SUMMARY
It remains unclear whether basophils and mast cells are derived from a common progenitor. Furthermore, how basophil versus mast cell fate is specified has not been investigated. Here, we have identified a population of granulocyte-macrophage progenitors (GMPs), which were highly enriched in the capacity to differentiate into basophils and mast cells while retaining a limited capacity to differentiate into myeloid cells. We have designated these progenitor cells “pre-basophil and mast cell progenitors” (pre-BMPs). STAT5 signaling was required for the differentiation of pre-BMPs into both basophils and mast cells and was critical for inducing two downstream molecules: C/EBPα and MITF. We have identified C/EBPα as the critical basophil transcription factor for specifying basophil cell fate and MITF as the crucial transcription factor for specifying mast cell fate. C/EBPα and MITF silenced each other’s transcription in a directly antagonistic fashion. Our study reveals how basophil and mast cell fate is specified.
INTRODUCTION
Basophils and mast cells share many common characteristics, such as the expression of a high-affinity immunoglobulin E (IgE)receptor (FcεR), and contain many ofthe same granules (Galli and Franco, 2008; Marone et al., 2002). Conversely, these cells also show notable differences. Basophils circulate in the blood stream, whereas mast cells reside in tissue. Mature basophils do not proliferate and have a short lifespan of approximately 60 hours (Ohnmacht and Voehringer, 2009), whereas mature mast cells can proliferate and have a much longer lifespan of up to several months (Galli et al., 2008). Functionally, both basophils and mast cells are the key effectors in type-2 immunity that cause allergic disease and provide protection against parasitic infections. Accumulated evidence supports the non-redundant role of basophils in immune regulation, protective immunity, allergy, and autoimmunity (Karasuyama et al., 2011). Recent success in using anti-IgE antibody to treat various allergic disorders in humans supports the importance of FcεR-expressing basophils and mast cells in human diseases (Busse et al., 2011; Holgate et al., 2005). Thus, a more comprehensive understanding of the developmental pathway for basophils and mast cells is of substantial value.
The hematopoietic hierarchy consists of various stem cells and progenitors. Long-term repopulating hematopoietic stem cells (HSCs) are at the top of the hematopoietic hierarchy. These cells possess the capacity for self-renewal and the potential to give rise to all types of blood cells. Long-term HSCs can generate short-term repopulating HSCs, which then give rise to multiple potential progenitors (MPPs). MPPs, in turn, can give rise to both common lymphoid progenitors and common myeloid progenitors (CMPs). CMPs can differentiate into granulocyte-monocyte progenitors (GMPs) (Kondo et al., 2003). GMPs give rise to eosinophil lineage-restricted progenitors (Iwasaki et al., 2005), basophil lineage-restricted progenitors (BaPs), neutrophils and macrophages (Arinobu et al., 2005).
The origin of basophils and mast cells has been a long-standing, unsolved, and important issue in hematology. By using colony formation assays, two groups have claimed that basophils develop from a common basophil and eosinophil progenitor (Denburg et al., 1985; Leary and Ogawa, 1984). Whether basophils and mast cells are derived from a common progenitor remains a controversial issue. Galli and colleagues found mast cell lineage-restricted progenitors (MCPs) in the bone marrow and proposed that MCPs were derived from multiple potential progenitors (MPPs) instead of CMPs or GMPs (Chen et al., 2005). Alternatively, Akashi and colleagues showed that both basophils and mast cells were derived from CMPs and GMPs (Arinobu et al., 2009); they further showed that basophil-mast cell progenitors (BMCPs) found in the spleen gave rise to both basophils and mast cells (Arinobu et al., 2005). However, the validity of BMCPs as authentic bi-potential basophil-mast cell progenitors has recently been challenged by a study in which Galli and colleagues demonstrated that BMCPs only gave rise to mast cells (Mukai et al., 2012).
Furthermore, the mechanisms by which basophil cell fate versus mast cell fate is specified remains undetermined. Regulatory networks containing primary and secondary determinants of cell fate have been shown to be critical in making T cell, B cell, macrophage, and neutrophil cell fate choices in the hematopoietic system (Laslo et al., 2008). For instance, Singh and colleagues demonstrated that a high dose of a transcription factor from the ETS family, PU.1, drove GMPs to differentiate into macrophages (Laslo et al., 2006), whereas a high C/EBPα /PU.1 ratio directed the differentiation of GMPs into neutrophils (Dahl et al., 2003). PU.1 induced the secondary determinants Egr1,2 and Nab-2 to suppress neutrophil cell fate, whereas C/EBPα induced Gfi to suppress macrophage cell fate. The actions of Egr1,2 and Nab-2 and Gfi were found to be directly antagonistic to one another (Laslo et al., 2006). Despite the previous identification of several factors involved in the differentiation of basophils and mast cells, it remains unclear how these factors relate to one another in specifying basophil versus mast cell fate. Thus, STAT5 (Shelburne et al., 2003), GATA1 (Migliaccio et al., 2003), GATA2 (Tsai and Orkin, 1997), and MITF (Kitamura et al., 2002; Takemoto et al., 2008) have each been demonstrated as critical for mast cell differentiation, whereas STAT5 (Ohmori et al., 2009), Runx1 (Mukai et al., 2012), GATA2 and C/EBPα (Iwasaki et al., 2006) have each been implicated to play imperative roles in basophil differentiation. It remains unknown which of the aforementioned factors are the master determinants for basophil versus mast cell fate.
In this study, we identified a population of granulocyte-macrophage progenitors (GMPs) that possessed highly enriched capacity to differentiate into basophils and mast cells while still retaining a limited capacity to differentiate into myeloid cells. We have designated these progenitors as pre-basophil and mast cell progenitors (pre-BMPs). Our analysis revealed that a regulatory network—composed of STAT5, C/EBPα and MITF—plays a critical role in determining basophil versus mast cell fate. Specifically, the upregulation of both C/EBPα and MITF in pre-BMPs was STAT5-dependent and expression of C/EBPα and MITF was mutually exclusive. Dominance of C/EBPα resulted in the differentiation of pre-BMPs into basophils, whereas dominance of MITF led to the differentiation of pre-BMPs into mast cells.
RESULTS
A Subset of GMPs in the Bone Marrow Acquires the Capacity to Produce Type 2 Cytokines
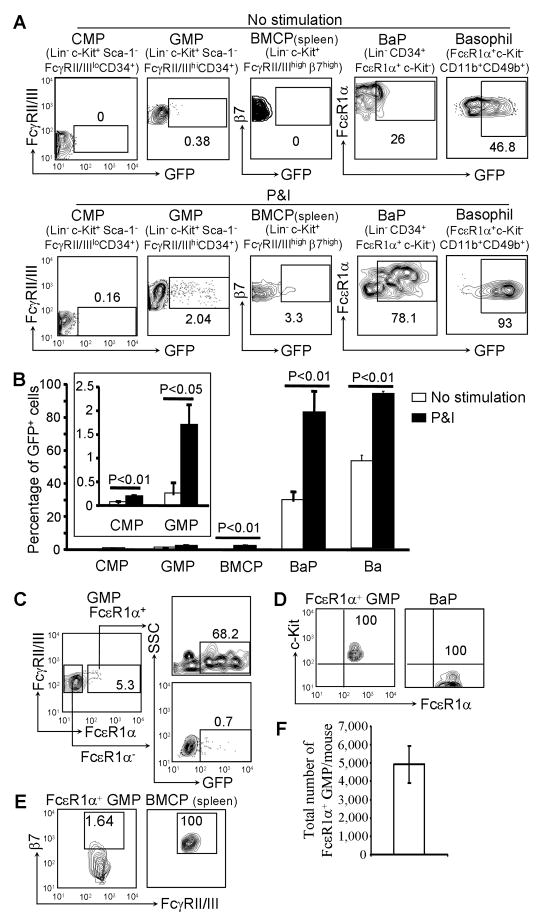
Mature basophils are known to be robust type 2 cytokine-producing cells. To determine at which developmental stage the bone marrow progenitor cells begin to acquire such capacity, we analyzed Il4-Gfp reporter gene expression in CMPs, GMPs, BaPs, and mature basophils from the bone marrow of heterozygous IL-4 reporter mice (Il4G4/+ mice) as well as in BMCPs from the spleen of Il4G4/+ mice (the reported BMCPs were not found in the bone marrow; thereafter, BMCPs refer to BMCPs in the spleen) (Arinobu et al., 2005). We found that CMPs did not express GFP regardless of whether they were stimulated with PMA and ionomycin (P&I), whereas a subset of GMPs expressed GFP only when stimulated by P&I (Figure 1A and 1B). BaPs and mature basophils expressed GFP without any P&I stimulation, but stimulation markedly increased the percentage of GFP+ BaPs and basophils (Figure 1A and 1B). A small percentage of BMCPs also expressed GFP with P&I stimulation (Figure 1A and 1B). We did not find any difference in the number of GFP+ GMPs between Il4G4/+ mice and Il4G4/G4 mice (data not shown). These data suggest that a small subset of GMPs acquires the capacity to transcribe the Il4 gene.
Figure 1. A Subset of GMPs Begins to Acquire the Capacity to Express the Il4 Reporter Gene.
(A) FACS analysis of GFP expression in progenitors of the bone marrow or the spleen prepared from Il4G4/+ mice. The numbers inside the FACS plots indicate the percentage within the gated populations. (B) The percentages of GFP+ cells within the gated populations (mean ± SD, n=3). (C) FACS sorted GMPs from the bone marrow of Il4G4/+ mice were stimulated with P&I for 6 hours and stained with APC-labeled anti-FcεR1α antibody. (D) c-Kit expression comparison between FcεR1α+GMPs and BaPs. (E) β7 expression comparison between FcεR1α+ GMPs and BMCPs in the spleen. (F) Total number of FcεR1α+ GMPs in two tibias and two femur bones per mouse (mean ± SD, n=6). Data are representative of three independent experiments with similar results. Also see Figure S1.
To search for a surface marker that identifies IL-4 competent GMPs, we analyzed FcεR1α expression of the GFP+ GMPs because FcεR1α has been associated with IL-4-producing basophils and mast cells. We found that about 70% of FcεR1α+ GMPs expressed GFP when stimulated with P&I (Figure 1C), whereas virtually no FcεR1α− GMPs expressed GFP when stimulated (Figure 1C). Thus, FcεR1α served as an appropriate surface marker for identifying GFP+ GMPs. To determine if the FcεR1α+ GMPs represented a novel population of progenitors, we phenotypically compared them with BaPs or BMCPs. The identified FcεR1α+ GMPs differed from BaPs by the expression of c-Kit (Figure 1D). FcεR1α+ GMPs were distinguishable from BMCPs because the majority of FcεR1α+ GMPs did not express β7 integrin—only 1.6% of FcεR1α+ GMPs expressed low amounts of β7 integrin, whereas BMCPs expressed high amounts of β7 integrin (Figure 1E). In fact, β7 integrin was a useful marker for enriching mast cells but not basophils (Figure S1). An average of 4,929 FcεR1α+ GMPs cells were found in two tibia and two femur bones per mouse (Figure 1F).
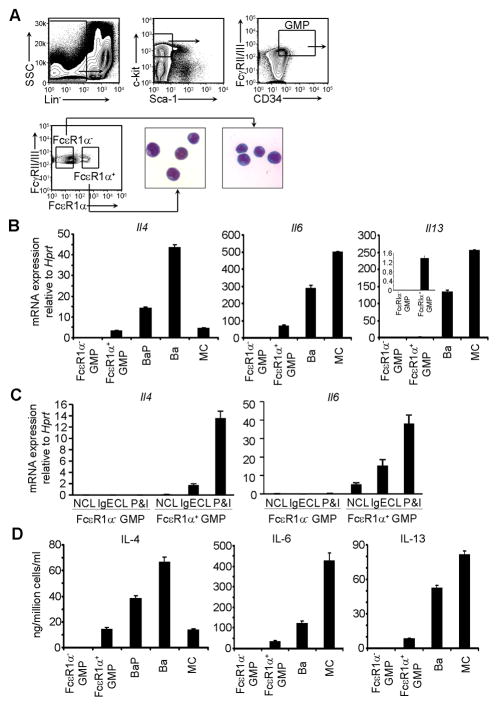
Mature basophils are known to produce IL-4, IL-5, IL-6, and IL-13 (Karasuyama et al., 2011). We FACS-sorted FcεR1α+ GMPs using the sorting gates shown in Figure 2A. Morphologically, the FcεR1α+ GMPs resembled immature progenitor cells rendering them indistinguishable from the morphology of FcεR1α− GMPs (Figure 2A). We showed that the sorted FcεR1α+ GMPs, but not the FcεR1α− GMPs, expressed Il4, Il6, and Il13 mRNA with P&I stimulation (Figure 2B) or IgE cross-linking of FcεR (Figure 2C). These early progenitors also produced small amounts of IL-4, IL-6, and IL-13 proteins (Figure 2D). The capacity of FcεR1α+ GMPs to express IL-4, IL-6, and IL-13 was low compared to that of BaPs and mature basophils (Figure 2B and 2D) and the capacity to express IL-6 and IL-13 was even lower relative to that of mast cells (Figure 2B and 2D). In contrast, the capacity to express IL-4 was comparable to that of mast cells (Figure 2B and 2D). However, IL-5 mRNA or protein remained undetectable (data not shown). These data demonstrate that FcεR1α+ GMPs represent the earliest hematopoietic progenitors to acquire the capacity to express type 2 cytokines.
Figure 2. FcεR1α+ GMPs Possess the Ability to Express IL-4, IL-6, and IL-13.
(A) The gates for sorting FcεR1α− and FcεR1α+ GMPs. The sorted FcεR1α− and FcεR1α+ GMPs were stained with May-Grunwald Giemsa. (B) The sorted cells were stimulated with P&I for 4 hours. mRNA expression was measured by qPCR (mean ± SD, triplicates). Ba=basophils; MC=BMMC. (C) The sorted FcεR1α− and FcεR1α+ GMPs were not cross-linked (NCL) with IgE, cross-linked with IgE (IgECL) or stimulated with P&I for 4 hours. (D) The sorted cells were stimulated with P&I overnight. IL-4, IL-6, and IL-13 proteins were measured by ELISA (mean ± SD, triplicates). Data represent two independent experiments with similar results.
FcεR1α+ GMPsContain Highly Enriched Common Basophil and Mast Cell Progenitors
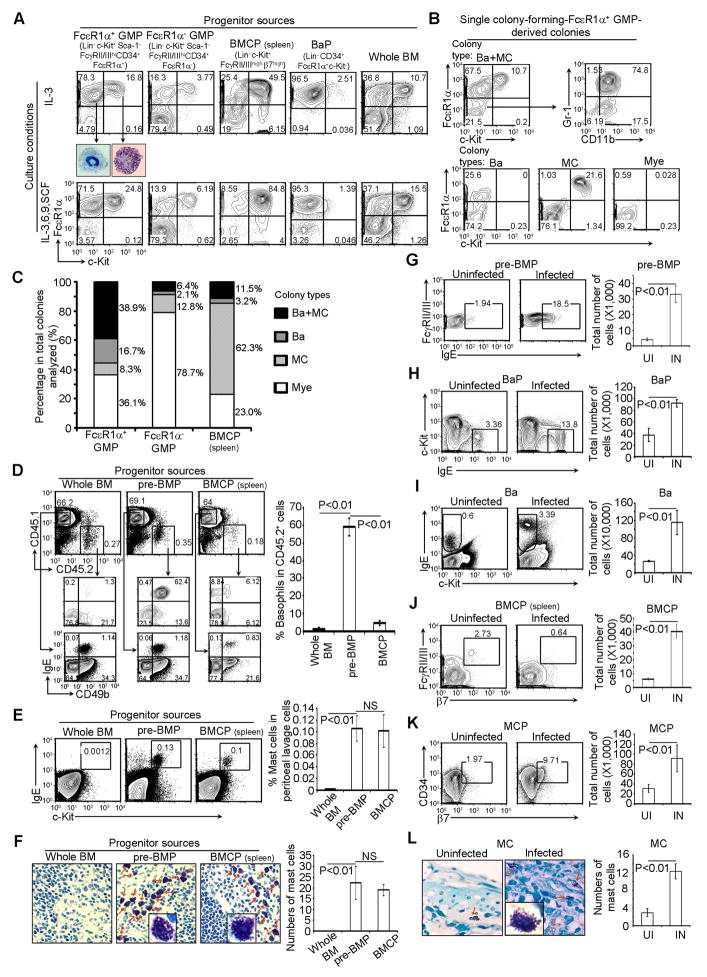
Because FcεR1α+ GMPs phenotypically belong to a subset of GMPs, it is possible that they are the progenitor cells of BaPs, MCPs or BMCPs. To test these possibilities, we differentiated the FACS-sorted FcεR1α+ GMPs in the presence of IL-3 for 1 day or 3 days and then reanalyzed the FcεR1α+ GMPs-derived cells. We showed that FcεR1α+ GMPs gave rise to BaPs and MCPs but not BMCPs (Figure S2A). Indeed, after 5 days of culture in the presence of IL-3 alone or IL-3 in combination with IL-6, IL-9, and SCF, FcεR1α+ GMPs differentiated exclusively into basophils and mast cells under both culture conditions (Figure 3A), BaPs gave rise exclusively to basophils, and BMCPs gave rise mostly to mast cells (Figure 3A). The IL-3, IL-6, IL-9, and SCF combination did not enhance basophil differentiation from BMCPs, a result that is consistent with a recent report (Mukai et al., 2012). SCF alone was sufficient for FcεR1α+ GMPs to differentiate into mast cells although it was much less sufficient for FcεR1α+ GMPs to differentiate into basophils (Figure S2D).
Figure 3. pre-BMP Populations Contain Highly Enriched Common Basophil-Mast Cells Progenitors.
(A) FcεR1α+ GMPs, FcεR1α− GMPs, and BaPs FACS-sorted from BM cells, BMCPs FACS-sorted from the spleen, and whole BM cells were cultured under the indicated conditions for 5 days. Basophils and mast cells sorted from FcεR1α+ GMPs cultured in IL-3 were stained by May-Grunwald Giemsa (40x). (B) Single colony-forming-FcεR1α+ GMP-derived colonies were analyzed by FACS. (C) Colony composition. Each type of colony was calculated as the percentage of total colonies analyzed. (D) FACS analysis of CD45.2+ basophils in the BM of pre-BMP-reconstituted CD45.1 recipient mice (mean ± SD, n = 3). (E) FACS analysis of mast cells from the peritoneal cavity of reconstituted Kitw–sh/w–sh mice (mean ± SD, n = 3). (F) Toluidine blue staining of spleen sections from reconstituted Kitw–sh/w–sh mice (40x, insert, 100x). Mast cells are indicated by red arrows. The right panel shows the average number of mast cells in five different fields (40x) randomly selected from the sections of spleens (mean ± SD, n = 3). (G-K) FACS analysis of pre-BMPs, BaPs, basophils, and MCPs in the bone marrow or BMCPs in the spleen (J) of mice uninfected (UI) or infected (IN) withS. mansoni for 7 weeks (left), and the total number of cells in two femur bones per mouse or in the spleen (right). Data represent the mean ± SD (n=5). (L) Mast cells in the large intestine of uninfected or infected mice were stained with toluidine blue (left, 40x, insert, 100x) and shows the average number of mast cells in five different fields randomly selected from the sections of large intestine (mean ± SD, n=3). Data represent three independent experiments with similar results. Also see Figure S2.
Because the highly purified FcεR1α+ GMP populations still contained heterogeneous progenitors, we determined if both basophils and mast cells could be derived from a single FcεR1α+ GMP. Using a limiting dilution analysis assay, the colony-forming efficiency for FcεR1α+ GMPs was determined to be 1 out of every 133 FcεR1α+ GMPs; the colony-forming efficiency for BMCPs was determined to be 1 out of every 64 BMCPs (Figure S2B). In order to grow single colony-forming progenitor-derived colonies, we seeded the sorted progenitors at a density below the colony-forming frequency (see the Supplemental Experimental Procedures for further explanation). We analyzed single colony-forming-FcεR1α+ GMP-derived colonies (Figure 3B) or single colony-forming-BMCP-derived colonies (Figure S2C) by FACS and found that 39% of single colony-forming-FcεR1α+ GMP-derived colonies contained both basophils and mast cells, 17% contained only basophils and 8% contained only mast cells (Figure 3C). The number of single FcεR1α+ GMP-derived colonies that contained both basophils and mast cells represented a 6-fold enrichment compared with single colony-forming FcεR1α− GMP (regular GMPs)-derived colonies (Figure 3C). Compared with BMCPs, the FcεR1α+ GMP population contained nearly 4 times the number of single progenitors capable of producing both basophils and mast cells (Figure 3C). We found that BMCPs contained highly enriched mast cell-lineage restricted progenitors (62 % of single BMCPs gave rise to mast cells, Figure 3C). Within the single colony-forming-FcεR1α+ GMP-derived colonies or single colony-forming-BMCP-derived colonies, we noted the presence of cells that stained negative for FcεR1α and positive for CD11b and (or) Gr-1 (Figure 3B), suggesting that single common basophil-mast cell progenitors in the FcεR1α+ GMP population also retained the capacity to differentiate into macrophages and neutrophils when cultured in semi-solid culture media in the presence of IL-3.
FcεR1α+ GMPs showed a reduced capacity to form colonies. The colony-forming efficiency of FcεR1α+ GMPs reduced to 26.9% and 56.2% of regular GMPs and splenic BMCPs, respectively (Figure S2B). We observed that FcεR1α+ GMPs formed colonies in the presence of IL-3 and GM-CSF and failed to form colonies in the presence of G-CSF, M-CSF, or IL-5 (Figure S2E). Furthermore, we did not find that FcεR1α+ GMPs to be responsive to IL-25 (data not shown). Altogether, because it was determined that FcεR1α+ GMPs were more mature than GMPs and because FcεR1α+ GMPs possessed great potential to differentiate into basophils and mast cells but had not yet fully committed into bi-potential basophil-mast cell potential progenitors, we named FcεR17α+ GMPs pre-basophil-mast cell progenitors (pre-BMPs).
Pre-BMPs Give Rise to Basophils and Mast Cells in Vivo
To determine if pre-BMPs can give rise to basophils in vivo, we injected FACS-sorted pre-BMPs into irradiated CD45.1 mice. Three days later, we found that over 60% of the pre-BMPs-derived cells in the bone marrow of reconstituted CD45.1 mice were basophils (Figure 3D). The percentage of the pre-BMPs-derived basophils decreased to approximately 8% six days after reconstitution (Figure S2F) and no pre-BMPs-derived basophils were detected at one month after reconstitution (Figure S2G). We did not find CD45.2+ T cells or B cells in the CD45.1 mice reconstituted with pre-BMP cells at one month after reconstitution (Figure S2G). These data were consistent with the notion that pre-BMPs do not possess the ability to self-renew and that basophils have a short lifespan. The basophil potential of pre-BMPs assessed using the in vivo method was similar to that measured using the in vitro method (Figure 3A and 3D), and this potential as measured by the in vivo method was 12-fold higher than that of BMCPs and 55-fold higher than that of unsorted bone marrow cells (Figure 3D).
To determine if pre-BMPs can give rise to mast cells, we injected FACS-sorted pre-BMPs into irradiated, mast cell-deficient mice (KitW–sh/W–sh), whose progenitors are unable to differentiate into mast cells, allowing more efficient pre-BMP-derived mast cell reconstitution. We found that pre-BMPs effectively reconstituted mast cells in both the peritoneal cavity and in the spleen of the reconstituted KitW–sh/W–sh mice within 5 to 6 weeks (Figure 3E and 3F). The mast cell reconstitution efficiency of pre-BMPs was comparable to that of BMCPs, and was 73-fold higher than that unsorted bone marrow cells (Figure 3E and 3F).
To determine the physiological relevance of pre-BMPs, we infected mice with Schistosoma mansoni cercaria, a type of parasite that induces a strong type-2 immune response, and observed a 10-fold expansion of pre-BMPs after 7 weeks of infection (Figure 3G). Consistently, BaPs, basophils, MCPs, and mast cells were also markedly expanded (Figure 3H, 3I, 3K, and 3L). The percentage of BMCPs in the spleens of infected mice decreased, yet the total numbers of BMCPs still markedly increased because the spleens of infected mice were extremely enlarged (Figure 3J). These data also support the notion that pre-BMPs are the in vivo progenitors in the production of basophils and mast cells.
STAT5 Is Imperative for the Differentiation of pre-BMPs into Both Basophils and Mast cells but not Myeloid Cells
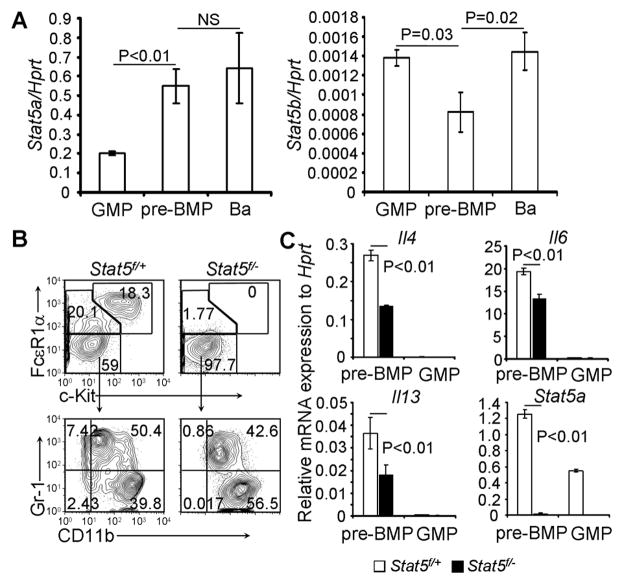
STAT5 is a signaling molecule that can be activated by many cytokines and growth factors (Leonard and O’Shea, 1998). We previously demonstrated that STAT5 deficiency impaired the development of multiple lineages, including basophils (Ohmori et al., 2009). However, the precise role of STAT5 in the differentiation of pre-BMPs into basophils has not yet been determined. We showed that Stat5a mRNA was upregulated in pre-BMPs compared with regular GMPs (Figure 4A). To examine the role of STAT5 in the differentiation of pre-BMPs into basophils and mast cells, we treated inducible STAT5 knockout mice (Stat5f/− RosaYFP/YFP Mx1-cre mice) and control mice (Stat5f/+RosaYFP/YFP Mx1-cre mice) with i.p. injection of poly I-C and then waited 21 to 30 days before analysis. Poly I-C treatment effectively deleted the Stat5a/b gene (Figure S3A). STAT5 deficiency had no effect on the number of T cells, B cells, neutrophils or macrophages in the spleen (Figure S3B and S3C) and caused a 50% reduction in the number of pre-BMPs, BaPs and basophils in the bone marrow (Figure S3D and S3E). Because STAT5 can be deleted in any cell at any developmental stage in the in vivo inducible gene deletion system, the effects of in vivo STAT5 deficiency on the number of pre-BMPs, BaPs, and basophils might not reflect the precise role of STAT5 in pre-BMP differentiation. To address this issue, we combined the in vivo inducible gene deletion system with prospective FACS sorting. We demonstrated that the FACS-sorted YFP+ pre-BMPs, prepared from poly IC-treated Stat5f/− RosaYFP/YFP Mx1-cre mice but not from poly I-C-treated Stat5f/+RosaYFP/YFP Mx1-cre mice, failed to differentiate into either basophils or mast cells, but they did differentiate into myeloid cells (Figure 4B). We did not find evidence of STAT5 haploid insufficiency in the differentiation of pre-BMPs into basophils or mast cells (Figure S3F and S3G). We showed that STAT5 was imperative for upregulating Il4, Il6, and Il13 mRNA expression in pre-BMPs (Figure 4C). The role of STAT5 in the differentiation of basophils and mast cells was further confirmed using a different Stat5 inducible gene deletion system (Figure S3H). These results demonstrate that STAT5 signaling is required for the differentiation of pre-BMPs into both basophils and mast cells in vitro and is critical in the acquisition of type 2 cytokine-expressing capacity in pre-BMPs.
Figure 4. STAT5 Is Imperative for the Differentiation of pre-BMPs into Both Basophils and Mast Cells.
(A) Stat5a/b mRNA expression in GMPs (FcεR1α− GMPs), pre-BMPs and basophils was measured by qPCR (mean ± SD, triplicates). Data represent two independent experiments. (B) YFP+ pre-BMPs were FACS sorted from bone marrow cells of poly IC-treated mice and cultured in methylcellulose containing medium in the presence of IL-3 for 9 days. (C) YFP+ pre-BMPs were FACS sorted from poly I-C-treated mice and stimulated with P&I for 4 hours. mRNA was measured by qPCR (mean ± SD, triplicates). Data represent two (B) or three (C) independent experiments with similar results. Also see Figure S3.
C/EBPα Is Required for the Differentiation of pre-BMPs into Basophils and Is Necessary for Maintaining Basophil Identity
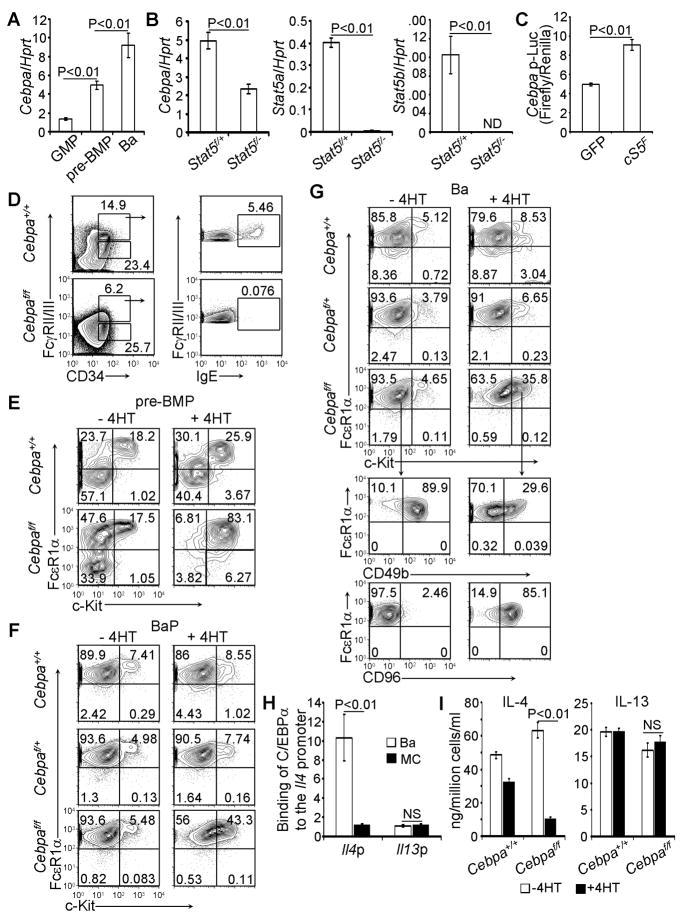
To search for downstream transcription factors that direct the differentiation of pre-BMPs into basophils, we examined the potential role of C/EBPα because it has been implicated in promoting basophil differentiation (Iwasaki et al., 2006). We observed that Cebpa mRNA in pre-BMPs was upregulated compared with that of regular GMPs (Figure 5A). We further showed that upregulation of Cebpa mRNA in pre-BMPs was STAT5-dependent (Figure 5B) and that overexpression of a constitutively active Stat5a mutant, cS5F (Moriggl et al., 2005), transactivated the Cebpa promoter (Figure 5C). C/EBPα has been established as a crucial transcription factor in the developmental transition from CMPs to GMPs (Zhang et al., 2004). To determine whether or not C/EBPα is required for the development of pre-BMPs, we treated both inducible Cebpa knockout mice (Cebpaf/f RosaYFP/creER mice) and control mice (Cebpa+/+Rosa YFP/creER mice) with tamoxifen and found that the development of pre-BMPs was abolished in the bone marrow of Cebpaf/f RosaYFP/creER mice but not Cebpa+/+Rosa YFP/creER mice (Figure 5D), indicating that C/EBPα is required for pre-BMP development.
Figure 5. C/EBPα Is Required for the Differentiation of pre-BMPs into Basophils and Is Necessary for Maintaining Basophil Identity.
(A) Cebpa mRNA expression (mean ± SD, triplicates). (B) Cebpa and Stat5a/b mRNA expression in YFP+pre-BMPs of poly I-C-treated mice as indicated (mean ± SD, triplicates). (C) Constitutively active Stat5 (cS5F) activated Cebpa promoter luciferase activity (mean ± SD, triplicates). (D) pre-BMPs in the bone marrow of the tamoxifen-treated mice were analyzed by FACS two weeks later after the treatment. YFP+ cells are shown. (E-G) pre-BMPs, BaPs, and basophils were FACS sorted from the untreated mice (Cebpaf/f= Cebpaf/f/RosaYFP/creER mice) and cultured in methylcellulose containing medium with or without the addition of 4HT for 9 days or complete medium in the presence of IL-3 with or without the addition of 4HT for 5 days (G). YFP+ cells are shown for 4HT-treated groups. (H) ChIP analysis of C/EBPα binding to the Il4 promoter in basophils (Ba) and mast cells (MC). (I) MACS sorted basophils were treated with or without 4HT for 5 days. ELISA analysis of IL-4 and IL-13 is shown (mean ± SD, triplicates). Data represent two (E, H, I) or three (A-D, F-G) independent experiments. Also see Figure S4.
To precisely determine the stage-specific, developmental role of C/EBPα in the differentiation of pre-BMPs into basophils, we FACS-sorted pre-BMPs from the bone marrow of Cebpaf/f RosaYFP/creER mice and Cebpa+/+Rosa YFP/creER control mice and cultured them in medium containing methylcellulose in the presence of both IL-3 and 4- hydroxytamoxifen (4HT), which induced deletion of the floxed Cebpa gene, for 9 days. In the absence of C/EBPα, pre-BMPs failed to differentiate into basophils (Figure 5E). Strikingly, we noted that Cebpa−/− pre-BMPs differentiated exclusively into mast cells (Figure 5E). To determine whether or not C/EBPα is required for BaPs to differentiate into basophils, we isolated BaPs from the bone marrow of Cebpaf/f RosaYFP/creER mice and either Cebpaf/+ RosaYFP/creER or Cebpa+/+Rosa YFP/creER control mice and cultured them in the presence of 4HT. We found that approximately 43 % of Cebpa−/− BaPs differentiated into c-Kit-expressing “mast cell-like” cells (Figure 5F). No dosage requirement was observed for C/EBPα in the differentiation of BaPs into basophils (Figure 5F). Even mature basophils converted into “mast cell-like cells”—i.e., reduced CD49b expression and increased CD96 expression—in the absence of C/EBPα (Figure 5G). A key difference between basophils and mast cells is that mast cells live much longer. We noticed that after two weeks of culture, WT basophils died whereas the majority of converted cells still lived. Phenotypically, Cebpa−/− basophils completely converted into mast cells after two weeks of culture (Figure S4). One of the major characteristics of basophils, compared with mast cells, is their ability to produce large quantities of IL-4. We previously reported that C/EBPα activated Il4 promoter activity (Qi et al., 2011). Thus, we assessed whether C/EBPα was required for maintaining the ability of basophils to produce IL-4. We showed that C/EBPα bound to the Il4 promoter, but not to the Il13 promoter, in primary basophils (Figure 5H) and that IL-4 production by basophils was dramatically reduced when the Cebpa gene was deleted following 4HT treatment (Figure 5I). C/EBPα did not appear to be required for IL-13 production (Figure 5I). These results demonstrate that C/EBPα is not only required for the differentiation of pre-BMPs into basophils, but is also required for maintaining the identity of basophils.
C/EBPα Promotes Basophil Molecular Programming and Simultaneously Represses Mast Cell Molecular Programing by Directly Inhibiting Mitf Gene Transcription
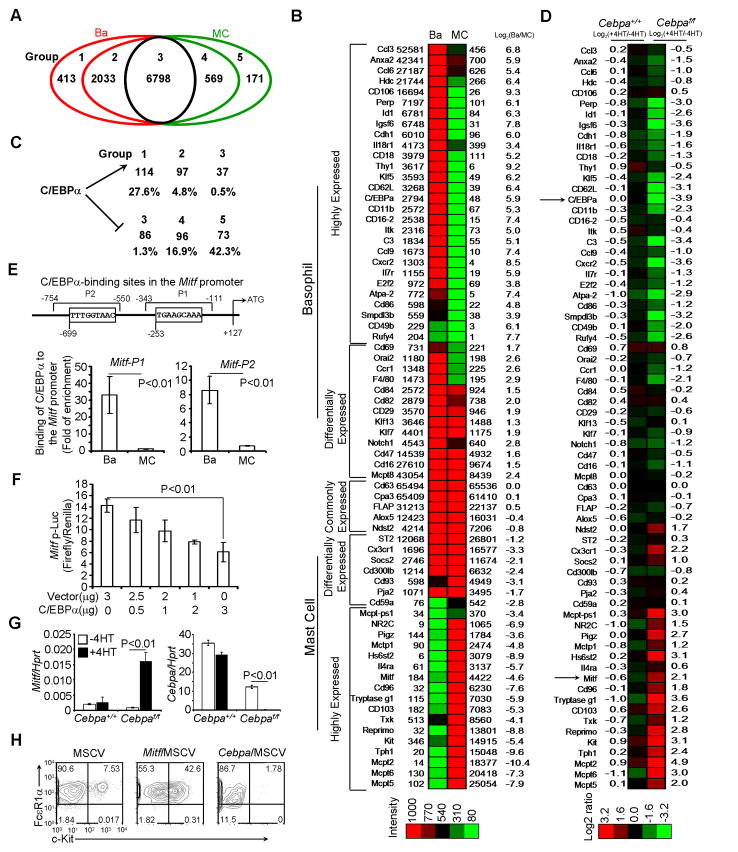
To investigate the mechanisms by which C/EBPα drives basophil differentiation and maintains basophil identity, we examined whether C/EBPα promotes basophil molecular programming and simultaneously represses mast cell molecular programming. We performed genome-wide gene expression profiling on basophils and mast cells and found that 6798 genes were shared by both mast cells and basophils; 2033 genes were expressed 2 to 10-fold (log2 1–3.3) higher in basophils (differentially expressed in basophils, group 2 of Figure 6A); and 413 genes were expressed greater than 10-fold (log2 3.3) higher in basophils (highly expressed in basophils, Group 1 of Figure 6A). Conversely, we found that 569 genes were expressed 2–10-fold (log2 −1 to −3.3) higher in mast cells and 171 genes were highly expressed in mast cells [greater than 10 fold (log2 −3.3)] (Group 4 and 5 of Figure 6A, respectively). Genes that were known to be critical in mediating basophil or mast cell function were represented in the heat map (Figure 6B). Expression profiles of the 20 genes that were highly expressed in either basophils or mast cells were verified by qPCR (Figure S5A) or by FACS (Figure S5B).
Figure 6. C/EBPα Promotes Basophil Molecular Programming and Simultaneously Represses Mast Cell Molecular Programming.
(A) Genome-wide gene expression profiling was performed on basophils and mast cells using microarray. Five groups of genes are shown in the Venn diagram. (B) The representative genes in each of five groups are shown in the heat map. (C) Numbers and percentages of genes in each group that were C/EBPα–dependent (indicated by arrow) or C/EBPα–repressed (indicated by bar). (D) The representative genes that were C/EBPα dependent (the log2 +4HT/−4HT ratio < −1) or C/EBPα repressed (the log2 +4HT/−4HT ratio > 1). (E) C/EBPα-binding sites and ChIP analysis of C/EBPα binding to the Mitf promoter (mean ± SD, triplicates). (F) C/EBPα suppressed Mitf promoter luciferase activity (mean ± SD, triplicates). (G) Re-expression of Mitf mRNA in Cebpaf/f or Cebpa+/+basophils treated with or without 4HT for 5 days (mean ± SD, triplicates). The right panel indicates a complete deletion of the Cebpa gene by 4HT treatment. (H) Purified BaPs were infected with GFP, C/EBPα or MITF virus and analyzed 5 days after infection. GFP+ cells are shown. Data (E-H) represent two independent experiments. Also see Figure S5 and Table S1.
Next, we examined which genes depended on C/EBPα for their expression and which genes were repressed by C/EBPα. We treated purified basophils prepared from Cebpaf/f RosaYFP/creER mice and Cebpa+/+ RosaYFP/creER control mice with or without 4HT treatment for 5 days. Gene expression in the treated basophils was analyzed using microarray analysis. Overall, deletion of Cebpa in basophils resulted in a reduction of mRNA expression for 248 genes and led to an increase in mRNA expression for 255 genes (Figure 6C). The majority of the C/EBPα-regulated genes were either differentially or highly expressed in basophils or mast cells (Figure 6C and 6D). The C/EBPα–dependency of genes shown in Figure 6B is presented in Figure 6D. Green represents a reduction in mRNA expression and red represents an increase in mRNA expression. Among C/EBPα-repressed genes, many were highly expressed in mast cells, such as Mitf, Mcpt, Kit, and Tph (Figure 6B and 6D). In the absence of C/EBPα, we did not observe enhanced expression of macrophage or neutrophil genes that were already expressed in basophils (Cd11b, Gr1 and F4/80), re-expression of new macrophage (Mmp12, Mpg-1, and Msr1) or neutrophil genes (Ela2, Prtn3, and Lactotransferrin) or re-expression of T cell, B cell or eosinophil surface markers (Table S1), indicating that C/EBPα represses mast cell programming, but not other cell-fate programming, in committed basophil progenitors and mature basophils.
C/EBPα could directly repress many mast cell-specific genes or indirectly by suppressing a master mast cell transcription factor that promotes transcription of mast cell-specific genes. Among C/EBPα-repressed genes, Mitf was of particular interest because it has been shown to play a critical role in mast cell differentiation. We found two C/EBPα-binding sites in the Mitf gene promoter region and demonstrated that C/EBPα bound to the Mitf promoter in basophils but not in mast cells (Figure 6E). C/EBPα inhibited Mitf promoter-driven luciferase activity (Figure 6F). Further, when the Cebpa gene was deleted in basophils, the Mitf gene was re-expressed (Figure 6D and 6G). To determine if re-expression of MITF in basophils is sufficient to promote mast cell-specific gene expression, we overexpressed MITF in BaPs and found that overexpression of MITF alone redirected committed BaPs to differentiate into mast cells (Figure 6H). These data demonstrate that C/EBPα represses mast cell programming in committed basophils by directly suppressing Mitf gene transcription (Figure S7).
MITF Directs the Differentiation of pre-BMPs into Mast Cells and Is Required for Repressing Basophil Programming by Directly Inhibiting Cebpa Gene Transcription
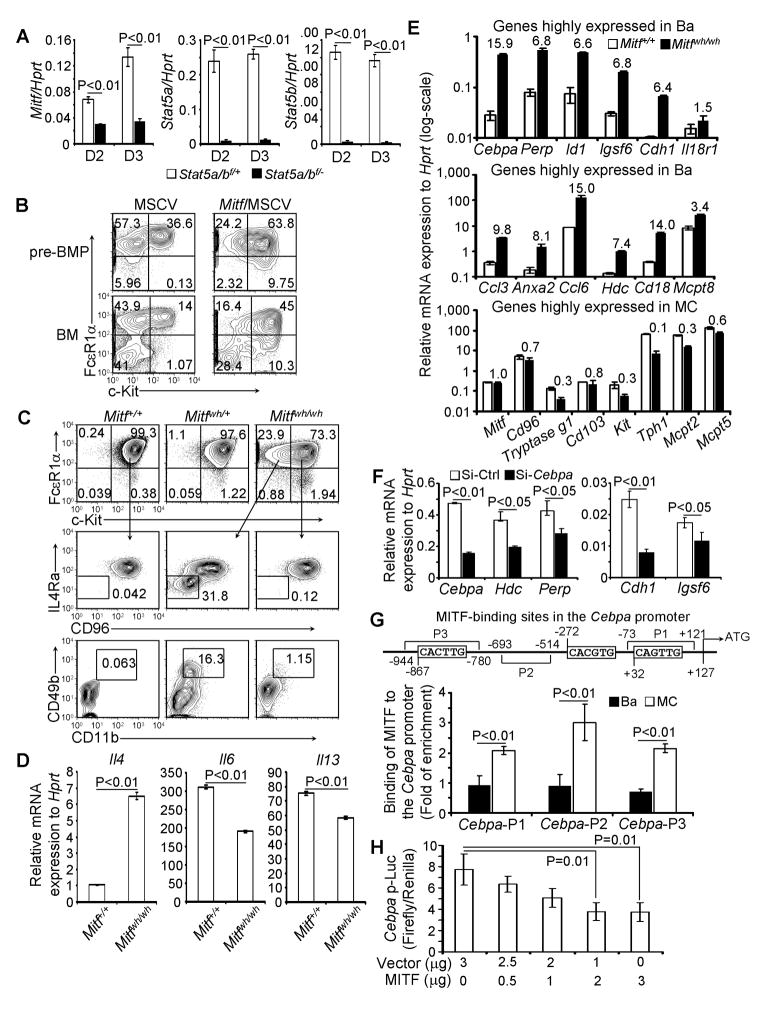
We found that Mitf is one of the downstream STAT5 genes—upregulation of Mitf was STAT5-dependent (Figure 7A). Overexpression of MITF was sufficient to drive pre-BMPs to differentiate into mast cells (Figure 7B). To test if Mitf is required for maintaining mast cell identity, we examined whether or not a Mitfmu tant would lead to the acquisition of basophil characteristics in Mitf mutant mast cells. We chose a Mitf mutant that has a mild effect on mast cell development so that we could observe any potential “mast cell to basophil” switch; specifically, we selected the Mitfwh/wh mutant. Mitfwh/wh mice harbor two copies of the Mitf gene with a single amino acid mutation at the basic domain (DNA binding domain), which results in deficient but demonstrable DNA binding on mast cell specific genes (Kim et al., 1999). We observed that Mitfwh/wh mast cells switched to “basophil-like” cells—i.e., lost c-Kit and IL-4 receptor α chain expression but gained CD49b expression—after 28 days of culture (Figure 7C). Mitfwh/wh mast cells also expressed more Il4 but less Il6 and Il13 mRNA than did WT mast cells (Figure 7D). We profiled the mRNA expression of Cebpa and other basophil-enriched genes—identified using genome wide transcription analysis—and found that Mitfwh/wh mast cells re-expressed high amounts of Cebpa mRNA and other basophil-enriched genes (Figure 7E). Mitfwh/wh mast cells did not re-express macrophage- or neutrophil-specific genes, nor did they re-express T cell, B cell or eosinophil surface markers (Figure S6), indicating that MITF represses basophil molecular programming, but not other cell-fate programming, in mast cells. Knockdown of Cebpa in Mitfwh/wh mast cells using Cebpa SiRNA inhibited the re-expression of C/EBPα-dependent genes (Figure 7F).
Figure 7. MITF Directs the Differentiation of pre-BMPs into Mast Cells and Is Required for Repressing Basophil Programming.
(A) The sorted YFP+ GMPs from poly I-C treated mice were cultured for 2 or 3 days in the presence of IL-3. Mitf and Stat5a/b mRNA were measured by qPCR (mean ± SD, triplicates). (B) pre-BMPs were infected with GFP or MITF virus. Cells were analyzed 5 days after infection. GFP+ cells are shown. (C) BM cells from Mitf wh/wh and C57BL/6 mice were cultured in complete IDMEM in the presence of IL-3 for 28 days. (D) WT and Mitf wh/wh BMMCs were stimulated with P&I for 4 hours. Il4, Il6 and Il13 mRNA was measured by qPCR (mean ± SD, triplicates). (E) mRNA expression of Cebpa and a set of basophil and mast cell genes in the cultured BMMC were analyzed by qPCR (mean ± SD, triplicates). The numbers indicate fold of difference in mRNA expression between WT and Mitfwh/wh mast cells. (F) qPCR measurements of mRNA expression in Mitfwh/wh BMMC transfected with Cebpa siRNA (mean ± SD, triplicates). (G) MITF-binding sites and ChIP analysis of MITF binding to the Cebpa promoter (mean ± SD, triplicates). (H) MITF suppressed Cebpa promoter luciferase activity (mean ± SD, triplicates). Data represent two independent results. Also see Figure S6 and S7.
To examine whether MITF directly suppresses Cebpa transcription, we searched and found three MITF binding sites (E-box) in the Cebpa promoter (Figure 7G). We showed that MITF was recruited into the Cebpa promoter in mast cells but not in basophils (Figure 7G). Co-expression of MITF significantly inhibited Cebpa promoter-driven luciferase gene transcription activity (Figure 7H), indicating that MITF represses basophil molecular programming in committed mast cells by directly suppressing the critical basophil transcription factor, C/EBPα (Figure S7).
DISCUSSION
In this study, we have identified a population of GMPs that contain highly enriched common basophil and mast cell progenitors. These cells are phenotypically distinct from BMCPs, first reported to be common basophil-mast cell progenitors, found in spleen. A recent study reported that BMCPs only give rise to mast cells (Mukai et al., 2012). Our data demonstrate that BMCPs predominantly give rise to mast cells in vitro and in vivo, whereas pre-BMPs contain highly enriched common basophil-mast cell progenitors. Upon in vitro culture, pre-BMPs fail to express β7 integrin (a marker for both BMCP and MCP). This finding does not support the conjecture that pre-BMPs are precursors for BMCPs. It is interesting that pre-BMPs retain some capacity to differentiate into myeloid cells under semi-solid culture conditions. We also note that pre-BMPs show reduced growth potential compared with conventional GMPs, suggesting that pre-BMPs are more differentiated than regular GMPs. Thus, we propose that pre-BMPs are the precursors of both BaPs and MCPs in the bone marrow. Consistent with this proposal, we note that upon infection with Schistosoma mansoni, the number of pre-BMPs increases dramatically. In parallel, the number of BaPs and MCPs also increases markedly. Although our in vitro data do not support that pre-BMPs give rise to splenic BMCPs, we cannot rule out whether pre-BMPs give rise to some splenic BMCPs in vivo. We think it is unlikely that pre-BMPs (a subset of GMPs) exist in the spleen because GMPs do not normally reside in the spleen. The precise developmental relationship among pre-BMPs, BaPs, MCPs, and BMCPs can be further defined by lineage-tracking experiments.
Recently, a novel type of innate cell that can produce a large amount of IL-13 in response to IL-25, IL-33, and parasitic infection has been identified (Moro et al., 2010; Neill et al., 2010). They can be identified by flow cytometry as Lin− Sca-1+, c-Kit+, − or lo, ST2+, IL-7R+, IL17RB+, CD25+, CD44+ (Brickshawana et al., 2011; Moro et al., 2010; Neill et al., 2010). Although innate type 2 lymphoid cells express stem-progenitor markers, they represent terminally differentiated effector cells and they do not possess progenitor activity. Another population of IL-25-elicited Lin− Sca-1+ cells in gut-associated lymphoid tissue, named MPPtype2, has been reported (Saenz et al., 2010). Pre-BMPs differ from the reported MPPtype2 cells in several aspects. First, MPPtype2 cells have been identified mainly in gut-associated lymphoid tissue, whereas pre-BMPs have been identified in the bone marrow. Second, pre-BMPs and MPPtype2 cells are phenotypically different. MPPtype2 cells contained two types of cells: one was defined as Lin− c-Kit+ Sca-1+IL-4-GFP− (Il4 gene reporter mice were used) and the other was defined as Lin− c-Kit+ Sca-1+IL-4-GFP+. Lin− c-Kit+ IL-4-GFP+ cells give rise to mast cells, whereas Lin− c-Kit+ IL-4-GFP− cells give rise to basophils, mast cells, and myeloid cells (basophil-mast cell-producing-MPP type2 cells). Pre-BMPs were identified as Lin− c-Kit+ Sca-1−FcεR1α+ IL-4-GFP+. The major phenotypical difference is that pre-BMPs are Sca-1 negative. It remains unclear whether a single MPP type2 cell can give rise to both basophils and mast cells in vitro and bulk MPP type2 cells can generate basophils and mast cells in vivo. Third, the major functional difference between pre-BMPs and basophil-mast cell-producing-MPP type2 cells is the ability of pre-BMPs to express IL-4. Finally, we found that pre-BMPs were not responsive to IL-25. We envision that one of the advantages of progenitors equipped with the capacity to produce type 2 cytokines could be that progenitors can rapidly differentiate into effectors at the site of inflammation.
How basophil versus mast cell fate is specifiedhas been a long-standing, unsolved issue. It appears that basophil cell fate and mast cell fate are mutually exclusive (e.g., under normal physiological conditions, the common basophil-mast cell progenitor differentiates into either a basophil or a mast cell but rather than a cell that displays both basophil and mast cell characteristics). Thus, we hypothesize that the master determinant for basophil cell fate must promote transcription of a set of basophil-specific genes that bestow basophil identity and function while simultaneously repressing transcription of a set of mast cell-specific genes that specify mast cell identity and function. Here, we develop an approach in which we combine prospective FACS sorting of defined progenitors with an inducible gene deletion within the defined progenitor population. This approach allows us to efficiently delete a gene of interest at a defined developmental stage. We have identified C/EBPα as the critical basophil transcription factor for specifying basophil cell fate. C/EBPα promoted a set of basophil-specific genes, while simultaneously repressing a set of mast cell-specific genes. Using a retroviral infection approach, Akashi and colleagues previously reported that C/EBPα was critical for the differentiation of GMPs into basophils (Iwasaki et al., 2006). However, they did not observe that C/EBPα was required for basophil maintenance (Arinobu et al., 2005). This could be due to the difficulty of deleting the Cebpa gene at a precise developmental stage using retrovirus.
We have also identified MITF as a crucial transcription factor for specifying mast cell fate and promoting a set of mast cell-specific genes, while simultaneously repressing a set of basophil-specific genes. Kitamura et. al. used various mutants at the Mitf locus to demonstrate the role of MITF in mast cell development. They have shown that MITF is required for the expression of many mast cell-specific genes, including cKit (Kitamura et al., 2002). We report here that MITF also suppresses many basophil-specific genes while having no effect on genes that govern T cell, B cell, eosinophil, neutrophil or macrophage development. Taken together, we have demonstrated that MITF is the key mast cell transcription factor.
Our experimental data also provide an explanation regarding how basophil cell fate and mast cell fate are mutually exclusive. We demonstrated that C/EBPα and MITF silence each other’s transcription in a directly antagonistic fashion by binding to the other’s promoter, thus suppressing its respective promoter activity. It is interesting to note that the molecular programing of basophil and mast cell is so closely intertwined that in the absence of C/EBPα or MITF, basophils could only re-express mast cell-specific genes or mast cells could only re-express basophil-specific genes, respectively, but not genes that govern other cell fates. This finding suggests that the suppression of other cell fate molecular programming might be mediated by transcriptional repressors that are commonly expressed in both basophils and mast cells. A shared molecular signature and a restricted, bidirectional convertibility between basophil and mast cell also strongly support that basophils and mast cells are progenies from a common progenitor. A better understanding of how C/EBPα and MITF specify basophil versus mast cell fate through direct mutual repression provides a model system to facilitate our understanding of cell fate determination in other cell types.
EXPERIMENTAL PROCEDURES
Mice
Mouse strains and sources are listed in the Supplemental Experimental Procedures. All animal experiments were approved by the National Jewish Health Institutional Animal Care and Use Committee.
FACS Analysis and Sorting
Cells prepared from various tissues or cell cultures were stained with fluorochrome-labeled antibodies. Stained cells were acquired by CyAN (DakoCytomation, Glostrup, Denmark) and analyzed using FlowJo software (Tree Star, Ashland, OR). Cell sorting was carried out using a Moflo machine (DakoCytomation). A more detailed description of cell surface markers used to define various progenitors is included in the Supplemental Experimental Procedures.
In Vitro Differentiation of Progenitors
Various progenitors were isolated by FACS sorting and seeded in semi-solid medium supplemented with IL-3 (20 ng/ml) for 7–9 days or in complete IDMEM in the presence of IL-3 (20 ng/ml) or IL-3 in combination with IL-6 (20 ng/ml), IL-9 (40 ng/ml), and SCF (50 ng/ml) for indicated periods of time. A more detailed procedure for semi-solid culture has been described in the Supplemental Experimental Procedures.
Generation of Chimeric Mice
For in vivo analysis of donor-derived basophils, FACS-sorted CD45.2+ progenitors were injected into lethally irradiated CD45.1 congenic recipient mice intravenously. Three days later, the bone marrow cells of the reconstituted mice were analyzed by FACS. For the in vivo analysis of donor-derived mast cells, FACS-sorted progenitors were injected into lethally irradiated KitW–sh/W–sh mice (Chen et al., 2005). Five to six weeks later, peritoneal cells and spleens of the reconstituted mice were analyzed.
Statistical Analysis
All of the error bars in this report represent SDs. For ELISA or qPCR analyses, Means ± SDs were derived from triplicate measurements of one experiment. Pooled data are indicated in the figure legends. The difference between two samples was analyzed with Student’s t-test.
Supplementary Material
Acknowledgments
We thank R. Moriggl for providing the plasmid of cS5F; D. Fisher of Harvard University for providing bone marrow cells of Mitfwh/wh mice; T. Ludwig of Columbia University for providing a breeding pair of RosacreERT2 mice; H. Goettle, D. Bohrer-Kunter, and J. D. Williams for technical assistance; H. Lei for the help of setting up the ChIP assay; M. Leyden for animal husbandry; S. Sobus and J. Looms for FACS analysis and cell sorting; H. Chu for assisting with microscopy; A. Gerber for sharing Luminomiter; and L. Lenz and W. Born for critical reading the manuscript. This study was supported by National Institutes of Health Grants RO1AI083986 (H. H.), RO1 AI05917 (H. H.), American Recovery and Reinvestment Act 3R01AI068083-04S1 (H. H.), R01 AI079087 (D. W.), PO1 HL44612 (D. W.), K08HL105536 (B.G) and Parker B. Francis Fellowship (B.G.).
Abbreviations used in this paper
- HSCs
hematopoietic stem cells
- MPPs
multiple potential progenitors
- CMPs
common myeloid progenitors
- GMPs
granulocyte-monocyte progenitors
- BaPs
basophil progenitors
- and pre-BMPs
pre-basophil and mast cell progenitors
Footnotes
ACCESSION NUMBERS
The microarray data have been deposited in the Gene Expression Omnibus(GEO) database under the accession number GSE41596.
Supplemental information includes seven figures, two tables, and Supplemental Experimental Procedures and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arinobu Y, Iwasaki H, Akashi K. Origin of basophils and mast cells. Allergol Int. 2009;58:21–28. doi: 10.2332/allergolint.08-RAI-0067. [DOI] [PubMed] [Google Scholar]
- Arinobu Y, Iwasaki H, Gurish MF, Mizuno S, Shigematsu H, Ozawa H, Tenen DG, Austen KF, Akashi K. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci U S A. 2005;102:18105–18110. doi: 10.1073/pnas.0509148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickshawana A, Shapiro VS, Kita H, Pease LR. Lineage Sca1+c-Kit CD25+ Cells Are IL-33–Responsive Type 2 Innate Cells in the Mouse Bone Marrow. The Journal of Immunology. 2011;187:5795–5804. doi: 10.4049/jimmunol.1102242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse WW, Morgan WJ, Gergen PJ, Mitchell HE, Gern JE, Liu AH, Gruchalla RS, Kattan M, Teach SJ, Pongracic JA, et al. Randomized trial of omalizumab (anti-IgE) for asthma in inner-city children. N Engl J Med. 2011;364:1005–1015. doi: 10.1056/NEJMoa1009705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Grimbaldeston MA, Tsai M, Weissman IL, Galli SJ. Identification of mast cell progenitors in adult mice. Proc Natl Acad Sci U S A. 2005;102:11408–11413. doi: 10.1073/pnas.0504197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, Simon MC. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4:1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- Denburg JA, Telizyn S, Messner H, Lim B, Jamal N, Ackerman SJ, Gleich GJ, Bienenstock J. Heterogeneity of human peripheral blood eosinophil-type colonies: evidence for a common basophil-eosinophil progenitor. Blood. 1985;66:312–318. [PubMed] [Google Scholar]
- Galli SJ, Franco CB. Basophils are back! Immunity. 2008;28:495–497. doi: 10.1016/j.immuni.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Galli SJ, Tsai M, Piliponsky AM. The development of allergic inflammation. Nature. 2008;454:445–454. doi: 10.1038/nature07204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holgate S, Casale T, Wenzel S, Bousquet J, Deniz Y, Reisner C. The anti-inflammatory effects of omalizumab confirm the central role of IgE in allergic inflammation. The Journal of allergy and clinical immunology. 2005;115:459–465. doi: 10.1016/j.jaci.2004.11.053. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, Gurish MF, Takatsu K, Akashi K. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J Exp Med. 2005;201:1891–1897. doi: 10.1084/jem.20050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasuyama H, Mukai K, Obata K, Tsujimura Y, Wada T. Nonredundant roles of basophils in immunity. Annu Rev Immunol. 2011;29:45–69. doi: 10.1146/annurev-immunol-031210-101257. [DOI] [PubMed] [Google Scholar]
- Kim DK, Morii E, Ogihara H, Lee YM, Jippo T, Adachi S, Maeyama K, Kim HM, Kitamura Y. Different Effect of Various Mutant MITF Encoded by mi, Mior, or Miwh Allele on Phenotype of Murine Mast Cells. Blood. 1999;93:4179–4186. [PubMed] [Google Scholar]
- Kitamura Y, Morii E, Jippo T, Ito A. Regulation of mast cell phenotype by MITF. Int Arch Allergy Immunol. 2002;127:106–109. doi: 10.1159/000048178. [DOI] [PubMed] [Google Scholar]
- Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA, Weissman IL. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- Laslo P, Pongubala JM, Lancki DW, Singh H. Gene regulatory networks directing myeloid and lymphoid cell fates within the immune system. Seminars in immunology. 2008;20:228–235. doi: 10.1016/j.smim.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Leary AG, Ogawa M. Identification of pure and mixed basophil colonies in culture of human peripheral blood and marrow cells. Blood. 1984;64:78–83. [PubMed] [Google Scholar]
- Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- Marone G, Galli SJ, Kitamura Y. Probing the roles of mast cells and basophils in natural and acquired immunity, physiology and disease. Trends in immunology. 2002;23:425–427. doi: 10.1016/s1471-4906(02)02274-3. [DOI] [PubMed] [Google Scholar]
- Migliaccio AR, Rana RA, Sanchez M, Lorenzini R, Centurione L, Bianchi L, Vannucchi AM, Migliaccio G, Orkin SH. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1 low mouse mutant. The Journal of experimental medicine. 2003;197:281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriggl R, Sexl V, Kenner L, Duntsch C, Stangl K, Gingras S, Hoffmeyer A, Bauer A, Piekorz R, Wang D, et al. Stat5 tetramer formation is associated with leukemogenesis. Cancer Cell. 2005;7:87–99. doi: 10.1016/j.ccr.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J-i, Ohtani M, Fujii H, Koyasu S. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- Mukai K, Benbarak MJ, Tachibana M, Nishida K, Karasuyama H, Taniuchi I, Galli SJ. Critical role of P1-Runx1 in mouse basophil development. Blood. 2012;120:76–85. doi: 10.1182/blood-2011-12-399113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TKA, Bucks C, Kane CM, Fallon PG, Pannell R, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori K, Luo Y, Jia Y, Nishida J, Wang Z, Bunting KD, Wang D, Huang H. IL-3 induces basophil expansion in vivo by directing granulocyte-monocyte progenitors to differentiate into basophil lineage-restricted progenitors in the bone marrow and by increasing the number of basophil/mast cell progenitors in the spleen. J Immunol. 2009;182:2835–2841. doi: 10.4049/jimmunol.0802870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht C, Voehringer D. Basophil effector function and homeostasis during helminth infection. Blood. 2009;113:2816–2825. doi: 10.1182/blood-2008-05-154773. [DOI] [PubMed] [Google Scholar]
- Qi X, Nishida J, Chaves L, Ohmori K, Huang H. CCAAT/enhancer-binding protein alpha (C/EBPalpha) is critical for interleukin-4 expression in response to FcepsilonRI receptor cross-linking. J Biol Chem. 2011;286:16063–16073. doi: 10.1074/jbc.M110.213389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz SA, Siracusa MC, Perrigoue JG, Spencer SP, Urban JF, Jr, Tocker JE, Budelsky AL, Kleinschek MA, Kastelein RA, Kambayashi T, et al. IL25 elicits a multipotent progenitor cell population that promotes TH2 cytokine responses. Nature. 2010;464:1362–1366. doi: 10.1038/nature08901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne CP, McCoy ME, Piekorz R, Sexl V, Roh KH, Jacobs-Helber SM, Gillespie SR, Bailey DP, Mirmonsef P, Mann MN, et al. Stat5 expression is critical for mast cell development and survival. Blood. 2003;102:1290–1297. doi: 10.1182/blood-2002-11-3490. [DOI] [PubMed] [Google Scholar]
- Takemoto CM, Lee YN, Jegga AG, Zablocki D, Brandal S, Shahlaee A, Huang S, Ye Y, Gowrisankar S, Huynh J, McDevitt MA. Mast cell transcriptional networks. Blood Cells, Molecules, and Diseases. 2008;41:82–90. doi: 10.1016/j.bcmd.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, Shigematsu H, Levantini E, Huettner CS, Lekstrom-Himes JA, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.