Abstract
Objective
A considerable fraction of prostate cancers harbor a gene fusion between the androgen-regulated TMPRSS2 and ERG, one of the most frequently over-expressed proto-oncogenes in prostate cancer. Here, we investigated if inherited genetic variation upstream of ERG alters prostate cancer risk and survival.
Methods
We genotyped 21 haplotype tagging SNPs (htSNPs) covering 123 kb of 5′UTR DNA including exon 3 of ERG in 2,760 incident prostate cancer cases and 1,647 controls from a population-based Swedish case–control study (CAPS). Individual SNPs and haplotypes were tested for association with prostate cancer risk and survival.
Results
One haplotype—′CTCGTATG′ located 100 kb upstream of ERG—was associated with lethal prostate cancer (HR, 1.36; 95% CI, 1.2–1.9, p = 0.006). Carriers of the variant ‘T’ allele of rs2836626 were diagnosed with higher TNM-stage (p = 0.009) and had an increased risk of prostate cancer-specific death (HR = 1.3; 95% CI, 1.1–1.7, p = 0.009). However, this association did not remain statistically significant after adjusting for multiple testing. We found overall no association between ERG variation and prostate cancer risk.
Conclusions
Genetic variation upstream of ERG may alter prostate cancer stage and ultimately prostate cancer-specific death but it is unlikely that it plays a role in prostate cancer development.
Keywords: Prostate cancer, ERG, Haplotype, Polymorphism, Survival
Introduction
Recent studies show that a substantial fraction of prostate cancers harbor a gene fusion between the androgen-regulated TMPRSS2 (21q22.3) and an ETS family transcription factor, most commonly ERG (21q22.3) [1–5]. ERG, located 2.8 Mb downstream from TMPRSS2, is recognized as a proto-oncogene frequently over-expressed in prostate cancer [6]. It has been suggested that the juxtaposition with TMPRSS2 triggers ERG over-expression [1] causing epigenetic reprogramming, WNT signaling and down-regulation of cell death pathways [4]. It appears as TMPRSS2-ERG fusion prostate cancers have a more aggressive natural history and they have been associated with clinical features including Gleason score [7, 8], tumor stage [2, 9] and prostate cancer-specific death [10]. However, results have been conflicting [11–17] and it is still unclear what role the ERG:TMPRSS2 fusion plays in prostate cancer.
The TMPRSS2:ERG fusion is most often induced by a ∼ 2.8 Mb deletion on chromosome 21q22.2–3 merging the two genes together. However, parts of the genes may also switch positions without any loss of intermediate genomic DNA [2, 4, 16, 17]. Rearrangement through deletion has been associated with high tumor stage and metastases to pelvic lymph nodes [2] implying that invasion regulators may reside between the two genes.
We hypothesize that germline DNA variation located 5’ of ERG affects prostate cancer-specific death. Germline variation may alter ERG expression either directly through regulatory mechanisms or by affecting the presence of the TMPRSS2:ERG fusion and thereby ERG expression. We genotyped 21 htSNPs covering 123 kb of the upstream genomic DNA of ERG in 2,760 prostate cancer cases and 1,647 controls. Individual SNPs and haplotypes were assessed for association between polymorphisms and prostate cancer-specific death. As a corollary, we also assessed the genotyped SNPs with respect to prostate cancer risk.
Subjects and methods
Case–control study
CAPS (CAncer Prostate in Sweden) is a population-based prostate cancer case–control study described earlier [18]. All men living in the northern and central parts of Sweden under the age of 80, and all men living in the Stockholm region and south-eastern part of Sweden under the age of 65 years constituted the study base. We invited all men with a newly diagnosed prostate cancer between March 2001 and October 2003. In total, 3,648 patients were identified and of them, 3,161 (87%) agreed to participate. We obtained information about clinical characteristics (Table 1) from the Swedish National Prostate Cancer Register (http://www.roc.se).
Table 1.
Characteristic for prostate cancer cases and controls enrolled in the CAPS study
| Characteristics1 | Cases | Controls | ||
|---|---|---|---|---|
|
|
|
|||
| n = 2,760 | % | n = 1,722 | % | |
| Age (years) | ||||
| ≤59 | 551 | 20.0 | 282 | 16.4 |
| 60–69 | 1329 | 48.2 | 733 | 42.6 |
| ≥70 | 880 | 31.9 | 707 | 41.0 |
| PSA levels2, ng/ml | ||||
| <4 | 141 | 5.2 | 1,418 | 82.0 |
| 4–9.99 | 950 | 35.4 | 237 | 13.8 |
| 10–19.99 | 635 | 23.6 | 41 | 2.4 |
| 20–49.99 | 439 | 16.3 | 20 | 1.2 |
| 50–99.99 | 217 | 8.1 | 3 | 0.2 |
| ≥100 | 304 | 11.3 | 2 | 0.1 |
| T stage | ||||
| T0/TX | 77 | 2.8 | – | – |
| T1 | 1,035 | 37.5 | – | – |
| T2 | 861 | 31.2 | – | – |
| T3 | 684 | 24.8 | – | – |
| T4 | 103 | 3.7 | – | – |
| N stage | ||||
| N0/NX | 2,667 | 96.6 | – | – |
| N1-N3 | 93 | 3.4 | – | – |
| M stage | ||||
| M0/MX | 2,497 | 90.5 | – | – |
| M1 | 263 | 9.5 | – | – |
| Gleason Score | ||||
| ≤4 | 98 | 3.9 | – | – |
| 5 | 280 | 11.1 | – | – |
| 6 | 944 | 37.4 | – | – |
| 7 | 761 | 30.1 | – | – |
| 8 | 242 | 9.6 | – | – |
| 9 | 176 | 7.0 | – | – |
| 10 | 24 | 1.0 | – | – |
| WHO Grade | ||||
| GI/GX | 1,883 | 68.2 | – | – |
| GII | 569 | 20.6 | – | – |
| GIII | 308 | 11.2 | – | – |
| Prostate cancer stage2 | ||||
| Localized | 1,583 | 57.4 | – | – |
| Advanced | 1,177 | 42.6 | – | – |
Characteristics were not available for all study participants
Case subjects were classified as advanced cases if they met at least one of the following criteria: T3/T4, N+, M+, Gleason score of 8–10 or PSA level ≥50 ng/ml
Control subjects were randomly selected from the Swedish Population Registry, frequency matched according to the expected age distribution of cases (groups of five-year interval) and geographical region (two regions, representing north and south of Sweden including Stockholm). Control subjects were recruited concurrent with case subjects. A total of 3,153 controls were invited and 2,149 (68%) agreed to participate.
For all participants, a blood sample and a questionnaire concerning risk factors for prostate cancer were collected. At time of this study, DNA was available for 2,760 cases and 1,722 controls. Each participant gave written informed consent. The research ethical committees at the Karolinska Institutet and Umeå University approved the study.
Follow-up
We collected information about prostate cancer-specific mortality for each case subject in CAPS. Each study participant is identified through his individually unique national registration number which includes the date of birth. Using this registration number, follow-up was achieved through record linkage to the Swedish Cause of Death Registry (www.socialstyrelsen.se). Subjects were followed until 1 March 2007. For individuals deceased after 31 December 2004, cause of death was established through review of death certificates by an experienced oncologist. We defined prostate cancer-specific death as those who had prostate cancer classified as the underlying cause of death. The average follow-up time was 4.3 years (range 0.33 to 6.5 years). A total of 576 (18%) individuals were deceased during follow-up and of those, 393 (12%) had prostate cancer classified as their underlying cause of death. At time for this study, DNA was available for 338 men who had died from prostate cancer.
SNP selection and tagging methodology
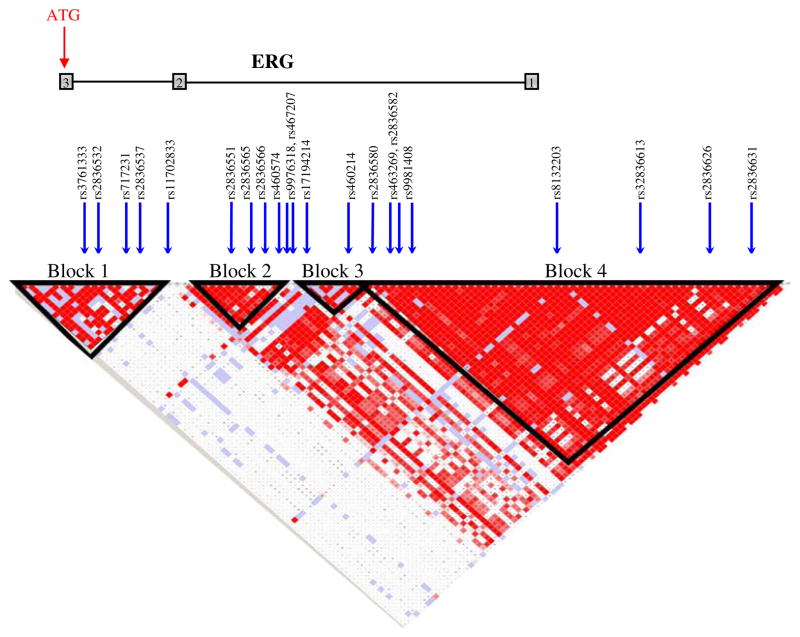
We determined the haplotype structure of ERG using publicly available genotype data from the CEPH population in the International HapMap project (www.hapmap.org). We only included SNPs with a minor allele frequency (MAF) above 5%. By including complete haplotype blocks as defined by Gabriel et al. [19], our target region spanned 121 kb upstream and 1.8 kb downstream from the start site of translation. This region included four distinct haplotype blocks which were separately tagged using the tagSNPS software [20]. htSNPs were selected to capture at least 95% of the haplotype diversity. In addition, we filled the gap between the blocks by tagging the whole region and chose possible additional SNPs needed to ensure a total coverage of at least 95% of the total common genetic variation. In total, 23 htSNPs were selected and genotyped (Fig. 1).
Fig. 1.
Linkage disequilibrium (LD) structure of ERG 5′UTR. Arrows depict selected and genotyped htSNPs in this study. Block limits are indicated by black triangles in the LD plot. Exons are indicated with gray squares
Genotyping
Genotyping details have been described earlier [18]. Shortly we used matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (SEQUENOM Inc., San Diego, California, USA) [21]. PCR assays and associated extension reactions were designed using the SpectroDESIGNER software (Sequenom Inc., San Diego, California). Primer sequences are available on request.
Statistical methods
We tested for Hardy–Weinberg equilibrium for each SNP using a replication method as implemented in the GENETICS package in the public available software R [22]. Association between prostate cancer risk and each SNP was assessed using a likelihood ratio test of a covariate equal to number of rare alleles (0, 1, or 2) based on an unconditional logistic regression model as implemented in R. We calculated per-allele odds ratios and corresponding 95% CI. We used the HAPLO.STATS package [23] in R to test for association between ERG haplotypes and prostate cancer risk. Haplotypes with a frequency <5% were pooled together. We adjusted all analyses for age and geographical region.
Follow-up began at the date of diagnosis and ended at the date of death or the last follow-up (1 March 2007). A likelihood ratio test of a covariate equal to number of rare alleles (0, 1, or 2) based on the Cox proportional hazards model was utilized to test for association between SNP and prostate cancer-specific death as implemented in R. The proportional hazards assumption was tested based on Schoenfeld residuals as implemented in Stata, version 8 [24]. Based on the number of SNPs tested, we performed a data simulation by randomly permuting genotype status and then reevaluate the Cox regression for each SNP. p-Values adjusted for multiple testing were then computed based on the empirical distribution of the maximum of the 21 test statistics obtained from Cox regression analysis for each permutation. A total of 10,000 replicas were run. We used the Kruskal-Wallis test to test for correlation between clinical characteristics and SNP genotypes. To estimate haplotypic effects on survival, we used the THESIAS software which allows analysis of censored data using a standard Cox proportional hazards formulation [25]. Hazard ratios and corresponding confidence intervals were estimated for each haplotype by comparison to a reference haplotype chosen as the most frequent one. A likelihood ratio test was used to perform a global test of association between haplotypes and prostate cancer death. Effects associated with rare haplotypes (frequency <0.05) were not estimated. All p-values are based on two-sided tests.
Results
Genotyping failed for two SNPs, rs464980 and rs2836542 but the high coverage of the genetic variation in the region was maintained (r2 = 0.94). The remaining SNPs had an average success rate of 98.3% (range: 96.3–99.4). All SNPs were in Hardy-Weinberg equilibrium among controls (p >0.01). The concordance rate between blinded duplicated samples (n = 330) was 99.93%.
Survival analysis
Two common SNPs, rs2836626 and rs2836582 located in block 4, were associated with prostate cancer survival (Table 2). Compared with carriers of the most common allele, carriers of the rs2836626 ‘T’ allele (MAF = 21%) were at increased risk to die from prostate cancer (HR: 1.28, 95% CI, 1.1–1.5, p = 0.009) whereas carriers of the rare ‘T’ allele (MAF = 23%) of rs2836582 had a significant better prognosis (HR = 0.80, 95% CI, 0.7–1.0, p = 0.02). No other of the 19 SNPs examined were associated with survival. Multivariate Cox regression analysis adjusted for TNM-stage, Gleason sum, and PSA levels at diagnosis did not alter the associations (HR = 1.27, p = 0.01 for rs2836626 and HR = 0.82, p = 0.05 for rs2836582) and neither did adjustment for treatment or family history (data not shown). We also tested the association between rs2836626 and rs2835582 and overall survival. Even though both SNPs were still significant (HR = 1.22, p = 0.02 and HR = 0.84, p = 0.05, respectively), the associations were attenuated. This is not surprising as the majority of deaths in this population (n = 576) are due to prostate cancer (n = 393). Therefore, the association with prostate cancer-specific death will also be observed in the overall survival analysis. Although rs2836626 and rs2835582 lies within the same haplotype block, their r2 is only 0.03 (D’ = 0.69). When both SNPs are included in the same model, rs2836626 remains significant (HR = 1.22, p = 0.04) while rs2836582 is only suggestively associated (HR = 0.82, p = 0.06). This suggests that both SNPs measure an indirect signal located within block 4. Carriers of the rs2836626 ‘T’ high-risk allele were also diagnosed with a significant higher TNM-stage than non-carriers (p = 0.009). Indeed, in survival analysis only adjusted for TNM-stage the association between rs2836626 and prostate cancer-specific survival disappeared (HR = 1.12, p = 0.22). However, when we adjusted our analyses for multiple testing using permutation tests, no association remained significant (p = 0.13 for rs2836626).
Table 2.
Association between ERG SNPs and prostate cancer risk and survival in CAPS. Prostate cancer risk was assessed with an unconditional logistic regression adjusted for age and geographical region. Hazard ratios and corresponding confidence intervals for survival analysis was performed with Cox regression
| SNP | Chromosomal position | Allele | Allele frequency (%) | Prostate cancer risk | Prostate cancer-specific survival | |||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| Cases (n = 2,760) | Controls (n = 1,647) | OR (95% CI) | p | HR (95% CI) | p | |||
| rs3761366 | 38870792 | G | 92.0 | 92.5 | 1.00 (ref) | 0.27 | 1.00 (ref) | 0.11 |
| A | 8.0 | 7.5 | 1.09 (0.9–1.3) | 0.79 (0.6–1.1) | ||||
| rs2836532 | 38870914 | A | 62.5 | 62.6 | 1.00 (ref) | 0.96 | 1.00 (ref) | 0.27 |
| T | 37.5 | 37.4 | 1.00 (0.9 –1.1) | 0.92 (0.8–1.1) | ||||
| rs717231 | 38873032 | G | 74.2 | 74.4 | 1.00 (ref) | 0.80 | 1.00 (ref) | 0.74 |
| T | 25.8 | 25.6 | 1.01 (0.9–1.1) | 1.03 (0.9–1.2) | ||||
| rs2836537 | 38873418 | A | 54.9 | 53.9 | 1.00 (ref) | 0.42 | 1.00 (ref) | 0.24 |
| C | 45.1 | 46.1 | 0.96 (0.9 –1.1) | 0.91 (0.8–1.1) | ||||
| rs11702833 | 38882034 | C | 66.8 | 65.9 | 1.00 (ref) | 0.43 | 1.00 (ref) | 0.26 |
| A | 33.2 | 34.1 | 0.96 (0.9–1.1) | 0.91 (0.8–1.1) | ||||
| rs2836551 | 38894247 | T | 59.3 | 60.0 | 1.00 (ref) | 0.35 | 1.00 (ref) | 0.67 |
| C | 40.7 | 40.0 | 1.04 (1.0–1.1) | 0.97 (0.8–1.1) | ||||
| rs2836565 | 38898571 | G | 74.4 | 73.2 | 1.00 (ref) | 0.32 | 1.00 (ref) | 0.35 |
| A | 25.6 | 26.3 | 0.95 (0.9–1.1) | 0.92 (0.8–1.1) | ||||
| rs2836566 | 38898599 | G | 78.7 | 77.7 | 1.00 (ref) | 0.45 | 1.00 (ref) | 0.12 |
| A | 21.3 | 22.3 | 0.96 (0.9–1.0) | 1.16 (1.0–1.4) | ||||
| rs460574 | 38908115 | C | 61.0 | 62.0 | 1.00 (ref) | 0.30 | 1.00 (ref) | 0.56 |
| T | 39.0 | 38.0 | 1.05 (1.0–1.1) | 0.95 (0.8–1.1) | ||||
| rs9976318 | 38912560 | A | 90.9 | 90.6 | 1.00 (ref) | 0.81 | 1.00 (ref) | 0.87 |
| G | 9.1 | 9.4 | 0.98 (0.8–1.1) | 0.98 (0.8–1.3) | ||||
| rs467207 | 38917952 | G | 78.0 | 79.0 | 1.00 (ref) | 0.27 | 1.00 (ref) | 0.71 |
| A | 22.0 | 21.0 | 1.06 (1.0–1.2) | 1.03 (0.9–1.2) | ||||
| rs17194214 | 38922657 | C | 80.9 | 79.6 | 1.00 (ref) | 0.26 | 1.00 (ref) | 0.21 |
| T | 19.1 | 20.4 | 0.94 (0.8–1.0) | 1.13 (0.9–1.4) | ||||
| rs460214 | 38927951 | T | 68.5 | 69.0 | 1.00 (ref) | 0.59 | 1.00 (ref) | 0.17 |
| C | 31.5 | 31.0 | 1.03 (0.9–1.1) | 1.12 (1.0–1.3) | ||||
| rs2836580 | 38932654 | C | 81.1 | 79.9 | 1.00 (ref) | 0.29 | 1.00 (ref) | 0.14 |
| G | 18.9 | 20.1 | 0.94 (0.8–1.1) | 1.15 (1.0–1.4) | ||||
| rs463269 | 38936307 | T | 77.8 | 77.6 | 1.00 (ref) | 0.83 | 1.00 (ref) | 0.34 |
| C | 22.2 | 22.4 | 0.99 (0.9–1.1) | 0.91 (0.8–1.1) | ||||
| rs2836582 | 38937178 | C | 77.3 | 77.4 | 1.00 (ref) | 0.65 | 1.00 (ref) | 0.02 |
| T | 22.7 | 22.6 | 1.02 (0.9–1.1) | 0.80 (0.7–1.0) | ||||
| rs9981408 | 38939316 | G | 74.0 | 73.3 | 1.00 (ref) | 0.48 | 1.00 (ref) | 0.12 |
| T | 26.0 | 26.7 | 0.97 (0.9–1.1) | 1.14 (1.0–1.4) | ||||
| rs8132203 | 38956590 | T | 94.5 | 94.1 | 1.00 (ref) | 0.68 | 1.00 (ref) | 0.77 |
| A | 5.5 | 5.9 | 0.96 (0.8–1.2) | 1.05 (0.8–1.4) | ||||
| rs2836613 | 38969046 | A | 76.5 | 76.7 | 1.00 (ref) | 0.84 | 1.00 (ref) | 0.53 |
| G | 23.5 | 23.3 | 1.01 (0.9–1.1) | 0.94 (0.8–1.1) | ||||
| rs2836626 | 38980308 | G | 79.5 | 78.2 | 1.00 (ref) | 0.46 | 1.00 (ref) | 0.009 |
| T | 20.5 | 21.2 | 0.96 (0.9–1.1) | 1.28 (1.1–1.5) | ||||
| rs2836631 | 38987776 | T | 56.0 | 55.2 | 1.00 (ref) | 0.59 | 1.00 (ref) | 0.69 |
| G | 44.0 | 44.8 | 0.98 (0.9–1.1) | 1.03 (0.9–1.2) | ||||
We observed borderline association between haplotypes and prostate cancer-specific death in block 4 (p global = 0.06, Table 3). Specifically, ′CTCGTATG′ carriers had a 36% increased risk of prostate cancer-specific death compared to the most common haplotype (95% CI, 1.1–1.7, p = 0.006). In addition, a single haplotype in block 3 (p = 0.03) and the ′GTCTTAGT′. haplotype in block 4(p = 0.04) were nominally associated with prostate cancer-specific survival (Table 3).
Table 3.
Association between ERG haplotypes and prostate cancer risk and survival in CAPS. The 5′ region of ERG can be divided in four distinct blocks with limited haplotype diversity
| Block | Haplotype1 | Frequency (%) | Prostate cancer risk | Prostate cancer-specific survival | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Cases | Controls | p | HR (95% CI) | p | ||
| 1 | – | – | – | 0.99 (global) | – | 0.34 (global) |
| GAGAC | 38 | 38 | 0.62 | 1.00 (Ref.) | – | |
| GAGAA | 15 | 14 | 0.96 | 1.06 (0.9–1.2) | 0.61 | |
| GTTCC | 13 | 12 | 0.81 | 1.22 (1.0–1.5) | 0.09 | |
| GTTCA | 7 | 7 | 0.87 | 0.96 (0.7–1.3) | 0.80 | |
| GTGCC | 5 | 5 | 0.97 | 0.79 (0.5–1.2) | 0.22 | |
| 2 | – | – | – | 0.08 (global) | – | 0.46 (global) |
| CGGT | 37 | 35 | 0.15 | 1.00 (Ref) | – | |
| TAGC | 25 | 26 | 0.35 | 0.96 (0.8–1.1) | 0.63 | |
| TGAC | 21 | 22 | 0.15 | 1.14 (0.9–1.4) | 0.20 | |
| TGGC | 10 | 9 | 0.10 | 1.02 (0.8–1.3) | 0.88 | |
| 3 | – | – | – | 0.31 (global) | – | 0.23 (global) |
| AGCT | 47 | 48 | 0.16 | 1.00 (ref) | – | |
| AACT | 22 | 21 | 0.32 | 1.11(0.9–1.3) | 0.27 | |
| AGCC | 12 | 10 | 0.05 | 1.09 (0.8–1.4) | 0.51 | |
| AGTC | 10 | 11 | 0.45 | 1.34 (1.0–1.7) | 0.02 | |
| GGTC | 9 | 9 | 0.91 | 1.09 (0.8–1.4) | 0.53 | |
| 4 | – | – | – | 0.80 (global) | – | 0.06 (global) |
| CTTGTAGG | 21 | 21 | 0.69 | 1.00 (ref) | – | |
| CTCGTATG | 19 | 19 | 0.99 | 1.36 (1.1–1.7) | 0.006 | |
| GTCTTAGT | 18 | 18 | 0.45 | 1.27 (1.0–1.6) | 0.04 | |
| CCCGTGGT | 16 | 15 | 0.70 | 1.04 (0.8–1.3) | 0.76 | |
| CTCTTAGT | 8 | 8 | 0.97 | 1.09 (0.8–1.5) | 0.58 | |
| CTCGTAGT | 6 | 5 | 0.17 | 0.87 (0.6–1.3) | 0.50 | |
| CCCGAGGT | 5 | 6 | 0.64 | 1.16 (0.8–1.6) | 0.38 | |
SNPs, block 1: rs3761366, 2836532, 717231, rs2836537, rs11702833; SNPs, block 2: rs2836551, rs2836565, rs2836566, rs460574; SNPs, block 3: rs9976318, rs467207, rs17194214, rs460214; SNPs, block 4: rs2836580, rs463269, rs2836582, rs9981408, rs8132203, rs2836613, rs2836626, rs2836631
To explore how representative included cases are for Swedish prostate cancer, we compared survival outcome and clinical characteristics between participating cases with DNA available for analysis and participating cases that did not provide DNA. There was no difference in prostate cancer-specific death between those who had participated with a blood sample and those who had not (p = 0.61). Moreover, we observed no difference regarding T stage (p = 0.33), N stage (p = 0.78), M stage (p = 0.92), Gleason score (p = 0.42), or PSA levels at time for diagnosis (P = 0.54), indicating that cases in the CAPS population with DNA available for analysis are representative for Swedish prostate cancer cases.
Association analysis
We observed no association between ERG SNPs and prostate cancer risk (p-values ranging from 0.26 to 0.96, Table 2). The genomic structure of the ERG promoter region includes four distinct haplotype blocks (Fig. 1). There was no correlation between ERG haplotypes and prostate cancer risk (Table 3). These data suggest that if common genetic variation at the ERG locus has a notable influence on risk of developing prostate cancer, it would be found outside our target region and demonstrate no LD with SNPs and haplotypes examined in this study.
To explore if case-misclassification (control subjects with undiagnosed prostate cancer) affected the results, we repeated association analysis between ERG genetic variants and prostate cancer risk excluding control subjects with a PSA level above four (n = 303). However, neither ERG SNPs nor ERG haplotypes showed significant association with disease risk in this restricted analysis (data not shown).
Discussion
This is the first study to evaluate if polymorphisms located in the promoter region of ERG are involved in prostate cancer development or in the progression to a lethal phenotype. We found evidence that genetic variation in this region alters prostate cancer-specific survival because individual SNPs and haplotypes in block 4 were associated with prostate cancer-specific death. Block 4, located 100 kb upstream of ERG, spans a region of 57 kb. Specifically, carriers of the variant rs2836626 ‘T’ allele were at 30% higher risk to die from prostate cancer and carriers of the ′CTCGTATG′ haplotype were at 36% increased risk. The variant ‘T’ allele of rs2836626 is only present at the ‘CTCGTATG’ haplotype indicating that this haplotype harbors a yet untyped causal marker showing strong linkage disequilibrium (LD) with rs2836626. We also observed significant association between rs2836626 and higher TNM-stage (p = 0.009). However, there are no known regulatory sequences in this region making it difficult to validate our findings from a biological point of view. In contrast, we observed no association between genetic variation and prostate cancer risk. There has been no implication of linkage to this region in Caucasians, although a linkage signal has been observed at 21q22.13–22.3 among eight pedigrees of African-American ancestry [26]. However, when we adjusted for multiple testing, no association remained statistically significant and considering the number of SNPs tested, we cannot rule out the possibility of chance findings.
There are both strengths and weakness with this study. CAPS is a well-powered population-based case–control study in a homogenous population. As PSA screening was limited in Sweden at the time for study recruitment [27]. CAPS includes predominantly symptomatic prostate cancers. The high proportion of clinically significant cancers is manifested by the high number of prostate cancer deaths, (12%) despite the relative short time of follow-up. Because we had no access to tumor samples, SNPs and haplotypes could not be studied in relation to fusion status or ERG expression. Another limitation is the restricted genetic region investigated. We cannot exclude that genetic variation outside the investigated region plays a significant role in prostate cancer.
Given that the fusion may be important in the natural history of prostate cancer, SNPs in this region may affect prognosis by induce/inhibit fusion. TMPRSS2:ERG rearrangement through genomic loss between the genes has been observed in a higher proportion of cancers with high-tumor stage and metastases to pelvic lymph nodes [2]. As we observed a high proportion of high grade tumors related to rs2836626 genetic variation in this region might favor fusion through deletion. Alternatively, genetic structure located in the 5′ UTR has a direct effect on ERG activation.
The TMPRSS2:ERG fusion is to date the most common chromosomal rearrangement in epithelial cancers. Its identification and implicated role in clinical prostate cancer provides new insight into the molecular mechanisms of prostate cancer. Understanding what drives this genetic rearrangement and its consequences may pave the way toward new areas of disease management.
In this study, we identified a borderline association between inherited genetic variation upstream of ERG and prostate cancer-specific survival. Unfortunately, we were not able to interpret our results in relation to TMPRSS2:ERG fusion status. If confirmed in other large studies, our results might help understanding the mechanisms behind the most common chromosomal rearrangement in prostate cancer to this date and further clarify the importance of ERG activation in the outcome of prostate cancer.
Acknowledgments
The authors thank all study participants in the CAPS study and all urologists who included their patients in the CAPS study. We also thank the Regional Cancer Registries and the CAPS-steering committee including Drs. Jan-Erik Johansson and Eberhart Varenhorst. We thank the personnel at the Medical Biobank in Umeå for skilfully handling the blood samples and the personnel at Mutation Analysis Facility (MAF) at Karolinska Insititutet for excellent genotyping. This work was supported by the Swedish Cancer Society and partially supported by the National Cancer Institute CA 105055 (Jianfeng Xu).
Contributor Information
Sara Lindström, Email: slindstr@hsph.harvard.edu, Department of Radiation Sciences, Oncology, Umeå University, Umeå, Sweden. Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Nobels Väg, P.O. Box 281, 17177 Stockholm, Sweden.
Hans-Olov Adami, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Nobels Väg, P.O. Box 281, 17177 Stockholm, Sweden. Department of Epidemiology, Harvard School of Public Health, Boston, MA, USA.
Katarina Bälter, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Nobels Väg, P.O. Box 281, 17177 Stockholm, Sweden.
Jianfeng Xu, Center for Human Genomics, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
S. Lilly Zheng, Center for Human Genomics, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
Jielin Sun, Center for Human Genomics, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
Pär Stattin, Department of Surgical and Perioperative Sciences, Urology and Andrology, Umeå University, Umeå, Sweden.
Henrik Grönberg, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Nobels Väg, P.O. Box 281, 17177 Stockholm, Sweden.
Fredrik Wiklund, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Nobels Väg, P.O. Box 281, 17177 Stockholm, Sweden.
References
- 1.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2205;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 2.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66(17):8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 3.Cerveira N, Ribeiro FR, Peixoto A, et al. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8(10):826–832. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iljin K, Wolf M, Edgren H, et al. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res. 2006;66(21):10242–10246. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66(17):8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 6.Petrovics G, Liu A, Shaheduzzaman S, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24(23):3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 7.Rajput AB, Miller MA, De Luca A, et al. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60(11):1238–1243. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnel AD, Lafargue CJ, Vollmer RT, Corcos J, Bismar TA. TMPRSS2-ERG fusion is frequently observed in gleason pattern 3 prostate cancer in a canadian cohort. Cancer Biol Ther. 2009;8(2):125–130. doi: 10.4161/cbt.8.2.7134. [DOI] [PubMed] [Google Scholar]
- 9.Mehra R, Tomlins SA, Shen R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20(5):538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 10.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26(31):4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 11.Jhavar S, Brewer D, Edwards S, et al. Integration of ERG gene mapping and gene-expression profiling identifies distinct categories of human prostate cancer. BJU Int. 2008 Nov 28; doi: 10.1111/j.1464-410X.2008.08200.x. in press. [DOI] [PubMed] [Google Scholar]
- 12.FitzGerald LM, Agalliu I, Johnson K, et al. Association of TMPRSS2-ERG gene fusion with clinical characteristics and outcomes: results from a population-based study of prostate cancer. BMC Cancer. 2008;8:230. doi: 10.1186/1471-2407-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saramäki OR, Harjula AE, Martikainen PM, Vessella RL, Tammela TL, Visakorpi T. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14(11):3395–3400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- 14.Mehra R, Tomlins SA, Yu J, et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent meta-static prostate cancer. Cancer Res. 2008;68(10):3584–3590. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam RK, Sugar L, Yang W, et al. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97(12):1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27(3):253–263. doi: 10.1038/sj.onc.1210640. doi:10.1038/sj.onc.12 10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer Res. 2006;66(22):10658–10663. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- 18.Lindstrom S, Wiklund F, Adami HO, Balter KA, Adolfsson J, Gronberg H. Germ-line genetic variation in the key androgen-regulating genes androgen receptor, cytochrome P450, and steroid-5-alpha-reductase type 2 is important for prostate cancer development. Cancer Res. 2006;66(22):11077–11083. doi: 10.1158/0008-5472.CAN-06-3024. [DOI] [PubMed] [Google Scholar]
- 19.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 20.Stram DO, Haiman CA, Hirschhorn JN, et al. Choosing haplotype-tagging SNPS based on unphased genotype data using a preliminary sample of unrelated subjects with an example from the Multiethnic Cohort Study. Hum Hered. 2003;55(1):27–36. doi: 10.1159/000071807. [DOI] [PubMed] [Google Scholar]
- 21.Jurinke C, van den Boom D, Cantor CR, Koster H. Automated genotyping using the DNA MassArray technology. Methods Mol Biol. 2002;187:179–192. doi: 10.1385/1-59259-273-2:179. [DOI] [PubMed] [Google Scholar]
- 22.R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2006. [Google Scholar]
- 23.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.StataCorp. Stata Statistical Software: release 8. StataCorp LP; College Station, TX: 2005. [Google Scholar]
- 25.Tregouet DA, Tiret L. Cox proportional hazards survival regression in haplotype-based association analysis using the Stochastic-EM algorithm. Eur J Hum Genet. 2004;12(11):971–974. doi: 10.1038/sj.ejhg.5201238. [DOI] [PubMed] [Google Scholar]
- 26.Schaid DJ, McDonnell SK, Zarfas KE, et al. Pooled genome linkage scan of aggressive prostate cancer: results from the international consortium for prostate cancer genetics. Hum Genet. 2006;120(4):471–485. doi: 10.1007/s00439-006-0219-9. [DOI] [PubMed] [Google Scholar]
- 27.Stattin P, Johansson R, Damber JE, et al. Non-systematic screening for prostate cancer in Sweden—survey from the National Prostate Cancer Registry. Scand J Urol Nephrol. 2003;37(6):461–465. doi: 10.1080/00365590310015778. [DOI] [PubMed] [Google Scholar]