 ± 0.06 | 0.67 | 67.00 |
| W119L€ | No | 900(R) | Neg(R) | 95%(R) | 0.88 | 0.33 ±0.15 | 2.18 | 7.51 |
| Y34D | n.a. | 100(R) | Neg(R) | 78%(R) | 20.58 | 0.85 ± 0.32 | 1.42 | 0.08 |
| L85P€ | n.a. | 900(R) | Neg(R) | 90%(R) | * | 4.39 ± 1.75 | 0.96 | |
| Y64D€ | No | ND | Pos(S) | 100%(R) | 57.83 | 0.35 ±0.17 | 3.90 | 0.19 |
| L116P | No | 100 | Neg(R) | 46%(R) | 50.15 | 0.22 ± 0.02 | 4.39 | 0.40 |
| WT3 (H37Rv) | No | 50(S) | Pos(S) | 1%(S) | 38.4 | 1(ref) | 15.98 | 0.42 |
| WT14 | No | 900(R) | Neg(R) | 69%(R) | Iden. | 0.06 ± 0.02 | 1.45 | 0.63 |
| WT15 | No | 900(R) | Neg(R) | 63(R) | Iden. | 0.43 ± 0.25 | 3.07 | 0.19 |
| WT16 | Yes (−11) | ND | Neg(R) | 88(R) | Iden. | 0.22 ±0.10 | 0.25 | 0.03 |
| WT4 | No | 50(S) | Pos(S) | 1(S) | Iden. | 2.2 ±0.41 | 6.91 | 0.08 |
| WT5 | No | 50(S) | Pos(S) | 1(S) | Iden. | 0.46 ± 0.07 | 14.42 | 0.82 |
| WT6 | No | ND | Pos(S) | 68(R) | Iden. | 6.87 ±2.15 | 2.04 | 0.01 |
| WT7 | No | ND | Pos(S) | 32(R) | Iden. | 0.23 ±0.01 | 2.58 | 0.29 |
| WT8 | No | 800(R) | Neg(R) | 53(R) | Iden. | 0.003 ± 0.002 | 1.76 | 18.33 |
| WT9 | Yes (−11) | ND | Neg(R) | 53(R) | Iden. | 0.20 ±0.10 | 5.92 | 0.77 |
WT: pncA wild type strain; ND: non-determined; Iden: PZAse activity Identical to H37RV; S: PZA susceptible; and R: PZA resistant; and PZA-MIC is measured in PZA μg/ml, lower than 100 μg/ml is considered PZA susceptible.
PZAse activity is measured in μmol POA min−1 mg−1 PZAse.
VPOA is measured in nmol POA mg−1 protein min−1. The pncA expression level represents the average of two biological replicates and the total range of variation.
PZAse expressed as an insoluble protein without detectable activity.
New strain reported.
2.2. PZA susceptibility testing
PZA resistance was determined by Bactec, which is considered as the gold standard. In addition, PZA susceptibility was also tested by Wayne activity, and 7H9 culture, as previously described.19 Briefly, mycobacterial growth was estimated by measuring the release of 14CO2 under PZA inhibition (100 μg/ml) at pH 6.0. The Bactec-growth index (BGI) estimated the PZA resistance level by the radioactive-growth index, which is the ratio of radioactivity of the media containing the bacteria and PZA, and the media containing the bacteria without PZA, expressed as a percentage. Less than 20% was considered as an indicator of a PZA susceptible strain.
The 7H9 culture broth enriched with ADC (albumin, dextrose and catalase) at pH 6.0 was used to determine the PZA minimum inhibitory concentration (MIC). A MIC less than 100 μg/ml of PZA was considered as an indicator of a PZA susceptible strain.9
The Wayne activity, a qualitative colorimetric method, detects POA released by the bacilli into the culture media.28 The presence of a reddish color indicates PZAse activity and was considered as an indicator of PZA susceptibility, whereas negative reaction was considered as an indicator of PZA resistance.29
2.3. Cloning of the M. tuberculosis pncA gene
Expression vectors containing pncA gene were used to express PZAse from previous studies.19–21 L4S, P54L, P62L, Q10P, R29P, L85P and Y64D PZAses were obtained by cloning the pncA gene according to Sheen et al.19 Briefly, The entire pncA gene was amplified by PCR using oligo-nucleotides with restriction sites for NcoI and XhoI (5′ CCC CCA TGG GCC GGG CGT TGA TCA TC 3′ and 5′ CCC CTC GAG GGA GCT GCA AAC CAA CTC 3′). The purified pncA gene amplification (575 bp) was double digested and inserted in the pET28a plasmid using T4 DNA ligase (NEBioLabs, Ipswich, MA). For purification, a hexahistidine tag was added to the carboxy-terminal end. Escherichia coli Novablue cells (Novagen, San Diego, CA) were transformed using heat shock protocol. Plasmid DNA was extracted and sequenced to ensure that the pncA gene is in-frame with the appropriate hexahistidine tag.
2.4. Expression and purification of the recombinant PZAses
The recombinant PZAses were expressed and purified as described previously.19 Briefly, the PZAses were expressed in E. coli BL21(DE3)pLysS cells (Novagen) under induction of 1 mM isopropyl ß-d-thiogalactoside for 4h at 37 °C. The cells were harvested by centrifugation at 4830×g at 4 °C for 10 min. The pellet was resuspended in 20 ml of binding buffer (20 mM imidazole, 0.5 M NaCl and 20 mM phosphate buffer pH 7.4) and lysed by repeated freeze-thaw cycles followed by sonication. After centrifugation at 17,572×g at 4 °C for 30 min the supernatant was loaded on a 5 ml His-bind column (Pharmacia, Piscataway, NJ) and washed with 40 mM imidazole, 0.5 M NaCl and 20 mM phosphate buffer, pH 7.4. The bound protein was eluted with 60 mM imidazole, 0.5 M NaCl and 20 mM phosphate buffer, pH 7.4. Aliquots of the fractions obtained were analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The purified protein was concentrated and then washed three times with 20 mM Tris–HCl, pH 7.9 by ultrafiltration at 4 °C. The protein concentration was determined according to the method of Bradford.20
2.5. PZAse enzymatic activity
PZAse activity was measured as previously described.19 Briefly, different concentrations of PZA (0–4 mM) were hydrolyzed with 1 μM of the recombinant PZAse. POA production was evidenced with addition of 20% FeNH4(SO4)2 and the reaction was stopped with 0.1 M glycine–HCl (pH 3.4) after 1 min reaction. The absorbance was measured at 450 nm and OD was adjusted to a standard curve of known concentrations. Each recombinant PZAse was tested at least three times. The enzymatic activity was estimated as the amount of POA produced in a 1 min reaction divided by the total amount of enzyme.
2.6. POA efflux rate and PZA flux rate
Both POA efflux and PZA flux rates were measured as previously described.20 In brief, approximately 2 mg of bacterial dry weight was incubated with 1 mM of PZA at 37 °C and for different times (12, 24, 36 and 48 h). The extracellular fraction was recovered after centrifugation and heated at 100 °C for 20 min to deactivate any remaining bacteria. The bacterial sediment was suspended in 10 mM citrate buffer (pH 6.2) and heated at 100 °C for 20 min to lyse bacteria (intracellular fraction). Samples were then centrifuged and supernatants were stored at 4 °C. The protein concentration in the supernatant was measured by the Bradford method. Initial POA was measure in both fractions using FeNH4(SO4)2, and color absorbance was read at 450 nm. PZA from both fractions was converted to POA by incubation with 1 μM PZAse (final concentration) at 37 °C for 1 h, and then final POA was measured. Each measurement was done in duplicate. The difference between final POA and initial POA represents PZA concentration. The rate at which PZA enters the bacteria and the rate at which POA is released from the bacteria (POA efflux rate) were estimated by the linear regression coefficient of a second order linear regression curve of POA concentration vs. time. Rates were estimated in nmol of POA or PZA per milligram of protein per minute.
2.7. Efficiency of the POA efflux pump system
The intrinsic efficiency of the POA efflux pump system (EffPOApump) was estimated as the POA efflux rate (VPOA) normalized by the PZAse activity (ActivityPZase) and the pncA mRNA level (mRNApncA), which is a close estimate of the intracellular PZAse concentration. Being defined in this way, the efficiency of the POA efflux pump is a non-dimensional parameter (i.e. it does not have a specific unit).
This estimation is based under the assumption that the POA efflux rate is directly proportional to the PZAse activity, the PZAse intracellular concentration, and the intrinsic efficiency of the POA efflux pump system. Moreover, we consider that the mRNA level is a suitable indicator of the intracellular concentration of the PZAse by assuming that no bias occurs during pncA translation.
2.8. pncA RNA isolation
In order to minimize alterations of the pncA expression level we tested all strains in the same growth phase. M. tuberculosis strains were reactivated in 7H10 agar for three weeks. One aliquot (loop) was harvested to a tube containing 7H9 medium and incubated for one week. The cultures were adjusted to McFarland 2 in a 10 ml-bacterial solution and the bacteria were separated by centrifugation at 1500×g for 15 min. Total RNA was extracted from two independent biological replicates using FastRNA Pro Blue kit in a FastPrep-24 instrument (Applied Biosystems) accordingly to the instructions provided by the manufacturer. The RNA quality and integrity were confirmed by agarose gel electrophoresis standardized RNA, in TBE-1% agarose gel. Quantitation of RNA was done by spectrophotometry using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). To avoid contamination with genomic DNA and in order to reduce the noise in the qRT-PCR, the extracted RNA was treated with DNase I (RNase-Free) in two steps. First, 200 ng of RNA was treated with 0.4 U DNAse/μg of RNA in a 100 μl reaction volume containing 1× DNAse buffer and DECP treated water, followed by retreatment under the same conditions but in half the reaction volume. Both steps were carried out at 37 °C for 30 min followed by DNAse inactivation at 75 °C for 5 min.
2.9. Reverse transcription of pncA
The reverse transcription (RT) was carried out independently using specific primers to pncA and 16S housekeeping genes using 200 ng of RNA in separate tubes in a PTC-100 Programmable Thermal Controller (MJ Research, Inc). Reaction was carried out with TaqMan Reverse Transcription Reagents (Applied Biosystems), in 20 μl containing 4 μg RNA, 1× RT-buffer, 5.5 mM MgCl2, 2 mM dNTPs, 0.5 μM pncA primer (pncA R: 5′ CCG TCT GGC GCA CAC AAT G 3′), 0.4 μM 16S primer (16S R: 5′ CGC TCGC ACC CTA CGT ATT AC 3′), 0.4 U/μl RNAse inhibitor, 3 U/μl Multi Scribe reverse transcriptase and DECP treated water. With the following thermal parameters: 1 cycle at 48 °C for 45 min, 1 cycle at 95 °C for 5 min, and the stable phase at 5 °C for 5 min. To control for DNA contamination, a reaction without reverse transcriptase was added.
2.10. Quantitative real-time RT-PCR of pncA
Each cDNA was amplified in a StepOnePlus RT-PCR System using Taqman Universal PCR Master Mix (Applied Biosystems). Primers designed to get a product of 116 base pairs of pncA were: (pncA F: 5′ CGA GAA CGG CAC GCC ACT G 3′, pncA R: 5′ CCG TCT GGC GCA CAC AAT G 3′, pncA probe: 5′ FAM-TTG GCT GCG GCA ACG CGG CGT CGA T-BHQ1 3′ and for 16S housekeeping gene were (16S F: 5′ TTC TCT CGG ATT GAC GGT AGG T 3′,16SR: 5′ CGC TCG CAC CCT ACG TAT TAC 3′, 16S probe 5′ FAM-AGC ACC GGC CAA CTA CGT GCC AG-BHQ1 3′). Reactions were carried out in 20 μl containing 0.5 μM for each primer, 0.2 μM and 3 μl of cDNA. With following thermal cycler parameters: 1 cycle at 50 °C for 2 min, 1 cycle at 95 °C for 10 min, and 40 cycles each at 95 °C for 20 s and a combined annealing and extension step at 55 °C for 1 min. The threshold cycle (Ct) was calculated by the instrument's Step One Plus v2.1 software (Applied Biosystems). qRT-PCR for each gene was done in duplicate, and for each reaction the entire experiment was repeated three times on RNA samples extracted from two independent cultures grown. The relative quantification of pncA expression was done with relative standard curve method, which required a standard curve for each gene. pncA expression was normalized against the 16S housekeeping. To calculate the threshold cycle (Ct) for each sample, it was normalized to the Ct of the 16S amplified. For this study, the Ct was fixed in the exponential phase of the PCR and was determined taking into account the smallest difference between the Ct for the same sample. Quantitative analysis of the PCR products was performed using StepOne Software v2.1 (Applied Biosystems).
2.11. Data analysis
Associations between the three PZA susceptibility tests: Bactec-growth index, Wayne activity and PZA-MIC were calculated with the Fisher exact test. Unpaired Student's t-test was used to test significant difference of resistance level between PZA susceptible and resistant strains. Linear and logistic regressions were used to test the association of the molecular and biochemical factors with PZA susceptibility. The adjusted effect of each molecular/biochemical factor on the PZA susceptibility was assessed with a multiple regression analysis after adjusting for potential confounding. Given that the POA efflux rate is directly related to the PZAse activity and the level of pncA expression, we created and tested a new parameter equal to the product of PZAse activity and the level of pncA expression (AEx). Every analysis was corrected for multivariate outliers with the test of Hadi30 and nested regression models were compared with the likelihood ratio test. All statistical analyses were conducted with 5% significance.
3. Results
3.1. PZA susceptibility and pncA sequencing
The BGI, PZA-MIC and Wayne activity of the studied strains are shown in Table 1. PZA susceptible strains (BGI lower than 20%) showed a BGI (N = 5, mean = 1.20%, SD = 1.09) significantly lower than PZA resistant strains (N = 24, mean = 71.46%, SD = 23.36), (P < 0.0001, Student's t-test). According to the PZA-MIC, 5 strains were susceptible (PZA-MIC lower than 100 μg/ml) and 17 resistant. Eight strains were not processed due to limited specimen availability. Nine strains were Wayne positive and 21 strains were, Wayne negative. Both the MIC and Wayne activity were significantly associated with BGI (R = 0.64, P = 0.001; and R = 0.48, P = 0.009 for a Pearson correlation, respectively). H37Rv strain, the PZAse mutated D136G strain, and two of the pncA wild type strains, were susceptible for all tests. We reported the D136G mutated strain before as a controversial strain because of its low PZAse activity but PZA susceptible phenotype.20,21
From the 30 strains for which the pncA promoter was sequenced, 2 PZA resistant strains, WT9 and WT16 with pncA wild type sequence, showed a mutation in the promoter region at nucleotide −11 bp.
3.2. pncA expression
The level of pncA expression of strain H37Rv was considered as reference (equal to 1), and other values indicated the folds of expression compared to it. Overall, the level of pncA expression showed variability (Table 1), ranging from 0.003 fold to 8.07 fold. The two biological replicates showed very similar levels of pncA expression. The average and the range of variation (minimum, maximum) are shown (Table 1). The reaction without reverse transcriptase, showed a difference of 8 CT (data not shown) confirming that DNA contamination is negligible.
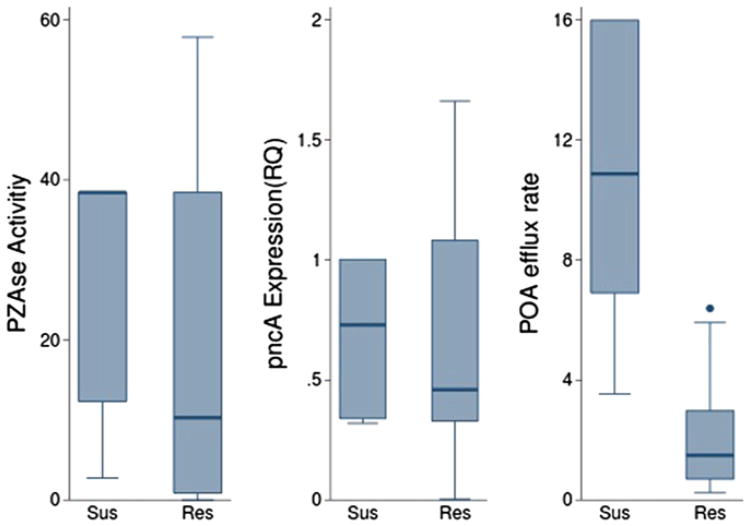
According to Bactec we found no significant difference in the level of pncA expression between susceptible strains (0.86, SD = 0.80), and resistant strains (1.36, SD = 2.15) (P = 0.61, Student's t-test, Table 2) (Figure 2). Interestingly, the two pncA wild type strains with mutations in the pncA promoter region, WT9 and WT16, showed only one-fifth of the reference level of pncA expression and had PZA resistant phenotype. Moreover, there was no significant difference in the level of pncA expression between strains with mutations in the PZAse coding region and PZAse wild type strains (P = 0.59, Student's t-test). Remarkably, one PZA resistant strain (PZAse mutation H51R, which impairs the PZAse activity; Table 1) showed a level of pncA expression of 8-fold of the reference strain. In contrast, seven PZA resistant strains (PZAse wild-types WT14, WT16, WT7, WT8 and WT9, and PZAse mutants L116P and Y64D) had normal to high PZAse activity but low level of pncA expression (Table 1).
Table 2.
Comparison of biochemical parameters between PZA susceptible and PZA resistant strains according to different susceptibility tests.
| Mycobacterium tuberculosis biochemical parameters | |||
|---|---|---|---|
|
| |||
| PZA susceptibility test | pncA expression P-value | PZAse activity P-value | VPOA P-value |
| Bactec-growth index | 0.614 | 0.257 | <0.001 |
| PZA-MIC | 0.392 | 0.039 | 0.002 |
| Wayne activity | 0.862 | 0.047 | <0.001 |
Means of biochemical parameters of PZA susceptible and resistant strains determined by Bactec-growth index, PZA-MIC and Wayne activity were compared using one-sided Student's T-test to estimate the P-value.
Figure 2.
Distribution of biochemical parameters by PZA susceptibility determined by Bactec and 7H9 culture. Graph box compares POA efflux rate, PZAse activity and level of pncA expression between PZA susceptible and PZA resistant strains, according to Bactec (a) and 7H9 culture (b). S, PZA susceptible; and R, PZA resistant.
3.3. PZAse enzymatic activity
The H37Rv reference strain had a PZAse activity of 38.4 μmol POA min−1 mg−1 PZAse. The mean PZAse activity of the MIC/7H9 susceptible strains was significantly higher than MIC/7H9 resistant strains (P = 0.04). The mean PZAse activity of Wayne positive strains was significantly higher than Wayne negative strains (P = 0.04, Table 2).
The mean PZAse activity of Bactec susceptible strains (26.06 μmol POA min−1 mg−1 PZAse, SD = 17.23) showed a trend of higher values than the corresponding to the Bactec resistant strains (20.09 μmol POA min−1 mg−1 PZAse, SD = 18.60), although the difference was non significant (P = 0.257, Student's t-test; Table 1, Figure 2). Two PZA resistant and PZAse mutated strains had higher PZAse activity than H37Rv wild type reference strain (PZAse mutants L116P and Y64D: 50.15 and 57.83 μmol POA min−1 mg−1 PZAse, respectively). L116P was resistant to PZA in all three tests, while Y64D was resistant in Bactec but susceptible in Wayne. Both also had low level of pncA expression of one-fifth and one-third of the reference level, respectively. PZAse with mutation L85P had non-detectable PZAse activity because it was insoluble when expressed recombinantly. It is possible that the lack of solubility could be due to the structure-disruptive effect of the presence of a proline in the alpha helix where L85 belongs.
3.4. POA efflux rate and PZA flux rate
The H37Rv reference strain showed the highest POA efflux rate, (15.98 nmol POA mg−1 protein min−1). According to Bactec, the mean POA efflux rate for susceptible strains (9.63 nmol POA mg−1 protein min−1, SD = 5.32), was significantly higher than the corresponding to resistant strains (2.15 nmol POA mg−1 protein min−1, SD = 1.75), (P < 0.0001, Student's t-test, Table 2 and Figure 2).
As shown in Table 1, F94L, Y34D, and WT15 strains with acceptable levels of PZAse activity and pncA expression were still PZA resistant strains and showed low POA efflux rates, probably due to a failure in their POA efflux pump system. Remarkably is the presence of the pncA wild type susceptible strain, WT5, that despite having half (0.46) of the level of pncA expression of H37Rv reference strain, its POA efflux rate was still similar to H37Rv (14.42 nmol POA mg−1 protein min−1). This suggests a more efficient POA efflux pump system probably associated to compensating mutations. As shown in previous findings,20 the PZA flux rate was uniformly distributed across the analyzed strains, and no significant difference was observed between PZA resistant and PZA susceptible strains, confirming that PZA flux occurs by passive diffusion.
3.5. Integral analysis and relevance of predictive factors
3.5.1. Correlation between parameters
The POA efflux rate was significantly correlated with the PZAse activity (determination coefficient of linear regression R2 = 0.39; P = 0.03), but was not significantly correlated with the level of pncA expression (R2 = −0.19; P = 0.31). This means that PZAse activity explains about 39% of the variability of the POA efflux rate.
3.5.2. Prediction of PZA susceptibility
POA efflux rate was the best predictor and significantly explained the variability of PZA resistance according to the BGI, 7H9/MIC, and Wayne activity when treated as a continuous or dichotomous variable (Table 3A and B). In the logistic regression, PZAse activity was associated with PZA resistance according to 7H9/MIC and Wayne activity with borderline significance.
Table 3.
Relationship of PZA susceptibility with specific biochemical parameters by considering the susceptibility as: (A), a continuous variable (linear regression); and (B), a dichotomous variable (logistic regression).
| (A) Linear regression to model PZA susceptibility PZA susceptibility | ||||
|---|---|---|---|---|
|
| ||||
| Predictor | Bactec-growth index | PZA-MIC | ||
|
|
|
|||
| R2 (P-value) | Reg. coef. | R2 (P-value) | Reg. coef. | |
| pncA expression | 0.06 (0.199) | 4.26 | 0.008 (0.693) | 15.72 |
| PZAse activity | 0.06 (0.177) | −0.48 | 0.018 (0.548) | −2.71 |
| VPOA | 0.51 (0.000) | −6.341 | 0.238 (0.021) | −44.53 |
| (B) Logistic regression to model PZA susceptibility PZA susceptibility | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Predictor | Bactec-growth index | PZA-MIC | Wayne activity | |||
|
|
|
|
||||
| OR | P-value | OR | P-value | OR | P-value | |
| pncA expression | 1.195 | 0.611 | 1.412 | 0.409 | 0.964 | 0.856 |
| PZAse activity | 0.981 | 0.502 | 0.949 | 0.089 | 0.960 | 0.102 |
| VPOA | 0.396 | 0.032 | 0.461 | 0.021 | 0.652 | 0.042 |
R2: determination coefficient, OR: Odds ratio, Reg. coef.: regression coefficient, VPOA: POA efflux rate. After testing for a multiple linear regression to model Bactec, MIC, and Wayne, we found that any other variable with exception of VPOA showed to be significant. This is expected to occur because the VPOA absorbs the variability of both the PZAse activity and the pncA expression level. PZA-MIC is measure in PZA μg/ml. PZA-MIC larger than 100 μg/ml is considered PZA resistant.
Although low pncA expression explained the specific cases of PZA resistance in pncA wild type strains WT8, WT9, WT14 and WT16, neither the level of pncA expression nor AEx were significantly associated with PZA resistance. However, AEx showed a trend to explain a higher variability of PZA resistance than the level of pncA expression only. POA efflux rate had the highest determination coefficient (R2 = 0.51) to predict PZA resistance defined as the BGI in a linear regression (P < 0.001). This means that about 51% of the variability of PZA resistance is being explained by the POA efflux rate. The adjusted multiple linear regression model to predict PZA resistance confirmed that POA efflux rate was the only significant predictor.
3.5.3. Efficiency of the POA efflux pump system
The efficiency of the POA efflux pump system for each strain is shown in Table 1. The POA efflux pump efficiency showed a large variability among the tested strains, ranging from 0.01 to 67, and its value for the H37Rv reference strain was 0.42. According to BGI, the POA efflux pump efficiency of the susceptible strains (mean 1.39; SD = 1.49) was not significantly different than the corresponding to the resistant strains (mean 6.11; SD = 14.72) (P = 0.76), however, a trend of lower values among the PZA susceptible strains was observed.
4. Discussion
This study is the first to measure the level of pncA expression concurrently with the PZAse activity and the POA efflux rate, and estimate their adjusted effects on the PZA susceptibility in M. tuberculosis. As discussed in a previous study,20 the POA efflux rate depends on at least three independent factors: (1) the PZAse activity, (2) the intracellular concentration of PZAse, which was estimated by the level of pncA mRNA, and (3) the efficiency of the POA efflux pump system. Although individual factors were found significantly associated with PZA resistance (with the exception of the level of pncA mRNA), POA efflux rate was the only significant factor after adjusting for the other factors for each of the three PZA susceptibility tests. This finding supports the fact that the POA efflux rate incorporates the information carried by the PZAse activity and the level of pncA expression. The concentration of the POA efflux pump in the cellular membrane should also affect the POA efflux rate, however, the gene encoding for this system is still unknown and it is not possible to be estimated.
Our results confirmed that the POA efflux rate was the best predictor of PZA resistance followed by PZAse activity, while pncA expression was not significantly associated. However, it is remarkable that in specific pncA wild type strains, the cause of resistance to PZA is likely to be the low level of pncA expression, which causes a limited intracellular PZAse activity that reduces the POA efflux rate, resulting in resistance as previously discussed.20 This is the first time that a reduction of pncA expression has been shown to be associated with PZA resistance in pncA wild type strains. Studies in other enzymes have shown that the intracellular overall enzymatic activity depends not only on the mutations in the coding gene, and the structure/function of the enzyme, but also on the intracellular concentration of the enzyme.31,32 Therefore, a different phenotype could result from a lack of expression of the particular enzyme with a wild type coding gene. Remarkably, one PZA resistant strain showed a pncA transcription level of 8-fold the H37Rv reference strain, which was the highest level of pncA mRNA observed. This strain exhibited the PZAse mutation H51R, which reduces significantly the PZAse activity because of the alteration of the metal binding site.19 It is possible that in this strain, the increase of pncA transcription rate could have occurred as a compensatory mechanism against the reduction of PZAse activity. This kind of compensatory mechanism has been observed before in other organisms.35,36
Although mutations in the pncA promoter region caused the pncA mRNA level to be reduced to one-fifth of the H37Rv reference strain, this study also shows evidence that strains without mutations in the promoter have a notable variation in the levels of pncA mRNA. This variability could be explained by other reasons, like variations in the expression of transcription factors, probably due to mutations, as to what has been previously found on another enzymes in different organisms.33,34 The lack of significant correlation between the POA efflux rate and the level of pncA expression found in this study, suggests that the kinetics of the POA efflux may not depend linearly on the PZAse intracellular concentration, but possibly has a non-linear correlation in which after reaching a certain level of PZAse concentration, the POA efflux rate is close to an equilibrium range.
Gene expression patterns may be affected by many environmental factors, including bacterial growth phase. In order to minimize alterations of the pncA expression level we tested all strains in the same growth phase, however we are assuming that any other factor is affecting the pncA expression. The expression of PZAse protein is dependent both on the number of pncA transcripts and the stability of those transcripts. It is possible that pncA mutations near the 3′ or 5′ of transcripts might affect mRNA stability and therefore protein expression levels. This should be considered in future studies. Furthermore, PZA efficacy might be affected by other factors including ATP concentration that may alter the POA efflux pump, or the redox state that mediates the reduction of HPOA to POA. Further studies are required in order to control and adjust for these factors and to understand their effects.
The estimation of the intrinsic efficiency of the POA efflux pump system as the POA efflux rate divided by the PZAse activity and the PZAse intracellular concentration (estimated by the pncA transcription level), showed an important variability. This suggests that variations in the structure-function of the POA efflux pump are occurring and possibly originated by mutations in its coding gene. In addition, mutations in the promoter region may affect the POA efflux pump gene expression, altering the concentration of pumps in the cell membrane.
5. Conclusion
This study shows that the variation in pncA expression is likely to give rise to PZA resistance in M. tuberculosis pncA wild type strains by reducing the concentration of intracellular PZAse. In addition our results confirmed that the POA efflux rate was the best predictor of PZA resistance followed by the PZAse activity, which is consistent with the fact that the POA efflux rate contains the information carried by the PZAse activity and the level of pncA expression. This makes the POA efflux rate a more reliable predictor of PZA resistance. The estimation of the efficiency of the POA efflux pump system revealed an important variability, suggesting that mutations in the associated gene(s) or promoter(s) might be present.
Acknowledgments
Funding: This research was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health US, under the terms of Award 1R01TW008669-01. PS and MZ were supported by TMRC New Tools to Understand and Control Endemic Parasites # 1 P01 AI51976 and Global Research Training Grant # 3 D43 TW006581.
Footnotes
Conflict of interest: None declared.
Ethical approval: Not required.
References
- 1.Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, et al. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163(9):1009–21. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- 2.Bloom BR, Murray CJ. Tuberculosis: commentary on a reemergent killer. Science. 1992;257(5073):1055–64. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 3.WHO. Management of chronic and multidrug-resistant cases Treatment of tuberculosis: guidelines for national programmes. 3rd. Geneva: Jotto Associati s.a.s.- Biella-Italy; pp. 200343–5. [Google Scholar]
- 4.Konno K, Feldmann FM, McDermott W. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am Rev Respir Dis. 1967;95(3):461–9. doi: 10.1164/arrd.1967.95.3.461. [DOI] [PubMed] [Google Scholar]
- 5.Mitchison DA. The action of antituberculosis drugs in short-course chemotherapy. Tubercle. 1985;66(3):219–25. doi: 10.1016/0041-3879(85)90040-6. [DOI] [PubMed] [Google Scholar]
- 6.Steele MA, Des Prez RM. The role of pyrazinamide in tuberculosis chemotherapy. Chest. 1988;94(4):845–50. doi: 10.1378/chest.94.4.845. [DOI] [PubMed] [Google Scholar]
- 7.Vasquez-Campos L, Asencios-Solis L, Leo-Hurtado E, Quispe-Torres N, Salazar-Lindo E, Bayona J, et al. Drug resistance trends among previously treated tuberculosis patients in a national registry in Peru, 1994–2001. Int J Tuberc Lung Dis. 2004;8(4):465–72. [PubMed] [Google Scholar]
- 8.Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother. 2003;52(5):790–5. doi: 10.1093/jac/dkg446. [DOI] [PubMed] [Google Scholar]
- 9.Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, et al. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41(3):540–3. doi: 10.1128/aac.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Scorpio A, Nikaido H, Sun Z. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Bacteriol. 1999;181(7):2044–9. doi: 10.1128/jb.181.7.2044-2049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng SJ, Thibert L, Sanchez T, Heifets L, Zhang Y. pncA mutations as a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis: spread of a monoresistant strain in Quebec, Canada. Antimicrob Agents Chemother. 2000;44(3):528–32. doi: 10.1128/aac.44.3.528-532.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Mitchison D. The curious characteristics of pyrazinamide: a review. Int J Tuberc Lung Dis. 2003;7(1):6–21. [PubMed] [Google Scholar]
- 13.Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE, 3rd, et al. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science. 2011;333(6049):1630–2. doi: 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano K, Takahashi M, Kazumi Y, Fukasawa Y, Abe C. Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuber Lung Dis. 1997;78(2):117–22. doi: 10.1016/s0962-8479(98)80004-x. [DOI] [PubMed] [Google Scholar]
- 15.Hou L, Osei-Hyiaman D, Zhang Z, Wang B, Yang A, Kano K. Molecular characterization of pncA gene mutations in Mycobacterium tuberculosis clinical isolates from China. Epidemiol Infect. 2000;124(2):227–32. doi: 10.1017/s0950268899003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mestdagh M, Fonteyne PA, Realini L, Rossau R, Jannes G, Mijs W, et al. Relationship between pyrazinamide resistance, loss of pyrazinamidase activity, and mutations in the pncA locus in multidrug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1999;43(9):2317–9. doi: 10.1128/aac.43.9.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SK, Lee JY, Chang CL, Lee MK, Son HC, Kim CM, et al. pncA mutations in clinical Mycobacterium tuberculosis isolates from Korea. BMC Infect Dis. 2001;1:4. doi: 10.1186/1471-2334-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sreevatsan S, Pan X, Zhang Y, Kreiswirth BN, Musser JM. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997;41(3):636–40. doi: 10.1128/aac.41.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheen P, Ferrer P, Gilman RH, Lopez-Llano J, Fuentes P, Valencia E, et al. Effect of pyrazinamidase activity on pyrazinamide resistance in Mycobacterium tuberculosis. Tuberculosis. 2009;89(2):109–13. doi: 10.1016/j.tube.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimic M, Fuentes P, Gilman RH, Gutierrez AH, Kirwan D, Sheen P. Pyrazinoic acid efflux rate in Mycobacterium tuberculosis is a better proxy of pyrazinamide resistance. Tuberculosis. 2012;92(1):84–91. doi: 10.1016/j.tube.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimic M, Loli S, Gilman RH, Gutierrez A, Fuentes P, Cotrina M, et al. A new approach for pyrazinamide susceptibility testing in Mycobacterium tuberculosis. Microb Drug Resist. 2012 doi: 10.1089/mdr.2011.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wade MM, Zhang Y. Mechanisms of drug resistance in Mycobacterium tuberculosis. Front Biosci. 2004;9:975–94. doi: 10.2741/1289. [DOI] [PubMed] [Google Scholar]
- 23.Sun Z, Zhang Y. Reduced pyrazinamidase activity and the natural resistance of Mycobacterium kansasii to the antituberculosis drug pyrazinamide. Antimicrob Agents Chemother. 1999;43(3):537–42. doi: 10.1128/aac.43.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandler JL, Gholson RK. De novo biosynthesis of nicotinamide adenine dinucleotide in Escherichia coli: excretion of quinolinic acid by mutants lacking quinolinate phosphoribosyl transferase. J Bacteriol. 1972;111(1):98–102. doi: 10.1128/jb.111.1.98-102.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foster JW, Moat AG. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol Rev. 1980;44(1):83–105. doi: 10.1128/mr.44.1.83-105.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calbreath DF, Joshi JG. Inhibition of nicotinamidase by nicotinamide adenine dinucleotide. J Biol Chem. 1971;246(13):4334–9. [PubMed] [Google Scholar]
- 27.Jewett MW, Jain S, Linowski AK, Sarkar A, Rosa PA. Molecular characterization of the Borrelia burgdorferi in vivo-essential protein PncA. Microbiology. 2011;157(Pt 10):2831–40. doi: 10.1099/mic.0.051706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wayne LG. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am Rev Respir Dis. 1974;109(1):147–51. doi: 10.1164/arrd.1974.109.1.147. [DOI] [PubMed] [Google Scholar]
- 29.Trivedi SS, Desai SG. Pyrazinamidase activity of Mycobacterium tuberculosis – a test of sensitivity to pyrazinamide. Tubercle. 1987;68(3):221–4. doi: 10.1016/0041-3879(87)90058-4. [DOI] [PubMed] [Google Scholar]
- 30.Hadi AS, Jefferey S. Procedures for the identification of multiple outliers in linear models. J Am Stat Assoc. 1993;88(424):1264–72. [Google Scholar]
- 31.Takakura M, Kyo S, Kanaya T, Tanaka M, Inoue M. Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res. 1998;58(7):1558–61. [PubMed] [Google Scholar]
- 32.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27(11):2291–8. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsushita K, Kobayashi S, Kato M, Itoh Y, Okuyama K, Sakiyama S, et al. Reduced messenger RNA expression level of p21 CIP1 in human colorectal carcinoma tissues and its association with p53 gene mutation. Int J Cancer. 1996;69(4):259–64. doi: 10.1002/(SICI)1097-0215(19960822)69:4<259::AID-IJC3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 34.Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314(5807):1930–3. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 35.Lucassen M, Schmidt A, Eckerle LG, Portner HO. Mitochondrial proliferation in the permanent vs. temporary cold: enzyme activities and mRNA levels in Antarctic and temperate zoarcid fish. Am J Physiol Regul Integr Comp Physiol. 2003;285(6):R1410–20. doi: 10.1152/ajpregu.00111.2003. [DOI] [PubMed] [Google Scholar]
- 36.Seedorf U, Leberer E, Kirschbaum BJ, Pette D. Neural control of gene expression in skeletal muscle. Effects of chronic stimulation on lactate dehydrogenase isoenzymes and citrate synthase. Biochem J. 1986;239(1):115–20. doi: 10.1042/bj2390115. [DOI] [PMC free article] [PubMed] [Google Scholar]