The importance of peripheral arterial diseases
Broadly defined, peripheral vascular disease refers to disease of the extra-cardiac blood vessels, including diseases of the arteries, veins and lymphatics. Peripheral arterial disease (PAD) refers to disease affecting non-coronary arteries, but is most often used to describe disease of the arteries supplying the limbs. Peripheral arterial disease is most commonly due to atherosclerosis, but may also be secondary to cardiac or vascular embolism, vasculitis, hypercoagulopathy, vascular dissection, vascular compression syndromes, and other less common disorders. In addition, peripheral arterial diseases include those characterized by fixed or dynamic stenoses, as well as aneurysmal disease such as abdominal aortic aneurysm (AAA). In this review, we focus on atherosclerotic arterial occlusive disease affecting the vessels supplying blood flow to the lower extremities (PAD), and discuss our current understanding and the future directions of PAD genetics.
PAD is a significant public health problem, and a major source of morbidity and mortality that affects approximately 8 million Americans. PAD contributes to impaired quality of life (eg. intermittent claudication reducing mobility), morbidity (eg. non-healing ulcers and ischemic rest pain) and mortality (generally due to its association with coronary and carotid artery disease). PAD is responsible for approximately half a million hospitalizations and 100,000 angiograms annually 1, 2. Due in part to a general unfamiliarity with these diseases amongst the primary care community, PAD patients receive suboptimal treatment compared to patients with coronary artery disease (CAD), being prescribed therapeutic doses of statins, anti-hypertensive medicines, and antiplatelet agents less commonly than patients with CAD 3-6. Much remains unknown about the biological origins of this disease and how to effectively identify and treat affected individuals.
Using genetics to identify pathophysiological pathways and novel therapeutic targets
A greater understanding of how genetic variation influences susceptibility to PAD may inform the development of novel therapeutics. High-throughput, whole-genome technologies efforts have recently made inroads toward these goals (Table). The advent of genome-wide association studies (GWAS, discussed below) and cDNA microarrays (which measure the mRNA expression levels of all coding genes) has allowed for the unbiased detection of pathways which are differentially activated in affected versus unaffected individuals. Whereas these are the contemporary tools now driving genetic investigations of PAD, it is important to first put this recent work into a historical context.
Table.
Historical approaches to PAD genetics: The Candidate Gene Approach, Linkage Analysis, and Genome-Wide Association Studies.
| Candidate Gene Approach | Linkage Analysis | Genome Wide Association Study (GWAS) | |
|---|---|---|---|
| Description | Typically a case-control approach which searches for a statistical association between a specific genetic variant (i.e. a SNP) and a disease of interes | A family-based approach where the genome is scanned for pre-specified DNA markers that are known to be highly variable (i.e. microsatellites). Regions that are found more commonly in the diseased members of the family are said to be ‘linked’ to the causative gene, which is then pursued with fine mapping -Also known as ‘positional cloning’ |
A novel approach where single nucleotide polymorphisms (SNPs) are genotyped across the entire genome in subjects with and without a given disease. SNPs which differ in frequency between cases compared to controls are ‘associated’ with disease |
| Strengths |
|
|
|
| Weaknesses |
|
|
|
| Examples related to PAD | ∼ 20 identified to date, reviewed in 7, 8
|
Rare examples in the literature including: |
Earlier studies documenting the heritability of PAD
PAD is a complex disorder from a genetic standpoint. Unlike monogenic vascular syndromes such as Marfan and Loey's Dietz that manifest a Mendelian inheritance pattern14, atherosclerotic PAD likely results from dozens or hundreds of genes interacting with each other and the environment to cause disease7. Epidemiological studies suggest that over 50% of the population burden of PAD is attributable to classical risk factors such as smoking, diabetes mellitus, dyslipidemia and hypertension15. The remaining risk is thus accounted for by other unmeasured environmental or genetic components.
Several studies indicate a heritable component to PAD. In a study of patients with premature PAD (onset before age 50), half of the subjects' asymptomatic siblings had occult lower extremity atherosclerosis as determined by duplex ultrasonography16. A recent Swedish twin study17used discharge diagnosis to define PAD, and is likely more representative of patients with symptomatic claudication or critical limb ischemia. Genetic effects accounted for 58% (95% CI, 50% to 64%) and non-shared environmental effects for 42% (95% CI, 36% to 50%) of the phenotypic variance among twins in this report. Three studies conducted to date estimate that 21% of the inter-individual variation in ABI is attributable to inherited factors. These studies include the National Heart, Lung, and Blood Institute (NHLBI) Twin Study18, the Genetic Epidemiology Network Arteriopathy Study (GENOA)19, and a study performed in the Framingham Offspring Cohort20. Many of the participants in these studies had ABI values in or near the reference range. While sibling studies frequently overestimate attributable genetic risk because of shared environmental risk factors within families21, these reports each provided evidence that ABI is at least moderately inheritable, and justify the search for responsible gene variants. Historical methods included case-control approaches and linkage analyses.
Earlier studies to identify genetic determinants of PAD
Candidate Gene Studies
Using the candidate gene approach, one searches for an association between a specific variant in a specific gene (eg. a single nucleotide polymorphism, or SNP) and a clinical phenotype (generally defined by a low ABI in PAD patients). Such polymorphisms may alter a gene's expression by altering the binding of its required transcription factors, impairing the stability or intracellular trafficking of its mRNA transcript, or limiting its ability to be translated into a functional protein. Often, the polymorphism may be linked to another gene that is responsible for the disease. Collections of cases and controls are assembled and genotyped for variants in a suspected pathway 22. Driven by our understanding of atherosclerotic disease, most efforts thus far have focused on genes that are known or suspected to be related to modulating lipids, blood pressure, vasomotor tone, inflammation or thrombosis. Association studies have included genes related to coagulation and platelet aggregation (prothrombin, Factor V Leiden), leukocyte activation (IL-6, E-Selectin, ICAM), and endothelial function (NOS3), amongst others (reviewed in 7, 8, 23).
Unfortunately, these studies have shed little light onto the pathophysiology of PAD. Plagued by small sample sizes and inadequate statistical power, many of the originally positive studies have not been successfully replicated in independent cohorts, suggesting the original association was falsely positive 24. Concerns over inadequate matching of racial/ethnic groups (who are known to have different rates of PAD as well as different allele frequencies) are justified by the possibility that ‘population stratification’ may lead to spurious associations 25. Moreover, a significant proportion of the candidate gene studies reported to date do not conform to Hardy-Weinberg equilibrium (i.e. the genotype was not distributed across the population as predicted by classical genetics) suggesting systematic genotyping errors or selection bias 23. Perhaps most concerning, a number of these studies appear to have been un-blinded, additionally casting doubt on their conclusions.
Taken together, these candidate gene studies are inconclusive. Certain variants that have been identified appear promising and warrant additional investigation, such as polymorphisms in the homocysteine pathway regulating enzyme MTHFR26-29, the inflammatory cytokine IL-6 30, 31, and the vascular adhesion molecule ICAM30, 32. Finally, a major limitation of candidate gene studies is that they are not likely to uncover novel mechanisms, as the choice of gene variants is typically determined by previous observations indicating a putative role for the gene in atherosclerosis.
Linkage Studies
Family-based linkage studies have also been applied to understanding vascular diseases 33. In this approach, the genome is scanned at a low resolution (every 10 centimorgans) for markers known as microsatellites that are co-inherited with the phenotype of interest (typically several million base pairs between tags). Once a marker has been firmly associated within the affected members of the study families, the surrounding region of the genome is fine-mapped to identify the causal gene which is ‘linked’ to the microsatellite and the disease. This method is conceptually superior to the candidate gene approach in that it does not require an a priori hypothesis about which gene is responsible for the disease and it scans the entire genome. This approach has been powerful in a number of Mendelian diseases, both vascular (e.g. NOTCH3 signaling in the autosomal dominant stroke syndrome, CADASIL34, 35) and otherwise (e.g. sarcomeric proteins in hypertrophic cardiomyopathy 36).
In the realm of PAD, however, the results have been somewhat disappointing. To date, only one positive linkage study has been reported. This study of 116 extended Icelandic families (884 subjects) identified a locus on chromosome 1p31 that was associated with angiographically or surgically documented PAD 9. This locus, known as PAOD1, had a logarithm of odds (LOD) score of 3.93 (> 3 is significant). Moreover, this locus was independent of other vascular risk factors and the association was strengthened when stroke patients were removed. Together, these findings suggest that the associated gene may specifically predispose patients to vascular disease in the lower extremity vascular bed. Unfortunately, the causative gene on Chromosome 1 has not yet been identified and this association has not been replicated by other investigators. The only other sizable linkage study performed to date, which utilized only ∼ 1/3 the number of microsatellite makers studied in the preceding study, failed to definitively identify a significant genomic locus 19.
To be effective, linkage studies require extended family pedigrees and genes with large effect sizes 37. As PAD tends to affect older adults, it is difficult to compile large collections of affected families. Further, this disease is polygenic and results from a number of factors each with a modest effect on risk, rather than one dominant gene that will be easily detected. Together, it is not surprising that this approach has not been met with more success in PAD.
GWAS Studies
Recent major advances in human genetics promise to overcome each of these deficits 38. In 2001, the Human Genome Project was completed, fully codifying the 3.1 billion nucleotides that make up our genetic code. Perhaps even more importantly, the International HapMap project provided a ‘catalog’ of common polymorphisms across the genome in 2004, allowing us for the first time to study the natural variation that makes each individual unique. These tools, combined with technological advances that have enabled the relatively inexpensive genotyping of millions of ‘tag’ SNPs simultaneously, have revolutionized modern genomics research. Employing the genome-wide association study (GWAS) approach, researchers can now scan the full genomes of large cohorts of patients to identify variants that are over-represented in subjects with a given disease compared to unaffected controls (see review in 38, 39). Unlike the candidate gene approach, the GWAS platform allows for an agnostic approach where no prior knowledge is required to implicate novel biological pathways 40. Further, the genome-wide approach allows for the consideration of polymorphisms in so-called ‘gene-deserts’ that do not encode any of the known ∼20,000 protein coding genes. These areas may contain non-coding elements that can modify gene expression, such as long-noncoding RNAs.
Unlike conventional linkage analysis using microsatellites, the GWAS technology scans the genome at much higher resolution and with greater fidelity. Moreover, late-onset diseases with high mortality rates (such as PAD) can be studied without the need for extended family pedigrees, and genes with modest effect size (OR of 1.2) can be detected. One drawback is that this approach requires large numbers of subjects (thousands), and because of multiple comparisons, the level of significance must be very high for a variant to be reliably associated with disease. Nevertheless, this approach has already provided revolutionary insights in several fields, as typified by the implication of the complement pathway in macular degeneration, autophagy in Crohn's disease, and hedgehog signaling in human height 41-43.
Since 2006, multiple loci have been definitively associated with cardiovascular disease44. These lead SNPs have been replicated by several consortia in multiple racial ethnic groups. The strongest and most consistent association with cardiovascular disorders is with the intergenic portion of Chromosome 9p21 (p = 1.6 ×10−25) 11, which has been queried in well over 100,000 patients 45, 46. Variants at this locus have been reported to be responsible for as much as 21% of one's lifetime risk of myocardial infarction (MI) 47. Importantly, these same polymorphisms also correlate with risk in the periphery, including aneurysmal disease, stroke and arterial stiffness 12, 48. The link to peripheral vascular disorders is most robust for AAA and intracranial aneurysms (accounting for 26% of the attributable risk for these diseases), and persists even after removing patients with a history of MI. A follow up study on the representative 9p21 SNP rs1333049supported this association with PAD status as well as severity of ABI in a cohort of older individuals (mean age 76) 49. The association persisted after controlling for diabetes, smoking, lipid levels, prior MI and hypertension, suggesting a novel pathophysiological mechanism at play.
The fact that the 9p21 SNPs correlate with disease status independently of known traditional risk factors has increased the enthusiasm for understanding the biology mediated by these SNPs. It is notable that these same SNPs also predict risk of the intracranial non-atherosclerotic berry aneurysm 12. Taken together, we and others have postulated that 9p21 likely does not alter vessel wall inflammation, thrombosis or lipid accumulation- but rather regulates the structural integrity of the artery itself. Given the particularly strong link to aneurysmal disorders, it is likely that the dominant role of 9p21 variants in vivo will center around vascular smooth muscle cell physiology and cell-fate decision making 50.
While much work has focused on the 9p21 hits described above, it is worth highlighting that a number of other GWAS polymorphisms have been implicated as significant at the genome-wide level. As the majority of these localize to genes not previously implicated in vascular disease, they certainly warrant investigation and should also yield novel biological targets. For example, one of the most exciting leads comes from a recent GWAS which found an associated SNP within a cluster of genes that encode nicotinic acetylcholine (nACh) receptors 13. This variant not only correlated with PAD (10% of the attributable risk for disease), but also predicted nicotine dependence and number of cigarettes smoked per day. It may be that the link to PAD occurs indirectly (i.e. by increasing lifetime smoking burden) or possibly by directly modulating the effect of tobacco on the vasculature. In this regard, we have shown that nicotine can directly accelerate plaque neovascularization and atherosclerosis by stimulating vascular nAChreceptors 51, 52.
Whereas the genetic variations discovered by GWAS have been predictive of CAD as well as PAD, it is likely that genetic variations that are more specific for PAD ultimately will be uncovered. Indeed, it is already apparent that the conventional risk factors have different predictive value for PAD, with tobacco exposure and diabetes mellitus being stronger risk factors for PAD than is dyslipidemia 53, 54. Consistent with these differences in pathophysiological determinants is the observation of a different proteomic profile in patients with PAD and CAD (PAD/CAD), than those patients with CAD alone. Specifically, beta 2 microglobulin, cystatin C and C-reactive protein are each found in higher levels in PAD/CAD, and can be used together to stratify the risk of PAD in a susceptible population 55, 56.
Use of genetics in PAD detection and treatment
A greater understanding of the genetic underpinnings of PAD could enhance our capacity to detect those at risk for disease. PAD is underdiagnosed, and these patients are not receiving medication known to reduce morbidity and mortality. Only 10-30% of PAD patients manifest the ‘classic’ symptom of intermittent claudication 3, 57. While the ABI test, which measures the ratio of blood pressure in the lower and upper extremities, is a simple and useful office-based technique to detect PAD 58, evidence indicates that this test is underutilized. In fact, the PARTNERS (Peripheral Arterial Disease Detection, Awareness, and Treatment in Primary Care) ABI screening study of nearly 7,000 adult general medicine patients found that over half of those with PAD had been previously undetected 3. Even when widely implemented, the ABI does not correlate with functional status, and only poorly with disease progression 59, 60. A blood test that could identify those at greatest risk for PAD could focus additional screening toward those populations.
Currently, genetic screening to assess the risk or progression of PAD is far from being realized. However, the possibility of such screening in the future is foreshadowed by recent advances in cardiac transplantation. In cardiac transplant recipients, the AlloMap test (which measures the expression of 11 genes in peripheral blood samples) has been shown to accurately predict allograft rejection 61. This blood test has the potential of replacing the current screening methodology which is invasive and expensive (ie. frequent endomyocardial biopsies and echocardiograms). Clearly, standardized, off-the-shelf blood tests that will obviate the need for technical imaging studies would greatly enhance our ability to rapidly identify at-risk individuals, initiate therapies early in the course of disease, and prognosticate with greater accuracy. Given the fact that as many as 60 million Americans technically meet the guideline criteria for a screening ABI, it is clear that an effective PAD panel could focus health care resources on those most at risk 55.
More powerful tools, that will arise from our forthcoming ability to rapidly and cheaply sequence the entire genome, will certainly provide further insights, point out additional targets and allow for therapies tailored to an individual's personal genetic makeup. To this final point, the field of pharmacogenomics, which studies the role of genetic differences in the response to drug therapy, has been rapidly expanding. A clear example relevant to PAD comes from the discovery that carriers of the common loss-of-function cytochrome p-450 enzyme polymorphism,CYP2C19, metabolize clopidogrel (the most efficacious antiplatelet drug in PAD 62) significantly more slowly than unaffected controls (reviewed in 63). These patients have reduced platelet inhibition on standard clopidogrel doses and experience a higher risk of major adverse cardiovascular events. As such, the FDA has provided a “black-box” warning for this agent in so-called “poor-metabolizers”. We do not yet know if higher doses of clopidogrel will reduce the risk of future event (without increasing bleeding). However, it is clear that clinicians will soon need to become facile with the concept of pharmacogenomics when choosing a drug for a particular patient 64. Before long, we anticipate that a patient's genetic variations will be documented and will factor into drug selection similarly to other current considerations, such as age, weight and concurrent medications.
The future of PAD genetics
Gene-by-environment interactions
Gene-by-environment (GxE) interactions are hypothesized to play an important role in the expression of disease, and have spurred investigations into the pathophysiological synergy of genomic and environmental exposures. Recently, this topic was approached with the first Environment-Wide Association Study (“EWAS”) which identified the pesticide heptachlor epoxide as being associated with type II diabetes 65. Application of similar ‘environment-wide’ scans will likely also be useful in PAD.
Epigenetic factors
Additionally, we will become more sophisticated in our evaluation of noncoding RNAs and other epigenetic modifications that promote peripheral arterial disease. The microRNAs are diffusible ∼ 22 nucleotide single-stranded RNAs that target coding mRNA transcripts for degradation 66. Though discovered less than a decade ago, they are now known to regulate upwards of 50% of the entire genome. This remarkable pathway, which consists of only ∼ 1000 genes, has already been implicated in a number of pathways relevant to PAD including impaired diabetic neoangiogenesis, smooth muscle remodeling, and circulating endothelial progenitor cell number 67-70. The advent of microarrays that can analyze the small RNA subfraction will certainly identify other microRNAs which are relevant to peripheral arterial disease.
Beyond microRNAs, tools now also exist to measure other epigenetic factors in PAD, such as histone modification, chromatin remodeling and DNA methylation 71. These processes induce structural modifications to the DNA molecule, rather than alterations in the DNA sequence, itself. Changes in DNA methylation or chromatin modifications make a given gene more or less amenable to transcription, and thus can alter the expression of that gene. Epigenetic alterations may occur postnatally and reproducibly localize to certain regions of the chromosome in disease. Detecting these patterns can provide insights that will implicate causative genes. It is possible that epigenetic ‘signatures’ can even be inherited across families, and that traits acquired due to environmental exposure can be passed to offspring to contribute to the familial concentration of disease. While most of the cardiovascular epigenetic research completed to date has focused on pathways related generally to atherosclerosis and vascular biology 72, interesting studies are forthcoming in AAA disease 73 and should also prove informative in PAD.
Acquired mitochondrial genetic alterations
There is accumulating evidence for a mitochondriopathy in PAD that contributes to the impairment in functional capacity74. The mitochondriopathy may be due in part to acquired alterations in mitochondrial DNA (mtDNA). In the patient with PAD, intermittent claudication is associated with ischemia-reperfusion, a known stimulus for generation of reactive oxygen species75. Regular bouts of ischemia-reperfusion may damage mtDNA, which is particularly vulnerable due to its proximity to reactive oxygen species generated by the electron transport chain (ETC), and because of its lack of protective histones. Indeed, by comparison to limbs of age-matched subjects without PAD, the skeletal muscle mitochondria in the most affected limb of patients with PAD have almost 20-fold greater frequency of the 4977-bpmtDNA deletion76.These mitochondrial genetic abnormalities may explain in part the mitochondriopathy of PAD. Notably, the only ETC protein activity which seems unaffected in PAD skeletal muscle is that of complex II, which is encoded by nuclear, rather than mitochondrial, DNA74. However, similar mtDNA alterations can be seen in the unaffected limb in patients with unilateral PAD, suggesting that systemic oxidative stress or other factors may be contributing to the mtDNA abnormalities in PAD77. In any event, medical therapy to reduce mitochondrial injury or enhance mitochondrial function may represent an interesting therapeutic avenue in PAD78.
Comparative genomics
Known differences in susceptibility to hindlimb ischemia between mouse strains are also now being exploited to advance our understanding of peripheral arterial disease and angiogenesis. Using an elegant comparative genomics approach, Dokun and colleagues performed a linkage analysis on mice from 6 genetic backgrounds that had undergone femoral artery ligation 10. They identified a quantitative trait locus on chromosome 7 that influences wound healing in situations of tissue hypoxia and critical limb ischemia. Other investigators have found that this same locus also appears to control over 50% of the variability in infarct burden in a mouse model of ischemic stroke 79. While the molecular mechanisms downstream of this region have yet to be defined, this example highlights the power of leveraging the natural differences that occur due to genetic variation between inbred animal lineages.
Exomic and whole genome sequencing
The most important advances, however, are likely to occur to as a result of the even more powerful genetic tools on the horizon. While the GWAS platform has provided new insights that will reshape our approach to PAD genetics, this tool is not without limitations 38, 80, 81. High-density genotyping chips now can identify up to 2.5 million individual markers per subject, but rely on imputation to predict the remaining SNPs in our genome. Rare, ‘personal’ variants (i.e. occurring with a frequency of less than 1% across the population) are not accurately cataloged for consideration with the GWAS approach. Moreover, historical GWAS genotyping has had little if any power to detect so-called structural variants such as insertions, deletions and copy number variants - all of which have been implicated in vascular disease. The advent of exome and truly full genome sequencing, where all existing nucleotides are codified, will address these limitations. Companies such as Complete Genomics and Illumina now provide full genome sequencing services for as little as a few thousand dollars per patient (depending on order size 82), and can provide this information in a matter of days (versus the $3 billion and 10 years required to sequence the first genome in the Human Genome Project). As Moore's Law on the declining cost of computing continues to be outstripped by the falling price of sequencing, it will not be long until standard academic laboratories will have bench-top sequencers capable of scanning entire genomes for $1000 or less per genome83.
PAD collaborations and advanced informatics
Most importantly, this generation will see the creation of larger and more successful collaborations focused on the investigation of PAD. With superior phenotypic characterization and longer term follow-up, these international groups will organize studies specifically intended for combining datasets and optimizing future meta-analyses 23. Novel approaches will focus on collecting ‘genetically enriched’ subjects to examine the extremes of phenotypes. For example, our group has collected subjects with very early onset of atherosclerotic disease (<45 years old) in the ADVANCE study 84, and others have chosen to study the opposite end of the spectrum with subjects over 80 who have no medical comorbidities (the “Wellderly”).
In addition to the creation of consortia for conducting genetic studies of complex diseases such as PAD, there is considerable interest in leveraging the electronic medical record (EMR) for high throughput phenotyping to facilitate such studies. The Electronic Medical Records and Genomics (eMERGE) network (www.gwas.org) was established in 2007 to develop and implement methods for leveraging biorepositories linked to EMR systems for large-scale genomic research 85, 86. One of the participating sites in this network, Mayo Clinic, is conducting a GWAS of PAD that includes 1,648 cases and 1,675 controls87 recruited in the clinical setting and with linkage to the EMR. Mining of structured data from the EMR as well as natural language processing of unstructured text was used to obtain relevant covariates. 87
Conclusions
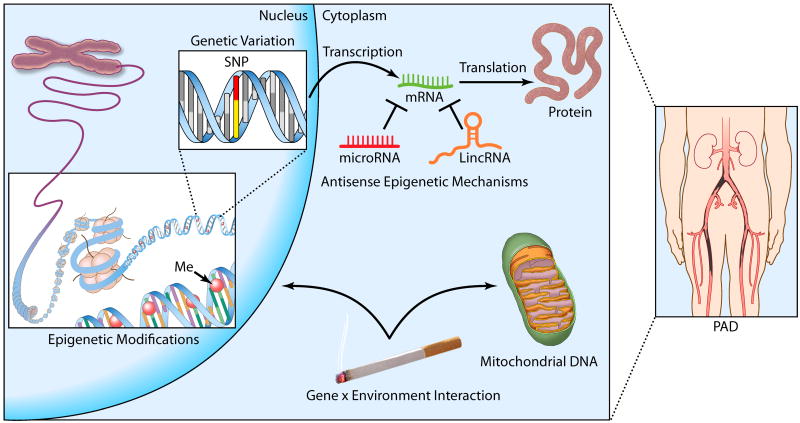
PAD is an important and highly prevalent condition with a heritable component. Vascular biologists and geneticists have begun to make inroads on this complex disease by applying increasingly sophisticated genetic approaches (Figure). We look forward to the next decade of research, as evolving technologies and interdisciplinary collaborations promise to finally provide insights on this often morbid condition.
Figure 1.
The Genetics of PAD. Factors associated with risk of peripheral arterial disease include variation in the genetic code (e.g. single nucleotide polymorphisms or SNPs) and epigenetic DNA modifications (e.g. cytosine methylation). Post-transcriptional regulation by non-coding RNAs may also alter expression of gene products relevant to vascular homeostasis. Finally, recently appreciated mitochondrial DNA variation and complex gene-by-environment interactions may also be involved in PAD pathogenesis, as discussed in the text.
Acknowledgments
Funding Sources: This work was supported by grants from the National Institutes of Health (K12HL087746and U01HL100397 to JPC and U0-1 HG-04599 to IJK) and the American Heart Association (10BGIA3290011 to NJL).
Footnotes
Conflict of Interest Disclosures: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gillum RF. Peripheral arterial occlusive disease of the extremities in the united states: Hospitalization and mortality. Am Heart J. 1990;120:1414–1418. doi: 10.1016/0002-8703(90)90257-x. [DOI] [PubMed] [Google Scholar]
- 2.Golomb BA, Dang TT, Criqui MH. Peripheral arterial disease: Morbidity and mortality implications. Circulation. 2006;114:688–699. doi: 10.1161/CIRCULATIONAHA.105.593442. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch AT, Criqui MH, Treat-Jacobson D, Regensteiner JG, Creager MA, Olin JW, Krook SH, Hunninghake DB, Comerota AJ, Walsh ME, McDermott MM, Hiatt WR. Peripheral arterial disease detection, awareness, and treatment in primary care. JAMA. 2001;286:1317–1324. doi: 10.1001/jama.286.11.1317. [DOI] [PubMed] [Google Scholar]
- 4.Gasse C, Jacobsen J, Larsen AC, Schmidt EB, Johannesen NL, Videbaek J, Sorensen HT, Johnsen SP. Secondary medical prevention among danish patients hospitalised with either peripheral arterial disease or myocardial infarction. Eur J Vasc Endovasc Surg. 2008;35:51–58. doi: 10.1016/j.ejvs.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Mechtouff L, Touze E, Steg PG, Ohman EM, Goto S, Hirsch AT, Rother J, Aichner FT, Weimar C, Bhatt DL, Alberts MJ, Mas JL. Worse blood pressure control in patients with cerebrovascular or peripheral arterial disease compared with coronary artery disease. J Intern Med. 2010;267:621–633. doi: 10.1111/j.1365-2796.2009.02198.x. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch AT, Murphy TP, Lovell MB, Twillman G, Treat-Jacobson D, Harwood EM, Mohler ER, 3rd, Creager MA, Hobson RW, 2nd, Robertson RM, Howard WJ, Schroeder P, Criqui MH. Gaps in public knowledge of peripheral arterial disease: The first national pad public awareness survey. Circulation. 2007;116:2086–2094. doi: 10.1161/CIRCULATIONAHA.107.725101. [DOI] [PubMed] [Google Scholar]
- 7.Knowles JW, Assimes TL, Li J, Quertermous T, Cooke JP. Genetic susceptibility to peripheral arterial disease: A dark corner in vascular biology. Arterioscler Thromb Vasc Biol. 2007;27:2068–2078. doi: 10.1161/01.ATV.0000282199.66398.8c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katwal AB, Dokun AO. Peripheral arterial disease in diabetes: Is there a role for genetics? Curr Diab Rep. 2011;11:218–225. doi: 10.1007/s11892-011-0188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudmundsson G, Matthiasson SE, Arason H, Johannsson H, Runarsson F, Bjarnason H, Helgadottir K, Thorisdottir S, Ingadottir G, Lindpaintner K, Sainz J, Gudnason V, Frigge ML, Kong A, Gulcher JR, Stefansson K. Localization of a gene for peripheral arterial occlusive disease to chromosome 1p31. Am J Hum Genet. 2002;70:586–592. doi: 10.1086/339251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dokun AO, Keum S, Hazarika S, Li Y, Lamonte GM, Wheeler F, Marchuk DA, Annex BH. A quantitative trait locus (lsq-1) on mouse chromosome 7 is linked to the absence of tissue loss after surgical hindlimb ischemia. Circulation. 2008;117:1207–1215. doi: 10.1161/CIRCULATIONAHA.107.736447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A genome-wide association study in europeans and south asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 12.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, Jaaskelainen JE, Kyo Y, Lenk GM, Sakalihasan N, Kostulas K, Gottsater A, Flex A, Stefansson H, Hansen T, Andersen G, Weinsheimer S, Borch-Johnsen K, Jorgensen T, Shah SH, Quyyumi AA, Granger CB, Reilly MP, Austin H, Levey AI, Vaccarino V, Palsdottir E, Walters GB, Jonsdottir T, Snorradottir S, Magnusdottir D, Gudmundsson G, Ferrell RE, Sveinbjornsdottir S, Hernesniemi J, Niemela M, Limet R, Andersen K, Sigurdsson G, Benediktsson R, Verhoeven EL, Teijink JA, Grobbee DE, Rader DJ, Collier DA, Pedersen O, Pola R, Hillert J, Lindblad B, Valdimarsson EM, Magnadottir HB, Wijmenga C, Tromp G, Baas AF, Ruigrok YM, van Rij AM, Kuivaniemi H, Powell JT, Matthiasson SE, Gulcher JR, Thorgeirsson G, Kong A, Thorsteinsdottir U, Stefansson K. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 13.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsater A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizuguchi T, Matsumoto N. Recent progress in genetics of marfan syndrome and marfan-associated disorders. J Hum Genet. 2007;52:1–12. doi: 10.1007/s10038-006-0078-1. [DOI] [PubMed] [Google Scholar]
- 15.Meijer WT, Grobbee DE, Hunink MG, Hofman A, Hoes AW. Determinants of peripheral arterial disease in the elderly: The rotterdam study. Arch Intern Med. 2000;160:2934–2938. doi: 10.1001/archinte.160.19.2934. [DOI] [PubMed] [Google Scholar]
- 16.Valentine RJ, Verstraete R, Clagett GP, Cohen JC. Premature cardiovascular disease is common in relatives of patients with premature peripheral atherosclerosis. Arch Intern Med. 2000;160:1343–1348. doi: 10.1001/archinte.160.9.1343. [DOI] [PubMed] [Google Scholar]
- 17.Wahlgren CM, Magnusson PK. Genetic influences on peripheral arterial disease in a twin population. Arterioscler Thromb Vasc Biol. 2011;31:678–682. doi: 10.1161/ATVBAHA.110.210385. [DOI] [PubMed] [Google Scholar]
- 18.Carmelli D, Fabsitz RR, Swan GE, Reed T, Miller B, Wolf PA. Contribution of genetic and environmental influences to ankle-brachial blood pressure index in the nhlbi twin study. National heart, lung, and blood institute. Am J Epidemiol. 2000;151:452–458. doi: 10.1093/oxfordjournals.aje.a010230. [DOI] [PubMed] [Google Scholar]
- 19.Kullo IJ, Turner ST, Kardia SL, Mosley TH, Jr, Boerwinkle E, de Andrade M. A genome-wide linkage scan for ankle-brachial index in african american and non-hispanic white subjects participating in the genoa study. Atherosclerosis. 2006;187:433–438. doi: 10.1016/j.atherosclerosis.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Murabito JM, Guo CY, Fox CS, D'Agostino RB. Heritability of the ankle-brachial index: The framingham offspring study. Am J Epidemiol. 2006;164:963–968. doi: 10.1093/aje/kwj295. [DOI] [PubMed] [Google Scholar]
- 21.Risch N. The genetic epidemiology of cancer: Interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev. 2001;10:733–741. [PubMed] [Google Scholar]
- 22.Kwon JM, Goate AM. The candidate gene approach. Alcohol Res Health. 2000;24:164–168. [PMC free article] [PubMed] [Google Scholar]
- 23.Zintzaras E, Zdoukopoulos N. A field synopsis and meta-analysis of genetic association studies in peripheral arterial disease: The cumagas-pad database. Am J Epidemiol. 2009;170:1–11. doi: 10.1093/aje/kwp094. [DOI] [PubMed] [Google Scholar]
- 24.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: An approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Sofi F, Lari B, Rogolino A, Marcucci R, Pratesi G, Dorigo W, Pratesi C, Gensini GF, Abbate R, Prisco D. Thrombophilic risk factors for symptomatic peripheral arterial disease. J Vasc Surg. 2005;41:255–260. doi: 10.1016/j.jvs.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Fowkes FG, Lee AJ, Hau CM, Cooke A, Connor JM, Lowe GD. Methylene tetrahydrofolate reductase (mthfr) and nitric oxide synthase (ecnos) genes and risks of peripheral arterial disease and coronary heart disease: Edinburgh artery study. Atherosclerosis. 2000;150:179–185. doi: 10.1016/s0021-9150(99)00366-4. [DOI] [PubMed] [Google Scholar]
- 28.Sabino A, Fernandes AP, Lima LM, Ribeiro DD, Sousa MO, de Castro Santos ME, Mota AP, Dusse LM, das Gracas Carvalho M. Polymorphism in the methylenetetrahydrofolate reductase (c677t) gene and homocysteine levels: A comparison in brazilian patients with coronary arterial disease, ischemic stroke and peripheral arterial obstructive disease. J Thromb Thrombolysis. 2009;27:82–87. doi: 10.1007/s11239-007-0172-z. [DOI] [PubMed] [Google Scholar]
- 29.Todesco L, Angst C, Litynski P, Loehrer F, Fowler B, Haefeli WE. Methylenetetrahydrofolate reductase polymorphism, plasma homocysteine and age. Eur J Clin Invest. 1999;29:1003–1009. doi: 10.1046/j.1365-2362.1999.00578.x. [DOI] [PubMed] [Google Scholar]
- 30.Flex A, Gaetani E, Angelini F, Sabusco A, Chilla C, Straface G, Biscetti F, Pola P, Castellot JJ, Jr, Pola R. Pro-inflammatory genetic profiles in subjects with peripheral arterial occlusive disease and critical limb ischemia. J Intern Med. 2007;262:124–130. doi: 10.1111/j.1365-2796.2007.01791.x. [DOI] [PubMed] [Google Scholar]
- 31.Flex A, Gaetani E, Pola R, Santoliquido A, Aloi F, Papaleo P, Dal Lago A, Pola E, Serricchio M, Tondi P, Pola P. The -174 g/c polymorphism of the interleukin-6 gene promoter is associated with peripheral artery occlusive disease. Eur J Vasc Endovasc Surg. 2002;24:264–268. doi: 10.1053/ejvs.2002.1711. [DOI] [PubMed] [Google Scholar]
- 32.Gaetani E, Flex A, Pola R, Papaleo P, De Martini D, Pola E, Aloi F, Flore R, Serricchio M, Gasbarrini A, Pola P. The k469e polymorphism of the icam-1 gene is a risk factor for peripheral arterial occlusive disease. Blood Coagul Fibrinolysis. 2002;13:483–488. doi: 10.1097/00001721-200209000-00002. [DOI] [PubMed] [Google Scholar]
- 33.Tromp G, Kuivaniemi H. Developments in genomics to improve understanding, diagnosis and management of aneurysms and peripheral artery disease. Eur J Vasc Endovasc Surg. 2009;38:676–682. doi: 10.1016/j.ejvs.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, Maciazek J, Vayssiere C, Cruaud C, Cabanis EA, Ruchoux MM, Weissenbach J, Bach JF, Bousser MG, Tournier-Lasserve E. Notch3 mutations in cadasil, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 35.Ducros A, Nagy T, Alamowitch S, Nibbio A, Joutel A, Vahedi K, Chabriat H, Iba-Zizen MT, Julien J, Davous P, Goas JY, Lyon-Caen O, Dubois B, Ducrocq X, Salsa F, Ragno M, Burkhard P, Bassetti C, Hutchinson M, Verin M, Viader F, Chapon F, Levasseur M, Mas JL, Delrieu O, et al. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy, genetic homogeneity, and mapping of the locus within a 2-cm interval. Am J Hum Genet. 1996;58:171–181. [PMC free article] [PubMed] [Google Scholar]
- 36.Franz WM, Muller OJ, Katus HA. Cardiomyopathies: From genetics to the prospect of treatment. Lancet. 2001;358:1627–1637. doi: 10.1016/S0140-6736(01)06657-0. [DOI] [PubMed] [Google Scholar]
- 37.Whittemore AS, Nelson LM. Study design in genetic epidemiology: Theoretical and practical considerations. J Natl Cancer Inst Monogr. 1999:61–69. doi: 10.1093/oxfordjournals.jncimonographs.a024228. [DOI] [PubMed] [Google Scholar]
- 38.Ding K, Kullo IJ. Genome-wide association studies for atherosclerotic vascular disease and its risk factors. Circ Cardiovasc Genet. 2009;2:63–72. doi: 10.1161/CIRCGENETICS.108.816751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manolio TA, Collins FS. The hapmap and genome-wide association studies in diagnosis and therapy. Annu Rev Med. 2009;60:443–456. doi: 10.1146/annurev.med.60.061907.093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hakonarson H, Grant SF. Planning a genome-wide association study: Points to consider. Ann Med. 2011 doi: 10.3109/07853890.2011.573803. [DOI] [PubMed] [Google Scholar]
- 41.Lee JC, Parkes M. Genome-wide association studies and crohn's disease. Brief Funct Genomics. 2011;10:71–76. doi: 10.1093/bfgp/elr009. [DOI] [PubMed] [Google Scholar]
- 42.Katta S, Kaur I, Chakrabarti S. The molecular genetic basis of age-related macular degeneration: An overview. J Genet. 2009;88:425–449. doi: 10.1007/s12041-009-0064-4. [DOI] [PubMed] [Google Scholar]
- 43.Ku CS, Loy EY, Pawitan Y, Chia KS. The pursuit of genome-wide association studies: Where are we now? J Hum Genet. 2010;55:195–206. doi: 10.1038/jhg.2010.19. [DOI] [PubMed] [Google Scholar]
- 44.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O'Donnell CJ, McPherson R, Erdmann J, Samani NJ. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 48.Bjorck HM, Lanne T, Alehagen U, Persson K, Rundkvist L, Hamsten A, Dahlstrom U, Eriksson P. Association of genetic variation on chromosome 9p21.3 and arterial stiffness. J Intern Med. 2009;265:373–381. doi: 10.1111/j.1365-2796.2008.02020.x. [DOI] [PubMed] [Google Scholar]
- 49.Cluett C, McDermott MM, Guralnik J, Ferrucci L, Bandinelli S, Miljkovic I, Zmuda JM, Li R, Tranah G, Harris T, Rice N, Henley W, Frayling TM, Murray A, Melzer D. The 9p21 myocardial infarction risk allele increases risk of peripheral artery disease in older people. Circ Cardiovasc Genet. 2009;2:347–353. doi: 10.1161/CIRCGENETICS.108.825935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heeschen C, Jang JJ, Weis M, Pathak A, Kaji S, Hu RS, Tsao PS, Johnson FL, Cooke JP. Nicotine stimulates angiogenesis and promotes tumor growth and atherosclerosis. Nat Med. 2001;7:833–839. doi: 10.1038/89961. [DOI] [PubMed] [Google Scholar]
- 52.Heeschen C, Weis M, Aicher A, Dimmeler S, Cooke JP. A novel angiogenic pathway mediated by non-neuronal nicotinic acetylcholine receptors. J Clin Invest. 2002;110:527–536. doi: 10.1172/JCI14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newman AB, Siscovick DS, Manolio TA, Polak J, Fried LP, Borhani NO, Wolfson SK. Ankle-arm index as a marker of atherosclerosis in the cardiovascular health study. Cardiovascular heart study (chs) collaborative research group. Circulation. 1993;88:837–845. doi: 10.1161/01.cir.88.3.837. [DOI] [PubMed] [Google Scholar]
- 54.Kennedy M, Solomon C, Manolio TA, Criqui MH, Newman AB, Polak JF, Burke GL, Enright P, Cushman M. Risk factors for declining ankle-brachial index in men and women 65 years or older: The cardiovascular health study. Arch Intern Med. 2005;165:1896–1902. doi: 10.1001/archinte.165.16.1896. [DOI] [PubMed] [Google Scholar]
- 55.Cooke JP, Wilson AM. Biomarkers of peripheral arterial disease. J Am Coll Cardiol. 2010;55:2017–2023. doi: 10.1016/j.jacc.2009.08.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson AM, Kimura E, Harada RK, Nair N, Narasimhan B, Meng XY, Zhang F, Beck KR, Olin JW, Fung ET, Cooke JP. Beta2-microglobulin as a biomarker in peripheral arterial disease: Proteomic profiling and clinical studies. Circulation. 2007;116:1396–1403. doi: 10.1161/CIRCULATIONAHA.106.683722. [DOI] [PubMed] [Google Scholar]
- 57.McDermott MM, Liu K, Greenland P, Guralnik JM, Criqui MH, Chan C, Pearce WH, Schneider JR, Ferrucci L, Celic L, Taylor LM, Vonesh E, Martin GJ, Clark E. Functional decline in peripheral arterial disease: Associations with the ankle brachial index and leg symptoms. Jama. 2004;292:453–461. doi: 10.1001/jama.292.4.453. [DOI] [PubMed] [Google Scholar]
- 58.Hiatt WR. Medical treatment of peripheral arterial disease and claudication. N Engl J Med. 2001;344:1608–1621. doi: 10.1056/NEJM200105243442108. [DOI] [PubMed] [Google Scholar]
- 59.Szuba A, Oka RK, Harada R, Cooke JP. Limb hemodynamics are not predictive of functional capacity in patients with pad. Vasc Med. 2006;11:155–163. doi: 10.1177/1358863x06074828. [DOI] [PubMed] [Google Scholar]
- 60.Long J, Modrall JG, Parker BJ, Swann A, Welborn MB, 3rd, Anthony T. Correlation between ankle-brachial index, symptoms, and health-related quality of life in patients with peripheral vascular disease. J Vasc Surg. 2004;39:723–727. doi: 10.1016/j.jvs.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 61.Pham MX, Teuteberg JJ, Kfoury AG, Starling RC, Deng MC, Cappola TP, Kao A, Anderson AS, Cotts WG, Ewald GA, Baran DA, Bogaev RC, Elashoff B, Baron H, Yee J, Valantine HA. Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362:1890–1900. doi: 10.1056/NEJMoa0912965. [DOI] [PubMed] [Google Scholar]
- 62.A randomised, blinded, trial of clopidogrel versus aspirin in patients at risk of ischaemic events (caprie). Caprie steering committee. Lancet. 1996;348:1329–1339. doi: 10.1016/s0140-6736(96)09457-3. [DOI] [PubMed] [Google Scholar]
- 63.Fuster V, Sweeny JM. Clopidogrel and the reduced-function cyp2c19 genetic variant: A limited piece of the overall therapeutic puzzle. JAMA. 2010;304:1839–1840. doi: 10.1001/jama.2010.1566. [DOI] [PubMed] [Google Scholar]
- 64.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364:1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patel CJ, Bhattacharya J, Butte AJ. An environment-wide association study (ewas) on type 2 diabetes mellitus. PLoS One. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Small EM, Frost RJ, Olson EN. Micrornas add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caporali A, Meloni M, Vollenkle C, Bonci D, Sala-Newby GB, Addis R, Spinetti G, Losa S, Masson R, Baker AH, Agami R, le Sage C, Condorelli G, Madeddu P, Martelli F, Emanueli C. Deregulation of microrna-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123:282–291. doi: 10.1161/CIRCULATIONAHA.110.952325. [DOI] [PubMed] [Google Scholar]
- 68.Leeper NJ, Cooke JP. Microrna and mechanisms of impaired angiogenesis in diabetes mellitus. Circulation. 2011;123:236–238. doi: 10.1161/CIRCULATIONAHA.110.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leeper NJ, Raiesdana A, Kojima Y, Chun HJ, Azuma J, Maegdefessel L, Kundu RK, Quertermous T, Tsao PS, Spin JM. Microrna-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011;226:1035–1043. doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao T, Li J, Chen AF. Microrna-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am J Physiol Endocrinol Metab. 2010;299:E110–116. doi: 10.1152/ajpendo.00192.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan MS, Matouk CC, Marsden PA. Epigenetics of the vascular endothelium. J Appl Physiol. 2010;109:916–926. doi: 10.1152/japplphysiol.00131.2010. [DOI] [PubMed] [Google Scholar]
- 72.Yla-Herttuala S, Glass CK. Review focus on epigenetics and the histone code in vascular biology. Cardiovasc Res. 2011;90:402–403. doi: 10.1093/cvr/cvr119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krishna SM, Dear AE, Norman PE, Golledge J. Genetic and epigenetic mechanisms and their possible role in abdominal aortic aneurysm. Atherosclerosis. 2010;212:16–29. doi: 10.1016/j.atherosclerosis.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 74.Pipinos II, Judge AR, Selsby JT, Zhu Z, Swanson SA, Nella AA, Dodd SL. The myopathy of peripheral arterial occlusive disease: Part 1. Functional and histomorphological changes and evidence for mitochondrial dysfunction. Vasc Endovascular Surg. 2007;41:481–489. doi: 10.1177/1538574407311106. [DOI] [PubMed] [Google Scholar]
- 75.Ramachandran A, Jha S, Lefer DJ. Review paper: Pathophysiology of myocardial reperfusion injury: The role of genetically engineered mouse models. Vet Pathol. 2008;45:698–706. doi: 10.1354/vp.45-5-698. [DOI] [PubMed] [Google Scholar]
- 76.Bhat HK, Hiatt WR, Hoppel CL, Brass EP. Skeletal muscle mitochondrial DNA injury in patients with unilateral peripheral arterial disease. Circulation. 1999;99:807–812. doi: 10.1161/01.cir.99.6.807. [DOI] [PubMed] [Google Scholar]
- 77.Brass EP, Wang H, Hiatt WR. Multiple skeletal muscle mitochondrial DNA deletions in patients with unilateral peripheral arterial disease. Vasc Med. 2000;5:225–230. [PubMed] [Google Scholar]
- 78.Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321:1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keum S, Marchuk DA. A locus mapping to mouse chromosome 7 determines infarct volume in a mouse model of ischemic stroke. Circ Cardiovasc Genet. 2009;2:591–598. doi: 10.1161/CIRCGENETICS.109.883231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baudhuin LM. Genetics of coronary artery disease: Focus on genome-wide association studies. Am J Transl Res. 2009;1:221–234. [PMC free article] [PubMed] [Google Scholar]
- 81.Moore JH, Asselbergs FW, Williams SM. Bioinformatics challenges for genome-wide association studies. Bioinformatics. 2010;26:445–455. doi: 10.1093/bioinformatics/btp713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.http://www.signonsandiego.com/news/2011/may/09/illumina-drops-human-sequencing-price-4000/
- 83.http://www.economist.com/node/16791936
- 84.Assimes TL, Knowles JW, Basu A, Iribarren C, Southwick A, Tang H, Absher D, Li J, Fair JM, Rubin GD, Sidney S, Fortmann SP, Go AS, Hlatky MA, Myers RM, Risch N, Quertermous T. Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic advance study. Hum Mol Genet. 2008;17:2320–2328. doi: 10.1093/hmg/ddn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, Struewing JP, Wolf WA. The emerge network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, Crane PK, Pathak J, Chute CG, Bielinski SJ, Kullo IJ, Li R, Manolio TA, Chisholm RL, Denny JC. Electronic medical records for genetic research: Results of the emerge consortium. Sci Transl Med. 2011;3:79re–71. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kullo IJ, Fan J, Pathak J, Savova GK, Ali Z, Chute CG. Leveraging informatics for genetic studies: Use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc. 2010;17:568–574. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]