Abstract
In this study, we characterized full-length hepatitis C virus (HCV) genome sequences for 11 genotype 2 isolates. They were isolated from the sera of 11 patients residing in Canada, of whom four had an African origin. Full-length genomes, each with 18–25 overlapping fragments, were obtained by PCR amplification. Five isolates represent the first complete genomes of subtypes 2d, 2e, 2j, 2m and 2r, while the other six correspond to variants that do not group within any assigned subtypes. These sequences had lengths of 9508–9825 nt and each contained a single ORF encoding 3012–3106 aa. Predicted amino acids were carefully inspected and unique variation patterns were recognized, especially for a 2e isolate, QC64. Phylogenetic analysis of complete genome sequences provides evidence that there are a total of 16 subtypes, of which 11 have been described here. Co-analysis with 68 partial NS5B sequences also differentiated 18 assigned subtypes, 2a–2r, and eight additional lineages within genotype 2, which is consistent with the analysis of complete genome sequences. The data from this study will now allow 10 assigned subtypes and six additional lineages of HCV genotype 2 to have their full-length genomes defined. Further analysis with 2021 genotype 2 sequences available in the HCV database indicated that the geographical distribution of these subtypes is consistent with an African origin, with particular subtypes having spread to Asia and the Americas.
Introduction
Determination of hepatitis C virus (HCV) genotype is of clinical relevance (Simmonds et al., 2005), which is shown by differences in the sensitivity to alpha interferon and ribavirin combination therapy among patients. Those infected by genotypes 2 and 3 have a higher rate of sustained virological response than those infected by genotypes 1 and 4 (Feld & Hoofnagle, 2005; Manns et al., 2001, 2006; Pearlman, 2004). In addition, genotype 3 has been linked to the development of more pronounced hepatocellular steatosis (Abid et al., 2005; Hourioux et al., 2007; Jackel-Cram et al., 2007; Negro, 2006; Rubbia-Brandt et al., 2000). Although the mechanisms involved are currently not understood, there is probably an association with the genetic variation of the virus.
Understanding the origin and nature of HCV genetic diversity is critical for defining preventive strategies and for the development of therapies and a vaccine. Such diversity is the source of information about the virus’s epidemic history prior to its discovery, which has not been well-documented in non-Western countries. Canada is a country encompassing many immigrants from non-Western countries, among which a variety of HCV genotypes and subtypes have been detected (Murphy et al., 2007). These include a number of genotype 2 isolates isolated in the Canadian province of Quebec. In addition to the globally prevalent 2a, 2b and 2c subtypes, other isolates representing less common subtypes (2d, 2e, 2i, 2j, 2k, 2m, 2r) and many novel variants were identified. This study focused on defining the full-length genome sequences for 11 of these strains, which have a limited spread, with the purpose of analysing their genetic diversity and organization. Our findings should add insights to HCV genomic studies and to clinical applications.
Although Africa is commonly thought to be the region where genotype 2 has originated, a whole spectrum of genotype 2 subtypes and unclassified variants have been identified outside this continent (Simmonds et al., 2005; Pouillot et al., 2008; Markov et al., 2009). This raises the question, could HCV genotype 2 have originated in another region such as Europe? In the current study, we addressed this question in association with the geographical distribution and genetic variation of all genotype 2 sequences available in the Los Alamos HCV database (http://hcv.lanl.gov/).
Results
Genome sequence and organization
Full-length genomic sequences were successfully obtained for 11 HCV genotype 2 isolates each with 18–25 overlapping fragments (see Fig. S1, available in JGV Online). These sequences had the lengths of 9508–9825 nt, starting from the extreme 5′ UTR termini through to the 3′ poly(U) tracts or the highly conserved 3′ X tails (Table 1). Four isolates QC64/2e, QC178/2m, QC302/2 and QC331/2 had their sequences determined up to the 3′ X tail while all the other isolates only had their sequences determined to the poly(U) tract. The genomes each contained a single ORF of 9036–9318 nt, with 10 protein-coding regions of the following sizes: core (573 nt/191 aa), E1 (576 nt/192 aa), E2 (1095–1101 nt/365–367 aa), p7 (189–390 nt/63–130 aa), NS2 (651 nt/217 aa), NS3 (1893 nt/631 aa), NS4A (162 nt/54 aa), NS4B (783 nt/261 aa), NS5A (1398–1413 nt/466–471 aa) and NS5B (1776 nt/591 aa). Among these regions, E2, p7 and NS5A were variable in length (Table 1).
Table 1. Patient information for the 11 genotype 2 isolates and the number of nucleotides/amino acids in each genomic region.
Bold entries indicate regions that are variable in length.
| ID | Age | Sex | Origin | Full | ORF | 5′ UTR | Core | E1 | E2 | p7 | NS2 | NS3 | NS4A | NS4B | NS5A | NS5B | 3′ UTR |
| H77* | – | – | – | 9646 | 9036/3011 | 341 | 573/191 | 576/192 | 1089/363 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1344/448 | 1776/591 | 269 |
| QC114 | 57 | F | Portuguese | 9508 | 9102/3033 | 340 | 573/191 | 576/192 | 1101/367 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1398/466 | 1776/591 | 66 |
| QC178 | 69 | M | Vietnam | 9618 | 9105/3034 | 340 | 573/191 | 576/192 | 1101/367 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1401/467 | 1776/591 | 173 |
| QC182 | 36 | F | Cameroon | 9519 | 9114/3037 | 340 | 573/191 | 576/192 | 1101/367 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1410/470 | 1776/591 | 65 |
| QC232 | 68 | M | Egypt | 9509 | 9102/3033 | 340 | 573/191 | 576/192 | 1101/367 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1398/466 | 1776/591 | 67 |
| QC259 | 38 | F | Benin | 9508 | 9102/3033 | 340 | 573/191 | 576/192 | 1101/367 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1398/466 | 1776/591 | 66 |
| QC283 | 53 | F | Haiti | 9512 | 9102/3033 | 340 | 573/191 | 576/192 | 1101/367 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1398/466 | 1776/591 | 70 |
| QC289 | 41 | F | Africa | 9514 | 9102/3033 | 340 | 573/191 | 576/192 | 1101/367 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1398/466 | 1776/591 | 72 |
| QC297 | 68 | F | Italy | 9510 | 9096/3031 | 340 | 573/191 | 576/192 | 1095/365 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1398/466 | 1776/591 | 74 |
| QC302 | 61 | F | Europe? | 9615 | 9102/3033 | 340 | 573/191 | 576/192 | 1101/367 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1398/466 | 1776/591 | 173 |
| QC331 | 75 | M | Canada? | 9628 | 9111/3036 | 341 | 573/191 | 576/192 | 1098/366 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1410/470 | 1776/591 | 176 |
| QC64 | 65 | M | Indonesia | 9825 | 9318/3105 | 340 | 573/191 | 576/192 | 1101/367 | 390/130 | 651/217 | 1893/631 | 162/54 | 783/261 | 1413/471 | 1776/591 | 167 |
The H77 genome (GenBank accession no. NC_004102), HCV genotype 1, is included for comparison.
Genotype 2 classification
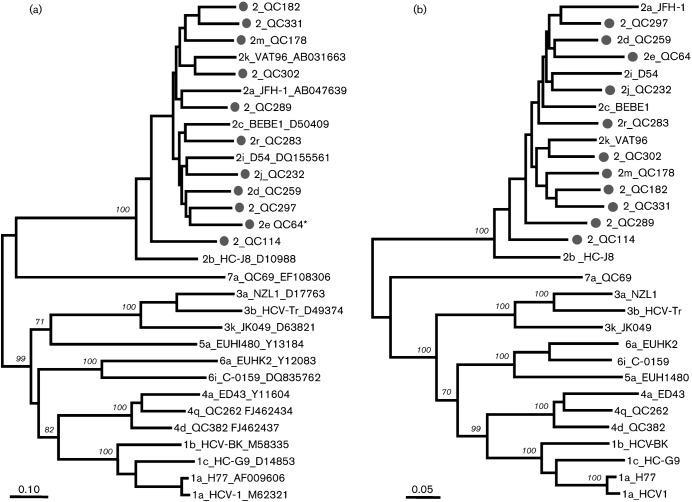
Fig. 1(a) shows the maximum-likelihood tree reconstructed with full-length genome sequences for all HCV genotypes. The tree displays seven major branches/clusters, each representing one genotype. Each major branch showed a bootstrap support of 100 % when a cluster was formed (i.e. genotypes with more than one subtype). Within genotype 2, the 11 isolates were distinct from subtypes 2a, 2b, 2c, 2i and 2k, for which full-length sequences were previously determined. Prior to our report, subtypes 2d, 2e, 2j, 2m and 2r were provisionally assigned only based on partial sequences. In the present study, these five subtypes were confirmed by their full-length genomes being determined. In addition, six isolates QC114, QC182, QC289, QC297, QC302 and QC331 showed evidence that they represent six new lineages. Fig. 1(b) shows the maximum-likelihood tree based on the amino acid sequences deduced for these isolates. A topology consistent with that in Fig. 1(a) was observed.
Fig. 1.
Maximum-likelihood phylogenies estimated from (a) full-length nucleotide sequences and (b) predicted amino acid sequences. Except for the 11 isolates from this study (•), reference sequences from subtypes 1a, 1b, 1c, 2a, 2b, 2c, 2i, 2k, 3a, 3b, 3k, 4a, 4d, 4q, 5a, 6a, 6i and 7a were also included. Each branch was led by an isolate named in the format: subtype_isolate_accession number. Isolate names that start with ‘2’ alone, without a letter, indicate that a subtype has not been assigned. Bootstrap supports are shown in italics. Bars, 0.10 nucleotide substitutions per site (a); 0.05 amino acid substitutions per site (b). *QC64 contains an extra p7 gene.
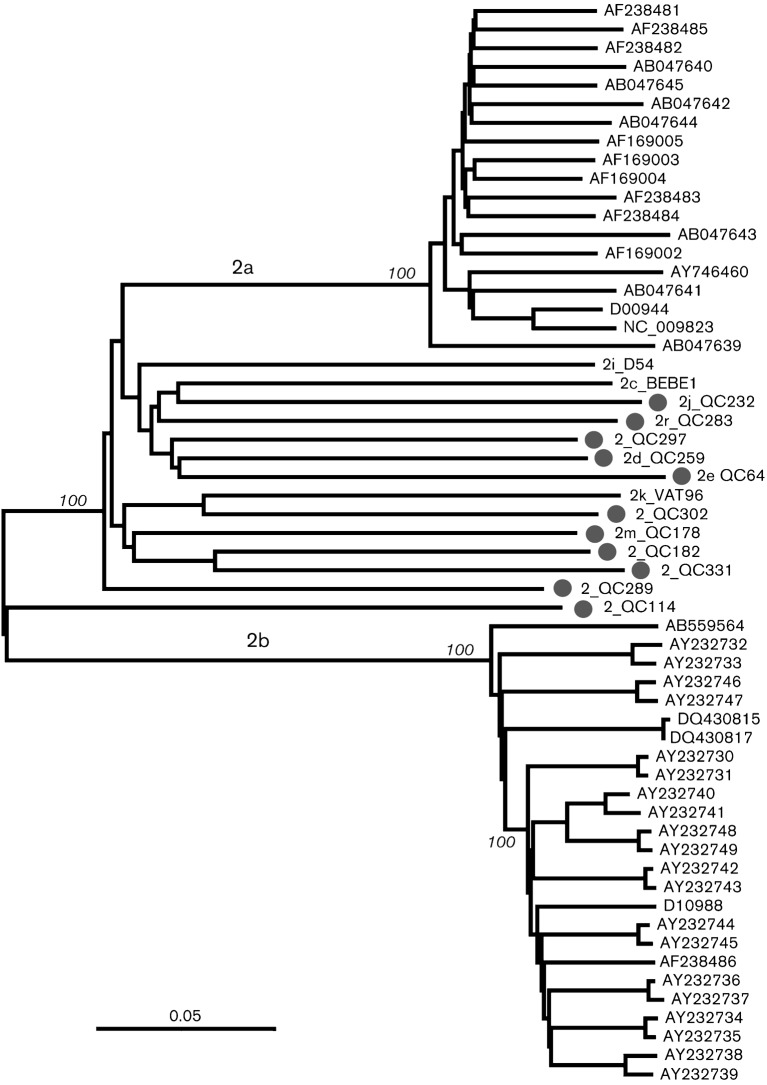
Fig. 2 shows a neighbour-joining tree reconstructed with full-length sequences for genotype 2. Besides the 11 isolates from the current study, the tree also included 47 sequences retrieved from the HCV database. Among them, 19 belonged to subtype 2a (12 from Japan, seven with unknown origin), 25 belonged to subtype 2b (23 from Japan and two from the USA), and one belonged to each of subtype 2c (from an Italian patient) (Nakao et al., 1996), subtype 2k (Samokhvalov et al., 2000) and subtype 2i (Noppornpanth et al., 2006). The tree showed that the 11 isolates were distinct from each other and from subtypes 2a, 2b, 2c, 2k and 2i. This provides evidence that the 11 isolates represent 11 distinct genotype 2 subtypes/lineages.
Fig. 2.
Neighbour-joining tree reconstructed with full-length genome sequences from 58 genotype 2 isolates. In addition to the 11 sequences from this study, the tree included 47 reference sequences. The latter contained 19 subtype 2a, 25 subtype 2b and one each of subtypes 2c, 2i and 2k. Subtypes 2a and 2b sequences are identified with their GenBank accession numbers. Sequences from this study are identified (•) and include the isolate name. Bar, 0.05 nucleotide substitutions per site.
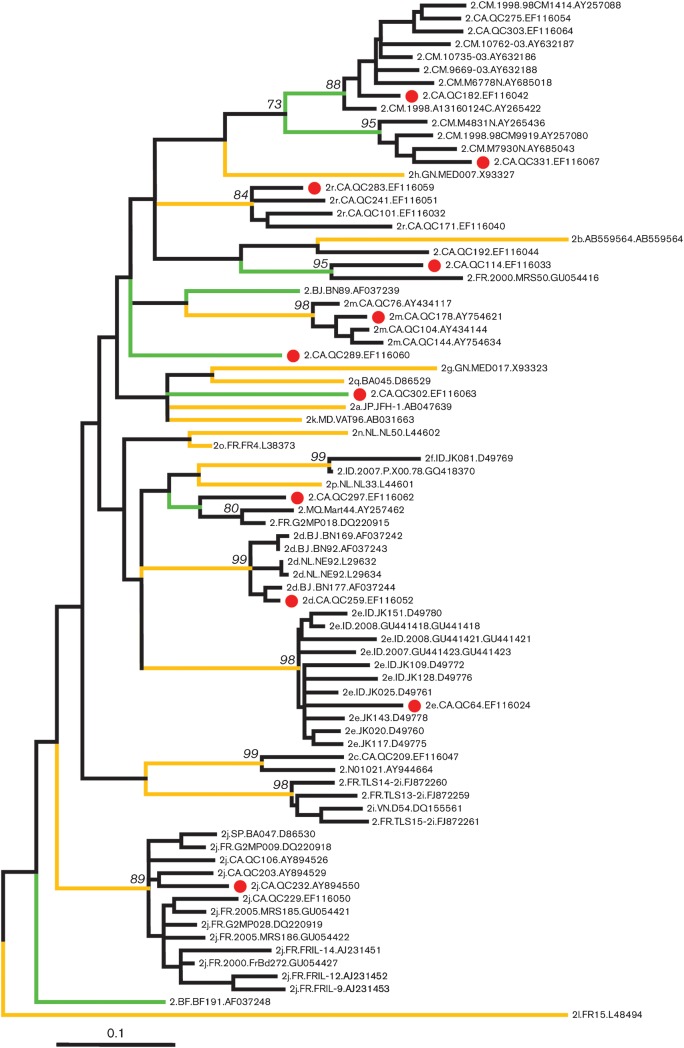
A segment of the NS5B region, corresponding to nt 8276–8615 in the numbering of the H77 genome (GenBank accession no. NC_004102), is reliable for differentiating HCV genotypes and subtypes. This region was further analysed using sequences from 68 genotype 2 isolates in addition to our 11 genotype 2 isolates. The tree (Fig. 3) shows 26 branches or clusters corresponding to assigned subtypes and unclassified lineages, congruent with those obtained for the complete genome sequences. Among them 18 (yellow-coloured branches) have been assigned subtypes 2a–2r and eight (green-coloured branches) have not been assigned to a subtype. QC259, QC64, QC232, QC178 and QC283 represent five of the 18 assigned while QC114, QC182, QC289, QC297, QC302 and QC331 represent six of the eight unassigned. QC259/2d grouped with five isolates, two from the Netherlands (Stuyver et al., 1994) and three from Africa (Jeannel et al., 1998); QC64/2e grouped with 10 isolates all from Indonesia (Tokita et al., 1996; Utama et al., 2010); QC232/2j grouped with nine isolates, one from Spain (BA047) (Holland et al., 1996) and eight from France (Thomas et al., 2007; Cantaloube et al., 2008; Le Pogam et al., 1998); QC178/2m and QC283/2r each grouped with three isolates, all from Canada; QC297 grouped with two isolates, one from the French Caribbean island of Martinique (Martial et al., 2004) and one from France (Thomas et al., 2007); QC331 grouped with three isolates all from Cameroon (Njouom et al., 2003a, 2005; Ndjomou et al., 2003a); QC182 grouped with eight isolates, two from Canada and six from Cameroon (Ndjomou et al., 2003; Pasquier et al., 2005; Njouom et al., 2003b). In contrast, QC302 and QC289 each represented an orphan isolate while QC114 grouped most closely with MRS50, a variant reported in France (Cantaloube et al., 2008).
Fig. 3.
Maximum-likelihood tree for 79 sequences of the NS5B region corresponding to nt 8276–8615 in the numbering of the H77 genome. Yellow branches mark the 18 assigned subtypes, 2a–2r. Green branches mark the eight unassigned lineages. All tips are named in the format: subtype, original country (for country codes see Table 2), sampling time (or missing), isolate ID, and GenBank accession number, each of which is separated by a dot. Bar, 0.1 nucleotide substitutions per site.
Geographical distribution of HCV genotype 2 isolates
In the Los Alamos HCV database (end of year 2010), a total of 4044 sequences have been classified in genotype 2. These sequences were filtered out and a total of 2021 individual isolates were distinguished that had their sampling countries recognized. The geographical distribution of these isolates is summarized in Tables 2 and 3. Subtypes 2a, 2b and 2c were ubiquitous; they were identified in 31, 21 and 19 countries/regions, respectively, accounting for 21.6, 24.7 and 17.8 % of the total 2021 isolates. This was followed by subtype 2k, which was found in nine countries in Europe, America, Africa, Asia and the Middle East, accounting for 3.27 % of isolates. In contrast, subtype 2i can be found only in four countries but in different continents; 2d, 2e, 2f and 2j each in three countries; 2h and 2l each in two countries; and the remaining seven subtypes (2g, 2m, 2n, 2o, 2p, 2q, 2r) each in one country. Diversity of reported genotype 2 isolates was the largest in Europe, especially in France, where a total of 17 subtypes were identified. However, 85.6 % (107 out of 125) of the African isolates were of unknown subtype, and 14.4 % (18 out of 125) belonged to six different subtypes (Table 3). The area of greatest diversity is presumably the region of origin, which therefore suggests Africa.
Table 2. Geographical distribution of HCV genotype 2 subtypes.
| Subtype | No. of countries in which subtype is detected | Total (%) |
| 2a | 31 | 437 (21.6) |
| 2b | 21 | 499 (24.7) |
| 2c | 19 | 360 (17.8) |
| 2k | 9 (AZ, CA, FR, MD, MG, MQ, RU, UZ, VN)* | 66 (3.27) |
| 2i | 4 (CA, DE, FR, VN) | 36 (1.78) |
| 2d | 3 (BJ, NL, CA) | 6 (0.300) |
| 2e | 3 (ID, CA, BN) | 22 (1.09) |
| 2f | 3 (DE, ID, CN) | 5 (0.247) |
| 2j | 3 (FR, CA, BA) | 9 (0.445) |
| 2h | 2 (DE, GN) | 2 (0.099) |
| 2l | 2 (FR, MQ) | 8 (0.396) |
| 2g | 1 (GN) | 10 (0.495) |
| 2m | 1 (CA) | 4 (0.198) |
| 2n | 1 (NL) | 1 (0.045) |
| 2o | 1 (FR) | 2 (0.099) |
| 2p | 1 (NL) | 1 (0.045) |
| 2q | 1 (BA) | 1 (0.045) |
| 2r | 1 (CA) | 4 (0.198) |
| Recomb. | 6 (IE, VN, PH, AR, RU, UZ) | 48 (2.38) |
| 2? | 31 | 500 (24.7) |
| Total | 56 | 2021 (100.0) |
Country codes officially assigned in ISO 3166-1: AR, Argentina; AZ, Azerbaijan; BA, Bosnia; BJ, Benin; BN, Brunei Darussalam; CA, Canada; CM, Cameroon; CN, China; DE, Germany; FR, France; GN, Guinea; ID, Indonesia; IE, Ireland; MD, Moldova, Republic of; MG, Madagascar; MQ, Martinique; NL, the Netherlands; PH, the Philippines; RU, Russian Federation; SP, Spain; UZ, Uzbekistan; VN, Vietnam (http://en.wikipedia.org/wiki/ISO_3166-1_alpha-2).
Table 3. Genotype 2 subtypes in different countries.
| Location | No. (subtypes)* | No. of isolates | Subtotal (%) |
| Europe | 17 (a, b, c, d, f, g, h, i, j, k, l, m, n, o, p, q, r) | 724 | 1020 (52.5) |
| (2?) | 296 | ||
| North America | 11 (a, b, c, d, e, i, j, k, l, m, r) | 207 | 254 (12.6) |
| (2?) | 47 | ||
| Asia | 9 (a, b, c, e, f, i, k, 1b/2k, 2i/6p) | 510 | 540 (26.7) |
| (2?) | 30 | ||
| Africa | 6 (b, c, d, g, h, k) | 18 | 125 (6.19) |
| (2?) | 107 | ||
| South America | 4 (a, b, c, a/c) | 53 | 69 (3.41) |
| (2?) | 16 | ||
| Middle East | 3 (a, b, k) | 5 | 9 (0.445) |
| 2? | 4 | ||
| Australia–Oceania | 2 (a, b) | 4 | 4 (0.198) |
| Total | 2021 |
The first row for each location shows the number of total assigned subtypes found in the location; the second row (2?) for each location represents isolates that are classified into genotype 2, but the subtype has not yet been assigned.
Pairwise comparison
Pairwise comparisons were performed between the 11 isolates and other isolates within genotype 2. The differences ranged from 16.9 % (QC182 vs QC331) to 23.6 % (2e_QC64 vs 2b_JPUT971017) at the nucleotide level and from 10.5 % (QC259 vs QC297) to 16.5 % (2e_QC64 vs 2b_JPUT971017) at the amino acid level. When compared with isolates of other genotypes, the differences ranged from 31.4 % (2r_QC283 vs 1a_H77) to 35.2 % (2_QC302 vs 3b_HCV-Tr) at the nucleotide level, and from 26.4 % (2d_259 vs 1a_H77) to 30.9 % (2e_QC64 vs 3b_Tr-kj) at the amino acid level. These results further support Fig. 1 and distinguish each isolate; they each belong to a different subtype.
To exclude the possibility of viral recombination, pairwise nucleotide similarity curves were plotted along HCV genomes. Comparing the 11 isolates against each other and against the 19 reference sequences representing different genotypes and subtypes shown in Fig. 1(a), no such evidence was revealed (data not shown).
Specific variations
E2 region.
QC297 comprises 365 aa and QC331 comprises 366 aa, while the remaining nine isolates each comprises 367 aa, the same size as other genotype 2 subtypes in this region. The E2 protein generally has 11 potential glycosylation sites. These sites were all conserved among the genotype 2 isolates, except for QC64 and BEBE1 (2c) which had a serine instead of an asparagine at the N5 site. All 16 sequences of genotype 2 (11 of our new isolates and five from the available database) showed 4 aa more in hypervariable region (HVR)2 or HVR3 than genotype 1. QC297 had a deletion of 2 aa and QC331 had a deletion of 1 aa in HVR1.
p7 sequences.
All but one of the isolates displayed a p7 sequence of 189 nt (63 aa). Surprisingly, QC64 had an insertion of 201 nt (67 aa) located directly after the p7 gene. Since the insertion appeared to be related to the p7 gene itself, a phylogeny was constructed for p7 sequences of all genotype 2 subtypes and representative sequences of the six other genotypes. For QC64, the 189 nt corresponding to the standard p7 sequence and the 201 nt corresponding to the insertion sequence were individually included in the analysis. Interestingly, this QC64 insertion was shown to cluster with the p7 sequence from QC64, and they both shared a nucleotide similarity of 78 %. The phylogenetic tree and the alignment of amino acid sequences are shown in Fig. 4.
Fig. 4.
Analysis of p7 sequences. (a) Phylogeny. p7 sequences from the 11 isolates in this study are marked (•). The insertion of isolate QC64 is indicated (○). Subtypes 1a, 1b, 1c, 2a, 2b, 2c, 2i, 2k, 3a, 3b, 3k, 4a, 4f, 5a, 6a, 6i and 7a were included as references. Bar, 0.1 nucleotide substitutions per site. (b) Multiple sequence alignment. The consensus sequence and distinct amino acids are shown in standard International Union of Pure and Applied Chemistry (IUPAC) codes. Dashes indicate gaps relative to the 2e_QC64 insertion sequence. The isolate names are listed to the right of the alignment and the QC64 insertion is indicated in grey.
NS5A.
Genotype 2 isolates show the longest NS5A region. For the 11 isolates, this region ranged between 1398 and 1413 nt (466–471 aa). Various insertions were observed in NS5A. To begin with, the interferon-sensitivity determining region (ISDR) of QC64 showed an insertion of 4 aa at amino acid numbering 2303–2306 (aa 261–264 in NS5A). Such an insertion is also observed in the BEBE1/2c isolate and in genotypes 1, 3, 5, 6 and 7 isolates, but not in genotype 4 isolates. An insertion was also observed in both QC64 and QC178 directly at the end of protein kinase R-binding domain (PKR-BD) region following the ISDR. They each had an insertion of 1 aa, corresponding to amino acid numbering 2349 of QC64 and 2278 of QC178 (aa 306 and 302 in NS5A, respectively). In the V3 region, QC182 and QC331 each displayed an insertion of 4 aa at amino acid numbering 2376–2379 and 2375–2378. Finally, when genotype 2 sequences as a whole were compared to the other HCV genotypes, they were found to contain an insertion in domain III of NS5A. The insertion occurred following the sequence SMPPLEGEPGDPDL (amino acid numbering 415–428 in the NS5A of H77) that is highly conserved in all HCV genotypes. At this position, genotype 2 isolates showed an insertion of 20 aa that was highly variable between subtypes with the median p-distance being 0.65 (0.30–0.90) (data not shown).
Discussion
In this study, full-length genome sequences were characterized for 11 genotype 2 isolates. Among them, QC259, QC64, QC232, QC178 and QC283 were previously assigned to subtypes 2d, 2e, 2j, 2m and 2r, respectively, based on partial C/E1 and NS5B sequences (Murphy et al., 2007). In the consensus paper, these five subtypes have been classified as ‘provisionally assigned subtypes’ (Simmonds et al., 2005) due to the absence of available full-length genome sequences. In this study, the full-length sequences of the five isolates all grouped independently from each other and from other genotype 2 isolates for which complete sequences are available. This now allows for the confirmed designations of subtypes 2d, 2e, 2j, 2m and 2r.
Full-length sequences were also obtained for six isolates designated QC114, QC182, QC289, QC297, QC302 and QC331. They also grouped independently from each other and from other genotype 2 complete sequences. However, we prefer not to assign them to new subtypes at the present moment. QC114, QC289 and QC302 do not group with a sufficient number of closely related isolates; they are not considered for a new subtype designation. Analysis of partial sequences in the NS5B region showed QC182, QC331 and QC297 each clustered with three or more isolates from the same geographical regions (Fig. 3). However, only a fraction of unclassified variants were included in Fig. 3. Inclusion of additional variants in the analysis did not allow for clear subtype separations (data not shown). They may represent continual variants resulting from endemic evolution. We would suggest that new subtype designations be given for these isolates only after their epidemiological significance is approved, the possibility of viral recombination is excluded, and if possible, more related candidates are entirely sequenced (Simmonds et al., 2005).
The Los Alamos HCV database contained 53 complete sequences of genotype 2. Among them, 38 had their geographical origins recognized. This included nine subtype 2a isolates from Asia, 23 subtype 2b isolates from both Asia and North America, one each of subtypes 2c, 2k and 1b/2k from Europe, and one each of subtypes 2i, 2i/6p and 2b/1b from Asia. Albeit 18 subtypes have been designated, the 53 full-length sequences only represent five of the genotype 2 subtypes (excluding recombinants), most of them belonging to subtypes 2a and 2b (Fig. 2). Our study now allows for 10 assigned subtypes (2a–e, 2i–k, 2m and 2r) and six unclassified and independently grouping variants to have their full-length genome sequences defined. Subtypes 2f, 2g, 2h, 2l, 2n, 2o, 2p and 2q remain to have their complete genome sequences determined – a condition essential for an accurate HCV nomenclature and for improving clinical strategies.
The recent consensus paper has provided updated criteria for classifying HCV isolates (Simmonds et al., 2005). Currently known HCV genotypes and subtypes are grouped into phylogenetic clusters that differ from each other by 31 % and 15 % of nucleotides, respectively. In this study, nucleotide differences of 31.4–35.2 % were observed for genotype 2 compared with other genotypes and 16.9–23.6 % among subtypes and unclassified variants. At the genotype level, the 11 isolates were closer to genotype 1 (31.4 % difference) but more different from genotype 3 (35.2 % difference). This is consistent with the fact that both genotypes 1 and 2 have a long-term local persistence in West Africa (Pybus et al., 2007; Markov et al., 2009; Ruggieri et al., 1996; Jeannel et al., 1998; Wansbrough-Jones et al., 1998; Candotti et al., 2003), while genotype 3 is endemic in South Asia that is far from Africa (Mellor et al., 1995). However, exceptions are observed: subtype 2e variants have been only identified in Indonesia (or immigrants from Indonesia) which is in Asia (Tokita et al., 1996; Utama et al., 2010), while subtype 3h isolates have been found exclusively in subjects originating from Somalia which is in East Africa (Abid et al., 2000). At the subtype level, the highest similarity was observed between QC182 and QC331, which differed by 16.9 % of nucleotides. Since the isolates clustering with QC182 and QC331 were all from West Africa, they may represent an ‘ancient’ lineage. In contrast, the largest subtypic difference was observed between QC64/2e and 2b_JPUT971017, which differed by 23.6 %, most probably due to their historical separation and geographical isolation.
Molecular epidemiology studies have revealed much greater diversity of HCV in certain regions of sub-Saharan Africa and in South and South-east Asia (Simmonds et al., 2005). Africa is where the diversity of genotype 2 isolates is thought to be the greatest. Table 3 showed that there are six subtypes found in Africa but more than 80 % genotype 2 sequences have not been classified into any subtypes. Although their origin in West Africa has been validated (Markov et al., 2009), genotype 2 viruses are widely distributed. Not only have they been found in other continents, particular subtypes have also shown unusual patterns of prevalence. The identification of 2e isolates all with an Indonesian origin is such an example. Another example is observed for 2m isolates, which are exclusively detected in Vietnam or in immigrants from Vietnam (unpublished data). Similarly, 2r isolates have been identified only in patients originating from the island of Hispaniola. Our speculation is that all genotype 2 subtypes had an ancestral origin from Africa but many had been later brought to Europe and European dominions by immigrants. This would be supported by the finding of a high number of distinct lineages in Africa. However, the number of lineages identified in Africa has been lower than that on other continents (Table 3). This could be due to two reasons. First, there has been limited sampling in Africa compared with that in the developed countries in Europe and in the other parts of the world. For example, 2i which is found in Europe, Vietnam and Canada has recently been found in Morocco in Africa (based on the updated database on March, 2011) (Qu et al., 1994; Cantaloube et al., 2003; Noppornpanth et al., 2006; Le Pogam et al., 1998; Murphy et al., 2007; Thomas et al., 2007;). Secondly, for many of the isolates in Africa only short segments of sequences are available (Markov et al., 2009). This has limited their analysis and hampered their comparison to other isolates. Greater efforts are needed for more extensive surveys in these African countries. Optimally, migration analyses will supply more accurate estimates.
One of the features observed among all genotype 2 sequences is the unique insertion of 20 aa near the end of the NS5A region. In this study, certain isolates also showed insertions in the ISDR and the PKR-BD. Although controversial, mutations in these latter regions have been associated with sensitivity to interferon treatment (Enomoto & Sato, 1995; Gale & Katze, 1998; Gale et al., 1998; Taylor et al., 1999). Another unique and striking insertion was found in the p7 region of the QC64 isolate, which contained 67 aa directly following the end of the p7 protein. Interestingly, the insertion and the p7 protein shared 78 % amino acid similarity and both were very different from all other isolates in the Los Alamos HCV database. A survey of additional isolates is required to see whether the insertion is common among other 2e viruses. The p7 protein is found to be critical for HCV infectivity and contains functionally important genotype-specific elements (Sakai et al., 2003). Repetition of this element may have the effect to increase viral infectivity and could be tested using the JFH1 cell culture model.
Methods
Samples.
Serum samples were obtained from 11 patients (four male and seven female) living in Quebec, Canada, between November 2000 and June 2006 as previously described (Murphy et al., 2007). Of these patients, four were Caucasians, four were African immigrants, two were Asians, and one was Haitian (Table 1). They were aged between 36 and 75 years old when sampled. HCV infection was confirmed by routine virological assays for clinical diagnosis.
PCR strategy.
Starting with 100 µl of serum, HCV genomic fragments were amplified using methods described previously (Li et al., 2006, 2009). In brief, RNA was extracted using TriPure (Roche), cDNA converted using random hexamers (Promega) and SuperScript III (Invitrogen) reverse transcriptase, and HCV fragments amplified using conventional PCR.
Using the BioEdit software (Hall, 1999), an alignment of full-length genomic sequences available for HCV genotype 2 subtypes 2a, 2b, 2c, 2k and 2i was made and degenerate primers were chosen according to the conserved regions in the alignment. A first set of fragments were amplified and sequenced using these degenerate primers. Specific primers were then designed based on the sequences of the fragments. These primers in combination with additional primers, degenerate or specific, were used to amplify the missing regions. Such a procedure continued until fragments covering the full HCV genome length were obtained. This strategy has been referred as ‘DNA walking through bridges on islands’ and has been successfully used to characterize many genotype 4 and 6 variants (Lu et al., 2007; Li et al., 2006, 2009). The degenerate primers used in this study are listed in Table S1 while the specific primers are not shown.
5′ and 3′ ends.
Because the extreme 5′ end of the HCV genome is highly conserved, the 25 nt at the beginning of the 5′ UTR of strain JFH1 were used as a primer in conventional PCR for amplifying the 5′ ends of all the 11 genotype 2 isolates. However, amplification of the 3′ ends employed a nested RACE PCR. The upstream primers were specific to the 3′ ends of the respective NS5B regions while the downstream primer (9587R, TTCACAGCTAGCCGTGACTAGG) was based on the highly conserved 98 nt 3′ X tail, corresponding to nt 9587–9565 in the H77 genome. For isolates failing in this amplification, poly A and nested universal primer (NUP) adaptor primers were used instead (Li et al., 2009). PCR was conducted for 35 cycles each consisting of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s except for the first cycle for which the denaturation step was conducted at 94 °C for 3 min. This was followed by a final cycle of extension at 72 °C for 7 min. FastStart Taq DNA Polymerase (Roche) was used as the reagent and standard procedures were taken to prevent false RT-PCR positives (Kwok & Higuchi, 1989).
Sequencing.
Amplicons were purified with the QIAquick PCR purification kit (Qiagen) according to the manufacturer’s protocol and directly sequenced in both directions to confirm the genetic heterogeneity observed at numerous positions throughout the genome. Amplicons obtained with degenerate primers were cloned with pGEM-T Easy Vector (Promega) and at least four clones were sequenced. The sequencing jobs were done using the ABI Prism BigDye 3.0 terminators and an appropriate primer on an ABI Prism 3137 genetic analyser (Applied Biosystems). All observed ambiguous nucleotides were recorded using standard IUPAC codes. ABI raw files were transferred onto a work station where chromatograms were visually inspected, manually edited and automatically assembled using the Seqman program of the Lasergene 8.1 software (DNAStar, Inc.). After the chromatograms of all amplicons successfully overlapped the whole HCV genome, the sequence job was cleared and the full-length consensus sequence was compiled.
Sequence analysis.
The 11 full-length HCV genomes were denoted according to the numbering of the H77 isolate from the extreme 5′ end to the 3′ X tail. Their amino acid sequences were deduced with the EditSeq program of the Lasergene 8.1 software. To analyse these sequences, many reference sequences were retrieved. They were contained in three datasets. The complete sequence dataset included 19 full-length genome sequences, not only from genotype 2, but also from genotypes 1, 3, 4, 5, 6 and 7. The complete sequence dataset for genotype 2 included 58 full-length sequences all from genotype 2. The partial NS5B sequence dataset included 68 partial NS5B sequences, spanning 340 nt and corresponding to nt 8276–8615 in the H77 genome. The three datasets of sequences were aligned using BioEdit software and checked with mega5 editor (Tamura et al., 2011) to display the genomic organization and genetic differences, with a particular interest in the amino acids of the interferon-sensitivity determining region (ISDR) (Enomoto & Sato, 1995), the RNA (dsRNA)-activated protein kinase (PKR) region, and additional domains in the 5′ UTR, E2 and NS5A regions (Gale & Katze, 1998; Gale et al., 1998; Taylor et al., 1999).
Recently beast software, in which a large number of evolutionary models and tree-based models can be implemented, has been widely used for Bayesian analysis of molecular sequences related by a phylogenetic tree. These models are suitable for analysing both within- and between-species sequence data. Possibly the most distinguishing feature is explicitly modelling the rate of molecular evolution on each branch in the phylogenetic tree rooted by an incorporated timescale. However, an unavoidable burden of this analysis is that, if no obvious prior distribution of a parameter exists, the prior distribution selected will intentionally influence the posterior distribution of the parameters of interest (Drummond & Rambaut, 2007). While the majority of genotype 2 sequences analysed here do not have their sampling dates recorded, in beast analysis we would have to include an estimateof a prior distribution, which would create uncertainties and bias. Therefore, phylogenetic trees in this study are better estimated by the traditional method.
Evolutionary analyses of viral ancestral relationships were conducted with mega5 software (Tamura et al., 2007). The evolutionary history was inferred by using the maximum-likelihood (ML) method based on the Hasegawa–Kishino–Yano nucleotide-substitution model (Hasegawa et al., 1985) under six categories of the gamma-distribution among-site rate variation (HKY+I+Г6) (Posada & Crandall, 1998). The transition/transversion ratio, proportion of invariable sites and gamma distribution shape parameter were estimated from the real sequence datasets. Base frequencies were adjusted to maximize the likelihood. Bootstrap resampling tests were performed 500 times.
To exclude recent virus recombination events (Kalinina et al., 2002, 2004; Colina et al., 2004; Legrand-Abravanel et al., 2007; Noppornpanth et al., 2006; Lee et al., 2010), the rdp3 software (Martin et al., 2005) was run with default settings modified as follows: (i) window size was 40 nt; (ii) the option of linear sequences was chosen; (iii) six different methods (rdp, geneconv, MaxChi, Bootscan, Chimaera and SiScan) were run simultaneously against the sequences listed; and (iv) only the events detected by more than two of the above methods were considered positive. This analysis was only performed for the complete genome sequence dataset.
Finally, to study the geographical distribution of genotype 2 viruses, a total of 4044 genotype 2 sequences were analysed. These sequences were retrieved from the Los Alamos HCV database (accessed at the end of 2010) and filtered with respect to their isolation time, sampling countries, genomic region and clone status, which distinguished 2021 individual isolates. To clarify their diversity with regard to geographical distributions, we tabled the information according to their subtypes, sampling countries and sampling continents.
Acknowledgements
This study was supported by a grant from the National Institute of Allergy and Infectious Diseases (5 R01 AI080734-03A). The funding agencies had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
A supplementary figure and table are available with the online version of this paper.
References
- Abid K., Quadri R., Veuthey A. L., Hadengue A., Negro F. (2000). A novel hepatitis C virus (HCV) subtype from Somalia and its classification into HCV clade 3. J Gen Virol 81, 1485–1493 [DOI] [PubMed] [Google Scholar]
- Abid K., Pazienza V., de Gottardi A., Rubbia-Brandt L., Conne B., Pugnale P., Rossi C., Mangia A., Negro F. (2005). An in vitro model of hepatitis C virus genotype 3a-associated triglycerides accumulation. J Hepatol 42, 744–751 10.1016/j.jhep.2004.12.034 [DOI] [PubMed] [Google Scholar]
- Candotti D., Temple J., Sarkodie F., Allain J. P. (2003). Frequent recovery and broad genotype 2 diversity characterize hepatitis C virus infection in Ghana, West Africa. J Virol 77, 7914–7923 10.1128/JVI.77.14.7914-7923.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantaloube J. F., Biagini P., Attoui H., Gallian P., de Micco P., de Lamballerie X. (2003). Evolution of hepatitis C virus in blood donors and their respective recipients. J Gen Virol 84, 441–446 10.1099/vir.0.18642-0 [DOI] [PubMed] [Google Scholar]
- Cantaloube J. F., Gallian P., Laperche S., Elghouzzi M. H., Piquet Y., Bouchardeau F., Jordier F., Biagini P., Attoui H., de Micco P. (2008). Molecular characterization of genotype 2 and 4 hepatitis C virus isolates in French blood donors. J Med Virol 80, 1732–1739 10.1002/jmv.21285 [DOI] [PubMed] [Google Scholar]
- Colina R., Casane D., Vasquez S., García-Aguirre L., Chunga A., Romero H., Khan B., Cristina J. (2004). Evidence of intratypic recombination in natural populations of hepatitis C virus. J Gen Virol 85, 31–37 10.1099/vir.0.19472-0 [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007). beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto N., Sato C. (1995). Clinical relevance of hepatitis C virus quasispecies. J Viral Hepat 2, 267–272 10.1111/j.1365-2893.1995.tb00040.x [DOI] [PubMed] [Google Scholar]
- Feld J. J., Hoofnagle J. H. (2005). Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436, 967–972 10.1038/nature04082 [DOI] [PubMed] [Google Scholar]
- Gale M., Jr, Katze M. G. (1998). Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol Ther 78, 29–46 10.1016/S0163-7258(97)00165-4 [DOI] [PubMed] [Google Scholar]
- Gale M. J., Jr, Korth M. J., Katze M. G. (1998). Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin Diagn Virol 10, 157–162 10.1016/S0928-0197(98)00034-8 [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41, 95–98 [Google Scholar]
- Hasegawa M., Kishino H., Yano T. (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22, 160–174 10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- Holland P. V., Barrera J. M., Ercilla M. G., Yoshida C. F., Wang Y., de Olim G. A., Betlach B., Kuramoto K., Okamoto H. (1996). Genotyping hepatitis C virus isolates from Spain, Brazil, China, and Macau by a simplified PCR method. J Clin Microbiol 34, 2372–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourioux C., Patient R., Morin A., Blanchard E., Moreau A., Trassard S., Giraudeau B., Roingeard P. (2007). The genotype 3-specific hepatitis C virus core protein residue phenylalanine 164 increases steatosis in an in vitro cellular model. Gut 56, 1302–1308 10.1136/gut.2006.108647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackel-Cram C., Babiuk L. A., Liu Q. (2007). Up-regulation of fatty acid synthase promoter by hepatitis C virus core protein: genotype-3a core has a stronger effect than genotype-1b core. J Hepatol 46, 999–1008 10.1016/j.jhep.2006.10.019 [DOI] [PubMed] [Google Scholar]
- Jeannel D., Fretz C., Traore Y., Kohdjo N., Bigot A., Pê Gamy E., Jourdan G., Kourouma K., Maertens G. & other authors (1998). Evidence for high genetic diversity and long-term endemicity of hepatitis C virus genotypes 1 and 2 in West Africa. J Med Virol 55, 92–97 [DOI] [PubMed] [Google Scholar]
- Kalinina O., Norder H., Mukomolov S., Magnius L. O. (2002). A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J Virol 76, 4034–4043 10.1128/JVI.76.8.4034-4043.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina O., Norder H., Magnius L. O. (2004). Full-length open reading frame of a recombinant hepatitis C virus strain from St Petersburg: proposed mechanism for its formation. J Gen Virol 85, 1853–1857 10.1099/vir.0.79984-0 [DOI] [PubMed] [Google Scholar]
- Kwok S., Higuchi R. (1989). Avoiding false positives with PCR. Nature 339, 237–238 10.1038/339237a0 [DOI] [PubMed] [Google Scholar]
- Le Pogam S., Dubois F., Christen R., Raby C., Cavicchini A., Goudeau A. (1998). Comparison of DNA enzyme immunoassay and line probe assays (Inno-LiPA HCV I and II) for hepatitis C virus genotyping. J Clin Microbiol 36, 1461–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. M., Lin H. J., Chen Y. J., Lee C. M., Wang S. F., Chang K. Y., Chen T. L., Liu H. F., Chen Y. M. (2010). Molecular epidemiology of HCV genotypes among injection drug users in Taiwan: full-length sequences of two new subtype 6w strains and a recombinant form_2b6w. J Med Virol 82, 57–68 10.1002/jmv.21658 [DOI] [PubMed] [Google Scholar]
- Legrand-Abravanel F., Claudinon J., Nicot F., Dubois M., Chapuy-Regaud S., Sandres-Saune K., Pasquier C., Izopet J. (2007). New natural intergenotypic (2/5) recombinant of hepatitis C virus. J Virol 81, 4357–4362 10.1128/JVI.02639-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Fu Y., Lu L., Ji W., Yu J., Hagedorn C. H., Zhang L. (2006). Complete genomic sequences for hepatitis C virus subtypes 6e and 6g isolated from Chinese patients with injection drug use and HIV-1 co-infection. J Med Virol 78, 1061–1069 10.1002/jmv.20663 [DOI] [PubMed] [Google Scholar]
- Li C., Lu L., Wu X., Wang C., Bennett P., Lu T., Murphy D. (2009). Complete genomic sequences for hepatitis C virus subtypes 4b, 4c, 4d, 4g, 4k, 4l, 4m, 4n, 4o, 4p, 4q, 4r and 4t. J Gen Virol 90, 1820–1826 10.1099/vir.0.010330-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Li C., Fu Y., Thaikruea L., Thongswat S., Maneekarn N., Apichartpiyakul C., Hotta H., Okamoto H. & other authors (2007). Complete genomes for hepatitis C virus subtypes 6f, 6i, 6j and 6m: viral genetic diversity among Thai blood donors and infected spouses. J Gen Virol 88, 1505–1518 10.1099/vir.0.82604-0 [DOI] [PubMed] [Google Scholar]
- Manns M. P., McHutchison J. G., Gordon S. C., Rustgi V. K., Shiffman M., Reindollar R., Goodman Z. D., Koury K., Ling M., Albrecht J. K. (2001). Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358, 958–965 10.1016/S0140-6736(01)06102-5 [DOI] [PubMed] [Google Scholar]
- Manns M. P., Wedemeyer H., Cornberg M. (2006). Treating viral hepatitis C: efficacy, side effects, and complications. Gut 55, 1350–1359 10.1136/gut.2005.076646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov P. V., Pepin J., Frost E., Deslandes S., Labbé A. C., Pybus O. G. (2009). Phylogeography and molecular epidemiology of hepatitis C virus genotype 2 in Africa. J Gen Virol 90, 2086–2096 10.1099/vir.0.011569-0 [DOI] [PubMed] [Google Scholar]
- Martial J., Morice Y., Abel S., Cabié A., Rat C., Lombard F., Edouard A., Pierre-Louis S., Garsaud P. & other authors (2004). Hepatitis C virus (HCV) genotypes in the Caribbean island of Martinique: evidence for a large radiation of HCV-2 and for a recent introduction from Europe of HCV-4. J Clin Microbiol 42, 784–791 10.1128/JCM.42.2.784-791.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. P., Williamson C., Posada D. (2005). rdp2: recombination detection and analysis from sequence alignments. Bioinformatics 21, 260–262 10.1093/bioinformatics/bth490 [DOI] [PubMed] [Google Scholar]
- Mellor J., Holmes E. C., Jarvis L. M., Yap P. L., Simmonds P., The International HCV Collaborative Study Group (1995). Investigation of the pattern of hepatitis C virus sequence diversity in different geographical regions: implications for virus classification. J Gen Virol 76, 2493–2507 10.1099/0022-1317-76-10-2493 [DOI] [PubMed] [Google Scholar]
- Murphy D. G., Willems B., Deschênes M., Hilzenrat N., Mousseau R., Sabbah S. (2007). Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5′ untranslated region sequences. J Clin Microbiol 45, 1102–1112 10.1128/JCM.02366-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao H., Okamoto H., Tokita H., Inoue T., Iizuka H., Pozzato G., Mishiro S. (1996). Full-length genomic sequence of a hepatitis C virus genotype 2c isolate (BEBE1) and the 2c-specific PCR primers. Arch Virol 141, 701–704 10.1007/BF01718327 [DOI] [PubMed] [Google Scholar]
- Ndjomou J., Pybus O. G., Matz B. (2003). Phylogenetic analysis of hepatitis C virus isolates indicates a unique pattern of endemic infection in Cameroon. J Gen Virol 84, 2333–2341 10.1099/vir.0.19240-0 [DOI] [PubMed] [Google Scholar]
- Negro F. (2006). Mechanisms and significance of liver steatosis in hepatitis C virus infection. World J Gastroenterol 12, 6756–6765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njouom R., Pasquier C., Ayouba A., Sandres-Sauné K., Mfoupouendoun J., Mony Lobe M., Tene G., Thonnon J., Izopet J., Nerrienet E. (2003a). Hepatitis C virus infection among pregnant women in Yaounde, Cameroon: prevalence, viremia, and genotypes. J Med Virol 69, 384–390 10.1002/jmv.10300 [DOI] [PubMed] [Google Scholar]
- Njouom R., Pasquier C., Ayouba A., Gessain A., Froment A., Mfoupouendoun J., Pouillot R., Dubois M., Sandres-Sauné K. & other authors (2003b). High rate of hepatitis C virus infection and predominance of genotype 4 among elderly inhabitants of a remote village of the rain forest of South Cameroon. J Med Virol 71, 219–225 10.1002/jmv.10473 [DOI] [PubMed] [Google Scholar]
- Njouom R., Pasquier C., Ayouba A., Tejiokem M. C., Vessiere A., Mfoupouendoun J., Tene G., Eteki N., Lobe M. M. & other authors (2005). Low risk of mother-to-child transmission of hepatitis C virus in Yaounde, Cameroon: the ANRS 1262 study. Am J Trop Med Hyg 73, 460–466 [PubMed] [Google Scholar]
- Noppornpanth S., Lien T. X., Poovorawan Y., Smits S. L., Osterhaus A. D., Haagmans B. L. (2006). Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J Virol 80, 7569–7577 10.1128/JVI.00312-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier C., Njouom R., Ayouba A., Dubois M., Sartre M. T., Vessière A., Timba I., Thonnon J., Izopet J., Nerrienet E. (2005). Distribution and heterogeneity of hepatitis C genotypes in hepatitis patients in Cameroon. J Med Virol 77, 390–398 10.1002/jmv.20468 [DOI] [PubMed] [Google Scholar]
- Pearlman B. L. (2004). Hepatitis C infection: a clinical review. South Med J 97, 364–373, quiz 374 10.1097/01.SMJ.0000118903.35704.30 [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K. A. (1998). modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Pouillot R., Lachenal G., Pybus O. G., Rousset D., Njouom R. (2008). Variable epidemic histories of hepatitis C virus genotype 2 infection in West Africa and Cameroon. Infect Genet Evol 8, 676–681 10.1016/j.meegid.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Pybus O. G., Markov P. V., Wu A., Tatem A. J. (2007). Investigating the endemic transmission of the hepatitis C virus. Int J Parasitol 37, 839–849 10.1016/j.ijpara.2007.04.009 [DOI] [PubMed] [Google Scholar]
- Qu D., Hantz O., Gouy M., Vitvitski L., Li J. S., Berby F., Tong S. P., Trépo C. (1994). Heterogeneity of hepatitis C virus genotypes in France. J Gen Virol 75, 1063–1070 10.1099/0022-1317-75-5-1063 [DOI] [PubMed] [Google Scholar]
- Rubbia-Brandt L., Quadri R., Abid K., Giostra E., Malé P. J., Mentha G., Spahr L., Zarski J. P., Borisch B. & other authors (2000). Hepatocyte steatosis is a cytopathic effect of hepatitis C virus genotype 3. J Hepatol 33, 106–115 10.1016/S0168-8278(00)80166-X [DOI] [PubMed] [Google Scholar]
- Ruggieri A., Argentini C., Kouruma F., Chionne P., D’Ugo E., Spada E., Dettori S., Sabbatani S., Rapicetta M. (1996). Heterogeneity of hepatitis C virus genotype 2 variants in West Central Africa (Guinea Conakry). J Gen Virol 77, 2073–2076 10.1099/0022-1317-77-9-2073 [DOI] [PubMed] [Google Scholar]
- Sakai A., Kaneko S., Honda M., Matsushita E., Kobayashi K. (1999). Quasispecies of hepatitis C virus in serum and in three different parts of the liver of patients with chronic hepatitis. Hepatology 30, 556–561 10.1002/hep.510300234 [DOI] [PubMed] [Google Scholar]
- Sakai A., Claire M. S., Faulk K., Govindarajan S., Emerson S. U., Purcell R. H., Bukh J. (2003). The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc Natl Acad Sci U S A 100, 11646–11651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samokhvalov E. I., Hijikata M., Gylka R. I., Lvov D. K., Mishiro S. (2000). Full-genome nucleotide sequence of a hepatitis C virus variant (isolate name VAT96) representing a new subtype within the genotype 2 (arbitrarily 2k). Virus Genes 20, 183–187 10.1023/A:1008182901274 [DOI] [PubMed] [Google Scholar]
- Simmonds P., Bukh J., Combet C., Deléage G., Enomoto N., Feinstone S., Halfon P., Inchauspé G., Kuiken C. & other authors (2005). Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42, 962–973 10.1002/hep.20819 [DOI] [PubMed] [Google Scholar]
- Stuyver L., van Arnhem W., Wyseur A., Hernandez F., Delaporte E., Maertens G. (1994). Classification of hepatitis C viruses based on phylogenetic analysis of the envelope 1 and nonstructural 5B regions and identification of five additional subtypes. Proc Natl Acad Sci U S A 91, 10134–10138 10.1073/pnas.91.21.10134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). mega4: Molecular Evolutionary Genetics Analysis (mega) software version 4.0. Mol Biol Evol 24, 1596–1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher J., Nei M., Kumar S. (2011). MEGAS: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. R., Shi S. T., Romano P. R., Barber G. N., Lai M. M. (1999). Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285, 107–110 10.1126/science.285.5424.107 [DOI] [PubMed] [Google Scholar]
- Thomas F., Nicot F., Sandres-Sauné K., Dubois M., Legrand-Abravanel F., Alric L., Peron J. M., Pasquier C., Izopet J. (2007). Genetic diversity of HCV genotype 2 strains in south western France. J Med Virol 79, 26–34 10.1002/jmv.20765 [DOI] [PubMed] [Google Scholar]
- Tokita H., Okamoto H., Iizuka H., Kishimoto J., Tsuda F., Lesmana L. A., Miyakawa Y., Mayumi M. (1996). Hepatitis C virus variants from Jakarta, Indonesia classifiable into novel genotypes in the second (2e and 2f), tenth (10a) and eleventh (11a) genetic groups. J Gen Virol 77, 293–301 10.1099/0022-1317-77-2-293 [DOI] [PubMed] [Google Scholar]
- Utama A., Tania N. P., Dhenni R., Gani R. A., Hasan I., Sanityoso A., Lelosutan S. A., Martamala R., Lesmana L. A. & other authors (2010). Genotype diversity of hepatitis C virus (HCV) in HCV-associated liver disease patients in Indonesia. Liver Int 30, 1152–1160 10.1111/j.1478-3231.2010.02280.x [DOI] [PubMed] [Google Scholar]
- Wansbrough-Jones M. H., Frimpong E., Cant B., Harris K., Evans M. R., Teo C. G. (1998). Prevalence and genotype of hepatitis C virus infection in pregnant women and blood donors in Ghana. Trans R Soc Trop Med Hyg 92, 496–499 10.1016/S0035-9203(98)90887-2 [DOI] [PubMed] [Google Scholar]