Abstract
The NEDD8 activating enzyme (NAE) is upstream of the 20S proteasome in the ubiquitin/proteasome pathway and catalyzes the first step in the neddylation pathway. NEDD8 modification of cullins is required for ubiquitination of cullin-ring ligases (CRLs), which regulate degradation of a distinct subset of proteins. The more targeted impact of NAE on protein degradation prompted us to study MLN4924, an investigational NAE inhibitor, in preclinical multiple myeloma (MM) models. In vitro treatment with MLN4924 led to dose-dependent decrease of viability (EC50=25–150nM) in a panel of human MM cell lines. MLN4924 was similarly active against a bortezomib-resistant ANBL-6 subline and its bortezomib-sensitive parental cells. MLN4924 had sub-μM activity (EC50 values <500nM) against primary CD138+ MM patient cells and exhibited at least additive effect when combined with dexamethasone, doxorubicin and bortezomib against MM.1S cells. The bortezomib-induced compensatory up-regulation of transcripts for ubiquitin/proteasome was not observed with MLN4924 treatment, suggesting distinct functional roles of NAE vs 20S proteasome. MLN4924 was well tolerated at doses up to 60mg/kg 2x daily and significantly reduced tumor burden in both a subcutaneous and an orthotopic mouse model of MM. These studies provide the framework for the clinical investigation of MLN4924 in MM.
Keywords: myeloma, cancer, NEDD8, NAE, microenvironment
Introduction
Following the successful clinical development of proteasome inhibitors, such as bortezomib (Bort) for injection, targeting protein homeostasis has become an attractive therapeutic strategy for multiple myeloma (MM) and other neoplasias. Cullin-ring ligases (CRLs) have recently become targets of interest because they regulate degradation of proteins more selectively than the 20S proteasome in the Ubiquitin/Proteasome protein degradation pathway(1). Specifically, NEDD8-activating enzyme (NAE) catalyzes the first step in the NEDD8 Neddylation pathway. NEDD8 modification of cullins is required for ubiquitination of CRLs and targeting NAE or Ubc12, the downstream enzyme in the pathway, allows for inhibition of the entire group of proteins degraded through the NEDDylation process(1, 2).
Preclinical and clinical data from our group and others have demonstrated the activity of proteasome inhibitors in plasma cell dyscrasias such as MM and Waldenstroms Macroglobulenemia (WM). This activity suggests that these cancers may also be sensitive to the inhibition of the Ubiquitin/Proteasome pathway at other levels, such as the NEDDylation component of protein degradation. To address this hypothesis we tested the preclinical activity of the NAE inhibitor MLN4924 against MM cells.
A panel of MM cell lines were treated with MLN4924 for 72hrs and compared to the colon carcinoma line HCT116. In addition, a longitudinal assessment of viability of MM.1S showed commitment to death <48hrs. We also observed that normal donor peripheral blood mononuclear cells (PBMCs) were less sensitive (EC50 >1000 nM) than the MM cell lines tested, suggest that this compound exhibits a rapid, tumor-selective effect at clinically relevant conditions. We also evaluated primary MM (CD138+) patient bone marrow cells and observed sub-μM activity by MLN4924. In addition, we tested a series of combinations of MLN4924 with dexamethasone, doxorubicin and bortezomib and observed additive activity or greater with MLN4924. Gene expression profiling revealed distinct signatures, as well as distinct patterns of gene expression changes which were induced by MLN4924 vs. Bort. Lastly, animal studies showed that MLN4924 was tolerated and active against MM xenografts in both subcutaneous and diffuse lesion mouse models of MM disease.
Material and Methods
Cell lines
We evaluated a panel of human MM cell lines Dox40 (kindly proved by Lori Hazlehurst; Lee H. Moffit Cancer Center, Tampa, FL;), H929 (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH; DSMZ), INA-6 (kindly provided by Renata Burger; University of Kiel; Kiel, Germany), JJN3 (DSMZ), KMS-18 (Japanese Collection of Research Bioresources; JCRB), KMS-34 (JCRB), MM.1S (Steve Rosen; Northwestern University), OPM2 (DSMZ) and the colon carcinoma cell line HCT116 (ATCC). The Bort-sensitive ANBL-6 cell line and their sub-line ANBL-6-V5R, which exhibits decreased response to Bort, were kindly provided by Dr Robert Orlowski (MD Anderson Cancer Center, Houston, TX). These lines were characterized by short tandem repeat (STR) profiling and compared with the known American Type Culture Collection database and German Collection of Microorganisms and Cell Cultures databases(3) and used within 6-months from receipt and/or characterization. Cells were grown in either RPMI-1640 (Mediatech Inc. Manassas, VA) or DMEM (Mediatech Inc.) media with 100 U/ml penicillin, 100 μg/ml streptomycin and 10% fetal calf serum (FCS) (GIBCO/BRL, Gaithersburg, MD), unless stated otherwise.
Compounds
MLN4924 and Bort (Fig. 1A) (4, 5), were provided by Millennium Pharmaceuticals (Cambridge, MA), resuspended in DMSO at 10mM and used at the concentrations indicated. Dex and doxo were purchased from Sigma, resuspended in DMSO and used at the concentrations indicated.
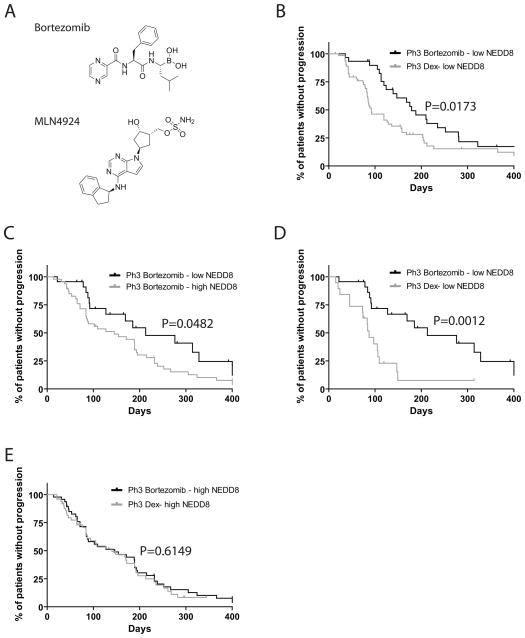
Figure 1. Levels of NEDD8 transcript in primary MM tumor cells correlate with clinical outcome.
Bort and MNL4924 structures are shown for these studies (A). Kaplan-Meier curves of progression-free survival for Bort-treated MM patients with low vs. high levels of NEDD8 transcript in the phase 2 SUMMIT (B, P=0.0173, log-rank test) and the phase 3 APEX trials (C, P=0.0482, log-rank test). Kaplan-Meier curves of progression-free survival for Bort- vs. dexamethasone-treated MM patients with low (D, P=0.0012, log-rank test) vs. high (E, P=0.6149, log-rank test) levels of NEDD8 transcript in the phase 3 APEX trial.
Correlation of Nedd8 transcript levels with clinical outcome
We evaluated the expression levels of NEDD8 transcript (201840_at) in the log2-transformed and median centered gene expression dataset of tumor cells from evaluable Bort-treated MM patients enrolled in phase II and III trials of relapsed/refractory MM (6, 7). Patients were classified as having high (top quartile of expression) vs. low (lower 3 quartiles of expression) levels of NEDD8 and Kaplan-Meier survival analyses comparing the 2 groups were performed using SPSS 17.0 and visualized with GraphPad Prism 5.0.
Cell viability assessment
Cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrasodium bromide (MTT; Chemicon International, Temecula, CA) colorimetric survival assay, as previously published (8) or CellTiterGlo (CTG; drug combination studies). In brief, MM cell lines were plated in 48-well plates at a density of 20,000 cells/well. MLN4924 was added at the concentrations indicated and compared to vehicle treated controls. Cultures were then incubated for 72 hrs in a 37°C incubator with 5% CO2. MTT was added to each well for the final 4 hours of treatment, followed by addition of isopropanol containing 0.04 N HCl to dissolve the crystals (at volume 1.5–3-fold of the total culture volume). Optical absorbance of the culture medium was then measured at 570/630 nm using a spectrophotometer (Molecular Devices Corp., Sunnyvale CA).
Peripheral blood samples from healthy donors were collected using heparinized tubes and processed by Ficoll (Amersham Biosciences, Piscataway, NJ)-density gradient centrifugation to isolate peripheral blood mononuclear cells (PBMCs), which were plated at 10,000 cells per well and pre-stimulated with 5 μg/mL phytohemagglutinin (PHA; Sigma-Aldrich, St Louis, MO) for 1hr. MLN4924 was administered at the doses indicated in the respective experiments for 72 hrs, followed by the addition of CTG and measurement as described above. Following consent, myeloma patient samples were collected, Ficoll gradient separated, and selected using CD138 micro-beads (Miltenyi Biotec; Bergisch Gladbach, Germany). Cells were cultured in RPMI-1640 (Mediatech, Inc.) with 100 U/ml penicillin, 100 μg/ml streptomycin and 20% fetal calf serum (FCS) (GIBCO/BRL) and treated as indicated in the figures. Viability was measured by CTG assay, as described above.
Cell death commitment assay
The minimum exposure of MM cells to MLN4924 that is required to commit them to death was evaluated by incubating cells in 24-well plates with MLN4924 (500nM) up to 72hrs. Following incubation, the cells were washed with drug-free medium to remove any residual drug, and then incubated in drug-free medium for an additional 3 days, resulting in equal length of incubation for all experimental conditions. MM cell survival was quantified by CTG and expressed as percentage of the value obtained from respective controls.
Stromal cell co-culture
The luciferase (luc)-expressing cell lines MM.1S-mCherry/luc and OPM2-mCherry/luc were generated by retroviral transduction and used for co-culture experiments with the luc-negative human stromal line HS-5, as described previously (9). Briefly, HS-5 stromal cells were plated in 384-well plates and allowed to attach overnight. MM cells were then plated and treated with MLN4924 for 48hrs at the doses indicated. Following incubation, luciferin substrate (Xenogen Corp, Alameda, CA) was added to the culture and the resulting bioluminescence signal was measured using an Envision luminometer (Perkin Elmer, Waltham, MA).
Osteoclast differentiation / co-culture
Peripheral blood mononuclear (PBMCs) were collected from collars of normal donors, ficoll gradient separated, resuspend in alpha-MEM media containing 10% FBS and Pen/Strep. Cells were cultured for 16 hrs to allow attachment and non-adherent cells were removed. Fresh media supplemented with MCSF (25ng/mL; R & D Systems # 216-MC-005) & RANKL (50ng/mL; Peprotech # 310-01) was added and cells were cultured for 3–4 weeks to differentiate mature osteoclasts. Half of media was changed 2X per week, containing fresh MCSF and RANKL. At 3–4 weeks a cells were trypsinized, replated and stained using TRAP stain to determine the differentiation of osteoclasts. Mature osteoclasts were plated in 96-well plates, allowed to attach overnight and used in co-culture experiments as indicated in the figure legends.
Animal Studies
The in vivo anti-MM activity of MLN4924 was evaluated in a subcutaneous xenograft model of RPMI8226/S MM cell line in CB.17-SCID mice. The anti-tumor activity and pharmacodynamic study in live mice was performed at Oncodesign S.A (Dijon, France). In brief, 2x107 RPMI 8226 cells were injected into the right flank of CB.17 SCID mice and studies were initiated once tumors had formed to approximately 200 mm3. MLN4924 was dosed subcutaneously at 10, 30 and 60 mg/kg BID for 21 days and tumor volume monitored. To evaluate the pharmacodynamic effects of MLN4924, a single subcutaneous dose of MLN4924 was administered and the effects on the NAE pathway were examined as described previously (4, 10).
The in vivo anti-MM activity of MLN4924 was evaluated in a previously established model of diffuse GFP/luc+ MM lesions in SCID/NOD mice. Briefly, male (6 to 8-week old) SCID/NOD mice were obtained from Charles River (Bar Harbor, ME); housed and monitored in the Animal Research Facility of the Dana-Farber Cancer Institute; gamma-irradiated (150 rads) using Cs137 γ-irradiator source; and received (24 hrs post-irradiation) tail i.v. injections of 106 MM.1S-GFP/luc cells per mouse. Mice were monitored regularly for changes in body weight, signs of infection or paralysis, and with weekly bioluminescence imaging(11).
Immunoblotting analysis
For immunoblotting analyses, MM.1S cells (10x106 cells per condition) were plated in RPMI-1640 medium with 10%FBS, penicillin, and streptomycin as previously described. MLN4924 was added at a concentration of 500nM for 0–72 hrs. Cell pellets were collected and treated with Triton X-100 lysis buffer containing 1 X PBS, Triton X-100 (1% v/v), sodium deoxycholate (0.5% w/v), SDS (0.1% w/v), EDTA (1 mmol/L), 1 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L sodium fluoride, 1 mmol/L sodium orthovanadate, 1 μg/mL aprotinin, 5 μg/mL leupeptin, and 5 μg/mL pepstatin A. The samples were cleared by centrifugation (14,000 rpm, 30 min, 4oC) and assessed for protein concentration by Bradford assay (Sigma). SDS-polyacrylamide gel electrophoresis (12%) was performed (30–50 μg of protein per lane), and proteins were electroblotted onto PVDF membranes. After 1 hr incubation in blocking solution (5% milk in TBS-T buffer), membranes were exposed to primary antibody overnight at 4oC. Following washing in TBS-T, the respective secondary horseradish peroxidase (HRP)-labeled antibody was added at 1:20,000 dilution for 1 hr at room temperature. The membrane was then washed with TBS-T for 45–60 min with multiple changes of the wash buffer, and the protein expression was visualized using the ECL technique. The primary antibodies used for immunoblotting were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), Upstate Biotechnologies (Lake Placid, NY) or Cell Signaling (Beverly, MA). Secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA).
Gene expression profiling of cells treated with MLN4924
Total RNA extraction and purification, cDNA synthesis and cRNA labeling, Affymetrix chip (human HG-U133 plus 2.0 array) hybridization and data analysis were performed as previously described (12–14). Briefly, MM.1S cells were treated with 250 nM MLN4924, Bort 10 nM or with DMSO as a control for the indicated time points; total RNA was then extracted and purified, cDNA synthesized and cRNA labeled prior to hybridization to the HG-U133 plus 2.0 arrays. Gene expression data have been deposited in Gene Expression Omnibus (GEO, GSE33577). Transcripts suppressed by MLN4924 were analyzed in publically available databases for relevance to clinical outcome and/or differential expression in various stages of plasma cell dyscrasias. We specifically analyzed the log2 transformed median centered values (derived from the Oncomine database) of different MLN4924-suppressed genes in the GEO (Gene Expression Omnibus, (15)) datasets GSE5900 (comparison of gene expression of bone marrow plasma cells (PCs)_ from healthy donors, MGUS, and smoldering myeloma (SMM) (16); GSE2658 (17); GSE2113 (comparison of gene expression profiles of purified PCs obtained from MGUS, newly diagnosed MM and from PCL patients) (18); GSE4452 (gene expression profiles of MM patients before treatment (19). Differential expression between normal PCs vs. MGUS vs. SMM; between MGUS vs. newly diagnosed MM vs. PCL patients; or between patients with differential patient outcome were evaluated using one-way analysis of variance (ANOVA) or unpaired T-test, as stated in the figure legends using Prism software (Graphpad).
Results
NEDD8 transcript levels correlates with clinical outcome in MM patients
We evaluated the expression levels of NEDD8 transcript (201840_at) in gene expression dataset of tumor cells from evaluable Bort-treated MM patients enrolled in phase II and III trials of relapsed/refractory MM (6, 7). We observed that patients with high expression of NEDD8 transcript had significantly shorter progression-free survival (PFS) compared to patients with low NEDD8 transcript levels. Among patients treated in the Phase III trial of Bort vs. Dex, those with low levels of NEDD8 transcript had significantly longer PFS with Bort than with Dex however, in patients with high levels of NEDD8 transcript there was no significant difference in patients treated with Bort vs. Dex (Fig. 1).
In vitro activity of MLN4924 against MM cell lines and primary patient samples
We treated a panel of MM cell lines with MLN4924 (0–1000nM) for 72hrs and evaluated viability using MTT colorimetric survival assays. The majority of cell lines had EC50 values <500nM (Fig. 2A). The most sensitive cells lines, such as OPM2, INA-6 and KMS34, exhibited EC50 values <100nM. In addition, we evaluated the impact of MLN4924 on the colorectal carcinoma cell line HCT116, previously reported to be MLN4924-responsive (4) for comparison.
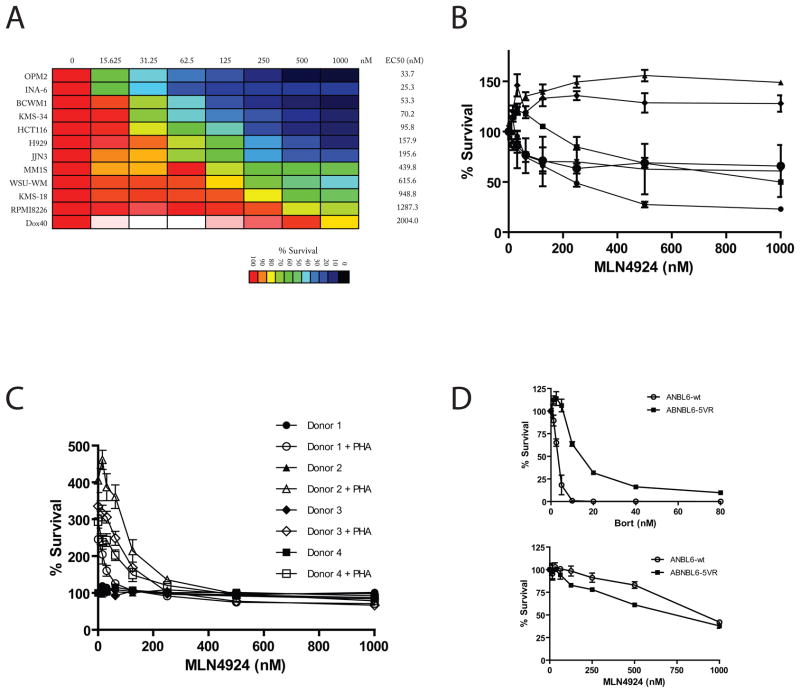
Figure 2. Activity of MLN4924 on MM cells.
MM (OPM2, INA-6, KMS-34, H929, JJN3, MM.1S, KMS-18 and Dox40), WM (BCWM1, WSU-WM) and colon carcinoma (HCT116) cell lines were treated with MLN4924 (0–1000nM for 72 hrs) and viability was assessed by MTT assay. The most sensitive cell lines (OPM2, INA-6) have EC50 values < 40 nM, whereas the most resistant MM cell line (Dox40) has an EC50 value > 2 μM (A). Bone marrow aspirates from MM patients were processed using Miltenyi anti-CD138 microbeads for purification of MM cells, which were then cultured in RPMI1640 media containing 20% serum. CD138+ selected cells were treated with increasing doses of MLN4924 for 72 hrs and viability was assessed by CellTiterGlo (B). To access the sensitivity of non-malignant cells, PBMCs were isolated from normal donors and screened in the presence and absence of PHA stimulation (5ng/mL) for 72hrs. The number of viable cells in each condition is expressed as percent of the non-PHA-stimulated drug-free control (C). The MM cell line ANBL6 and its sub-line ANBL6-V5R were tested for sensitivity to Bort and MLN4924. ANBL6-V5R was more resistant to Bort, but not MLN4924, compared to the parental cell line (D).
In addition to cell lines, CD138+ primary MM patient samples were evaluated for sensitivity to MLN4924. MM patient BM was collected, ficolled and then selected for CD138+ cells. Cell were then plated, and treated with increasing doses of MLN4924 for 72hrs. Of the 6 samples treated we observed activity of the drug against 4 samples with EC50 values <600nM. Two samples, however, remaining insensitive to the drug up to 1000nM of drug (Fig 2B).
For further understanding of the activity of MLN4924 against normal tissues, PBMCs from healthy donors were isolated, PHA-stimulated to induce cycling and proliferation, then treated with MLN4924 for 72hrs (Fig. 2C). The percentage of viable PBMCs was ≥90% following MLN4924 treatment in the absence of PHA stimulation. Sensitivity of PBMCs to MLN4924 was increased as a result of PHA-stimulation, but remained less than the most sensitive MM cell lines.
We also tested in vitro the activity of MLN4924 against the Bort-sensitive MM cell line ANBL6 and its sub-line ANBL6-V5R, which exhibits decrease response to Bort. We did not observe increased resistance of ANBL6-V5R cells to MLN4924 compared to the parental cell line (Fig. 2D).
Cell death commitment assays were performed on Dox40, MM.1S, OPM2 and KMS34 cells by exposing them to MLN4924 (500nM) for up to 72hrs, followed by drug washout and incubation in drug-free medium for an additional 72hrs. MLN4924 treatment for 48hrs resulted in commitment to death in OPM2, KMS34 and MM.1S (Suppl. Fig. 1), suggesting that molecular events initiating cell death can occur in less than 48hrs of MLN4924 treatment.
MLN4924 overcomes stroma and osteoclast protection of MM cells
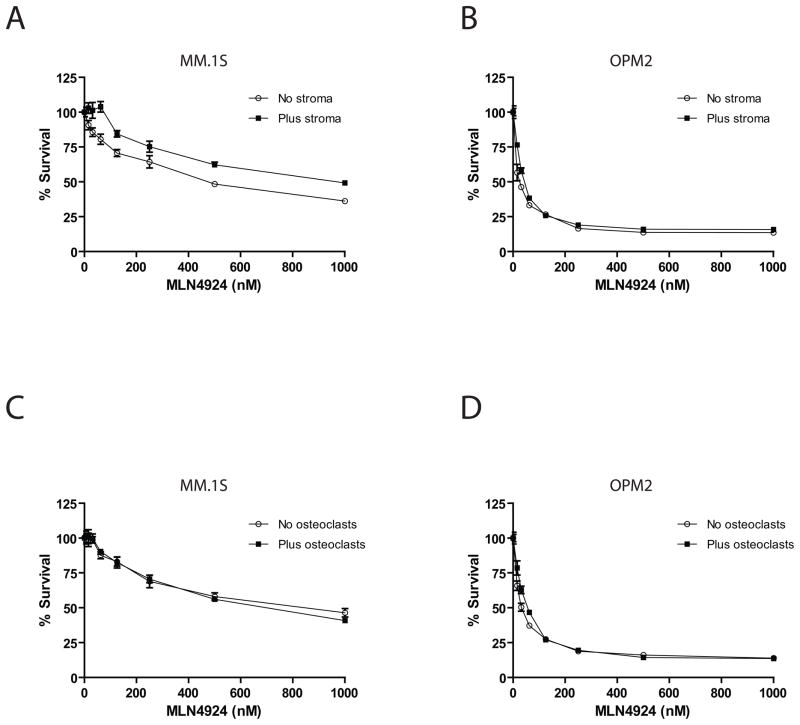
Luc+ MM.1S and OPM2 cells were cultured in the presence and absence of HS-5 stromal cells or differentiated osteoclasts for 72hrs. Cell viability was normalized to each respective drug-free control. Results show that MLN49424 was similarly active both in the presence and absence of HS-5 (Fig 3A, B) and presence and absence of osteoclasts (Fig. 3C, D), indicating that MLN4924 can overcome the protective effects of bone microenvironmental factors.
Figure 3. MLN4924 overcomes the protective effect of nonmalignant accessory cells of the bone microenvironment.
MM.1S and OPM2 MM cells were cultured in the presence and absence of HS-5 stromal cells (A, B) or differentiated osteoclasts (C, D) and treated with MLN4924 for 72 hrs. The resulting cell viability was normalized to each respective no drug control (n=4). Comparable activity was observed against MM.1S and OPM2 cells both in the presence or absence of these bone microenvironment cells with treatment with MLN4924.
Activity of MLN4924 in animal studies
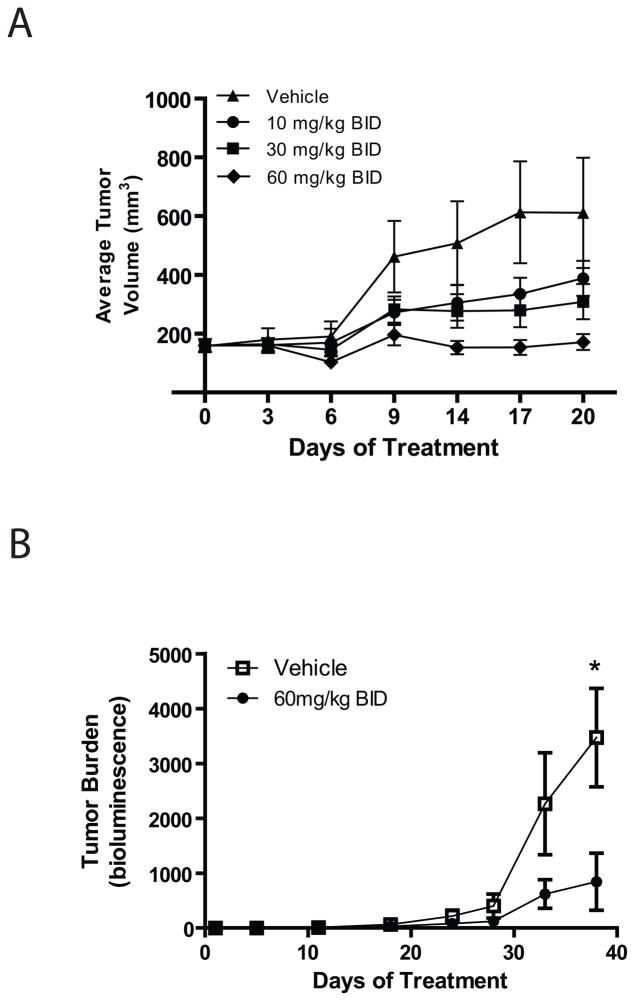
MLN4924 demonstrated dose-dependent anti-MM activity in the RPMI8226/S xenograft model in CB.17 SCID mice (Fig 4A). Complete inhibition of tumor growth was achieved at 60 mg/kg BID dose level which is below the MTD on this schedule. In addition, MLN4924 demonstrated dose-dependent inhibition of Nedd8-cullin levels with a concomitant increase in levels of two CRL substrates, Nrf-2 and pIKBa (Supplemental Figure 2). We also evaluated the in vivo anti-MM activity of MLN4924 in sublethally irradiated SCID/NOD mice injected i.v. with MM.1S-GFP/luc cells for establishment of diffuse MM bone lesions. MLN4924 treatment was associated with decrease in tumor burden (Fig. 4B) without a significant decrease in average body weight in the mice (data not shown).
Figure 4. In vivo anti-MM activity of MLN4924.
RPMI8226/S cells were injected subcutaneously in CB.17-SCID mice and treatment was initiated once tumors had reached approximately 200mm3. MLN4924 was dosed subcutaneously at 10, 30 and 60 mg/kg BID for 21 days and tumor volume monitored (N=12 per group). There was a statistically significant difference in tumor burden only when comparing the control with 60mg/kg group (p<0.05; 1-way ANOVA; A). In addition, an orthotopic model of MM was evaluated. Sublethally irradiated NOD/SCID mice injected I.V. with MM1.S MM cells were randomly assigned to receive MLN4924 treatment (60 mg/kg BID i.p.) or vehicle control. Tumor burden was evaluated by whole-body bioluminescence imaging weekly and tumor burden calculated using Living Image software (B). Treated mice had lower tumor burden compared to vehicle treated controls (p=0.1186 day 33; p=0.0269 day 38; N=6 per group).
Molecular sequelae of MLN4924 treatment
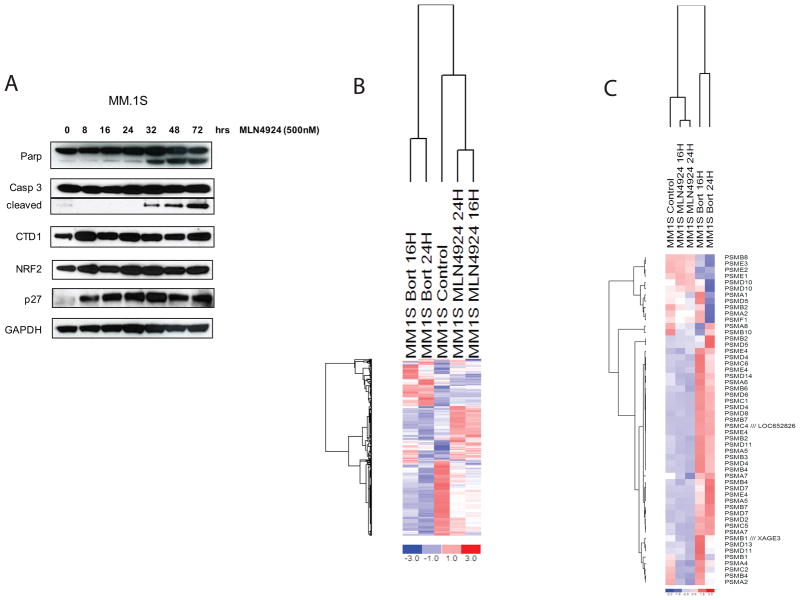
Immunoblotting of MLN4924-treated MM.1S showed upregulation (as early as 8hrs) of p27, CTD1 and NRF2, which are known targets of the NEDD8 pathway, followed by markers of cell death, such as cleavage of PARP and caspase-3 at later time-points (as early as 32 hours after initiation of treatment; Fig 5A). We also evaluated the gene expression profiling changes triggered in MM.1S cells treated with MLN4924 vs. Bort. We observed that the profile of MLN4924-triggered transcriptional changes was distinct from the one induced by Bort exposure (Fig. 5B). Indeed, known transcriptional responses to 20S proteasome inhibition, such as upregulation of proteasome subunits (Fig. 5C) or heat shock proteins (data not shown), were minimal or absent in the MLN4924-treated cells (Suppl. Table). In addition, we observed that several MLN4924-suppressed genes were associated with higher expression in MM patients with adverse clinical outcome or were expressed at higher levels in patients with PCL or MM compared to MGUS or normal PCs. For example, KLF2 (Suppl Fig 3A), PHACTR1 (Suppl Fig 3B), and FAM1985 (Suppl Fig 3C) were expressed at higher levels in smoldering myeloma and MGUS compared to normal plasma cells; FMA1985 was expressed higher in MM patients who were dead vs. alive at 5 years (Suppl Fig 3D); PSMC3IP was expressed at higher levels in patients who were dead vs. alive at 1 year after diagnosis (Suppl Fig 3E); and show that PSMC3IP was expressed at higher levels in PCL vs.MM vs. MGUS patients (Suppl Fig 3F). These results support the notion that the molecular sequelae of MLN4924 treatment include genes with putative roles in the progression and clinical outcome of MM patients.
Figure 5. Immunoblotting analysis and transcriptional profiles of MLN4924-treated MM cells.
MM.1S cells treated with MLN4924 show upregulation (as early as 8hrs) of p27, CTD1 and NRF2, which are known targets of the NEDD8 pathway, followed by markers of cell death, such as cleavage of PARP and caspase-3 at later time-points (as early as 32 hours after initiation of treatment) (A). The gene expression profiling changes triggered in MM.1S cells by MLN4924 vs. Bort were evaluated using the HG-U133plus2 oligonucleotide microarray. The profile of MLN4924-triggered transcriptional changes was distinct from the one induced by Bort exposure (B). Known transcriptional responses to 20S proteasome inhibition, such as upregulation of proteasome subunits (C) minimal or absent in the MLN4924-treated cells.
MLN4924 combinations with other anti-MM agents
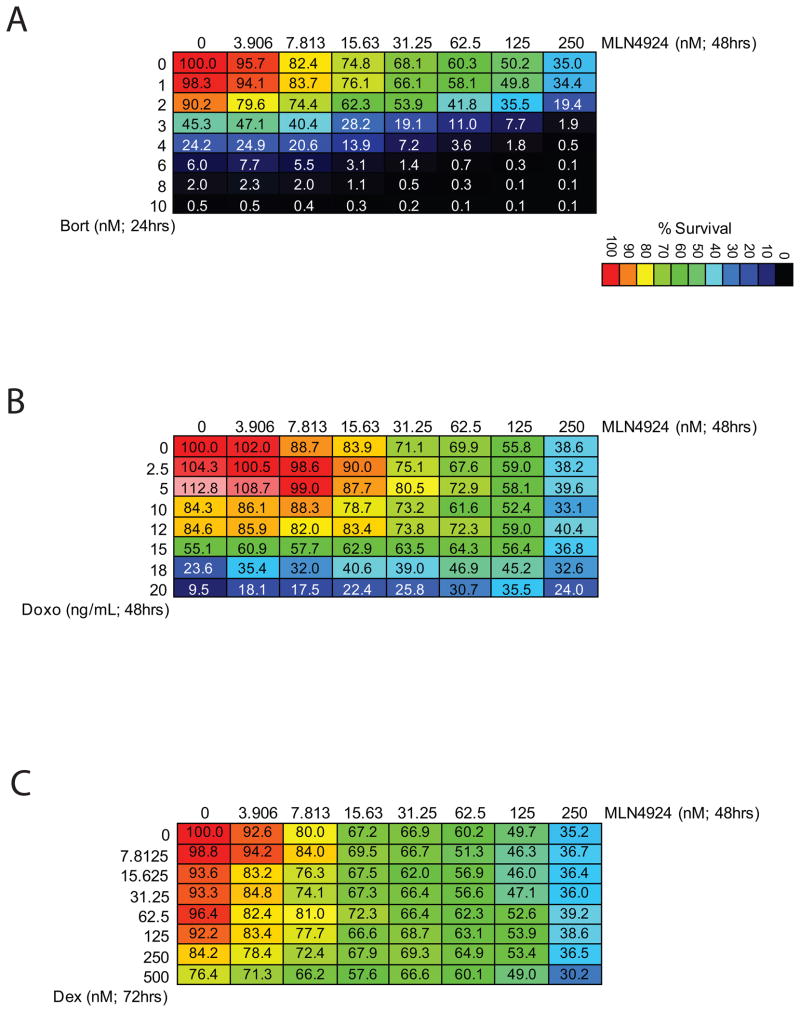
We next evaluated combinations of MLN4924 with agents used for clinical management of MM (Dex, Bort, doxorubicin). Although no synergy was observed between MLN4924 and Bort (Fig.6A), doxorubicin (Fig. 6B) or Dex (Fig. 6C), there also was no antagonism observed, suggesting that MLN4924 does not adversely impact the anti-MM activity of these approved agents.
Figure 6. Combinations of MLN4924 with established anti-MM agents.
MLN4924 treatment was combined with agents used for clinical management of MM, namely Bort (A), Doxo (B), and Dex (C). Activity was observed with these combinations at drug doses relevant to those achieved clinically in the treatment of patients.
Discussion
The preclinical and clinical development of the proteasome inhibitor Bort for the treatment of MM led to significant improvements in the clinical outcome of patients with this presently incurable plasma cell dyscrasia. The clinical success of Bort validated the notion that therapeutic targeting of ubiquitination and proteasome function is feasible without catastrophic toxicities. It also led to further efforts to identify other druggable targets in this pathway, particularly at molecular levels situated upstream of the proteasome. These targets have included the cullin-ring ligases (CRLs), which regulate degradation of proteins more selectively than the chymotryptic-like site of the 20S proteasome core. NEDD8-activating enzyme (NAE) has been the focus of particular attention because it catalyzes the first step in the NEDD8 neddylation pathway. NEDD8 modification of cullins is required for ubiquitination of CRLs and targeting NAE or Ubc12, the downstream enzyme in the pathway, allows for inhibition of the entire group of proteins degraded through the neddylation process.
Our interest on the therapeutic targeting of NAE was further enhanced by our observation that MM patients with high levels of NEDD8 transcript had shorter progression free survival upon treatment with Bort. We also observed that, in the randomized phase III APEX trial which compared Bort versus dexamethasone treatment in relapsed or refractory MM, Bort-treated patients with high levels of NEDD8 transcript did not have a significant advantage over dexamethasone-treated patients in terms of progression free survival. This suggests that perturbation of the NEDD8 cascade may characterize a subset of MM patients who do not receive maximum benefit from Bort-based therapies. For those subsets of patients, therapeutic targeting of the NEDD8 pathway could be useful.
We evaluated the anti-myeloma activity of the NAE inhibitor MLN4924 in our in vitro preclinical models and observed a dose-and time-dependent reduction in viability of MM cell lines as well as primary CD138+ MM tumor cells isolated from MM patients. The EC50 values of MLN4924 against MM cell lines and primary tumor cells was comparable to or lower than the values observed in preclinical studies of other neoplasias considered to be highly responsive to NAE inhibition(4, 10, 20). Importantly, this activity observed in vitro against MM cells could be achieved with exposure to MLN4924 for 48hrs or less. Furthermore, we observed that the response of MM cells to MLN4924 was similar when these cells were cultured alone vs. co-cultured with bone marrow stromal cells (BMSCs) or osteoclasts. This indicates that NAE inhibition can circumvent the proliferative / anti-apoptotic stimulation provided to MM cells by the interactions with nonmalignant accessory cells of the BM microenvironment. Consistent with this observation, MLN4924 significantly reduced the tumor burden of both subcutaneous and orthotopic MM lesions in immunocompromised mice.
Mechanistically, NAE inhibition with MLN4924 treatment was associated with early increase in protein levels of p27, CTD1 and NRF2. This observation is compatible with the known role of NAE in regulating intracellular levels of these proteins. A more open-ended analysis of the transcriptional profile of MLN4924-treated MM cells showed changes distinct from those triggered by Bort. For instance, while Bort triggers major increases in transcript levels for diverse heat shock proteins family members and proteasome subunits, MLN4924 triggered minimal, if any, such changes. This difference likely reflects the distinct functional consequences of inhibiting the beta 5 subunit of the 20S proteasome core vs. NAE. The beta 5 subunit of the proteasome core regulates the intracellular levels of a larger pool of ubiquitinated proteins. In contrast, NAE regulates the neddylation of a more restricted set of proteins. Because the targets of NAE regulation are enriched for proteins with critical roles in proliferation, survival or drug resistance, MLN4924 achieves anti-tumor responses, even though it does not induce the pronounced intracellular proteotoxic stress triggered by Bort. Interestingly, we observed that a myeloma cell line model with constitutive resistance to Bort and its isogenic Bort-sensitive parental line are both responsive to MLN4924. This suggests that resistance to inhibition of the beta 5 subunit of the 20S core by Bort does not necessarily confer cross-resistance to inhibition of upstream molecular targets, such as NAE.
Based on these observations, we further explored combination studies of MLN4924 with established anti-myeloma agents, such as bort, doxo and dex. These in vitro combinations showed no antagonism between the combined agents, suggesting that currently available regimens for MM treatment may be combined with MLN4924 in further preclinical and potentially clinical studies.
Our results indicate that NAE inhibition with MLN4924 has in vitro and in vivo activity in preclinical models of MM. The molecular impact of NAE inhibition is distinct from the sequelae triggered by Bort, as evidenced by the differential molecular profiles induced by MLN4924 vs. Bort, as well as the activity of MLN4924 against Bort-resistant cells. These observations provide a platform for further evaluation of NAE inhibitors to inform the design of clinical trials in MM.
Supplementary Material
Acknowledgments
Supported in part by the Chambers Medical Foundation (CSM, PGR), the Steven Cobb Foundation (D.W.M, C.S.M), the R.J. Corman Fund for Myeloma Research (PGR, CSM), the Stepanian Fund for Myeloma Research (PGR, CSM), the Honickman Fund (PGR, CSM) and National Institutes of Health Grants RO1-50947 (C.S.M and KCA), and PO-1-78378 (K.C.A.). C.S.M is supported by the “Dunkin Donuts Rising Stars” Program at the Dana-Farber Cancer Institute. We would like to thank Dr Robert Orlowski (MD Anderson Cancer Center, Houston, TX) for providing they ANBL-6 and ANBL-6-V5R cells
Footnotes
Author Contributions
D.W.M. conducted experiments, performed analysis and wrote the manuscript; J.D., H.J., L.B., Z.H., and J.U. conducted experiments, performed analysis; V.M. conducted experiments; P.G.S. performed analysis; P.G.R. provided patient samples; K.C.A. and S.P.T. participated in writing the manuscript: A.L.K. performed analysis; C.S.M. performed analysis, wrote manuscript and supervised the project.
Disclosure of Potential Conflicts of Interest
DWM is a founder and equity holder in Axios Biosciences. HMJ, JD, LB, ZH, VM, ALK have nothing to disclose. JY, PG are employees of Millennium Pharmaceuticals (The Takeda Oncology company); P.G.R. has received honoraria from Millennium, Celgene and Novartis; as well as research support from Johnson & Johnson and Bristol-Myers Squibb. K.C.A. has received research grants and honoraria from Millennium and Celgene, has been a consultant for Millennium, Celgene, Novartis, Bristol-Myers Squibb, Merck and Onyx and is a founder of Acetylon Pharmaceutials. SPT has received honoraria from and is consultant/member of advisory board for Millennium. CSM has received in the past Consultant honoraria from Millennium Pharmaceuticals, Novartis Pharmaceuticals, Bristol-Myers Squibb, Merck & Co., Kosan Pharmaceuticals, Pharmion, Johnson & Johnson and Amnis Therapeutics, licensing royalties from PharmaMar, and research funding from Amgen Pharmaceuticals, AVEO Pharma, EMD Serono, Sunesis, Gloucester Pharmaceuticals and Johnson & Johnson.
References
- 1.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15:3912–6. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 2.Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. The NEDD8 Conjugation Pathway and Its Relevance in Cancer Biology and Therapy. Genes Cancer. 2010;1:708–16. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.[cited; DSMZ]. Available from: http://www.dsmz.de/
- 4.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–6. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 5.Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, et al. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 1999;59:2615–22. [PubMed] [Google Scholar]
- 6.Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med. 2005;352:2487–98. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 7.Mulligan G, Mitsiades C, Bryant B, Zhan F, Chng WJ, Roels S, et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109:3177–88. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- 8.Mitsiades CS, Treon SP, Mitsiades N, Shima Y, Richardson P, Schlossman R, et al. TRAIL/Apo2L ligand selectively induces apoptosis and overcomes drug resistance in multiple myeloma: therapeutic applications. Blood. 2001;98:795–804. doi: 10.1182/blood.v98.3.795. [DOI] [PubMed] [Google Scholar]
- 9.McMillin DW, Delmore J, Weisberg E, Negri JM, Geer DC, Klippel S, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med. 2010;16:483–9. doi: 10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116:1515–23. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 11.Mitsiades CS, Mitsiades NS, Bronson RT, Chauhan D, Munshi N, Treon SP, et al. Fluorescence imaging of multiple myeloma cells in a clinically relevant SCID/NOD in vivo model: biologic and clinical implications. Cancer Res. 2003;63:6689–96. [PubMed] [Google Scholar]
- 12.Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Fanourakis G, Gu X, et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci U S A. 2002;99:14374–9. doi: 10.1073/pnas.202445099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci U S A. 2004;101:540–5. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Akiyama M, et al. Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell. 2004;5:221–30. doi: 10.1016/s1535-6108(04)00050-9. [DOI] [PubMed] [Google Scholar]
- 15.[cited; Gene Expression Omnibus]. Available from: http://www.ncbi.nlm.nih.gov/geo/
- 16.Zhan F, Barlogie B, Arzoumanian V, Huang Y, Williams DR, Hollmig K, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109:1692–700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattioli M, Agnelli L, Fabris S, Baldini L, Morabito F, Bicciato S, et al. Gene expression profiling of plasma cell dyscrasias reveals molecular patterns associated with distinct IGH translocations in multiple myeloma. Oncogene. 2005;24:2461–73. doi: 10.1038/sj.onc.1208447. [DOI] [PubMed] [Google Scholar]
- 19.Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–25. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 20.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.