Abstract
Locus Control Regions (LCR) are cis-acting gene regulatory elements with the unique, integration site-independent ability to transfer the characteristics of their locus-of-origin’s gene expression pattern to a linked transgene in mice. LCR activities have been discovered in numerous T cell lineage expressed gene loci. These elements can be adapted to the design of stem cell gene therapy vectors that direct robust therapeutic gene expression to the T cell progeny of engineered stem cells. Currently, transgenic mice provide the only experimental approach that wholly supports all the critical aspects of LCR activity. Herein we report manifestation of all key features of mouse T cell receptor (TCR)-α gene LCR function in T cells derived in vitro from mouse embryonic stem cells (ESC). High level, copy number-related TCRα LCR-linked reporter gene expression levels are cell type-restricted in this system, and upregulated during the expected stage transition of T cell development. We further report that de novo introduction of TCRα LCR linked transgenes into existing T cell lines yields incomplete LCR activity. Together, these data indicate that establishing full TCRα LCR activity requires critical molecular events occurring prior to final T-lineage determination. This study additionally validates a novel, tractable and more rapid approach for the study of LCR activity in T cells, and its translation to therapeutic genetic engineering.
Introduction
Locus control regions (LCR) have been discovered in numerous gene loci that are selectively active in T cells. An LCR is a cis-acting DNA element capable of transferring most aspects of the expression pattern of its gene locus of origin to a linked transgene in mice (1). These aspects include a predictable mRNA production level that also displays locus-of-origin appropriate developmental timing and tissue restriction. Furthermore, unlike most known cis-acting elements, an LCR can accomplish this at virtually any ectopic site of integration in the genome. Transgenic analyses of LCRs have clearly demonstrated their ability to overcome integration site-dependent position effects that can silence a transgene at a subset of ectopic genomic locations (2, 3). Thus, LCR driven transgene expression is present in the appropriate tissues of all transgene positive mice at levels that are roughly transgene copy number-dependent (4). The integration site-independent ability of the LCR to robustly and predictably regulate a linked heterologous transgene in time and space makes it a prime target in the search for DNA elements with the power to increase the specificity and robustness of therapeutic gene expression from lentiviral vectors. The number and variety of LCR activities that are active in T cells is unusually large. They are derived from functionally important gene loci that feature a diverse array of developmental expression patterns during T cell generation and function. These gene loci include human CD2 (5), human adenosine deaminase (6), mouse T cell receptor (TCR)-α (7), mouse interleukin-2 (8), mouse CD4 (9), human perforin (10) and the mouse TH2 cytokine cluster (11). Thus, the continued study of LCR activity is of particularly high significance to the understanding of T cell biology. In addition, these LCRs provide a potentially rich source of cis-acting DNA tools for creating vectors that can drive high level therapeutic cargo gene expression with developmentally directed characteristics in T cells.
T cells are a highly significant cell type to target for gene therapy. The αβ T cell receptor (TCR) complex is used by most circulating T cells to recognize antigen and initiate immune responses. T cells can be genetically modified to contain a specific, cloned TCR (12) or engineered chimeric antigen receptor (CAR) cDNAs (13) that encode receptors enabling them to initiate a desired immunotherapeutic response. Current efforts in this vein have treated hematologic malignancies by introducing CAR-encoding vectors directly into fully developed T cells (14). However, it is also possible, and desirable, to introduce therapeutic antigen receptor gene constructions into embryonic stem cells (ESC), induced pluripotential stem cells (iPSC) and hematopoietic stem cells (HSC) using lentiviral vectors. Such stem cell genetic engineering represents a promising approach for providing an individual with a longer-term source of T cells producing an introduced therapeutic antigen receptor gene product.
Naturally, ESC, iPSC and HSC populations all give rise to multiple cell lineages in addition to T cells, each of which executes a unique program of gene expression. The safest outcome of the above-described stem cell gene therapy approach would require restricting high-level production of the introduced TCR/CAR protein to the T cell progeny of the genetically engineered stem cells. Achieving this important goal will require major advances in the understanding of the cis-acting DNA sequence requirements for predictable spatiotemporal gene regulation in native chromatin during, and after, T cell differentiation from stem cells. It will further require increased knowledge of gene regulatory DNA with the capacity to insulate therapeutic genes from the silencing effects of the genome wide heterochromatin changes likely to accompany T cell development from stem cell precursors. The multiple functions of the LCRs active in T cells seem to hold the key to addressing both of these critical issues.
To date, the transgenic mouse is the only experimental model shown to support all aspects of an LCR’s complex activity at ectopic integration sites. While a powerful approach, experiments in transgenic mice are resource intensive and involve protracted timetables. Basic structure/function analyses of LCRs, and translation of this information to gene therapy vector design, can be greatly accelerated by the development of a cell culture model capable of supporting the many facets of LCR activity. However, early attempts to develop such a system for the β-globin LCR indicated that de novo introduction of LCR driven transgenes into differentiated cell lines does not support full LCR activity (15). Subsequent studies involving cell fusion have further suggested that the development of full LCR activity may require the LCR DNA to be present in the genome prior to cell lineage commitment (16).
We study the LCR present in the mouse TCRα gene locus (Figure 1A). The TCRα LCR was originally identified as a cluster of nine DNAse I hypersensitive sites (HS) (7) located in between the Cα constant region exons and the downstream Dad1 gene (17). It has been amply demonstrated, using multiple reporter transgenes, that the TCRα LCR drives copy number-related levels of linked transgene expression (18–20). Using randomly integrated transgenes in mice, we have identified five distinct functional sub-regions of this LCR. Two of these sub-regions are required for the LCRs spatiotemporal specificity (21). The others seem to provide a form of insulation capacity that prevents integration site-dependent position effects on TCRα LCR function (22, 23). Informed by our prior TCRα LCR studies in vivo, we sought to develop an experimental system to assay for complete TCRα LCR activity that was not dependent on transgenic mice.
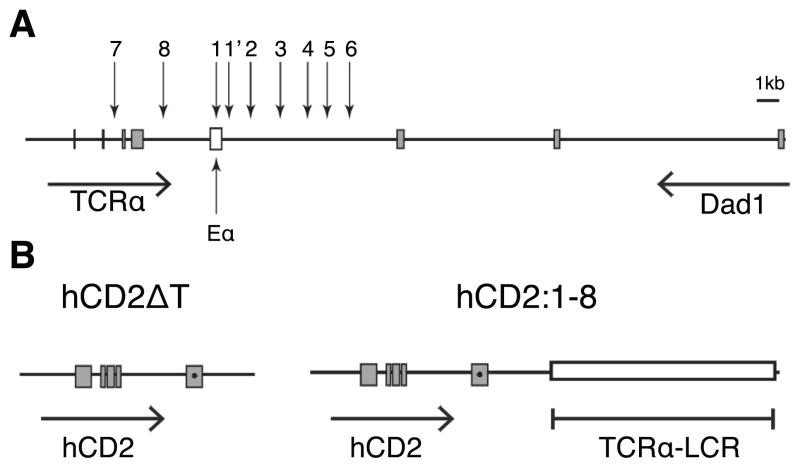
Figure 1. The TCRα LCR genomic region and reporter transgenes.
(A) Diagram of the TCRα/Dad1 locus. Vertical arrows depict DNase hypersensitive sites (HS)1-8 of the TCRα LCR. The open box marks the Eα classical transcriptional enhancer. All other boxes indicate exons of their respective genes. Horizontal arrows indicate the transcription orientation of the genes. Diagram is drawn to scale. (B) Depiction of the hCD2ΔT and hCD2:1-8 transgenes. A premature stop codon (●) was introduced in exon V prior to the codons of the cytoplasmic tail (24) In hCD2:1-8, the TCRα LCR cassette (18) (open box) containing an exon-free HS1-8 fragment is linked to hCD2ΔT gene fragment.
Here we report the finding of a cell culture model that supports all aspects of TCRα LCR activity observed in whole animals. The model involves transfection of mouse embryonic stem cells with a TCRα LCR linked reporter gene construct. Transfected embryonic stem cell clones are then induced to differentiate into T cells (and other hematopoietic progeny) in vitro. This approach permits the examination of reporter gene expression levels per transgene copy, as well as the developmental timing and cell type-restriction of reporter gene expression. As is observed in transgenic mice, TCRα LCR-linked reporter mRNA expression in this system correlates with integrated transgene copy number. Furthermore, high-level transgene expression is cell-type restricted, and is activated at the expected T cell developmental stage transition. These efforts validate in vitro ESC differentiation as an effective experimental model for the further study and translation of TCRα LCR activity. By comparing TCRα LCR activity in this system to that observed in directly transfected T cell lines, the present study also provides new evidence that the establishment of full LCR activity in lineage-committed cells requires molecular components acting prior to cell lineage differentiation.
Materials and Methods
Reporter gene constructs
The hCD2ΔT transgene (24) was excised from the pBluescript SK vector using Sal I and Bam HI. The hCD2:1-8 transgene (19) was excised using Kpn I and Not I. The SV40 promoter-driven Neomycin-G418 resistance cassette was excised from the pEYFP-C1 vector (Clontech) using Ssp I and EcoO109 I enzymes.
T cell line culture and transfection
T-cell lines VL3-3M2 (25) and C6VLB (26) were cultured in RPMI 1640 with 5% FBS and 10% FBS respectively, supplemented with 1% Penicillin-Streptomycin (Cellgro), 1% Glutagro (Cellgro) and 54μM β-mercaptoethanol (Sigma). Cells were transfected using a BioRad Gene Pulser (0.3 kV and 960 μF). Approximately 1 ×107 cells were re-suspended in 0.5 ml of Electroporation Buffer (Millipore) with 10 μg of hCD2:1-8 transgene fragment, or 5 μg of hCD2ΔT fragment. An equimolar amount of a Neomycin G418 resistance cassette was co-transfected with the reporter transgene. 24 hours post-transfection, Neomycin-G418 was added at a concentration of 0.4 mg/ml for VL3-3M2 and 0.35 mg/ml for C6VLB. Individual clones were obtained by serial dilution.
Embryonic stem cell (ESC) culture and transfection
The mouse ESR1 cell line was co-cultured with Mitomycin C arrested Mouse Embryonic Fibroblasts (MEFs) (Millipore) in Dulbeco’s Modified Eagle Media (DMEM) (Cellgro) supplemented with 20% FBS (Gemini), 1% Glutagro (Cellgro), 1% Penicillin/Streptomycin (Cellgro), 1% HEPES (Millipore), 1% Non Essential Amino Acids (Millipore), 0.1% Gentamycin (Life Technologies), 0.1% β-mercaptoethanol (Life Technologies), and 10 ng/ml of Leukemia Inhibitory Factor (LIF) (Millipore). Cells were transfected by BioRad Gene Pulser (0.24kV and 500μF). Approximately 1 ×107 ESCs were re-suspended in 0.5 ml of Electroporation Buffer (Millipore) with 15 μg of hCD2:1-8 transgene fragment, or 7.5 μg of hCD2ΔT fragment. An equimolar amount of a Neomycin-G418 resistance cassette was co-transfected with the reporter transgene. 48 hours post-transfection, G418 was added at a concentration of 0.18 mg/ml. Selection media was changed daily. Individual colonies were picked after 10 days and clonally propagated.
Both ESC and T cell transfectant clones were initially screened for transgene integration by PCR using primers complimentary to the hCD2 promoter region (Forward: 5′-GAGGAAAC CAACCCCTAAGATGAG-3′ Reverse: 5′-CGTAATCTCTTTGGAGACTGCACC-3′). Intact transgene copy number was subsequently determined via Southern blot using an 800 bp Bgl II probe from the HS6 region of the TCRα LCR, as described previously (19). Copy number estimates were determined by PhosphorImager analyses of at least three Southern blots for each set of clones. All clones directly compared in assays were analyzed for relative copy number on the same Southern blots. Enzymes and probes chosen enabled simultaneous detection of distinct sized fragments from both the endogenous TCRα locus and the transgene. Transgene signals were normalized to the endogenous signal within each sample.
In vitro ESC differentiation
The protocol for in vitro derivation of T cells, and other hematopoietic cell types, from mouse ESC was carried out as previously described (27). Emerging hematopoietic stem cells from day 8 co-cultures were harvested and transferred onto OP9-DL1 cell monolayers (to derive T-cells) (28) or OP9 cell monolayers (to derive monocytic, erythroid, or B-cells) (29). In a typical experiment, multiple, independent, transfected ESC clones were differentiated in parallel with a non-transfected ESR1 control co-culture. The ESR1 derived progeny were used as negative controls for the corresponding differentiation products of the multiple transfected ESC clones assessed in the same experiment. Cells were analyzed by flow cytometry on Day 12 of co-culture (to detect monocytic, erythroid, or early-stage developing T cells), Day 16 (to detect B-cells), and Day 18 (to detect later stage developing T cells). Co-cultures were harvested on Day 20 for RNA extraction, cDNA synthesis and Quantitative real time PCR (qRT-PCR) analysis.
Flow cytometry
FACSCalibur and FACSVantage devices were used. Antibodies used include Fluorescein isothiocyanate (FITC) conjugated, anti-hCD2 (clone S5.2), R-Phycoerythrin/cyanine dye 7 (PE-Cy7) conjugated anti-CD45 (Clone 30F-11), Allophycocyanin (APC) conjugated anti-CD44 (Clone IM7), PE-conjugated anti-CD25 (Clone 3C7), PE conjugated anti-CD8 (Clone 53–6.7), Anti CD16/32 (Clone 2.4G2), (all from BD Biosciences) and CD4 APC (Clone RM4-5), CD11b (RM2805), Ter119 (mTer04) and CD19 (RM7705) (Life Technologies). Dead Cell Discriminator (DCD) or 4′, 6-diamidino-2-phenylindole (DAPI) (Life Technologies) was used to label non-viable cells. Before staining, cells were pre-treated with anti CD16/32 antibody (to block Fc receptors). Cells were stained with fluorochrome-conjugated antibodies for 20 minutes and washed three times. For analyses, live cells were gated based on forward and side scatter and lack of DAPI or DCD signal. CD45 was additionally used to gate on white blood cell types derived in co-culture. FlowJo (Tree Star, Inc.) software was used for data analyses.
Quantitative real time PCR
On day 20 of ESC-OP9-DL1 co-culture, total RNA was extracted from one well of a 6 well plate per clone (Qiagen RNeasy micro kit). RNA for VL3-3M2 and C6VLB clones was extracted from 1 ×107 cells (Qiagen RNeasy mini kit). cDNA was synthesized from 1μg of each of these RNA samples (NEB Protoscript cDNA synthesis kit). qRT-PCR was performed using an Applied Biosystems 7500 device. Samples were prepared using the Dynamo SYBR green qPCR kit, (New England Biolabs). TCRα primers (20) were used to normalize for both loading and T cell content of the co-cultures. hCD2 primers were used to detect reporter gene expression (Forward: 5′-CCTTTCTGCTGGTGAACTTGTG-3′ Reverse: 5′-TCAACACAACCCTGACCTGTG-3′). Relative levels of hCD2 gene products were calculated as follows: Non-transfected ESR1 Ct (cycle threshold) value was used as the “calibrator”. Ct values observed using TCRα primers were used as our normalizing control, as TCRα is the “reference gene” in this assay. Ct values observed with hCD2 primers were all subtracted by the Ct values obtained for TCRα to obtain the normalized hCD2 ΔCt for each co-culture. Next, all ΔCt values were then subtracted by ΔCt of the calibrator to obtain ΔΔCt. Normalized relative hCD2 expression in T cells derived from each clone, was calculated by using the 2−ΔΔCT method. The resulting values were then divided by the transgene copy number determined for each clone. In an experiment, relative mRNA levels per transgene copy were determined from triplicate samples. The highest level observed in a given experiment was designated as 1.0. The relative mRNA levels observed in the three separate experiments were then averaged and graphed.
Results
Applying in vitro embryonic stem cell differentiation to the study of the TCRα LCR
Recent advances have enabled quantitative differentiation of various hematopoietic cell types from mouse embryonic stem cells (ESCs) in co-culture with a bone marrow derived feeder cell line (OP9) (29). ESCs-OP9 co-culture yields hematopoietic stem cells (HSCs) when the growth media is supplemented with fms-like tyrosine kinase receptor-3 ligand (Flt-3L) (30). Subsequent addition of interleukin (IL)-7 supports HSC differentiation into cells of monocytic (CD11bhi), erythroid (TER119+) and B lymphocyte (CD19+) lineages. HSCs can be driven towards development of T-lymphocytes by using OP9 cells transfected with a delta like ligand (DLL-1 or DLL-4), of the notch receptor (31). This model has proven valuable to the study of various aspects of T cell development that were formerly only approachable in whole animal models (32–34). We investigated if this in vitro ESC differentiation system would support the properties of the TCRα LCR that have been well demonstrated in transgenic mice. As in our previous work (19), here we use a reporter transgene fragment encoding a non-signaling variant of the human CD2 protein (hCD2ΔT) (24). Transgenic mice carrying the unlinked hCD2 reporter gene alone did not display detectable hCD2 reporter protein expression (35). Thus, this fragment was linked to HS 1-8 the TCRα LCR (Figure 1A) to create the hCD2:1-8 transgene construct that has been a suitable reporter for TCRα LCR activity (19, 20).
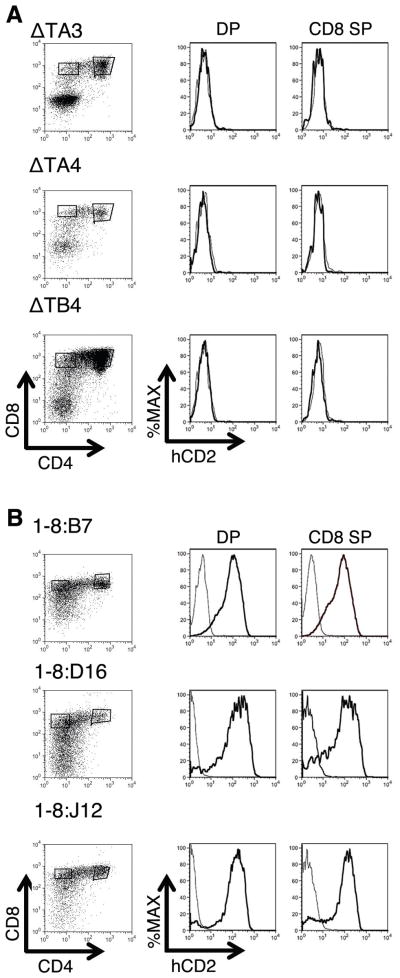
We generated ESC clones transfected with hCD2ΔT or hCD2:1-8 (Figure 1B). Each independently generated ESC clone represents an independent assay of a construct’s transcriptional activity. hCD2:1-8 and hCD2ΔT transfected ESC clones were differentiated into CD4, CD8 double positive (DP) and CD8 single positive (SP) T-lineage cells. All of the hCD2ΔT transfected clones yielded hCD2 negative DP and CD8 SP T-cells (Figure 2A). In contrast, all of the hCD2:1-8 clones produced hCD2 positive DP and CD8 SP T-cells (Figure 2B). Thus, hCD2 gene expression in this system is TCRα LCR-dependent and integration site-independent.
Figure 2. TCRα LCR-dependent hCD2 reporter protein expression in T-cells differentiated from ES cell clones.
(A) Flow cytometry analyses of hCD2 expression in three representative independent clones of ESC transfected with hCD2ΔT (ΔT) and subsequently differentiated into CD4,CD8 double positive (DP) and CD8 single positive (SP) T-cells. None of the clones produce T cells that are hCD2 positive (n=6). (B) DP and CD8 SP T cells derived from three representative independent ESC clones transfected with hCD2:1-8 are positive for hCD2 reporter protein. All hCD2:1-8 ESC clones (1-8:) produce T cells that express the hCD2 protein on their cell surface (n>12). Cell population gates are shown at left. hCD2 expression in transfected (dark curve) and non-transfected (light curve) gated cells is shown at right.
TCRα LCR does not drive consistent, high level hCD2:1-8 transgene expression in non-T lineage cell types derived in vitro
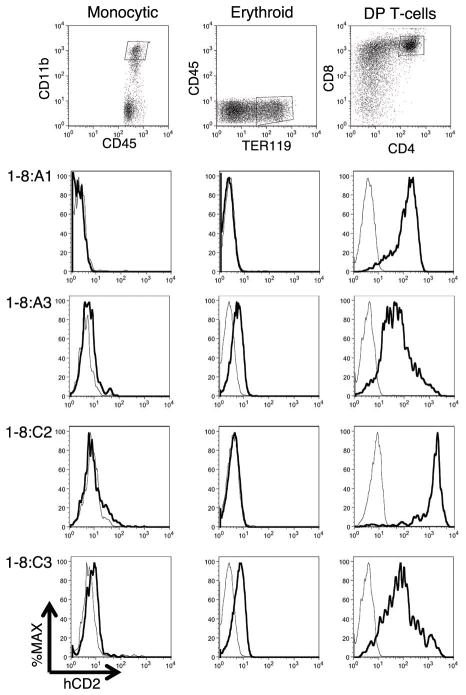
In transgenic mice, a reporter gene linked to the TCRα LCR is expressed primarily in lymphoid organs and not other tissues (18, 19). In the OP9 co-culture system, ESCs can be differentiated into various hematopoietic cell types. To test cell type distribution of TCRα LCR activity in the system, we differentiated the hCD2:1-8 transfected ESC clones into either TER119+ cells (indicative of erythroid lineage) or CD11bhi cells (monocytic lineage). These populations of cells were analyzed via flow cytometry for hCD2 expression. The hCD2 reporter gene signal was low to absent in these differentiation products after 12 days of co-culture on OP9 stroma (Figure 3). Longer co-culture time (16 days) did not result in hCD2 reporter gene upregulation in non-lymphoid cells (data not shown). We were able to generate B cells from five of the hCD2:1-8 transfected ESC clones after 16 days of OP9 co-culture. Congruent with the ectopic B-cell expression observed in hCD2:1-8 transgenic mice (19), reporter hCD2 expression was detected in three of the five clones (data not shown).
Figure 3. Cell type restriction on high-level hCD2:1-8 reporter transgene expression during in vitro hematopoiesis from ESCs.
Flow cytometric analysis of in vitro differentiated hematopoeietic progeny of four representative hCD2:1-8ESC clones (1-8:). Monocytic (CD11bhiCD45+) and Erythroid (TER119+CD45neg) cells were harvested on day 12 of OP9+ESC co-culture and were low to negative for hCD2. DP T-cells (CD4+CD8+) were harvested on Day 18 of OP9DL1+ESC co-culture and were strongly positive for hCD2. Representative target cell population gates are shown at top. hCD2 expression in gated transfected (dark curve) and non-transfected (light curve) cells is shown below in each column.
TCRα LCR linked reporter gene is expressed with endogenous TCRα gene-like kinetics during T-cell development in vitro
The endogenous TCRα gene is activated at or during the double negative (DN)3 (CD4−,CD8−, CD25+,CD44−) to DP stage transition of T-cell development (36). In transgenic mice, the hCD2:1-8 transgene is upregulated with similar kinetics (19). In contrast, the hCD2 reporter gene linked to its cognate LCR is activated at the DN1 stage of T cell development (37).
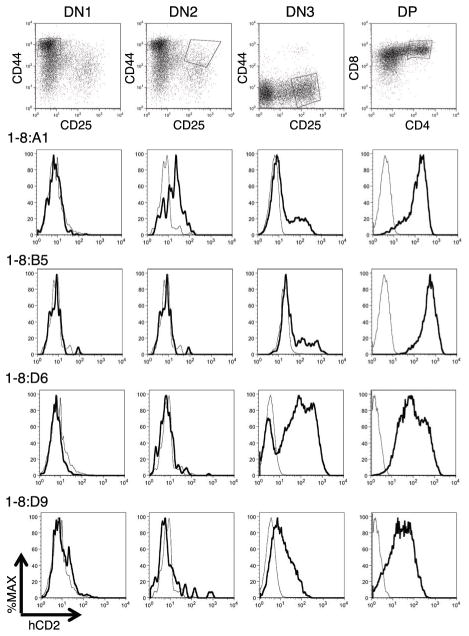
Representative hCD2:1-8 transfectant ESC clones were analyzed for hCD2 expression during the DN stages. Congruent with the expected timing of endogenous TCRα gene activation in vivo, we generally observed hCD2 reporter gene upregulation at DN3 or during the DN3 to DP transition in vitro (Figure 4). One clone (1-8:A1) produced DN2 stage cells displaying a low hCD2 signal reminiscent of the low activity occasionally observed for this transgene at the DN2 stage in vivo (19). Nevertheless, in the main, these data indicate that the TCRα LCR can confer the developmental timing of its locus of origin to a linked heterologous reporter gene during in vitro T cell differentiation.
Figure 4. Appropriate upregulation of the hCD2:1-8 reporter gene at DN3 or during the DN3 to DP stage transition of in vitro T cell development.
Flow cytometry analysis of T cell development from four representative hCD2:1-8ESC clones (1-8:) differentiated in the OP9DL1 co-culture system. Cells were harvested on day 12 of co-culture to detect CD4, CD8 double negative (DN)1 (CD44+CD25neg) and DN2 (CD44+CD25+) stage T cells, or day 18 to examine DN3 (CD44negCD25+) and DP (CD4+CD8+) cells. Note that clone 1-8:A1 also appears in Figure 3. Representative target cell population gates are shown at top. hCD2 expression in gated transfected (dark curve) and non-transfected (light curve) cells is shown below in each column.
The hCD2:1-8 transgene linked to TCRα LCR is expressed in a copy number dependent manner by T cells derived in vitro
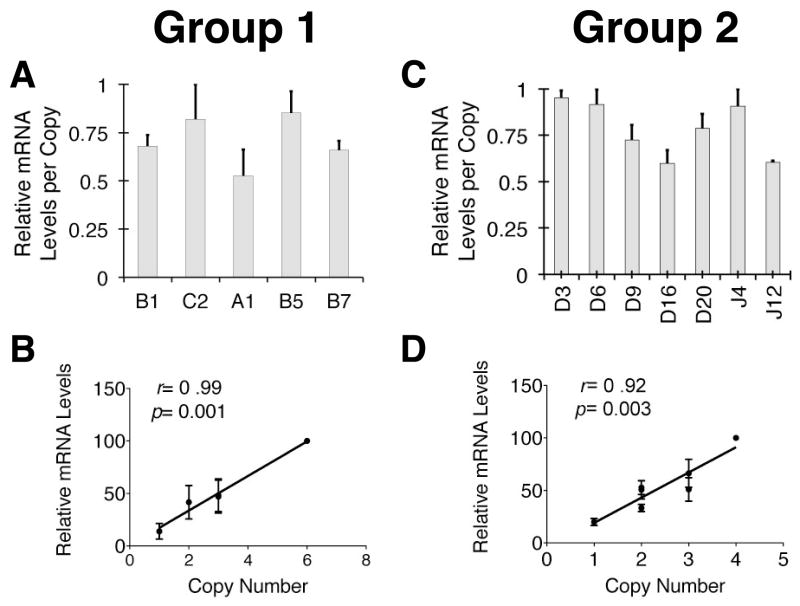
A hallmark manifestation of TCRα LCR activity in vivo is its ability to drive transgene copy number related mRNA levels from a linked reporter gene. To determine if the in vitro ESC differentiation system supported this aspect of TCRα LCR activity, we extracted total RNA from 20-day ESC-OP9-DL1 co-cultures that are rich in DP and CD8 single positive (SP) T-cells (27). The extracted RNA was analyzed for hCD2 expression levels using quantitative, real-time PCR (qRT-PCR). TCRα primers were used to normalize for loading variation and T cell content variability among the co-cultures. Figure 5 shows qRT-PCR data from analyses of two separate groups of TCRα-LCR linked reporter gene transfected ESC clones. Each group of clones was derived from independent transfections. RNA from T cells derived in vitro from each clone was analyzed in triplicate, and experiments on each of these two groups were repeated three times. The data demonstrate that reporter mRNA expression levels per copy vary within a very narrow 1.6-fold range in both sets of experiments (Figure 5A, 5C) and, thus, correlate strongly and significantly with the integrated transgene copy number (Figure 5B, 5D). These data are consistent with the full TCRα LCR activity observed in vivo. These data indicate that the TCRα LCR provides a degree of integration site-independence in this system comparable to that observed in transgenic mice. Taken together, the data described above indicate that the in vitro ESC differentiation system supports the full range of TCRα LCR activity seen in vivo.
Figure 5. TCRα LCR drives copy number-related hCD2 reporter mRNA levels in T cells derived in vitro from ESCs.
Quantitative (q)RT-PCR analysis of in vitro differentiated T cell progeny of representative hCD2:1-8ESC clones. Cells were harvested on day 20 of OP9DL1-ESC co-culture. (A) qRT-PCR results from T cells derived from five individual, independent hCD2:1-8ESC clones generated from an ESC transfection (Group 1). Copy number estimates are (L to R) 3, 6, 1, 2 and 3. Individual samples were run in triplicate in the qRT-PCR experiments. The y-axis signifies the relative mRNA levels detected in a given experiment with the highest level observed designated as “1.0”. Bars represent averages of three independent experiments (see Materials and Methods). The range of mRNA levels per transgene copy is 1.6-fold. (B) Graph of the correlation between relative mRNA level and transgene copy number. Prism (Graphpad) software was used to calculate x-y value correlation (r) and significance (p) noted on the graphs. The p-value was derived from an “F test” on the linear relationship between x and y values. (C) qRT-PCR results [analyzed and depicted as in (A)] from T cells derived from seven additional individual, independent hCD2:1-8ESC clones generated from an additional, independent ESC transfection (Group 2). Copy number estimates are (L to R) 2, 4, 1, 3, 3, 2 and 2. The range of mRNA levels per transgene copy is 1.6-fold. (D) Graph of correlation between relative mRNA level and transgene copy number [analyzed and depicted as in (B)].
Incomplete TCRα LCR activity after de novo introduction into lineage-committed T cell lines
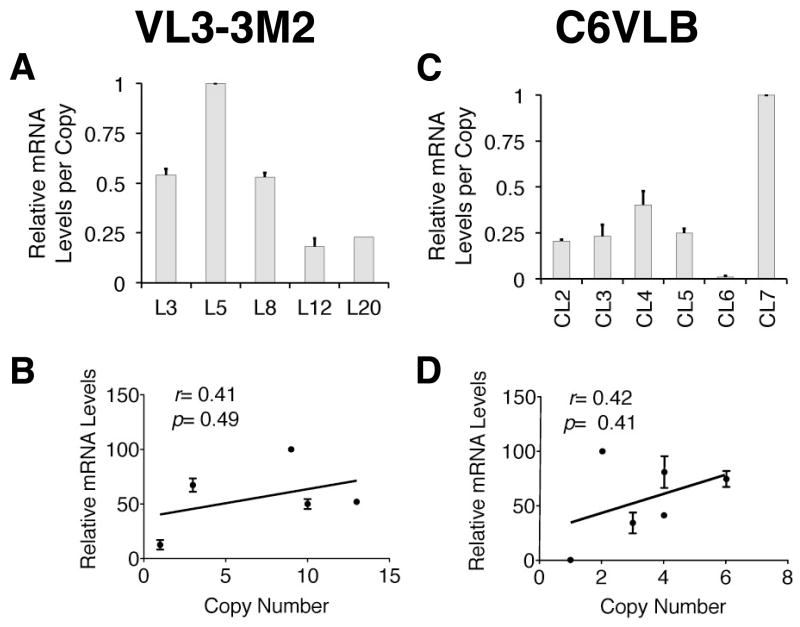
As mentioned previously, prior data had suggested that the development of full LCR activity may require the LCR DNA to be present in the genome of a cell prior to cell lineage commitment (16). We decided to directly test this hypothesis by assessing TCRα LCR activity after its de novo introduction into T cell lines. To do this, we used the identical reporter gene constructs and transfection/selection approach that yielded full TCRα LCR activity above in T cells derived in vitro from ESCs. Figure 6 shows qRT-PCR analyses of hCD2:1-8 reporter gene expression in transfected clones of two different T cell lines at distinct developmental stages. The VL3-3M2 (25) cell line represents a DP thymocyte stage, while the C6VLB (26) cell line represents a circulating CD4 SP stage. The graph depicts relative hCD2 reporter mRNA levels (analyzed in triplicate) per intact integrated transgene copy, as observed in three separate experiments for each cell line. In both cell lines, the expression levels per copy (5.5-fold for VL3 and 83.5-fold for C6VLB) fall outside the range of copy number-dependence that would be expected of full LCR activity (Figure 6A, 6C) (38). In sharp contrast to the strong single copy reporter gene expression seen in the T cells derived from transfected ESC clones (see Figure 5 clones A1 and D9), the single copy C6VLB clone (CL6) is highly sensitive to silencing. Even excluding the CL6 clone, the range of mRNA expression levels per transgene copy would be nearly 5-fold. While this might be indicative of “partial LCR” activity (38), this result would still fall outside the range characteristic of full LCR activity in vivo. Thus, in these cell lines, reporter expression does not correlate significantly with the integrated transgene copy number (Figure 6B, 6D). There are no material differences in the methods of transfection, selection and analyses of both the ESC and cell line clones used to generate TCRα LCR reporter gene positive T cells. Therefore, the data presented here support the above-stated hypothesis.
Figure 6. TCRα LCR driven reporter mRNA levels are not copy number-related after de novo transfection into established T-cell lines.
(A) qRT-PCR analysis of clones of a CD4, CD8 DP T cell line, VL3-3M2, transfected with the hCD2:1-8 reporter gene. Copy number estimates are (L to R) 8, 3, 1, 13 and 10. Individual samples were run in triplicate in the qRTPCR experiments. The y-axis shows relative mRNA levels detected in a given experiment with the highest level observed designated as “1.0”. Bars represent averages of three independent experiments (see Materials and Methods). The range of mRNA levels per copy is 5.5-fold. (B) Graph of the correlation between relative mRNA level and transgene copy number [analyzed and depicted as in Figure 5B, 5D]. (C) qRT-PCR results [analyzed and depicted as in (A)] of clones of C6VLB (a CD4+ T cell line) transfected with the hCD2:1-8 reporter gene. Copy number estimates are (L to R) 4, 3, 4, 6, 1, and 2. The range of mRNA levels per transgene copy is over 80-fold. (D) Graph of the correlation between relative mRNA level and transgene copy number [analyzed and depicted as in Figure 5B, 5D].
Discussion
The ability of an LCR to consistently establish an independently (and predictably) regulated gene locus at an ectopic site in the genome, has been linked to the prevention of heterochromatin-induced position effects (2, 22) and epigenetic modifications (39, 40) in multiple systems. At least three complete LCRs have been isolated and shown to dominantly and predictably regulate a linked, heterologous transgene when randomly integrated into the genome (18, 41, 42). As such, continued study of the unique properties of LCRs manifested at ectopic genomic locations is of high significance. LCR-driven transcription units provide valuable models for investigating the nature and activity of cis-acting gene regulatory DNA during development in a manner that is free of native locus-derived functional redundancy. They further provide critical opportunities to explore how these cis-acting DNA elements might be applied to the improvement of therapeutic transgenesis (43, 44).
A combination of somatic cell fusion (45, 46) and whole animal transgenic (4, 42) approaches identified the first described LCR in the human β-globin locus. Transgenic mice have been particularly important in the characterization of numerous LCRs and all the multiple dimensions of their regulatory activities [reviewed in (1)]. Nevertheless, the high cost and long timelines inherent in this approach inspired various efforts to develop additional models for the study of β-globin LCR activity. These ranged from “cell free” in vitro transcription (47) to somatic cell genetics (48) to direct, de novo introduction of LCR-driven reporter transgenes into established cell lines at pre-determined (49) or random (15) sites in the genome. The latter approach was proven unable to support the complete β-globin LCR activity seen in transgenic mice. In particular, integration-site independence was incomplete, as manifested by the absence of transgene copy number-related reporter gene expression levels (15). Introduction of β-globin LCR-driven reporter genes first into embryonic fibroblasts, followed by fibroblast cell fusion with erythroid cells seemed to rescue this deficiency (16). While this approach does not recapitulate normal erythroid cell development, these experiments suggested that β-globin LCR DNA must pre-exist in an undifferentiated environment before it can establish its full activity in differentiated cells. Pre-differentiation molecular “priming” events at the β-globin locus have been described that would support this notion (50–52). Epigenetic pre-priming of T cell expressed gene loci has also been discovered to occur as early as the ESC stage (53). These events may be critical to establishing proper transcriptional activation upon cellular differentiation (54) and may be part of the reason why differentiated cell lines seem unable to support full LCR activity de novo.
The development of protocols for in vitro differentiation of T cells from mouse ESC (27, 31) seemed to provide a system that, in principle, could incorporate the input of any early epigenetic priming events on gene regulation. We thus sought in this study to determine if complete TCRα LCR activity could be established in T cells derived via this technology. This approach was used once before to test the cell type specificity of the human Perforin gene and LCR contained in a bacterial artificial chromosome (10). We have now demonstrated here that the in vitro ESC to T cell differentiation system supports all known aspects of full TCRα LCR activity. The developmental timing, cell type-specificity and copy number-relatedness of TCRα LCR-driven reporter transgenes in this system are all comparable to that observed in transgenic mice. In contrast, the identical LCR-driven reporter constructs, transfection, selection and mRNA analysis approaches revealed that two distinct T cell lines were unable to support the full integration site-independence of TCRα LCR activity de novo. Together these data indicate that during the development of T cells from embryonic stem cell precursors, critical events prior to final T cell lineage differentiation are required for the establishment of complete TCRα LCR activity.
Studies of the human CD2 gene locus provided the first example of an LCR active in T cells (5). Both transgenic (2, 41, 55) and knock-in (56, 57) mouse studies of this LCR have demonstrated its impact on the ongoing establishment of chromatin and gene expression states during T cell development. A recent application of microcell mediated chromosome transfer to the study of selected aspects of Perforin LCR activity notwithstanding (10), the overwhelming majority of studies of the numerous other T cell-active LCRs have been similarly dependent on whole animal models. The present study now validates in vitro ESC to T cell differentiation as a novel experimental model for the study of LCR activity at ectopic sites. This system bears a close resemblance to normal cellular differentiation. Furthermore, it enables examination of all key aspects of LCR activity in T cells (integration site-independence, developmental timing, cell type-specificity) without the use of transgenic mice.
Combining TCRα LCR activity with in vitro T cell derivation from ESC can facilitate studies involving genetic manipulation of T cell development and function. It has been previously demonstrated that TCRα gene constructs containing the full TCRα LCR lead to robust cell surface transgenic TCR expression in mice with proper developmental kinetics (58). These mice were free of the T cell abnormalities that result from premature expression of the TCR during thymocyte development (59). The work described here indicates that it will be possible to generate similarly normal TCR transgenic T cells via transfection of ESCs followed by in vitro differentiation in OP9-DL1 co-culture. It has also recently been shown that CD8 T cells generated in vitro and adoptively transferred into syngeneic mice are able to generate antigen specific responses without graft-versus-host pathology (60). Thus, the present study also now augurs the feasibility of using TCRα LCR linked transgenes to create genetically modified CD8 T cells in vitro, whose activity can then be examined in vivo after transfer. Finally, in vitro hematopoiesis from ESC precursors should, in principle, support the further study of the numerous other LCRs previously identified in gene loci expressed in cells of the immune system. This approach will provide a rapid screening system for adapting the activity of the many T cell-active LCRs identified to gene therapy. These efforts should improve the efficacy and temporospatial specificity of therapeutic gene expression from vectors proposed for use in stem cell based approaches to T cell genetic engineering.
Acknowledgments
This work was initiated with the support of the New York State Department of Health via the Empire State Stem Cell Research Board (NYSTEM) grant C024302 (B.D.O.) and the Canadian Institutes of Health Research (J.C.Z.P.). This work was completed with the support of National Institutes of Health grant SC1-GM095402 (B.D.O.). A.L. acknowledges the support of a CUNY Doctoral Student Research Grant. A.L., M.K.-L. and J.L. are matriculated in the CUNY Doctoral Program in Biology. J.C.Z.P. is supported by a Canadian Research Chair in Developmental Immunology. The biomedical research infrastructure at Hunter College is supported in part by a Research Centers in Minority Institutions Program grant from the National Center for Research Resources (G12 RR003037) and the National Institute on Minority Health and Health Disparities (G12 MD007599) from the National Institutes of Health.
We thank Joon Kim for expert flow cytometry assistance, Rose Zamoyska for the hCD2ΔT transgene, Steve Smale for VL3-3M2 cells, Jim Allison for C6VLB cells, Paul Love for ESR1 cells, and Derek Sant’Angelo for helpful comments on the manuscript.
References
- 1.Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus Control Regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- 3.Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 4.Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 5.Lang G, Wotton D, Owen MJ, Sewell WA, Brown MH, Mason DY, Crumpton MJ, Kioussis D. The structure of the human CD2 gene and its expression in transgenic mice. EMBO J. 1988;7:1675–1682. doi: 10.1002/j.1460-2075.1988.tb02995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aronow BJ, Ebert CA, Valerius MT, Potter SS, Wiginton DA, Witte DP, Hutton JJ. Dissecting a locus control region: facilitation of enhancer function by extended enhancer-flanking sequences. Mol Cell Biol. 1995;15:1123–1135. doi: 10.1128/mcb.15.2.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz P, Cado D, Winoto A. A locus control region in the T cell receptor alpha/delta locus. Immunity. 1994;1:207–217. doi: 10.1016/1074-7613(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 8.Yui MA, Hernandez-Hoyos G, Rothenberg EV. A new regulatory region of the IL-2 locus that confers position-independent transgene expression. J Immunol. 2001;166:1730–1739. doi: 10.4049/jimmunol.166.3.1730. [DOI] [PubMed] [Google Scholar]
- 9.Adlam M, Siu G. Hierarchical interactions control CD4 gene expression during thymocyte development. Immunity. 2003;18:173–184. doi: 10.1016/s1074-7613(03)00021-9. [DOI] [PubMed] [Google Scholar]
- 10.Pipkin ME, Ljutic B, Cruz-Guilloty F, Nouzova M, Rao A, Zúñiga-Pflücker JC, Lichtenheld MG. Chromosome transfer activates and delineates a locus control region for perforin. Immunity. 2007;26:29–41. doi: 10.1016/j.immuni.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 11.Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 12.Papapetrou EP, Kovalovsky D, Beloeil L, Sant’angelo D, Sadelain M. Harnessing endogenous miR-181a to segregate transgenic antigen receptor expression in developing versus post-thymic T cells in murine hematopoietic chimeras. J Clin Invest. 2009;119:157–168. doi: 10.1172/JCI37216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davila ML, Brentjens R, Wang X, Riviere I, Sadelain M. How do CARs work?: Early insights from recent clinical studies targeting CD19. Oncoimmunology. 2012;1:1577–1583. doi: 10.4161/onci.22524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skarpidi E, Vassilopoulos G, Stamatoyannopoulos G, Li Q. Comparison of expression of human globin genes transferred into mouse erythroleukemia cells and in transgenic mice. Blood. 1998;92:3416–3421. [PubMed] [Google Scholar]
- 16.Vassilopoulos G, Navas PA, Skarpidi E, Peterson KR, Lowrey CH, Papayannopoulou T, Stamatoyannopoulos G. Correct function of the locus control region may require passage through a nonerythroid cellular environment. Blood. 1999;93:703–712. [PubMed] [Google Scholar]
- 17.Hong NA, Cado D, Mitchell J, Ortiz BD, Hsieh SN, Winoto A. A targeted mutation at the T-cell receptor alpha/delta locus impairs T- cell development and reveals the presence of the nearby antiapoptosis gene Dad1. Mol Cell Biol. 1997;17:2151–2157. doi: 10.1128/mcb.17.4.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortiz BD, Cado D, Chen V, Diaz PW, Winoto A. Adjacent DNA elements dominantly restrict the ubiquitous activity of a novel chromatin-opening region to specific tissues. EMBO J. 1997;16:5037–5045. doi: 10.1093/emboj/16.16.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrow F, Ortiz BD. The TCRalpha locus control region specifies thymic, but not peripheral, patterns of TCRalpha gene expression. J Immunol. 2005;175:6659–6667. doi: 10.4049/jimmunol.175.10.6659. [DOI] [PubMed] [Google Scholar]
- 20.Knirr S, Gomos-Klein J, Andino BE, Harrow F, Erhard KF, Kovalovsky D, Sant’Angelo DB, Ortiz BD. Ectopic T cell receptor-alpha locus control region activity in B cells is suppressed by direct linkage to two flanking genes at once. PLoS One. 2010;5:e15527. doi: 10.1371/journal.pone.0015527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortiz BD, Cado D, Winoto A. A new element within the T-cell receptor alpha locus required for tissue-specific locus control region activity. Mol Cell Biol. 1999;19:1901–1909. doi: 10.1128/mcb.19.3.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrow F, Amuta JU, Hutchinson SR, Akwaa F, Ortiz BD. Factors binding a non-classical Cis-element prevent heterochromatin effects on locus control region activity. J Biol Chem. 2004;279:17842–17849. doi: 10.1074/jbc.M401258200. [DOI] [PubMed] [Google Scholar]
- 23.Gomos-Klein J, Harrow F, Alarcón J, Ortiz BD. CTCF-independent, but not CTCF-dependent, elements significantly contribute to TCRalpha locus control region activity. J Immunol. 2007;179:1088–1095. doi: 10.4049/jimmunol.179.2.1088. [DOI] [PubMed] [Google Scholar]
- 24.Melton E, Sarner N, Torkar M, van der Merwe PA, Russell JQ, Budd RC, Mamalaki C, Tolaini M, Kioussis D, Zamoyska R. Transgene-encoded human CD2 acts in a dominant negative fashion to modify thymocyte selection signals in mice. Eur J Immunol. 1996;26:2952–2963. doi: 10.1002/eji.1830261222. [DOI] [PubMed] [Google Scholar]
- 25.Groves T, Katis P, Madden Z, Manickam K, Ramsden D, Wu G, Guidos CJ. In vitro maturation of clonal CD4+CD8+ cell lines in response to TCR engagement. J Immunol. 1995;154:5011–5022. [PubMed] [Google Scholar]
- 26.Allison JP, McIntyre BW, Bloch D. Tumor-specific antigen of murine T-lymphoma defined with monoclonal antibody. J Immunol. 1982;129:2293–2300. [PubMed] [Google Scholar]
- 27.Holmes R, Zúñiga-Pflücker JC. The OP9-DL1 system: generation of T-lymphocytes from embryonic or hematopoietic stem cells in vitro. Cold Spring Harb Protoc. 2009;2009:pdb prot5156. doi: 10.1101/pdb.prot5156. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 29.Kodama H, Nose M, Niida S, Nishikawa S. Involvement of the c-kit receptor in the adhesion of hematopoietic stem cells to stromal cells. Exp Hematol. 1994;22:979–984. [PubMed] [Google Scholar]
- 30.Cho SK, Webber TD, Carlyle JR, Nakano T, Lewis SM, Zúñiga-Pflücker JC. Functional characterization of B lymphocytes generated in vitro from embryonic stem cells. Proc Natl Acad Sci USA. 1999;96:9797–9802. doi: 10.1073/pnas.96.17.9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt TM, de Pooter RF, Gronski MA, Cho SK, Ohashi PS, Zúñiga-Pflücker JC. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol. 2004;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- 32.de Pooter RF, Schmitt TM, de la Pompa JL, Fujiwara Y, Orkin SH, Zúñiga-Pflücker JC. Notch signaling requires GATA-2 to inhibit myelopoiesis from embryonic stem cells and primary hemopoietic progenitors. J Immunol. 2006;176:5267–5275. doi: 10.4049/jimmunol.176.9.5267. [DOI] [PubMed] [Google Scholar]
- 33.Watarai H, Rybouchkin A, Hongo N, Nagata Y, Sakata S, Sekine E, Dashtsoodol N, Tashiro T, Fujii S, Shimizu K, Mori K, Masuda K, Kawamoto H, Koseki H, Taniguchi M. Generation of functional NKT cells in vitro from embryonic stem cells bearing rearranged invariant Valpha14-Jalpha18 TCRalpha gene. Blood. 2010;115:230–237. doi: 10.1182/blood-2009-04-217729. [DOI] [PubMed] [Google Scholar]
- 34.Arsov I, Adebayo A, Kučerová-Levisohn M, Haye J, MacNeil M, Papavasiliou FN, Yue Z, Ortiz BD. A role for autophagic protein beclin 1 early in lymphocyte development. J Immunol. 2011;186:2201–2209. doi: 10.4049/jimmunol.1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang G, Mamalaki C, Greenberg D, Yannoutsos N, Kioussis D. Deletion analysis of the human CD2 gene locus control region in transgenic mice. Nucleic Acids Res. 1991;19:5851–5856. doi: 10.1093/nar/19.21.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearse M, Wu L, Egerton M, Wilson A, Shortman K, Scollay R. A murine early thymocyte developmental sequence is marked by transient expression of the interleukin 2 receptor. Proc Natl Acad Sci USA. 1989;86:1614–1618. doi: 10.1073/pnas.86.5.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 38.Ellis J, Tan-Un KC, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human beta-globin locus control region. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- 39.Ho Y, Elefant F, Cooke N, Liebhaber S. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol Cell. 2002;9:291–302. doi: 10.1016/s1097-2765(02)00447-1. [DOI] [PubMed] [Google Scholar]
- 40.Santoso B, Ortiz BD, Winoto A. Control of organ specific demethylation by an element of the T-cell receptor alpha locus control region. J Biol Chem. 2000;275:1952–1958. doi: 10.1074/jbc.275.3.1952. [DOI] [PubMed] [Google Scholar]
- 41.Greaves DR, Wilson FD, Lang G, Kioussis D. Human CD2 3′-flanking sequences confer high-level, T cell-specific, position-independent gene expression in transgenic mice. Cell. 1989;56:979–986. doi: 10.1016/0092-8674(89)90631-4. [DOI] [PubMed] [Google Scholar]
- 42.Blom van Assendelft G, Hanscombe O, Grosveld F, Greaves DR. The beta-globin dominant control region activates homologous and heterologous promoters in a tissue-specific manner. Cell. 1989;56:969–977. doi: 10.1016/0092-8674(89)90630-2. [DOI] [PubMed] [Google Scholar]
- 43.May C, Rivella S, Callegari J, Heller G, Gaensler KM, Luzzatto L, Sadelain M. Therapeutic haemoglobin synthesis in beta-thalassaemic mice expressing lentivirus-encoded human beta-globin. Nature. 2000;406:82–86. doi: 10.1038/35017565. [DOI] [PubMed] [Google Scholar]
- 44.Lisowski L, Sadelain M. Locus control region elements HS1 and HS4 enhance the therapeutic efficacy of globin gene transfer in beta-thalassemic mice. Blood. 2007;110:4175–4178. doi: 10.1182/blood-2007-08-108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forrester WC, Takegawa S, Papayannopoulou T, Stamatoyannopoulos G, Groudine M. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 1987;15:10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forrester WC, Epner E, Driscoll MC, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 47.Armstrong JA, Emerson BM. NF-E2 disrupts chromatin structure at human beta-globin locus control region hypersensitive site 2 in vitro. Mol Cell Biol. 1996;16:5634–5644. doi: 10.1128/mcb.16.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. The locus control region is necessary for gene expression in the human beta-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng YQ, Warin R, Li T, Olivier E, Besse A, Lobell A, Fu H, Lin CM, Aladjem MI, Bouhassira EE. The human beta-globin locus control region can silence as well as activate gene expression. Mol Cell Biol. 2005;25:3864–3874. doi: 10.1128/MCB.25.10.3864-3874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vieira KF, Levings PP, Hill MA, Crusselle VJ, Kang SH, Engel JD, Bungert J. Recruitment of transcription complexes to the beta-globin gene locus in vivo and in vitro. J Biol Chem. 2004;279:50350–50357. doi: 10.1074/jbc.M408883200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bottardi S, Ross J, Pierre-Charles N, Blank V, Milot E. Lineage-specific activators affect beta-globin locus chromatin in multipotent hematopoietic progenitors. EMBO J. 2006;25:3586–3595. doi: 10.1038/sj.emboj.7601232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bottardi S, Aumont A, Grosveld F, Milot E. Developmental stage-specific epigenetic control of human beta-globin gene expression is potentiated in hematopoietic progenitor cells prior to their transcriptional activation. Blood. 2003;102:3989–3997. doi: 10.1182/blood-2003-05-1540. [DOI] [PubMed] [Google Scholar]
- 53.Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smale ST. Pioneer factors in embryonic stem cells and differentiation. Curr Opin Genet Dev. 2010;20:519–526. doi: 10.1016/j.gde.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hostert A, Tolaini M, Festenstein R, McNeill L, Malissen B, Williams O, Zamoyska R, Kioussis D. A CD8 genomic fragment that directs subset-specific expression of CD8 in transgenic mice. J Immunol. 1997;158:4270–4281. [PubMed] [Google Scholar]
- 56.Menzel U, Kosteas T, Tolaini M, Killeen N, Roderick K, Kioussis D. Modulation of the murine CD8 gene complex following the targeted integration of human CD2-locus control region sequences. J Immunol. 2011;187:3712–3720. doi: 10.4049/jimmunol.1100709. [DOI] [PubMed] [Google Scholar]
- 57.Menzel U, Ktistaki E, Tolaini M, Veiga-Fernandes H, Kioussis D. Replication allows inactivation of a knocked-in locus control region in inappropriate cell lineages. Proc Natl Acad Sci USA. 2010;107:16928–16933. doi: 10.1073/pnas.1010317107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kovalovsky D, Pezzano M, Ortiz BD, Sant’Angelo DB. A novel TCR transgenic model reveals that negative selection involves an immediate, Bim-dependent pathway and a delayed, Bim-independent pathway. PLoS One. 2010;5:e8675. doi: 10.1371/journal.pone.0008675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lacorazza HD, Tucek-Szabo C, Vasovic LV, Remus K, Nikolich-Zugich J. Premature TCR alpha beta expression and signaling in early thymocytes impair thymocyte expansion and partially block their development. J Immunol. 2001;166:3184–3193. doi: 10.4049/jimmunol.166.5.3184. [DOI] [PubMed] [Google Scholar]
- 60.Dervovic DD, Ciofani M, Kianizad K, Zúñiga-Pflücker JC. Comparative and functional evaluation of in vitro generated to ex vivo CD8 T cells. J Immunol. 2012;189:3411–3420. doi: 10.4049/jimmunol.1200979. [DOI] [PubMed] [Google Scholar]