Abstract
Human plasma chitotriosidase activity is a commonly used diagnostic and therapeutic biomarker for non-neuronopathic Gaucher disease. Chitotriosidase deficiency is common in non-African populations and is primarily caused by a 24 bp duplication in the encoding gene (CHIT1). Allele frequencies for the 24 bp duplication range from 20–50 % outside Africa. The present study found chitotriosidase deficiency to be rare in the South African Black population (1.6 %) and the otherwise common 24 bp duplication is absent in this African population. Instead, chitotriosidase deficiency is caused by a 4 bp deletion across the exon/intron 10 boundary (E/I-10_delGAgt) of the CHIT1 gene. The exact position of this mutation was found to differ from the previously reported location. Allele frequencies for six coding variants of CHIT1 (p.G102S, p.G354R, 24 bp duplication, E/I-10_delGAgt, p.A442V/G) were determined and the 4 bp deletion was found to be in complete linkage disequilibrium (LD) with two of the coding variants (p.G354R and p.A442V). The in silico assessments of the two missense mutations in LD predict a protein-damaging nature and functional studies are needed to clarify if one or both abolish the enzyme’s activity. Overall, the low frequency of chitotriosidase deficiency in South African Blacks makes chitotriosidase activity an excellent biomarker of choice in this population.
Electronic supplementary material:
The online version of this chapter (doi:10.1007/8904_2012_193) contains supplementary material, which is available to authorized users.
Introduction
Chitotriosidase (CHIT1) (EC 3.2.1.14) is a chitinase that hydrolyses chitin, a structural component found in fungi, nematodes, insects and shellfish. This enzyme is primarily secreted from human macrophages and is thought to play a role in the body’s defence against chitin-containing pathogens. Elevated plasma chitotriosidase levels are observed in several lysosomal storage disorders including Gaucher disease, as well as in β-thalassaemia and atherosclerosis (Barone et al. 2003a; Boot et al. 1999; Guo et al. 1995). This finding has led to the utilisation of chitotriosidase activity as a biomarker for conditions involving chitinase secretion from human macrophages (Renkema et al. 1998, 1995).
One limitation in the use of chitotriosidase activity as a biomarker is the presence of genetic variants in the chitotriosidase encoding gene CHIT1 which result in lack or reduced activity of this enzyme in some individuals. The most striking observation about this recessively inherited enzyme deficiency is its distinct difference in prevalence rates in various populations, as well as the dissimilar distribution of causative mutations. To date, three mutations that lead to deficient chitotriosidase expression (Boot et al. 1998; Grace et al. 2007) (Table 2) and three mutations that result in strongly reduced chitotriosidase expression in humans (Lee et al. 2007; Webb et al. 2006) (Table 2) are recorded in the Human Genome Mutation Database (www.hgmd.cf.ac.uk) (Stenson et al. 2003). The most common mutation reported in the literature is a 24bp duplication in exon 10 of the CHIT1 gene (OMIM 600031.0001) that results in aberrant splicing of the mRNA and subsequently leads to the diminished expression of an enzymatically inactive protein (Boot et al. 1998). Notably, chitotriosidase (chito) deficiency resulting from this 24bp duplication has been shown to be highly prevalent in Europeans and Asians, with allele frequencies reported to be ~ > 0.20 and > 0.50, respectively (Lee et al. 2007). A very different picture is observed in populations on the African continent, where chito deficiency is rare (0–2%) and the 24bp duplication is absent (Malaguarnera et al. 2003). Grace et al. (2007) screened 396 individuals from various ethnic groups with an unknown chito activity status for the presence of the 24bp duplication in the CHIT1 gene and found that 3/70 (4.3%) Black Americans and 3/64 (4.7%) Caribbean Blacks carry the 24bp duplication allele compared to carrier rates of 7/88 (8%) and 33/244 (13.5%) in the Dominican and Puerto Rican population, respectively.
Table 2.
Allele frequencies for six CHIT1 mutations in unrelated Black South Africans
| p.G102S (G>A) Exon 4 (rs2297950) |
p.G354R (G>A) Exon 10 (rs9943208) |
24bp duplication in Exon 10 | E/ I-10_delGAgt 4bp deletion |
p.A442V (C>T) Exon 12 (rs1065761) |
p.A442G (C>G) Exon 12 (rs1065761) |
|
|---|---|---|---|---|---|---|
| Mutant allele frequency | 0.257 (36/140) | 0.250 (5/20) | 0 (0/984) | 0.038 (5/130) | 0.049 (6/122) | 0.148 (18/122) |
| homozygote | 0.057 (4/70) | 0.200 (2/10) | 0 | 0.030 (2/65) | 0.032 (2/61) | 0.049 (3/61) |
| heterozygote | 0.286 (20/70) | 0.100 (1/10) | 0 | 0.015 (1/65) | 0.032 (2/61) | 0.197 (12/61) |
| Chitotriosidase activity of mutant alleles (assessed and reported by) | Reduced activity Lee et al. (2007) |
No stable protein Grace et al. (2007) |
Deficient Boot et al. (1998) |
Deficient Grace et al. (2007) |
Reduced activity Lee et al. (2007) |
Normal activity Lee et al. (2007) |
The study presented here was aimed at elucidating the overall prevalence of chito deficiency in the South African Black population and to investigate the underlying molecular mechanism. Those 10 individuals that showed either complete chito deficiency or low chito activity (≤10 nmol/h/ml) were earmarked for the analysis of six mutations: p.G102S (rs2297950) in exon 4, p.G354R (rs9943208) in exon 10, the 24bp duplication in exon 10, a 4bp deletion across the exon/intron-10 boundary (complex E/I-10) and a tri-allelic SNP p.A442V/G (rs1065761) in exon 12. In addition, estimates of the frequencies for these six CHIT1 variants were also ascertained in the general South African Black population.
Materials and Methods
Subjects
Peripheral blood samples from 492 unrelated Black South African Bantu of unknown disease status who presented for paternity testing at the National Health Laboratory Service (NHLS) were used in the present project. The study was conducted in two parts with two groups of subjects. In part one, we assessed chito activity in 280 participants (group 1) and tested for the presence of the 24bp duplication in all chito-deficient individuals (0 nmol/h/ml) that were found in this group.
In part two, we assessed chito activity in 212 participants (group 2). Genotype information was generated for all subjects with reduced or absent chito activity. A subset (70/212) of participants with normal chito activity was further investigated in order to obtain background CHIT1 allele frequency data for the Black South African Bantu population. Three out of six investigated loci harbour an allele that impacts on chito activity (Tables 1 and 2). Ethics clearance to undertake the study was obtained from the University of the Witwatersrand’s Human Research Ethics Committee (Medical) (protocol number M050706).
Table 1.
Chitotriosidase activity and CHIT1 genotype information for Chito-deficient and Chito-impaired individuals
| Chitotriosidase impaired individuals | Chito-activity [nmol/h/ml] |
p.G102S (G>A) Exon 4 (rs2297950) |
p.G354R (G>A) Exon 10 (rs9943208) |
c.1049_1072dup24 Exon 10 (24bp duplication) |
E/ I-10_delGAgt* (4bp deletion) |
p.A442V/G (C>T/G) Exon 12 (rs1065761) |
|---|---|---|---|---|---|---|
| Group 1 | ||||||
| 01 | 0 | ND | ND | WT / WT | ND | ND |
| 02 | 0 | ND | ND | WT / WT | ND | ND |
| 03 | 0 | ND | ND | WT / WT | ND | ND |
| 04 | 0 | ND | ND | WT / WT | ND | ND |
| 05 | 0 | ND | ND | WT / WT | ND | ND |
| 06 | 0 | ND | ND | WT / WT | ND | ND |
| Group 2 | ||||||
| 07 | 0 | G/G | A/A | WT / WT | Del / Del | T/T |
| 08 | 0 | G/G | A/A | WT / WT | Del / Del | T/T |
| 09 | 8.5 | G/G | G/A | WT / WT | WT / Del | C/T |
| 10 | 9.4 | G/G | G/G | WT / WT | WT / WT | C/C |
| 11 | 0.5 | G/G | G/G | WT / WT | WT / WT | C/G |
| 12 | 6.0 | G/G | G/G | WT / WT | WT / WT | C/C |
| 13 | 10.0 | G/G | G/G | WT / WT | WT / WT | C/C |
| 14 | 8.4 | G/G | G/G | WT / WT | WT / WT | C/C |
| 15 | 9.4 | G/G | G/G | WT / WT | WT / WT | C/C |
| 16 | 8.8 | G/G | G/G | WT / WT | WT / WT | C/C |
ND - not determined; WT- wild type
*c.1155delGA; 1156+1_1156+2delGT (see Fig. 1)
Methods
Plasma Chitotriosidase Assay
Plasma chitotriosidase activity was measured in 492 participants (group 1 and 2), using an adapted biochemical assay (Hollak et al. 1994).
CHIT1 Mutation Detection
Genomic DNA was extracted from peripheral blood samples, using the FlexiGene® DNA kit (Qiagen, USA) in accordance with the manufacturer’s protocol. DNA concentrations were spectrophotometrically assessed. Each sample was standardised to a working concentration of 100 ng/μL for further use.
The frequency of the 24bp duplication was assessed by subjecting genomic DNA to a PCR-based method described elsewhere (Boot et al. 1998). For group 2, ten sets of primers were designed to amplify the CHIT1 coding region, covering all 12 exons and the intron/exon boundaries. Genomic DNA in the 2 chito-deficient individuals was amplified and each amplicon was sequenced in both directions using BigDyeTM Terminator Ready Reaction Mix and the ABI PrismTM 3130XL genetic analyser (Applied Biosystems, USA). The SeqMan module from the Lasergene® software suite Lasergene® (DNASTAR Inc. Software, USA) was utilised for the evaluation of sequence variants compared to the published CHIT1 reference sequence (NM_003465).
Population Screening for CHIT1 Mutations
In order to obtain allele frequency data for the Black South African population, a subset of 70 (out of 212) group 2 participants was subjected to CHIT1 locus investigations. Five of the following six variants have been associated with reduced or deficient chito activity in the literature: p.G102S (rs2297950) in exon 4, p.G354R (rs9943208) in exon 10, the 24bp duplication in exon 10, a 4bp deletion across the exon/intron-10 boundary (complex E/I-10) and the tri-allelic SNP p.A442V (rs1065761) in exon 12 of the CHIT1 gene. The additional SNP allele p.A442G (rs1065761) has no adverse effect on chito activity (Table 2).
For p.G102S, a PCR assay followed by RFLP analysis was applied (Supplementary Table 1). The 259bp long PCR amplicon was subjected to restriction enzyme digestion with HpaII (10 units/10 μL PCR reaction) and the fragments were separated on a 4% Seakem agarose gel (FMC Bioproducts, USA) at 4°C. This restriction enzyme digest resulted in the presence of two fragments (240bp and 19bp) for the wild-type allele, whereas the p.G102S transversion abolishes the HpaII restriction site and the 259bp fragment remains uncleaved. In the heterozygote state, three fragments were visible on the gel.
PCR amplification, followed by direct DNA sequencing was used to examine exon 10 and 12 of the CHIT1 gene for the presence of p.G354R, the 24bp duplication, the 4bp deletion and p.A442V/G. PCR products were purified using DyeEx KitTM 2.0 kit (Qiagen, Hilden, Germany) and then sequenced in both directions using BigDyeTM Terminator Ready Reaction Mix and the ABI PrismTM 3130XL genetic analyser (Applied Biosystems, USA).
Evaluation of Missense Mutations
Variants detected in the coding region of CHIT1 were assessed using two web-based tools, PolyPhen (http://genetics.bwh.harvard.edu/pph) and SNPs3D (http://www.snps3d.org/). Briefly, PolyPhen computes the impact of an amino acid substitution on a human protein by analysing its structural and functional characteristics. The program generates a position-specific independent counts (PSIC) score for the two amino acid variants. If the difference between the calculated scores for these two amino acid variants (ΔPSIC) is ≤ 0.5, the impact of the amino acid substitution on the protein’s function is likely to be benign (Ramensky et al. 2002).
SNPs3D uses a machine learning technique for the in silico analysis of coding non-synonymous variants. The SNPs3D algorithm combines two support vector machine (SVM) methods and computes the impact of an amino acid change on the structure and stability of the protein. In addition, it incorporates evolutionary sequence information by comparing the mutated amino acid position to orthologue sequences. The output is an SVM score. A positive SVM score classifies the amino acid change as non-deleterious, while a negative score indicates a deleterious effect on the protein’s function. It has been noted that accuracy is significantly higher for SVM scores > 0.5 and < − 0.5 (Yue and Moult 2006).
Results
Plasma chito activities and CHIT1 genotyping
A mean chito activity of 68.77 nmol/h/ml (SD+/− 46.19) was observed in Black South African individuals (N = 210) with detectable enzyme activity (range 0.5–346.16 nmol/h/ml). In total, 8 of the 492 (1.6%) unrelated Black individuals that were tested biochemically showed no detectable chito activity (0 nmol/h/ml) and 8 out of 212 individuals (3.8%) showed low enzyme activity (<= 10 nmol/h/ml), indicative of bearing a mutant CHIT1 allele. The absence of the 24bp-duplication was confirmed in all individuals (N = 0/16) (Table 1).
Genotype analysis of ten chito-compromised probands from group 2 (N=212) revealed a 4bp deletion across the exon / intron 10 boundary (E/I-10_delGAgt) in homozygous state in Probands 7 and 8 and in heterozygous state in Proband 9 (Table 1). The exact position of this 4bp splice site deletion (E/I-10_delGAgt) and its impact on the mRNA splicing are shown in Fig. 1. All three individuals with the E/I-10_delGAgt allele also carried the p.G354R and the p.A442V variant. Based on our data, all three mutations appear to be in complete linkage disequilibrium (LD) with each other (Table 1). Since the three potentially damaging variants were present on each of the homozygote chito-deficient loci, we used online bioinformatic tools in conjunction with published information to assess the impact of mutations p.G354R and p.A442V independently (Table 3) (Ramensky et al. 2002; Yue and Moult 2006).
Fig. 1.
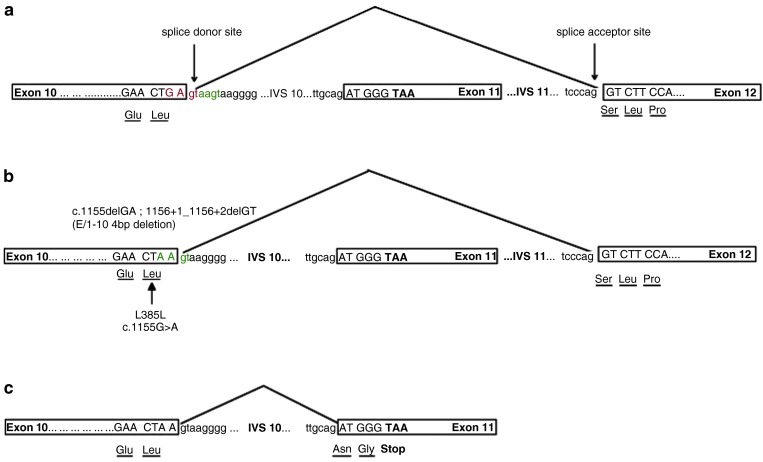
(a) The normal sequence and exon splicing of CHIT1 pre-mRNA, (exon 1-10 + exon 12) (Boot et al. 1998), with the 4bp deletion indicated in red. Flagged in green are the neighbouring nucleotides of the 4bp deletion. (b) As a result of the E/I-10 4bp deletion the pre-mRNA sequence of CHIT1 is altered. The occurring frame shift leads to silent mutation p.L385L as well as the creation of an alternative splice donor site. In the majority of splicing events (4/6 times) the splicosome recognises the alternative splice site and normal splicing occurs (exon 1-10 + exon 12) (Grace et al. 2007). (c) In 2/6 splicing events, the use of the alternative splice site leads to aberrant mRNA splicing. The inclusion of exon 11 in the mRNA leads to the introduction of a premature stop codon and no translation into protein occurs (Grace et al. 2007)
Table 3.
Computationally predicted functional impact of coding DNA variants on CHIT1 using two web-based tools
| Variant | PolyPhen ΔPSIC (http://genetics.bwh.harvard.edu/pph) |
SNP3D SVM score (http://www.snps3d.org/) |
Predicted effect on Chitotrosidase |
|---|---|---|---|
| G354R | 2.975 | –4.25 | Deleterious |
| A442V | 0.948 | –1.07 | Damaging |
| A442G | 0.867 | 1.55 | Inconclusive |
| ΔPSIC ≤ 0.5 = benign | SVM scores > +0.5= benign |
Discussion and Conclusion
The 1.6% overall prevalence of enzyme assay-based chitotriosidase deficiency detected in the Black SA Bantu population corroborates previous findings of notably lower prevalence of chito deficiency in African populations (0–2%) (Malaguarnera et al. 2003) in comparison with populations elsewhere in the world (Lee et al. 2007; Piras et al. 2007). In addition, our study revealed complete absence of the 24bp duplication in exon 10 of the CHIT1 gene as the causative mutation for chito deficiency in the Black Southern African population. This finding differs from investigations of chito-deficient alleles in subjects of African ancestry in previous studies (Grace et al. 2007; Lee et al. 2007) where the 24bp duplication was detected at appreciable frequencies (4.3% and 7%, respectively). However, it is likely that the 24bp duplication allele in African Americans, Caribbean Hispanics and Black individuals of self-reported ethnicity, all living outside of Africa, is a consequence of gene flow, if one considers the high frequency of the 24bp duplication in their respective surrounding populations. The observation of the 4bp splice site deletion (E/I-10_delGAgt) in homozygous state in our study allowed the precise localisation of the deletion, which is shifted by 2bp from the published position (Grace et al. 2007). The postulated functional impact of this 4bp deletion as described in the current study (Fig. 1) is strongly supported by previous CHIT1 mutation expression studies (Grace et al. 2007).
Mutation analysis in Black South African chito-deficient individuals shows that the 4bp deletion is in complete linkage disequilibrium (LD) with two additional coding variants (p.G354R and p.A442V) within the CHIT1 gene. It is not possible to accurately pinpoint the causal mutation that leads to the loss of chito activity in Black South Africans, since two of the three detected mutations (p.G354R, E/I-10_delGAgt) result in chito deficiency and the remaining one (p.A442V) results in reduced activity (Lee et al. 2007). It would be interesting to investigate allelic variants that occur at the CHIT1 locus in other African populations and establish if these three mutations are, firstly, present and, secondly, in LD with each other.
To date, the answers as to why the global population differences in chitotriosidase deficiency emerged and what the exact biological function of this enzyme is remain controversial (Barone et al. 2003b; Chien et al. 2005; Choi et al. 2001; Gordon-Thomson et al. 2009; Hall et al. 2007; Hise et al. 2003). In populations living outside Africa, chitotriosidase deficiency might confer a selective advantage against atopy by modulating the immune response (van Eijk et al. 2005) and, more recently, it was reported that heterozygosity for the 24bp duplication is associated with longevity in Mediterranean populations (Malaguarnera et al. 2010).
In conclusion, regardless of the driving force behind the emergence of chitotriosidase deficiency, we have shown that the prevalence of chitotriosidase deficiency in Black South Africans is low and it is therefore a conducive biomarker.
Supplementary Table 1.
Primer information for the amplification of the CHIT1 gene
| Exon | Primer sequence 5’ to 3’ | Annealing Temperature TA°C | PCR product size (bp) |
|---|---|---|---|
| 1 F | GACAGGGTGGCCAGATAAAA | 60 | 183 |
| 1 R | GGTAGCAAGTGGTCCCTGAA | ||
| 2 F | GAAGGTGAGAATGGCAGCAT | 58 | 152 |
| 2 R | GGGAAGGTGTTTTGGGACAT | ||
| 3 F | CCTGCTGACAGCTATCCCTTT | 60 | 296 |
| 3 R | TTATCTGTCACCCCACCACA | ||
| 4 F | CTGTTGTTCCTCCCCACTGT | 58 | 187 |
| 4 R | CCATCCAGGAGCTTTACCAC | ||
| 5 + 6 F | TGTTTTCTTGCCTGCTTCTG | 58 | 688 |
| 5 + 6 R | CTACCCATGGGAAAGCTCAG | ||
| 7 F | CCAATCCCTTGTTTCTCACC | 58 | 237 |
| 7 R | GCTCAAAAGAAGCCACCAAA | ||
| 8 + 9 F | CTCCTAGCTCTGCCTCTGGA | 62 | 762 |
| 8 + 9 R | TCACTAGGACCACCCCTCTG | ||
| 10 F | CCTGTCCAGAAGAGGTAGCC | 58 | 250 |
| 10 R | ACCTCCAAATTCCACCACTG | ||
| 11 +12 F | AGAAAGCCTCCCCTTAGCC | 62 | 844 |
| 11 + 12 R | CTCTGACCTGCGGATGTTTT | ||
| RFLP PCR 4F | CCATCGGAGGCTGGAATTCC | 56°C | 259 |
| RFLP PCR 4R | CTGGCAAGACTGGATCTGA |
Acknowledgements
This study was supported by the South African National Health Laboratory Service Research Trust. We would like to thank Jennifer Kromberg from the Mellon Research Foundation at the University of the Witwatersrand, and M. Ramsay and A. Nebel for useful comments on the manuscript.
Footnotes
Competing interests: None declared
References
- Barone R, Malaguarnera L, Angius A, Musumeci S. Plasma chitotriosidase activity in patients with beta-thalassemia. Am J Hematol. 2003;72(4):285–6. doi: 10.1002/ajh.10294. [DOI] [PubMed] [Google Scholar]
- Barone R, Simpore J, Malaguarnera L, Pignatelli S, Musumeci S. Plasma chitotriosidase activity in acute Plasmodium falciparum malaria. Clin Chim Acta. 2003;331(1–2):79–85. doi: 10.1016/S0009-8981(03)00089-5. [DOI] [PubMed] [Google Scholar]
- Boot RG, Renkema GH, Verhoek M, Strijland A, Bliek J, de Meulemeester TM, Mannens MM, Aerts JM. The human chitotriosidase gene Nature of inherited enzyme deficiency. J Biol Chem. 1998;273(40):25680–5. doi: 10.1074/jbc.273.40.25680. [DOI] [PubMed] [Google Scholar]
- Boot RG, van Achterberg TA, van Aken BE, Renkema GH, Jacobs MJ, Aerts JM, de Vries CJ. Strong induction of members of the chitinase family of proteins in atherosclerosis: chitotriosidase and human cartilage gp-39 expressed in lesion macrophages. Arterioscler Thromb Vasc Biol. 1999;19(3):687–94. doi: 10.1161/01.ATV.19.3.687. [DOI] [PubMed] [Google Scholar]
- Chien YH, Chen JH, Hwu WL. 2005. Plasma chitotriosidase activity and malaria. Clin Chim Acta 353(1–2):215; author reply 217. [DOI] [PubMed]
- Choi EH, Zimmerman PA, Foster CB, Zhu S, Kumaraswami V, Nutman TB, Chanock SJ. Genetic polymorphisms in molecules of innate immunity and susceptibility to infection with Wuchereria bancrofti in South India. Genes Immun. 2001;2(5):248–53. doi: 10.1038/sj.gene.6363767. [DOI] [PubMed] [Google Scholar]
- Gordon-Thomson C, Kumari A, Tomkins L, Holford P, Djordjevic JT, Wright LC, Sorrell TC, Moore GP. Chitotriosidase and gene therapy for fungal infections. Cell Mol Life Sci. 2009;66(6):1116–25. doi: 10.1007/s00018-009-8765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace ME, Balwani M, Nazarenko I, Prakash-Cheng A, Desnick RJ. Type 1 Gaucher disease: null and hypomorphic novel chitotriosidase mutations-implications for diagnosis and therapeutic monitoring. Hum Mutat. 2007;28(9):866–73. doi: 10.1002/humu.20524. [DOI] [PubMed] [Google Scholar]
- Guo Y, He W, Boer AM, Wevers RA, de Bruijn AM, Groener JE, Hollak CE, Aerts JM, Galjaard H, van Diggelen OP. Elevated plasma chitotriosidase activity in various lysosomal storage disorders. J Inherit Metab Dis. 1995;18(6):717–22. doi: 10.1007/BF02436762. [DOI] [PubMed] [Google Scholar]
- Hall AJ, Quinnell RJ, Raiko A, Lagog M, Siba P, Morroll S, Falcone FH. Chitotriosidase deficiency is not associated with human hookworm infection in a Papua New Guinean population. Infect Genet Evol. 2007;7(6):743–7. doi: 10.1016/j.meegid.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hise AG, Hazlett FE, Bockarie MJ, Zimmerman PA, Tisch DJ, Kazura JW. Polymorphisms of innate immunity genes and susceptibility to lymphatic filariasis. Genes Immun. 2003;4(7):524–7. doi: 10.1038/sj.gene.6364015. [DOI] [PubMed] [Google Scholar]
- Hollak CE, van Weely S, van Oers MH, Aerts JM. Marked elevation of plasma chitotriosidase activity A novel hallmark of Gaucher disease. J Clin Invest. 1994;93(3):1288–92. doi: 10.1172/JCI117084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Waalen J, Crain K, Smargon A, Beutler E. Human chitotriosidase polymorphisms G354R and A442V associated with reduced enzyme activity. Blood Cells Mol Dis. 2007;39(3):353–60. doi: 10.1016/j.bcmd.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaguarnera L, Ohazuruike LN, Tsianaka C, Antic T, Di Rosa M, Malaguarnera M. Human chitotriosidase polymorphism is associated with human longevity in Mediterranean nonagenarians and centenarians. J Hum Genet. 2010;55(1):8–12. doi: 10.1038/jhg.2009.111. [DOI] [PubMed] [Google Scholar]
- Malaguarnera L, Simpore J, Prodi DA, Angius A, Sassu A, Persico I, Barone R, Musumeci S. A 24-bp duplication in exon 10 of human chitotriosidase gene from the sub-Saharan to the Mediterranean area: role of parasitic diseases and environmental conditions. Genes Immun. 2003;4(8):570–4. doi: 10.1038/sj.gene.6364025. [DOI] [PubMed] [Google Scholar]
- Piras I, Melis A, Ghiani ME, Falchi A, Luiselli D, Moral P, Varesi L, Calo CM, Vona G. Human CHIT1 gene distribution: new data from Mediterranean and European populations. J Hum Genet. 2007;52(2):110–6. doi: 10.1007/s10038-006-0086-1. [DOI] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkema GH, Boot RG, Au FL, Donker-Koopman WE, Strijland A, Muijsers AO, Hrebicek M, Aerts JM. Chitotriosidase, a chitinase, and the 39-kDa human cartilage glycoprotein, a chitin-binding lectin, are homologues of family 18 glycosyl hydrolases secreted by human macrophages. Eur J Biochem. 1998;251(1–2):504–9. doi: 10.1046/j.1432-1327.1998.2510504.x. [DOI] [PubMed] [Google Scholar]
- Renkema GH, Boot RG, Muijsers AO, Donker-Koopman WE, Aerts JM. Purification and characterization of human chitotriosidase, a novel member of the chitinase family of proteins. J Biol Chem. 1995;270(5):2198–202. doi: 10.1074/jbc.270.5.2198. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21(6):577–81. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- van Eijk M, van Roomen CP, Renkema GH, Bussink AP, Andrews L, Blommaart EF, Sugar A, Verhoeven AJ, Boot RG, Aerts JM. Characterization of human phagocyte-derived chitotriosidase, a component of innate immunity. Int Immunol. 2005;17(11):1505–12. doi: 10.1093/intimm/dxh328. [DOI] [PubMed] [Google Scholar]
- Webb EL, Rudd MF, Sellick GS, El Galta R, Bethke L, Wood W, Fletcher O, Penegar S, Withey L, Qureshi others M, et al. Search for low penetrance alleles for colorectal cancer through a scan of 1467 non-synonymous SNPs in 2575 cases and 2707 controls with validation by kin-cohort analysis of 14 704 first-degree relatives. Hum Mol Genet. 2006;15(21):3263–71. doi: 10.1093/hmg/ddl401. [DOI] [PubMed] [Google Scholar]
- Yue P, Moult J. Identification and analysis of deleterious human SNPs. J Mol Biol. 2006;356(5):1263–74. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]


