Abstract
The structural microheterogeneity of heparin and heparan sulfate is one of the major reasons for the multifunctionality exhibited by this class of molecules. In a physiological context, these molecules primarily exert their effects extracellularly by mediating key processes of cellular cross-talk and signaling leading to the modulation of a number of different biological activities including development, cell proliferation, and inflammation. This structural diversity is biosynthetically imprinted in a nontemplate-driven manner and may also be dynamically remodeled as cellular function changes. Understanding the structural information encoded in these molecules forms the basis for attempting to understand the complex biology they mediate. This chapter provides an overview of the origin of the structural microheterogeneity observed in heparin and heparan sulfate, and the orthogonal analytical methodologies that are required to help decipher this information.
Keywords: Heparan sulfate, Heparin, Microheterogeneity, Biosynthesis, Structural analysis, Conformation, Mass spectrometry, Nuclear magnetic resonance, Structure–function relationships
1 Introduction
Heparin and heparan sulfate are complex, linear, acidic polysaccharides belonging to the glycosaminoglycan (GAG) family. In higher organisms, they can be found primarily on the cell surface or in the extracellular matrix, attached to a protein core. The structural diversity of heparin and heparan sulfate (HS) lies at the core of the varied range of physiological processes these molecules tend to modulate. Heparin is a well-known anticoagulant drug and is extensively used in medical practice (Wardrop and Keeling 2008). Heparin is isolated from animal organs, predominantly porcine intestinal mucosa, and goes through an extensive process of purification before it can be used for pharmaceutical purposes (Linhardt and Gunay 1999; Liu et al. 2009). The molecular basis for the anticoagulant function of heparin was elucidated in the early 1980s when a distinct pentasaccharide sequence within heparin chains was identified as being crucial for binding and activating antithrombin, leading to accelerated inhibition of the coagulation cascade (Lindahl et al. 1979; Rosenberg and Lam 1979). Identification of this unique structure–function relationship further strengthened the hypothesis that the variable sulfation within heparin and heparan sulfate encodes information that forms the basis for regulating other physiological activities as well.
Over the last 20 years, heparin and heparan sulfate have been shown to interact with a large number of important proteins thereby regulating a range of biological activities including cell proliferation, inflammation, angiogenesis, viral infectivity and development. Due to the structural diversity exhibited by these molecules, it is believed that possibly unique (in some cases) or most likely an ensemble of structural motifs might be responsible for different interactions. Therefore, it has become increasingly important to interpret the structural information represented in these complex molecules in order to enable a better understanding of their structure–function relationships.
The structural microheterogeneity is predominantly biosynthetically imprinted in a nontemplate-driven manner, as chains of heparin or heparan sulfate are elongated in the Golgi (Salmivirta et al. 1996). Furthermore, the purification and manufacture of heparin also leads to the introduction of additional chemical heterogeneity. Structural heterogeneity, coupled with a lack of ex vivo tools to amplify specific structures, makes the structural analysis of heparin/heparan sulfate extremely challenging. It is becoming increasingly clear that efficient, accurate structural analysis requires orthogonal analytical approaches to help decipher the information encoded in the saccharide sequences. As such, this chapter focuses on understanding the structural microheterogeneity of heparin and heparan sulfate; how it arises, and methods that have been developed to analyze this structural diversity.
2 Biosynthesis of Heparin: Encoding Microheterogeneity
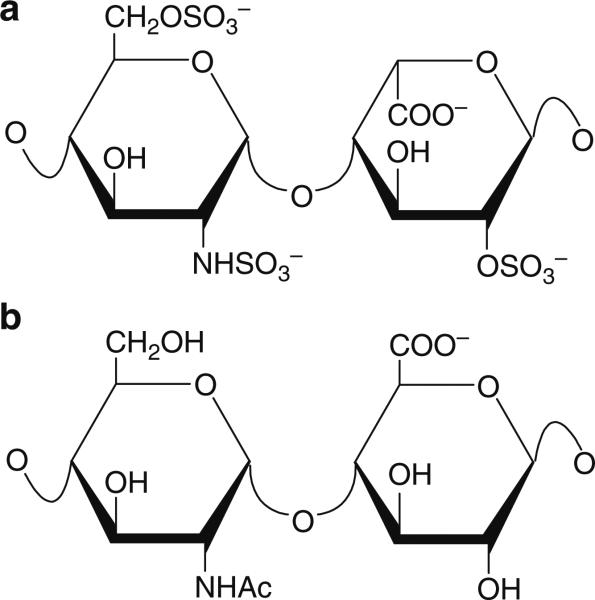
Heparin and heparan sulfate are built up of linear chains of repeating disaccharide units consisting of a glucosamine and uronic acid. The initial disaccharide unit that constitutes the growing chain during biosynthesis has a d-glucuronic acid β1 → 4 linked to a d-N-acetylglucosamine. These units are linked to each other via an α1 → 4 linkage. The subsequent modifications proceed in a sequential manner, beginning with the N-deacetylation and N-sulfonation of glucosamine residues within the chains. This is followed by epimerization of the glucuronic acid to iduronic acid and O-sulfonation at either the C-2 of the uronic acid or the C-6 of the glucosamine. The final modification step in this pathway is the O-sulfonation at the C-3 of the glucosamine. Each of these biosynthetic reactions is dependent on the previous modification to some extent, as products of one step can often act as substrates for subsequent steps. Another aspect of this pathway that has an important bearing on saccharide structural diversity is that each of these biosynthetic modification steps likely does not proceed to completion. Thus, the resulting chain can be differentially modified in various regions, accounting for a significant component of the structural heterogeneity observed in these molecules (Salmivirta et al. 1996; Sugahara and Kitagawa 2002). The structure of the basic disaccharide sequence in heparin and heparan sulfate along with the sites of variable sulfation is shown in Fig. 1. The average heparin disaccharide contains ~2.7 sulfate groups, whereas heparan sulfate contains ≥1 sulfate group per disaccharide (Toida et al. 1997). Also, while l-iduronic acid predominates in heparin, the d-glucuronic acid epimer represents the majority of the uronic acid present in heparan sulfate (Lindahl et al. 1998). Heparin is often referred to as the more completely modified version of heparan sulfate and also possesses the highest negative charge density of any known biological macromolecule. While heparan sulfate contains all of the structural variations found in heparin, the frequency of occurrence of the minor sequence variations is greater in HS; thus, the extent of structural heterogeneity observed in heparan sulfate is usually greater than that observed in heparin. However, both heparin and heparan sulfate chains are polydisperse, with a broad molecular weight distribution. Heparan sulfate chains are generally longer than heparin chains, and have an average molecular weight of ~30 kDa as compared to ~15 kDa for heparin. This structural variability at multiple levels makes heparin and heparan sulfate very challenging molecules to characterize.
Fig. 1.
The major repeating disaccharide unit in (a) heparin and (b) heparan sulfate. Structural heterogeneity arises due to the variable presence of either acetyl or sulfo groups at the N – position of the glucosamine, sulfation at the 2-O – position on the uronic acid or 6-O – and 3-O – positions on the glucosamine, and epimerization at the C-5 of the uronic acid
In addition to the structural heterogeneity, heparin and heparan sulfate also exhibit conformational flexibility due to the presence of iduronic acid in their linear sequence (Ferro et al. 1990; Sanderson et al. 1987). This provides an additional dimension through which chains of heparin and heparan sulfate can alter the spatial orientation of their sulfate groups to allow for productive binding to various proteins. Co-crystal structures of a heparin hexasaccharide with basic fibroblast growth factor demonstrate this phenomenon (Canales et al. 2005; Faham et al. 1996). One of the sulfated iduronic acid residues in the saccharide is observed to contact the protein in a 1C4 conformation. This conformation is typically not favored by the residue in aqueous solution.
3 Purification and Isolation of Heparin
Heparin is one of the most widely used carbohydrate drugs, and has been used as a pharmaceutical product for several decades. Clinically, heparin is used as a prophylactic agent to prevent the formation of thrombi, as well as for their initial treatment. Heparin is a natural product that is isolated from animal tissues. Currently, heparin is sourced almost exclusively from a single source, viz., porcine intestine, though other animal sources of heparin have been reported (Casu et al. 1995, 1996). Heparin from bovine sources, especially bovine lung, had been employed previously, but current use is not widespread due to concerns of BSE contamination. There are efforts ongoing to create heparin through defined chemical sulfonation of the appropriate saccharide backbone, coupled with in vitro enzymatic modification of chains through the use of recombinantly expressed enzymes of the biosynthetic pathway (Kuberan et al. 2003; Liu et al. 2010).
As behooves the fact that it is derived biosynthetically, the purification of heparin is a multistep involved process. Several basic steps are used in the initial purification of heparin from porcine intestinal mucosa to form crude heparin. Preparation of raw or crude heparin is typically performed outside of cGMP (Good Manufacturing Practices) and involves the physical separation of mucosa from the gut lining, solubilization through the addition of proteases, multiple precipitation and resolubilization steps, followed by a final ethanolic precipitation step (Liu et al. 2009). At this stage, crude heparin is typically a mixture of many components. Besides residual biomaterials, such as proteins, nucleic acids, and lipids, crude heparin contains a mixture of glycosaminoglycan complex polysaccharides, such as hyaluronic acid, chondroitin sulfate, as well as the two predominant components – heparin and dermatan sulfate.
Crude heparin is then transported into a cGMP facility for additional purification, testing and release. Depending on the manufacturer, different purification strategies are employed; broadly these involve additional precipitations, ion capture and exchange, and chemical steps to decolorize and depyrogenate heparin. Upon completion of final polishing steps, heparin is a fairly well-defined material, predominantly composed of glycosaminoglycan chains with some degree of polydispersity, but an average molecular weight that consistently is within the range of 12–20 kDa. It also has a fairly defined composition, primarily consisting of repeating units of the trisulfated disaccharide, i.e., a 2-O-sulfonated iduronic acid 1 → 4 linked to a 6-O, N-sulfonated glucosamine (Fig. 1).
Differential precipitation steps used in the purification of heparin are partly the reason for unfractionated heparin being one of the most highly sulfated glycosaminoglycans. Studies looking at waste material, so-called “side stream” heparin, indicate that undersulfated and/or lower molecular weight material is removed from the final product, yielding heparin chains with an average sulfate-to-carboxylate ratio, a measure of the degree of sulfation per average disaccharide, of 2.4-2.6 (Liu et al. 2009). Furthermore, this purification strategy likely limits the amount of undersulfated materials, including porcine-based heparan sulfate, which can be present within a sample. Porcine heparan sulfate, which may be created biosynthetically and characterized by a sulfate-to-carboxylate ratio of ~0.3 (Toida et al. 1997), is probably removed early on in the purification process, if even present at appreciable levels. Indeed, examination of crude heparin preparations indicates few, if any, of the signs of the presence of heparan sulfate. Additionally, considering that, at later polishing stages, precipitation results in the removal of heparin material with a higher degree of sulfation than HS, it is highly unlikely that appreciable levels of “true” heparan sulfate are present within heparin preparations. Conversely, dermatan sulfate, a higher sulfated glycosaminoglycan that contains a high degree of iduronic acid, has been shown to be an impurity in heparin preparations.
Apart from the inherent structural variability among the heparin chains, both in terms of sulfation and chain length, there are structural variations that are introduced as a function of the purification process, especially chemical treatment to decolor, deodorize, and depyrogenate heparin samples. Because a variety of chemical agents can be employed, including peroxide treatment under alkaline conditions, permanganate, hypochlorite, and ozone, the type and extent of modifications that can occur are variable and can be indicative of the manufacturing process employed.
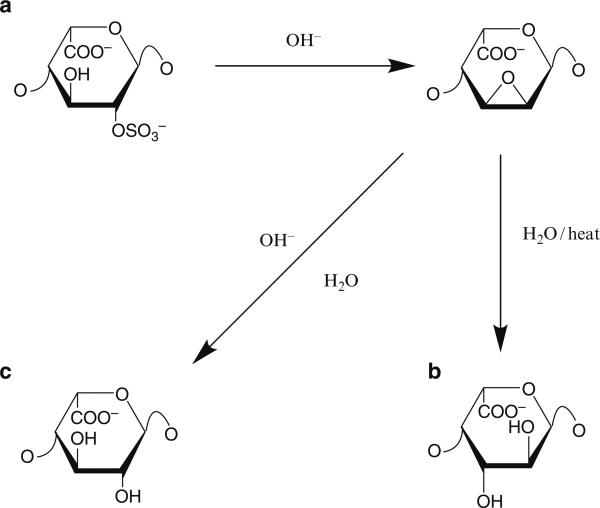
Under basic conditions, it is clear that some amount of 2-O sulfonated iduronic acid can be converted to an intermediate 2, 3-epoxide, followed by conversion to galacturonic acid (Fig. 2). We and others have found that this can constitute up to ~2–3% of the total disaccharide content of heparin (Rej et al. 1989). Additionally, under basic conditions, 6-O sulfonated glucosamine can undergo cyclization to form a 1,6 anhydro ring. Finally, peeling reactions via Millard chemistry can also occur, resulting in loss of a monosaccharide and the production of “odd-numbered” chains.
Fig. 2.
Sensitivity of the 2-O-sulfated iduronic acid in heparin to manufacturing conditions. Conversion of 2-O-sulfated iduronic acid in heparin to (a) 2,3 epoxide upon treatment with base. Subsequently, the 2,3 epoxide can convert to either (b) a galacturonic acid species on heat treatment, or (c) an iduronic acid (at higher pH)
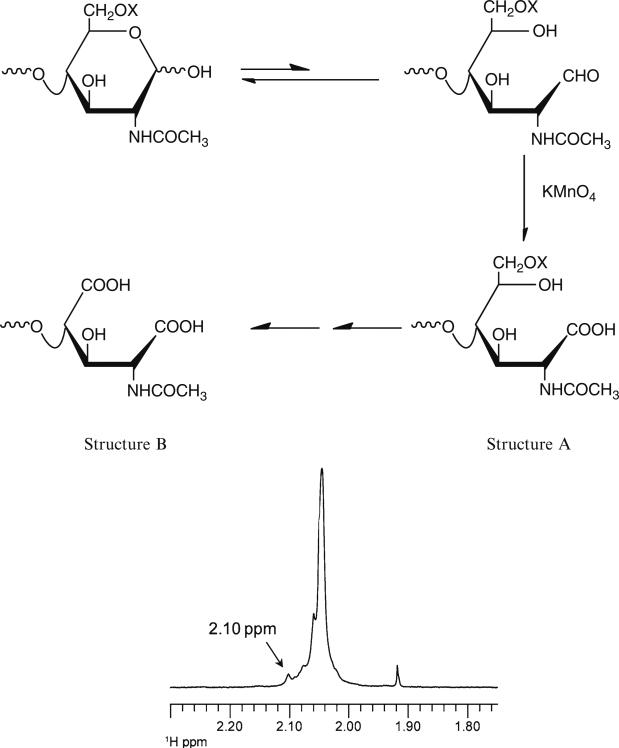
In addition to these reactions, which can occur to an appreciable extent, there are other minor modifications that can occur. Several groups have identified the fact that addition of oxidizing agents, such as peracetic acid, can result in transient O-acetylation. Potassium permanganate-based oxidation can extend to modification of N-acetylglucosamine moieties at the reducing ends of the chain resulting in formation of an N-acetylglucosaminic acid (Fig. 3). Presence of such a species has recently been shown to produce a characteristic signal in the 1H-NMR spectrum at 2.10 ppm, and is detected in heparin from several manufacturers (Beccati et al. 2010).
Fig. 3.
Potassium permanganate oxidation of the reducing end N-acetylglucosamine to N-acetylglucosaminic acid. The formation of Structure A causes a shift in the N-acetyl protons to 2.10 ppm, as shown in the 1 H-NMR spectrum. If X=H in Structure A, then the formation of Structure B may also be possible due to further oxidation
Taken together, it is clear that the production and purification of heparin starts with a diverse set of polysaccharide chains, with some degree of sequence variability. This variability is both reduced through purification of the final heparin product, which results in a narrowing of the polydispersity and a homogenization of the sulfation pattern of individual chains, and is increased, through introduction of purification process-specific alterations.
4 Structural Analysis of Heparin and Heparan Sulfate
Due to differential sulfation, domain structure (i.e., nonuniform distribution of disaccharides), and variable polysaccharide length, the structure and sequence of heparan sulfate (HS) is highly variable. This structural variability arises mostly from the fact that HS is synthesized in a nontemplate manner, through the concerted action of biosynthetic enzymes, including the O-sulfotransferases, the N-deactylase/N-sulfotransferase, and the C5-epimerase (Salmivirta et al. 1996). However, this variability, while significant, is controlled, through the tissue/cell-specific expression of enzyme isoforms. This has been confirmed experimentally by examining HS composition across different tissue preparations (Guimond et al. 2009; Shi and Zaia 2009) where there are primarily six major disaccharides (Table 1), with the most abundant being a nonsulfated uronic acid linked to an N-acetylglucosamine (I/G → HNAc). In this context, heparin can be thought of as a “specialized” HS, one that is more highly sulfated, and hence contains less variability. Especially in the context of HS, the challenge of detailed structural analysis is exacerbated by the fact that, because synthesis is nontemplate-driven, strategies employed for other template-driven biopolymers, such as DNA or proteins, are not amenable to sequencing complex polysaccharides. Thus, new glycan-specific techniques must either be developed de novo or existing techniques (e.g., proteomic-based techniques) must be substantially adapted.
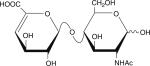
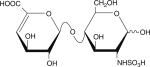
Table 1.
Six disaccharides that constitute the major building blocks of heparan sulfate chains
| Disaccharide structure | Structural notation |
|---|---|
|
ΔU-HNAc |
|
ΔU-HNS |
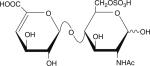
|
ΔU-HNAc6S |
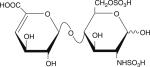
|
ΔU-HNS6S |
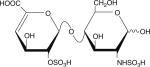
|
ΔU2S-HNS |
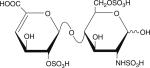
|
ΔU2S-HNS6S |
Numerous efforts have been made to facilitate the sequencing of heparin and HS chains, primarily to determine structure/function relationships for important protein-binding saccharides. In recent years, these efforts have been enhanced through the development of sophisticated analytical tools, including mass spectrometry and nuclear magnetic resonance (Kuberan et al. 2002; Yates et al. 1996; Saad et al. 2005). Additionally, heparin-degrading enzymes, primarily the bacterially derived heparinases, but also the mammalian enzyme heparanase, have proven to be important tools for sequencing of heparin and HS (Bisio et al. 2007).
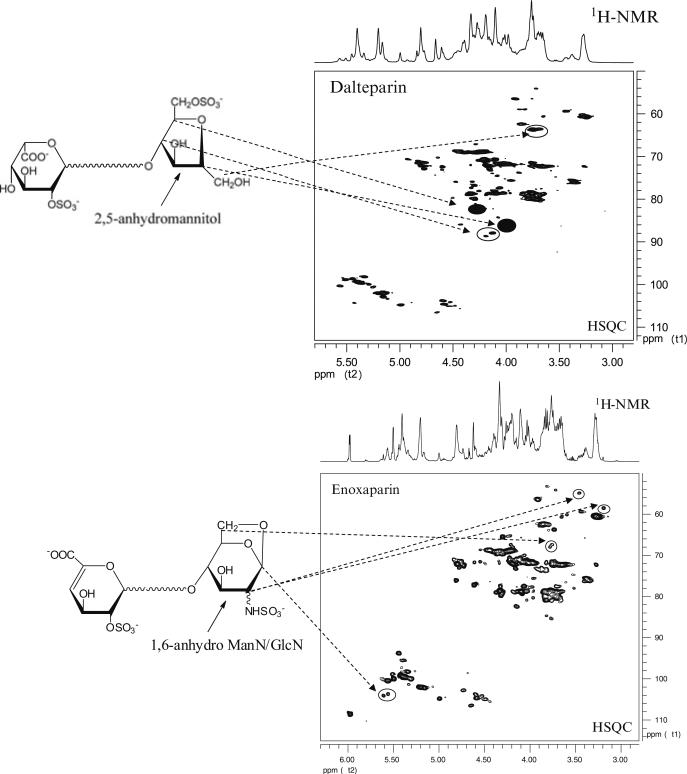
Broadly, efforts to provide structural information for heparin and HS include both top-down approaches, where key structural information is obtained on intact chains, and bottom-up approaches, where either partial or complete digestion of the chains is carried out, followed by identification /quantification of the resulting fragments. In terms of top-down approaches, monodimensional (1H and 13C) and multidimensional NMR have been employed. Proton and 13C NMR have been used to differentiate animal source and key structural motifs of heparin and HS (Guerrini et al. 2001, 2005). Additionally, for shorter heparin fragments, up to tetradecasaccharides, multidimensional NMR can be used to obtain complete sequence information. In the case of complex mixtures, including intact heparin and low-molecular-weight heparin, multidimensional NMR can be used to identify mono- and disaccharide constituents as well as unusual building blocks produced in low-molecular-weight heparins as a function of the process (Fig. 4) (Guerrini et al. 2007). Notably, multidimensional NMR was the key experimental technique to identify the structure of the heparin contaminant, oversulfated chondroitin sulfate (Guerrini et al. 2008).
Fig. 4.
HSQC spectra of low-molecular-weight heparins. 2D-NMR spectra of dalteparin and enoxaparin exhibit unique cross peaks arising from signature structures. These structures result from the specific chemistry used to depolymerize unfractionated heparin into lower molecular weight chains. The 2,5 anhydromannitol in dalteparin, and the 1,6-anhydro structure in enoxaparin, along with their associated signature cross peaks are shown
In addition to recent advances in NMR analysis of heparin/HS, mass spectrometry has been used to examine the fine structure of heparin and HS. While information from intact analysis of heparin chains is limited, in-line placement of high-resolution gel permeation chromatography has been successfully used to determine composition of low-molecular-weight heparin chains up to octadecasaccharide (Henriksen et al. 2004). Additionally, such an approach has recently been used to determine detailed structural information for the chondroitin sulfate proteoglycan, bikunin (Chi et al. 2008).
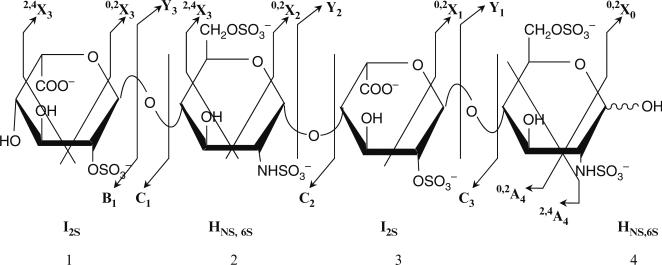
Coupled with the above advances, progress has also been made in strategies to fragment HS chains. Historically, application of high energy sufficient for fragmentation (and even ionization) has typically resulted in desulfation and the production of low information content fragments. These limitations have been overcome through several strategies, including formation of noncovalent complexes with cationic buffers or basic peptides (Juhasz and Biemann 1994; Rhomberg et al. 1998), application of gentle ionization conditions, as well as alternative strategies for fragmentation, including electron detachment dissociation (EDD) (Wolff et al. 2007). These advances have led to strategies where informative ions, including B and Y ions (Fig. 5) can be formed and detected. Such approaches have been used to sequence heparin-derived saccharides. However, there are limitations with these approaches; while sequence information has been obtained for sequences up to hexasaccharides, sequencing longer fragments still suffers from loss of structural information due to substantial sulfate loss during fragmentation.
Fig. 5.
Sequencing of heparin oligosaccharides by MS fragmentation. Several researchers have focused on developing MS-based sequencing procedures for heparin oligosaccharides. Shown here is different potential fragments for a representative tetrasaccharide: I2SHNS,6SI2SHNS,6S. In general, “cross-ring” cleavages, such as those that form fragments such as 0,2X0,1,2,3 and/or 2,4X 0,1,2,3 are more informative of sequence. However, fragments resulting from glycosidic bond cleavage are generally more prevalent. Additionally, fragments are often detected with some loss of sulfate (and water), resulting in sometimes ambiguous assignments; for example, the inability to distinguish B and Z ions from C and Y ions (the most common fragments). Finally, from published reports, more fragments and/or cross-ring fragmentations result from hexosamine (H) units than uronic (I/G) units
Bottom-up approaches, involving either partial or complete degradation of the heparin/HS chains and determination of the monosaccharide constituents of the fragments has been largely perfected. Quantification is typically completed through separating the resulting fragments by ion exchange or ion-pairing reverse phase HPLC or via capillary electrophoresis. Identification is typically completed either through co-injection with reference standards or through MS and MSn analysis by online coupling, especially for ion-pairing HPLC where labile pairing agents, such as dibutylamine, have simplified detection (Kuberan et al. 2002). An alternative strategy that has been employed is direct infusion of a digested mixture and detection and quantification by MS (Saad and Leary 2003, 2005).
In terms of digestion strategies, chemical means such as nitrous acid have been successfully used to determine disaccharide composition and have the added advantage that such strategies maintain the epimeric state of the uronic acid. However, the fact that no chromophore is generated and that detection typically must be completed through labeling (most often using radioactive sodium borohydride) has limited the adoption of this technique. An alternative strategy, employing bacterially derived heparinases, has been more widely adopted. Here, addition of one or more heparin-degrading enzymes, each with distinct, but overlapping substrate specificity, results in cleavage of heparin or HS (Ernst et al. 1995). Both complete digestion down to di- and tetrasaccharides, and partial digestion have been completed (Linhardt et al. 1988, 1990; Karamanos et al. 1997). Since these enzymes are lyases, digestion results in the formation of a Δ4,5 double bond on the uronic acid. Such species absorb UV light strongly, providing a tag for detection. Furthermore, structural information, specifically the presence or absence of an N-acetyl moiety, can be determined. Also of benefit is that the digestion can be completed at room temperature at pH of around 7, conditions conducive with the retention of labile sulfates. Alternatively, detection can be achieved through labeling the reducing end via reductive amination. Such approaches have dramatically increased sensitivity, especially in conjunction with laser-induced fluorescence, enabling detection of femtomoles of material.
Finally, given their high charge density and isomeric configurations (as well as the presence of α and β isoforms), often saccharide components will co-elute/co-migrate with one another. This greatly complicates both identification and quantification. While MS can be employed online to differentiate isomeric states, an alternative, easily implemented strategy is the use of additional bacterial or mammalian degradation enzymes, including the Δ4,5 glycuronidase (Myette et al. 2002), and the 2-O and 6-O sulfatases (Myette et al. 2003, 2009). Concerted or stepwise addition of these enzymes has been used to resolve di- and oligosaccharides, enabling accurate quantification and determination of mass balance.
Given the structural complexity of heparin and HS, as well as the increasingly sophisticated insight that we are obtaining into biological processes, it is likely that integration of several independent approaches, including both top-down and bottom-up approaches will prove important. Thus, while some methodologies, such as those outlined above, have provided superior information content, dataset integration enables more complete and rapid assessment of sequence and structure–function than any individual method used in isolation. As such development of frameworks to enable integration of datasets from multiple techniques will continue to prove important for the sequencing of heparin and HS (Venkataraman et al. 1999). As one example of such an integrative approach, results from NMR-based analysis of heparin fragments has been complemented and extended by CE-based disaccharide analysis after digestion with a cocktail of enzymes. In this manner, use of both datasets in an iterative and integrative manner enables one to “walk” through the saccharide sequence (Guerrini et al. 2002).
While the above approach has been successfully applied to isolated saccharides, or simple mixtures, for more complex mixtures, such as what has been isolated from cell surface HS, use of multiple approaches, or constraints, will likely be necessary; however, the approach is still valid. Depending on the complexity of the mixture, use of multiple, orthogonal constraints will likely be necessary, including both top-down and bottom-up approaches. Indeed, recently such logic was employed by the U.S. FDA in the approval of the first generic low-molecular-weight heparin.
5 Addressing Structure–Function Relationships for Heparan Sulfate
Heparan sulfate is known to interact with a wide variety of proteins and modulate their activity (Capila and Linhardt 2002; Lindahl 2007). A partial list of proteins, the role of HS, and structural motifs (i.e., length, sulfation pattern, etc.) that they recognize is listed in Table 2. The first (and still best) example of a defined protein-binding motif within heparin was the discovery of a discrete pentasaccharide sequence within heparin required for binding antithrombin. This pentasaccharide sequence is rare, occurring in only about one-third of heparin chains, and with even less frequency in heparan sulfate. Its most distinguishing feature is the presence of an unusual 3-O-sulfo group on an internal GlcNpS6S residue, which is absolutely essential for its high affinity binding to antithrombin. Notably, 3-O-sulfonation likely occurs late in the biosynthesis of heparin and heparan sulfate, and there are multiple 3-O-sulfotransferase isoforms having different substrate specificities. Given its relative rarity as well as the cloning and characterization of the biosynthetic enzymes, 3-O sulfonation has been used as a marker to identify sequences that are important in several biological systems, including viral entry and bridging of growth factor-receptor complexes, among others (Liu et al. 2002; Shukla et al. 1999).
Table 2.
Physiological and pathophysiological role of heparin and heparan sulfate
| Physiology/pathology | Binding proteins | Characteristics of heparin/HS binding |
|---|---|---|
| Anticoagulation | Antithrombin Thrombin |
Unique 5-mer sequence; requires 3-O-sulfonation Trisulfated disaccharide repeat |
| Cell proliferation and metastasis | FGF-2 FGF-1 Selectins |
4-mer to 6-mer; IdoA2S-GlcNS 4-mer to 6-mer; IdoA2S-GlcNS6S ≥4-mer; requires highly sulfated sequences, with presence of free amine residues |
| Inflammation | IL-8 SDF1α |
18-mer to 20-mer; requires alternating domains of high sulfation-low sulfation-high sulfation 12-mer to 14-mer; requires highly sulfated domains |
| Viral infectivity | HSV gD HIV gp120 Dengue virus envelope protein |
8-mer; requires a unique 3-O-sulfated, glucosamine at the reducing end 10-mer; highly sulfated sequences 10-mer; highly sulfated sequences |
| Development | HB-GAM | 16-mer to 18-mer; requires highly sulfated domains |
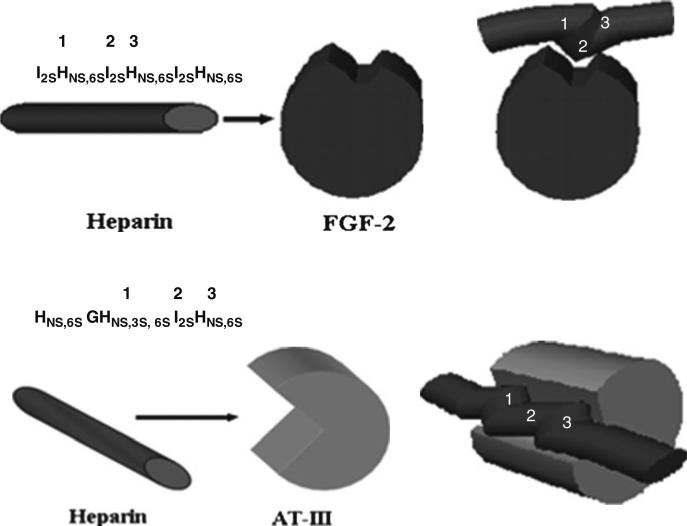
In addressing structure–function relationships for heparin/HS, reference to the antithrombin-heparin system provided a set of biochemical and analytical tools to identify high affinity sequences in heparin. With this system, the presence of a 3-O sulfate resulted in a ~100-fold increase in affinity, almost representing an “on/off switch”. Unlike this situation, many heparin/HS–protein interactions are often gradated, where sequence matters, but strength of binding to various structures depends on more than primary sequence. Addressing the structure–function relationships in these instances not only requires sequencing of HS from biologically relevant sources, but it also requires an integration of information from complementary approaches that permit a move from the biological to the structural space in an iterative and transitive manner. Approaching the problem in this way enables a fuller appreciation of all aspects of high-affinity interactions between soluble factors, including the key role that conformation plays in the high-affinity interaction between HS/heparin and growth factors. Introduction of conformational considerations has led to the introduction of the “kink” concept, where binding of growth factors to particular sequences induces a local distortion in the secondary structure of the polysaccharide chain, aligning sulfate groups for strong interaction with the protein, particularly basic residues (Fig. 6) (Raman et al. 2003). Thus, beyond primary sequence, conformational considerations, as well as the positioning of protein binding sites within the chain (thus modulating protein oligomerization) are essential to fully understanding structure–function relationships for this class of molecules.
Fig. 6.
Specificity of binding to heparin is governed by sequence and topology. Two examples of this “kink” modulating specificity include the FGF system where binding to a trisaccharide spanning kink [HNS,6SI2SHNS,6S] is enhanced by an internal iduronate residue adopting a particular conformation (1C4). Similarly, with the antithrombin system, a separate “kink”, namely a trisaccharide of the sequence HNS,3S,6SI2SHNS,6S influences “overwinding” of the helical axis, mediated in part through the 2S0 conformation of iduronate
6 Conclusions
Heparin, used clinically in both the treatment and prophylaxis of thrombosis, is one of the most widely used drugs. Given that the manufacture of heparin has been widely optimized, and given that a large number of proteins of physiological and pathophysiological importance interact with heparin and HS, raises the potential for the use of heparin/HS in a range of potential therapeutic applications, ranging from cancer to neurological disorders to inflammatory diseases. The major limitations in utilizing heparin/HS-based oligosaccharides for a wide range of disorders are (1) derivation of definitive structure–function relationships; and (2) the source and manufacturing of heparin has been optimized for high anticoagulant potency, which in essence becomes a side-effect, for nonanticoagulant applications. For the first consideration, the development of analytical technologies for the sequencing of heparin/HS as well as structural tools to understand high affinity binding in the context of protein structure have enabled and will continue to enable development of robust structure-activity understanding. For the second consideration, the introduction of low-molecular-weight heparins and the preparation of heparin oligosaccharides and synthetic analogs devoid of anticoagulant activity may open up a wide variety of new potential therapeutic applications in the treatment of cancer, and viral and bacterial infection, among others.
Acknowledgement
The authors thank Dr. Daniela Beccati for providing Fig. 4.
Contributor Information
Zachary Shriver, 77 Massachusetts Avenue, Building E25-Room 519, Cambridge, MA 02139, USA; Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Ishan Capila, Momenta Pharmaceuticals Inc., 675 West Kendall Street, Cambridge, MA 02142, USA.
Ganesh Venkataraman, Momenta Pharmaceuticals Inc., 675 West Kendall Street, Cambridge, MA 02142, USA.
Ram Sasisekharan, 77 Massachusetts Avenue, Building E25-Room 519, Cambridge, MA 02139, USA; Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA rams@mit.edu.
References
- Beccati D, Roy S, Yu F, Gunay NS, Capila I, Lech M, Linhardt RJ, Venkataraman G. Identification of a novel structure in heparin generated by potassium permanganate oxidation. Carbohydr Polymer. 2010;82:699–705. doi: 10.1016/j.carbpol.2010.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisio A, Mantegazza A, Urso E, Naggi A, Torri G, Viskov C, Casu B. High-performance liquid chromatographic/mass spectrometric studies on the susceptibility of heparin species to cleavage by heparanase. Semin Thromb Hemost. 2007;33:488–495. doi: 10.1055/s-2007-982079. [DOI] [PubMed] [Google Scholar]
- Canales A, Angulo J, Ojeda R, Bruix M, Fayos R, Lozano R, Gimenez-Gallego G, Martin-Lomas M, Nieto PM, Jimenez-Barbero J. Conformational flexibility of a synthetic glycosylaminoglycan bound to a fibroblast growth factor. FGF-1 recognizes both the (1)C(4) and (2)S(O) conformations of a bioactive heparin-like hexasaccharide. J Am Chem Soc. 2005;127:5778–5779. doi: 10.1021/ja043363y. [DOI] [PubMed] [Google Scholar]
- Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Casu B, Guerrini M, Naggi A, Torri G, De-Ambrosi L, Boveri G, Gonella S, Ronzoni G. Differentiation of beef and pig mucosal heparins by NMR spectroscopy. Thromb Haemost. 1995;74:1205. [PubMed] [Google Scholar]
- Casu B, Guerrini M, Naggi A, Torri G, De-Ambrosi L, Boveri G, Gonella S, Cedro A, Ferro L, Lanzarotti E, Paterno M, Attolini M, Valle MG. Characterization of sulfation patterns of beef and pig mucosal heparins by nuclear magnetic resonance spectroscopy. Arzneimittelforschung. 1996;46:472–477. [PubMed] [Google Scholar]
- Chi L, Wolff JJ, Laremore TN, Restaino OF, Xie J, Schiraldi C, Toida T, Amster IJ, Linhardt RJ. Structural analysis of bikunin glycosaminoglycan. J Am Chem Soc. 2008;130:2617–2625. doi: 10.1021/ja0778500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst S, Langer R, Cooney CL, Sasisekharan R. Enzymatic degradation of glycosaminoglycans. Crit Rev Biochem Mol Biol. 1995;30:387–444. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- Ferro DR, Provasoli A, Ragazzi M, Casu B, Torri G, Bossennec V, Perly B, Sinay P, Petitou M, Choay J. Conformer populations of L-iduronic acid residues in glycosaminoglycan sequences. Carbohydr Res. 1990;195:157–167. doi: 10.1016/0008-6215(90)84164-p. [DOI] [PubMed] [Google Scholar]
- Guerrini M, Bisio A, Torri G. Combined quantitative (1)H and (13)C nuclear magnetic resonance spectroscopy for characterization of heparin preparations. Semin Thromb Hemost. 2001;27:473–482. doi: 10.1055/s-2001-17958. [DOI] [PubMed] [Google Scholar]
- Guerrini M, Raman R, Venkataraman G, Torri G, Sasisekharan R, Casu B. A novel computational approach to integrate NMR spectroscopy and capillary electrophoresis for structure assignment of heparin and heparan sulfate oligosaccharides. Glycobiology. 2002;12:713–719. doi: 10.1093/glycob/cwf084. [DOI] [PubMed] [Google Scholar]
- Guerrini M, Naggi A, Guglieri S, Santarsiero R, Torri G. Complex glycosaminoglycans: profiling substitution patterns by two-dimensional nuclear magnetic resonance spectroscopy. Anal Biochem. 2005;337:35–47. doi: 10.1016/j.ab.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Guerrini M, Guglieri S, Naggi A, Sasisekharan R, Torri G. Low molecular weight heparins: structural differentiation by bidimensional nuclear magnetic resonance spectroscopy. Semin Thromb Hemost. 2007;33:478–487. doi: 10.1055/s-2007-982078. [DOI] [PubMed] [Google Scholar]
- Guerrini M, Beccati D, Shriver Z, Naggi A, Viswanathan K, Bisio A, Capila I, Lansing JC, Guglieri S, Fraser B, Al-Hakim A, Gunay NS, Zhang Z, Robinson L, Buhse L, Nasr M, Woodcock J, Langer R, Venkataraman G, Linhardt RJ, Casu B, Torri G, Sasisekharan R. Oversulfated chondroitin sulfate is a contaminant in heparin associated with adverse clinical events. Nat Biotechnol. 2008;26:669–675. doi: 10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimond SE, Puvirajesinghe TM, Skidmore MA, Kalus I, Dierks T, Yates EA, Turnbull JE. Rapid purification and high sensitivity analysis of heparan sulfate from cells and tissues: toward glycomics profiling. J Biol Chem. 2009;284:25714–25722. doi: 10.1074/jbc.M109.032755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen J, Ringborg LH, Roepstorrf P. On-line size-exclusion chromatography/mass spectrometry of low molecular mass heparin. J Mass Spectrom. 2004;39:1305–1312. doi: 10.1002/jms.723. [DOI] [PubMed] [Google Scholar]
- Juhasz P, Biemann K. Mass spectrometric molecular-weight determination of highly acidic compounds of biological significance via their complexes with basic polypeptides. Proc Natl Acad Sci USA. 1994;91:4333–4337. doi: 10.1073/pnas.91.10.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanos NK, Vanky P, Tzanakakis GN, Tsegenidis T, Hjerpe A. Ion-pair high-performance liquid chromatography for determining disaccharide composition in heparin and heparan sulphate. J Chromatogr A. 1997;765:169–179. doi: 10.1016/s0021-9673(96)00930-2. [DOI] [PubMed] [Google Scholar]
- Kuberan B, Lech M, Zhang L, Wu ZL, Beeler DL, Rosenberg RD. Analysis of heparan sulfate oligosaccharides with ion pair-reverse phase capillary high performance liquid chromatography-microelectrospray ionization time-of-flight mass spectrometry. J Am Chem Soc. 2002;124:8707–8718. doi: 10.1021/ja0178867. [DOI] [PubMed] [Google Scholar]
- Kuberan B, Beeler DL, Lech M, Wu ZL, Rosenberg RD. Chemoenzymatic synthesis of classical and non-classical anticoagulant heparan sulfate polysaccharides. J Biol Chem. 2003;278:52613–52621. doi: 10.1074/jbc.M305029200. [DOI] [PubMed] [Google Scholar]
- Lindahl U. Heparan sulfate-protein interactions – a concept for drug design? Thromb Haemost. 2007;98:109–115. [PubMed] [Google Scholar]
- Lindahl U, Backstrom G, Hook M, Thunberg L, Fransson LA, Linker A. Structure of the antithrombin-binding site in heparin. Proc Natl Acad Sci USA. 1979;76:3198–3202. doi: 10.1073/pnas.76.7.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl U, Kusche-Gullberg M, Kjellen L. Regulated diversity of heparan sulfate. J Biol Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Gunay NS. Production and chemical processing of low molecular weight heparins. Semin Thromb Hemost. 1999;25(Suppl 3):5–16. [PubMed] [Google Scholar]
- Linhardt RJ, Rice KG, Kim YS, Lohse DL, Wang HM, Loganathan D. Mapping and quantification of the major oligosaccharide components of heparin. Biochem J. 1988;254:781–787. doi: 10.1042/bj2540781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhardt RJ, Loganathan D, al-Hakim A, Wang HM, Walenga JM, Hoppensteadt D, Fareed J. Oligosaccharide mapping of low molecular weight heparins: structure and activity differences. J Med Chem. 1990;33:1639–1645. doi: 10.1021/jm00168a017. [DOI] [PubMed] [Google Scholar]
- Liu J, Shriver Z, Pope RM, Thorp SC, Duncan MB, Copeland RJ, Raska CS, Yoshida K, Eisenberg RJ, Cohen G, Linhardt RJ, Sasisekharan R. Characterization of a heparan sulfate octasaccharide that binds to herpes simplex virus type 1 glycoprotein D. J Biol Chem. 2002;277:33456–33467. doi: 10.1074/jbc.M202034200. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang Z, Linhardt RJ. Lessons learned from the contamination of heparin. Nat Prod Rep. 2009;26:313–321. doi: 10.1039/b819896a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Xu Y, Chen M, Weiwer M, Zhou X, Bridges AS, DeAngelis PL, Zhang Q, Linhardt RJ, Liu J. Chemoenzymatic design of heparan sulfate oligosaccharides. J Biol Chem. 2010;285:34240–34249. doi: 10.1074/jbc.M110.159152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myette JR, Shriver Z, Kiziltepe T, McLean MW, Venkataraman G, Sasisekharan R. Molecular cloning of the heparin/heparan sulfate delta 4,5 unsaturated glycuronidase from Flavobacterium heparinum, its recombinant expression in Escherichia coli, and biochemical determination of its unique substrate specificity. Biochemistry. 2002;41:7424–7434. doi: 10.1021/bi012147o. [DOI] [PubMed] [Google Scholar]
- Myette JR, Shriver Z, Claycamp C, McLean MW, Venkataraman G, Sasisekharan R. The heparin/heparan sulfate 2-O-sulfatase from Flavobacterium heparinum. Molecular cloning, recombinant expression, and biochemical characterization. J Biol Chem. 2003;278:12157–12166. doi: 10.1074/jbc.M211420200. [DOI] [PubMed] [Google Scholar]
- Myette JR, Soundararajan V, Shriver Z, Raman R, Sasisekharan R. Heparin/heparan sulfate 6-O-sulfatase from Flavobacterium heparinum: integrated structural and biochemical investigation of enzyme active site and substrate specificity. J Biol Chem. 2009;284:35177–35188. doi: 10.1074/jbc.M109.053801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman R, Venkataraman G, Ernst S, Sasisekharan V, Sasisekharan R. Structural specificity of heparin binding in the fibroblast growth factor family of proteins. Proc Natl Acad Sci USA. 2003;100:2357–2362. doi: 10.1073/pnas.0437842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rej R, Jaseja M, Perlin AS. Importance for blood anticoagulant activity of a 2-sulfate group on L-iduronic acid residues in heparin. Thromb Haemost. 1989;61:540. [PubMed] [Google Scholar]
- Rhomberg AJ, Ernst S, Sasisekharan R, Biemann K. Mass spectrometric and capillary electrophoretic investigation of the enzymatic degradation of heparin-like glycosaminoglycans. Proc Natl Acad Sci USA. 1998;95:4176–4181. doi: 10.1073/pnas.95.8.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg RD, Lam L. Correlation between structure and function of heparin. Proc Natl Acad Sci USA. 1979;76:1218–1222. doi: 10.1073/pnas.76.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad OM, Leary JA. Compositional analysis and quantification of heparin and heparan sulfate by electrospray ionization ion trap mass spectrometry. Anal Chem. 2003;75:2985–2995. doi: 10.1021/ac0340455. [DOI] [PubMed] [Google Scholar]
- Saad OM, Leary JA. Heparin sequencing using enzymatic digestion and ESI-MSn with HOST: a heparin/HS oligosaccharide sequencing tool. Anal Chem. 2005;77:5902–5911. doi: 10.1021/ac050793d. [DOI] [PubMed] [Google Scholar]
- Saad OM, Ebel H, Uchimura K, Rosen SD, Bertozzi CR, Leary JA. Compositional profiling of heparin/heparan sulfate using mass spectrometry: assay for specificity of a novel extracellular human endosulfatase. Glycobiology. 2005;15:818–826. doi: 10.1093/glycob/cwi064. [DOI] [PubMed] [Google Scholar]
- Salmivirta M, Lidholt K, Lindahl U. Heparan sulfate: a piece of information. FASEB J. 1996;10:1270–1279. doi: 10.1096/fasebj.10.11.8836040. [DOI] [PubMed] [Google Scholar]
- Sanderson PN, Huckerby TN, Nieduszynski IA. Conformational equilibria of alpha-L-iduronate residues in disaccharides derived from heparin. Biochem J. 1987;243:175–181. doi: 10.1042/bj2430175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Zaia J. Organ-specific heparan sulfate structural phenotypes. J Biol Chem. 2009;284:11806–11814. doi: 10.1074/jbc.M809637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- Sugahara K, Kitagawa H. Heparin and heparan sulfate biosynthesis. IUBMB Life. 2002;54:163–175. doi: 10.1080/15216540214928. [DOI] [PubMed] [Google Scholar]
- Toida T, Yoshida H, Toyoda H, Koshiishi I, Imanari T, Hileman RE, Fromm JR, Linhardt RJ. Structural differences and the presence of unsubstituted amino groups in heparan sulphates from different tissues and species. Biochem J. 1997;322(Pt 2):499–506. doi: 10.1042/bj3220499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Sequencing complex polysaccharides. Science. 1999;286:537–542. doi: 10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- Wardrop D, Keeling D. The story of the discovery of heparin and warfarin. Br J Haematol. 2008;141:757–763. doi: 10.1111/j.1365-2141.2008.07119.x. [DOI] [PubMed] [Google Scholar]
- Wolff JJ, Amster IJ, Chi L, Linhardt RJ. Electron detachment dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2007;18:234–244. doi: 10.1016/j.jasms.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates EA, Santini F, Guerrini M, Naggi A, Torri G, Casu B. 1H and 13C NMR spectral assignments of the major sequences of twelve systematically modified heparin derivatives. Carbohydr Res. 1996;294:15–27. doi: 10.1016/s0008-6215(96)90611-4. [DOI] [PubMed] [Google Scholar]