Abstract
Circadian rhythms have evolved to anticipate metabolic needs across the 24-hour light/dark cycle. This is accomplished by circadian expression of metabolic genes orchestrated by transcription factors through chromatin remodeling and histone modifications. Our recent genome-wide study on histone deacetylase 3 (HDAC3) in mouse liver provides novel insights into the molecular link between circadian rhythm and hepatic de novo lipogenesis. We found that liver-specific knockout of HDAC3 in adult mouse display severe hepatic steatosis associated with enhanced de novo lipogenesis and increased expression of lipogenic genes. Genome-wide analysis (ChIP-seq) revealed a pronounced circadian pattern of HDAC3 occupancy on genes involved in lipid metabolism, which is inversely related to histone acetylation and RNA polymerase II recruitment at these sites. The cistromes of HDAC3 and its binding partner, nuclear receptor co-repressor (NCoR), significantly overlap with that of Rev-erbα, a nuclear receptor directly involved in the core circadian machinery. Knockout of Rev-erbα in mouse also leads to hepatic steatosis and enhanced de novo lipogenesis. Collectively, these data suggest that the circadian epigenomic remodeling controlled by HDAC3, and largely directed by Rev-erbα, is essential for homeostasis of the lipogenic process in liver.
INTRODUCTION
Living organisms on Earth have a circadian clock built in by evolution through millions of years of living in a light/dark cycle of 24 hours, which allows anticipation of metabolic needs in a feedforward manner, rather than mere response to environment in a feedback way (Arble et al. 2010; Bass and Takahashi 2010; Huang et al. 2011). This is accomplished by coordinating circadian expression of genes involved in metabolism. Transcriptome profiling revealed that about 2–15% of all expressed genes are circadian, most of which participate in energy and xenobiotics metabolism, highlighting the inherent link between circadian rhythm and metabolism (Panda et al. 2002; Hughes et al. 2009; Asher and Schibler 2011).
The circadian expression of metabolic genes is dictated directly or indirectly by the core circadian clock machinery composed of several transcription factors and cofactors that form autoregulatory negative feedback loops through both transcriptional and posttranslational mechanisms (Green et al. 2008). In mammals, there is a central circadian clock residing in the central nervous system and multiple peripheral circadian clocks in peripheral tissues such as liver, heart, lung, adipose and pancreas. Although composed of similar molecular components, the central clock is mainly entrained by light, while the peripheral clocks receive systemic signals independently from both the central clock and other inputs such as feeding behavior (Green et al. 2008). Most rhythmic genes in liver rely on the liver peripheral clock rather than the central clock for the establishment of their circadian rhythm (Kornmann et al. 2007).
Disruption of circadian rhythm causes metabolic derangement, a notion supported by several lines of evidence from studies on both humans and animals. Epidemiology studies have found higher incidence of metabolic derangements such as obesity, diabetes and cardiovascular diseases in shift workers (De Bacquer et al. 2009; Scheer et al. 2009; Pietroiusti et al. 2010). Likewise, daytime feeding of nocturnal animals or simulation of shift work results in metabolic disorders (Arble et al. 2010; Salgado-Delgado et al. 2010). Genome-wide association studies (GWAS) have found association between metabolic derangement and genes Bmal1 and Clock that constitute the core circadian clock machinery). Animals bearing mutations or deletions on these genes also display abnormalities in carbohydrate and lipid metabolism (Turek et al. 2005; Lamia et al. 2008; Yang et al. 2009; Zhang et al. 2010; Marcheva et al. 2010).
Transcriptional regulation plays a central role in circadian rhythm, as most circadian genes characterized so far show circadian rhythm at the transcriptional level and the core circadian clock machinery is mainly composed of transcription activators and repressors. Transcriptional activation and repression is a complex biological process involving extensive epigenomic remodeling that leads to chromatin relaxation or condensation so that genomic DNA can be accessed by or restrained from the general transcription machinery. Covalent modifications on chromatin histones at different sites through phosphorylation, acetylation and methylation either alone or in combination, guide such remodeling processes in a language-like manner termed “histone code” (Strahl and Allis 2000; Berger 2007; Lee et al. 2010).
Histone acetylation at the promoter regions of circadian genes were found to fluctuate during the light/dark cycle in parallel with rhythmic expression of these genes, suggesting a role of histone acetylation in regulating the circadian rhythm (Crosio et al. 2000; Etchegaray et al. 2003; Curtis et al. 2004; Yoshihisa Naruse et al. 2004). Histone acetylation levels are controlled by the opposing actions of histone acetyltransferases (HATs) and histone deacetylases (HDACs). HATs and HDACs usually function as part of large multiprotein coactivator and corepressor complexes, and are recruited to active and silent genes respectively by sequence-specific DNA binding proteins. Clock, the founding member of the molecular clock machinery, was shown to have intrinsic HAT activity (Doi et al. 2006). And SIRT1, a class III HDAC, was found to interact with Clock and regulate and the amplitude of the circadian rhythm (Nakahata et al. 2008). Previous work from our lab has suggested that HDAC3 plays an important role in the maintenance of normal circadian rhythm in peripheral tissues (Alenghat et al. 2006).
HDAC3, AN EPIGENOMIC MODIFIER THAT KEEPS HEPATIC LIPOGENESIS IN CHECK
HDAC3 is the third HDAC identified in mammals by sequence homology to previously identified HDAC1 and HDAC2 (Yang et al. 1997). Biochemical purification studies by our laboratory and others have established that HDAC3 exists in transcription co-repressor complexes containing two homologous proteins: nuclear receptor co-repressor (NCoR) and silencing mediator for retinoid and thyroid receptors (SMRT) (Guenther et al. 2000; Li et al. 2000; Wen et al. 2000). As their names suggest, NCoR and SMRT are recruited to the chromatin by several nuclear hormone receptors and are responsible for transcriptional repression of these nuclear receptors under unliganded conditions. HDAC3 is the primary deacetylase in the NCoR/SMRT complex (Fischle et al. 2002). Moreover, enzymatic activity of HDAC3 is not fully activated until it binds to the deacetylase activating domain (DAD) of NCoR or SMRT (Guenther et al. 2001). Mice bearing a point mutation in NCoR DAD domain (N-DADm) that disrupts NCoR-HDAC3 interaction exhibit abnormal energy expenditure and a phase shift in the circadian expression of several metabolic genes in peripheral tissues, thus accentuating the important function of HDAC3 in metabolism and circadian rhythm (Alenghat et al. 2006).
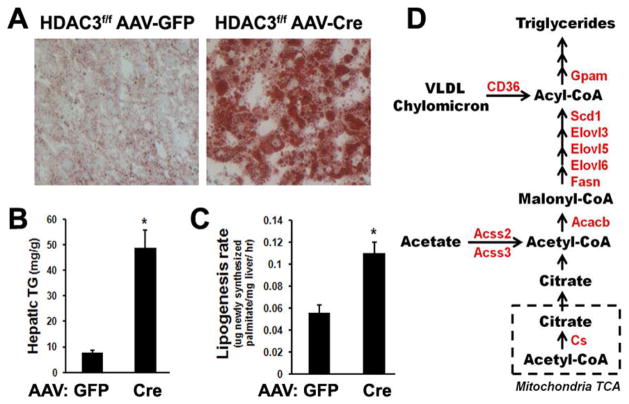
To further characterize tissue-specific functions of HDAC3, we have generated C57BL/6J mice with HDAC3 exon 4 to 7 flanked by loxP sites (HDAC3f/f mice). Liver-specific deletion of HDAC3 in adult mice was accomplished by intravenous injection of recombinant adenovirus-associated virus expressing Cre recombinase under a hepatocyte-specific thyroxin-binding globulin (TBG) promoter. Efficient liver-specific deletion of HDAC3 was confirmed at 1-week post-injection (Feng et al. 2011). At 2-weeks post-injection, the liver was significantly enlarged with massive accumulation of neutral lipid (Fig. 1A). Hepatic triglyceride content was increased 2–3 fold at 1-week post-injection and 5–10 fold at 2-weeks post-injection (Fig. 1B and data not shown).
Figure 1.
HDAC3 suppresses hepatic lipogenesis. (A) Oil Red O staining of neutral lipid in liver from 12-week-old HDAC3f/f mice 2weeks after intravenous injection of adenovirus-associated virus (AAV) expressing either green fluorescence protein (GFP) or Cre recombinase under a hepatocyte-specific thyroxin-binding globulin (TBG) promoter. (B) Hepatic triglyceride (TG) levels were measured by enzymatic colorimetric assay. Error bars indicate S.E.M. * P < 0.05. (C) Hepatic de novo lipogenesis (DNL) rate was measured in 12-week-old HDAC3f/f mice 1 week after injection with AAV-Cre in the light cycle, by tracing synthesis of 2H-labeled hepatic palmitate from 2H2O injected intraperitoneally. Error bars indicate S.E.M. * P < 0.05. (D) Transcription of genes (in red) involved in fatty acid synthesis was upregulated in the light cycle in liver from HDAC3f/f mice at 1 week after AAV-cre injection. Cs, citrate synthase; Acacb, acetyl-CoA carboxylase 2; Fasn, fatty acid synthase; Elovl, fatty acid elongase; Scd1, stearoyl-CoA desaturase 1; Gpam, glycerol-3-phosphate acyltransferase 1, mitochondrial; Acss, acyl-CoA synthetase; CD36, fatty acid translocase.
Enhanced fatty acid synthesis is associated with hepatic steatosis (Browning and Horton 2004). To test whether HDAC3 deletion enhances de novo lipogenesis, deuterated water was used as a tracer and newly synthesized 2H-labeled hepatic palmitate in HDAC3f/f mice at 1-week after AAV injection was analyzed by gas chromatography-electron impact ionization mass spectrometry (GC/MS). Hepatic de novo lipogenesis rate was significantly increased when HDAC3 was deleted (Fig. 1C), suggesting that HDAC3 suppressed lipogenesis. Transcriptome analysis by microarray showed that many lipid metabolic genes are upregulated in liver in the absence of HDAC3. Among the most prominently upregulated genes are those encoding key enzymes in lipogenesis such as fatty acid elongases (Elovls), stearoyl-CoA desaturase 1 (Scd1), fatty acid synthase (Fasn), acetyl-CoA carboxylase beta (Acacb), and glycerol-3-phosphate acyltransferase 1, mitochondrial (Gpam) (Fig. 1D). These findings are consistent with a previous report on development of fatty liver in mice when hepatic HDAC3 is deleted in utero (Knutson et al. 2008). Thus, HDAC3 is an epigenomic modifier that keeps hepatic lipogenesis in check under normal physiological conditions.
CHROMATIN OCCUPANCY OF HDAC3 IS CIRCADIAN AND ANTI-PHASE TO HEPATIC LIPOGENESIS
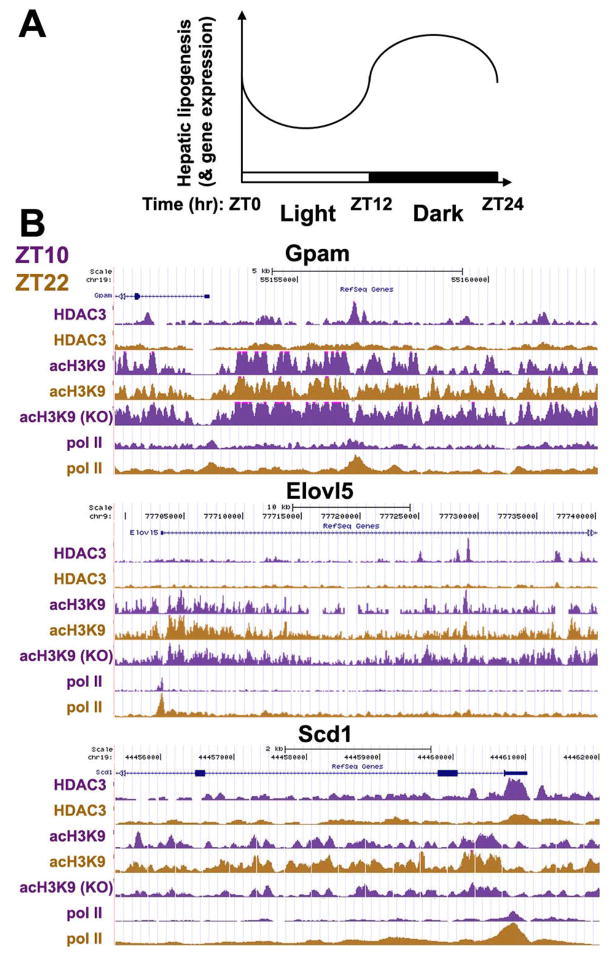
Hepatic lipogenesis is enhanced during feeding to allow conversion of excess nutrient metabolites to lipid for secretion from the liver and storage at the adipose tissue. Conversely, hepatic lipogenesis is suppressed during fasting when there are neither substrates nor metabolic needs for such processes. In nocturnal animals, feeding happens at night and fasting occurs during the day. Therefore hepatic lipogenesis shall presumably have a circadian pattern that is active at night and quiescent during the day. Indeed, metabolic flux tracing studies have clearly showed such pattern of circadian rhythm in hepatic lipogenesis in mice and rats, which is illustrated in Fig. 2A (Kimura et al. 1970; Hems et al. 1975; Cornish and Cawthorne 1978; Cincotta and Meier 1984).
Figure 2.
Chromatin occupancy of HDAC3 is circadian in anti-phase to hepatic lipogenesis. (A) Hepatic lipogenesis and expression of lipogenic genes is circadian with a zenith in the night cycle and a nadir in the light cycle in nocturnal animals such as mouse and rat. ZT: Zeitgeber time that marks the light/dark cycle -- light on at ZT0, light off at ZT12. (B) Circadian rhythm of hepatic HDAC3 chromatin occupancy on lipogenic genes is inversely associated with rhythmic acetylation at lysine 9 on histone 3 (acH3K9) and RNA polymerase II (Pol II) recruitment. Deletion of HDAC3 causes constitutive histone acetylation. Occupancy of HDAC3, acH3K9, and Pol II on the genome was determined by ChIP-seq and viewed by the UCSC genome browser. The y-axis indicates relative signal intensities that were normalized to reads per million (rpm) and put into the same scale between ZT10 and ZT22 for the same factor for easy comparison. Gpam, glycerol-3-phosphate acyltransferase 1, mitochondrial; Elovl5, fatty acid elongase 5; Scd1, stearoyl-CoA desaturase 1.
More interestingly, and less obviously, transcription of hepatic lipogenic genes such as multiple Elovls, Gpam, Fasn, acetyl-Coenzyme A carboxylase α (Acaca), and ATP-citrate lyase also display the same pattern of circadian rhythm, as illustrated in Fig. 2A (Zardoya et al. 1994; Panda et al. 2002; Brolinson et al. 2008; Hughes et al. 2009). This seems to suggest that the body has the ability to anticipate metabolic need by coordinately upregulating lipogenic enzymes during peak feeding times when they are most needed, while suppressing them during fasting period when they are not needed.
What is the molecular switch underlying this circadian rhythm? Given the ability of HDAC3 to both modify the epigenome and suppress hepatic lipogenesis, we set out to test if HDAC3 occupancy on the genome is circadian. Chromatin immunoprecipitation with an HDAC3-specific antibody was performed in mouse liver during the day at ZT10 (“Zeitgeber time” marks the light/dark cycle; light on at ZT0, light off at ZT12) and at night at ZT22, followed by massively parallel DNA sequencing (ChIP-seq). HDAC3 displays a prominent rhythmic occupancy on the genome with a zenith during the day and a nadir at night, as evidenced by both total number of binding sites (14578 sites during the day versus 120 sites at night) and average signal intensity at the center of the sites (1.6 signal per million total reads [RPM] during the day versus 0.4 RPM at night) (Feng et al. 2011). HDAC3 binding was enriched on genes involved in lipid metabolism, especially fatty acid synthesis, such as Gpam, Scd1 and Elovl5 (Fig. 2B). The circadian occupancy of HDAC3 is inversely associated with the genome-wide histone acetylation and RNA polymerase II recruitment at the same sites, suggesting that HDAC3 orchestrate a circadian epigenomic remodeling that leads to transcriptional repression of hepatic lipogenic genes during the day but allows transcriptional activation of these genes at night (Fig. 2B). Loss of this circadian rhythm results in constitutively high levels of expression of lipogenic genes that promote fatty acid synthesis and thus the hepatic steatosis found in HDAC3-liver specific knockout mice.
REV-ERBα REGULATES CIRCADIAN RECRUITMENT OF HDAC3 TO THE GENOME
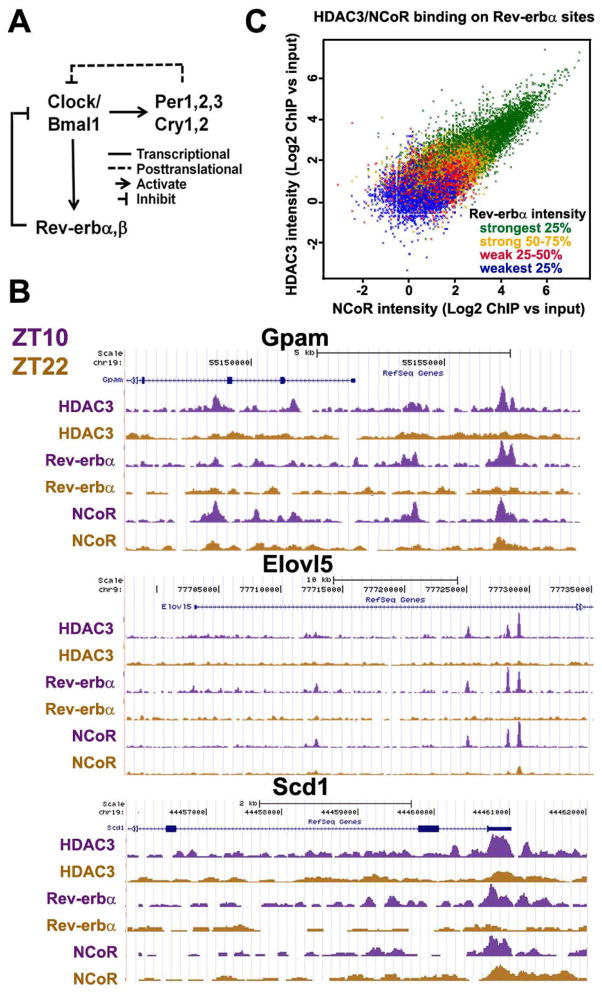
What is the DNA-sequence specific transcription factor(s) that dictate circadian recruitment of HDAC3 to the genome? Motif analysis showed that the most enriched motif among all HDAC3 binding sites is AGGTCA, a sequence known to be bound by many nuclear hormone receptors (Yang et al. 2007). Among all nuclear receptors, Rev-erbα and β are core members of the circadian clock machinery (Fig. 3A). Rev-erbα and β are direct target genes of Clock/Bmal1 and can in turn repress transcription of Clock/Bmal1. Rev-erb-mediated transcriptional repression of Clock/Bmal1 and Per/Cry-mediated posttranslational inhibition of Clock/Bmal1 are the two negative feedback loops within the core circadian clock machinery (Fig. 3A) (Preitner et al. 2002; Asher and Schibler 2011). Rev-erb proteins exhibit prominent circadian expression in mouse liver that is in-phase with genomic recruitment of HDAC3 (Balsalobre et al. 1998; Preitner et al. 2002).
Figure 3.
Rev-erbα controls circadian recruitment of HDAC3 to the genome. (A) The core molecular circadian clock machinery. Clock/Bmal1 heterodimer transcriptionally activate Period (Per 1, 2, 3), Cryptochrome (Cry 1, 2), and Rev-erbα&β. Per and Cry interferes with the activity of Clock/Bmal1 proteins, while Rev-erb represses transcription of Clock/Bmal1 genes. (B) Circadian occupancy of HDAC3, NCoR, and Rev-erbα on hepatic lipogenic genes as viewed by the UCSC genome browser. Gpam, glycerol-3-phosphate acyltransferase 1, mitochondrial; Elovl5, fatty acid elongase 5; Scd1, stearoyl-CoA desaturase 1. (C) For all sites occupied by Rev-erbα at ZT10 in mouse liver, ChIP-seq signal intensities for Rev-erbα, NCoR, and HDAC3 were normalized to reads per million (rpm) and computed as log2 of the ratio of ChIP rpm to input rpm. To analyze the 3-way correlation of binding affinities of the 3 factors, scatter plot with HDAC3 and NCoR signal intensities was color-coded by the quartile of signal intensity for Rev-erbα.
Previous work from our laboratory has revealed that Rev-erbα recruits NCoR/HDAC3 to the Bmal1 promoter in cultured liver cells and that HDAC3 is required for transcriptional repression of Bmal1 by Rev-erbα (Yin and Lazar 2005). It is perceivable that Rev-erbα may be responsible for the circadian chromatin recruitment of HDAC3 in liver on a genome-wide scale. To test this hypothesis, ChIP-seq experiments were performed with antibodies specific for Rev-erbα and NCoR in mouse liver at ZT10 and ZT22. As expected, there is a robust circadian rhythm in genome occupancy of Rev-erbα and NCoR that is in phase with HDAC3 binding (Fig. 3B). The genomic binding sites of Rev-erbα significantly overlap with those of HDAC3 and NCoR, especially on genes involved in fatty acid synthesis (Fig. 3B). More importantly, a close correlation exists between signal intensities of Rev-erbα binding and those of NCoR/HDAC3 at the same sites (Fig. 3C), and the circadian HDAC3 binding at several sites are lost in Rev-erbα null mice (Feng et al. 2011). Taken together, these data strongly support the notion that Rev-erbα recruits NCoR and HDAC3 to the genome and thus accounts for the circadian rhythmicity of their genomic occupancy.
If the circadian rhythm of HDAC3 occupancy on the genome is critical for keeping hepatic lipogenesis in check in the light cycle, loss of this rhythm in Rev-erbα null mice would promote hepatic de novo lipogenesis and precipitate hepatic steatosis in a similar manner as was found in the HDAC3 deficient liver. Indeed, Rev-erbα null mice display elevated intracellular lipid content in liver (Fig. 4A) and the hepatic triglyceride levels are nearly two fold higher than control wild-type mice (Fig. 4B). Isotope tracing studies showed that hepatic de novo lipogenesis is elevated in the light cycle in Rev-erbα null mice (Feng et al. 2011), indicating that the circadian epigenomic remodeling events dictated by Rev-erbα/NCoR/HDAC3 is important in suppression of hepatic lipogenesis in the light cycle.
Figure 4.
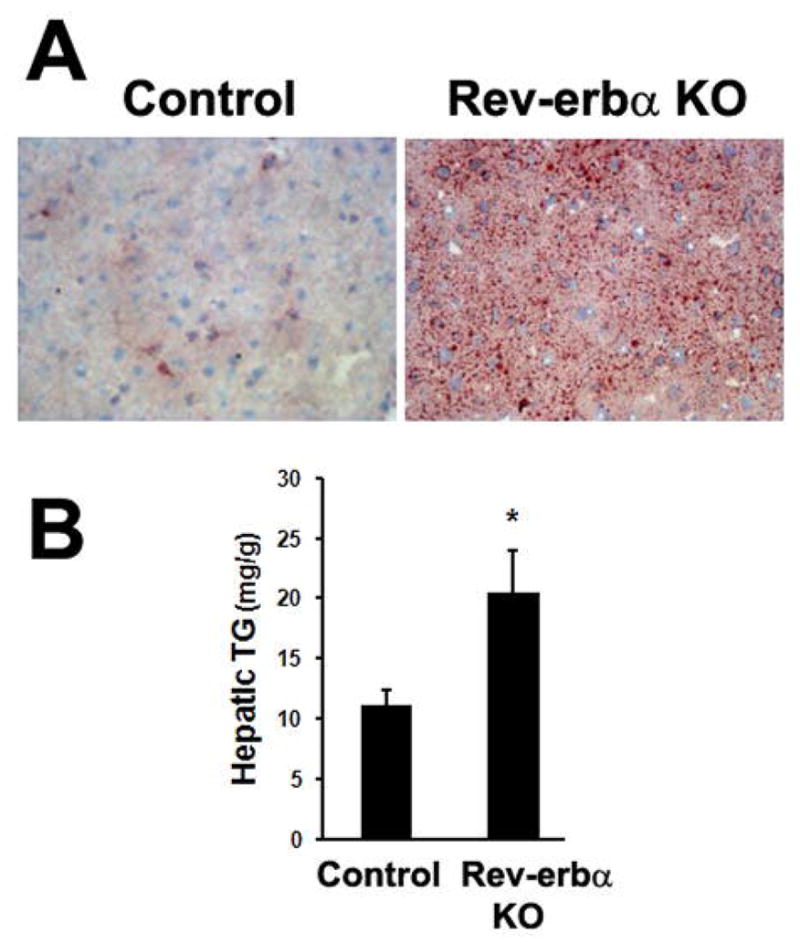
Loss of Rev-erbα-mediated circadian rhythm enhanced hepatic lipogenesis. (A) Oil Red O staining of neutral lipid in livers from 9-week-old Rev-erbα knockout (KO) mice and control wild-type mice. (B) Hepatic triglyceride (TG) levels were measured by enzymatic colorimetric assay. Error bars indicate S.E.M. * P < 0.05.
CONCLUSIONS AND PERSPECTIVES
In mammals, there is a circadian rhythm of hepatic lipogenesis activity that increases in the feeding/active phase and decreases in the fasting/sleep phase. The ability to anticipate metabolic needs are accomplished by coordinated upregulation of hepatic lipogenic genes in the dark cycle and downregulation in the light cycle in nocturnal animals. Here we show that circadian expression of hepatic lipogenic genes is orchestrated by HDAC3 through rhythmic epigenomic remodeling and histone deacetylation. Rev-erbα, a core component of the circadian clock, is largely responsible for circadian recruitment of HDAC3 and NCoR to the genome. Loss of the circadian epigenomic remodeling in either HDAC3 knockout or Rev-erbα knockout mouse liver results in abnormally elevated hepatic de novo lipogenesis in the light cycle that leads to hepatic steatosis. Our findings provide possible molecular explanation of the higher incidence of lipid metabolism derangements observed in shifting workers (De Bacquer et al. 2009; Esquirol et al. 2009; Pietroiusti et al. 2010) and the association between fatty liver diseases and genetic variations in the Clock gene in humans (Sookoian et al. 2007; Scott et al. 2008).
Deletion of HDAC3 from mouse liver results in 5–10-fold increase of hepatic triglyceride content, while knockout of Rev-erbα results in only a 2-fold increase. The relatively modest steatosis in Rev-erbα knockout mice implies that other transcription factor(s), such as Rev-erbβ, may contribute to chromatin recruitment of HDAC3 to repress expression of lipogenic genes. It is also possible that long-term whole-body knockout of Rev-erbα may foster compensatory responses that ameliorate the hepatic steatosis phenotype. Recent genome-wide studies have revealed that different transcription factors have unexpectedly high frequency of chromatin co-occupancy, suggesting the existence of extensive cooperative crosstalk between transcription factors in epigenomic remodeling (Lefterova et al. 2008; Nielsen et al. 2008; MacArthur et al. 2009; He et al. 2011; Siersbæk et al. 2011). It will be of great interest to understand how Rev-erbα/NCoR/HDAC3-mediated epigenomic remodeling can influence the activity of other transcription factors involved in lipogenesis such as the sterol regulatory element-binding proteins (SREBPs) and peroxisome proliferator-activated receptor γ (PPARγ).
Many small molecules that can manipulate activities of both HATs and HDACs are being developed as potential “epigenetic drugs” and some have shown promise in clinical studies in treatment of various diseases such as cancer (Carafa et al. 2011; Rekowski and Giannis 2010). The present study suggests potential side effects of these small molecules due to disruption of circadian rhythm and derangement of metabolism. In line with this notion, HDAC inhibitors have been shown to alter expression of circadian genes (Repouskou et al. 2010; Sanchis-Segura et al. 2009). On the other hand, these epigenetic drugs could also be useful in manipulating the circadian rhythm in a beneficial way (Perreau-Lenz et al. 2007).
Acknowledgments
Work is supported by the Cox Institute for Medical Research, NIH DK45586, DK43806, and RC1DK08623 (to M.A.L.)
References
- Alenghat T, Yu J, Lazar MA. The N-CoR complex enables chromatin remodeler SNF2H to enhance repression by thyroid hormone receptor. EMBO J. 2006;25:3966–3974. doi: 10.1038/sj.emboj.7601280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arble DM, Ramsey KM, Bass J, Turek FW. Circadian disruption and metabolic disease: findings from animal models. Best Pract Res Clin Endocrinol Metab. 2010;24:785–800. doi: 10.1016/j.beem.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- De Bacquer D, Van Risseghem M, Clays E, Kittel F, De Backer G, Braeckman L. Rotating shift work and the metabolic syndrome: a prospective study. Int J Epidemiol. 2009;38:848–854. doi: 10.1093/ije/dyn360. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Brolinson A, Fourcade S, Jakobsson A, Pujol A, Jacobsson A. Steroid hormones control circadian Elovl3 expression in mouse liver. Endocrinology. 2008;149:3158–3166. doi: 10.1210/en.2007-1402. [DOI] [PubMed] [Google Scholar]
- Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114:147–152. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafa V, Nebbioso A, Altucci L. Histone deacetylase inhibitors: recent insights from basic to clinical knowledge & patenting of anti-cancer actions. Recent Pat Anticancer Drug Discov. 2011;6:131–145. doi: 10.2174/157489211793980088. [DOI] [PubMed] [Google Scholar]
- Cincotta AH, Meier AH. Circadian rhythms of lipogenic and hypoglycaemic responses to insulin in the golden hamster (Mesocricetus auratus) Journal of Endocrinology. 1984;103:141–146. doi: 10.1677/joe.0.1030141. [DOI] [PubMed] [Google Scholar]
- Cornish S, Cawthorne MA. Fatty acid synthesis in mice during the 24hr cycle and during meal-feeding. Horm Metab Res. 1978;10:286–290. doi: 10.1055/s-0028-1093416. [DOI] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Curtis AM, S-beom Seo, Westgate EJ, Rudic RD, Smyth EM, Chakravarti D, FitzGerald GA, McNamara P. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J Biol Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Paolo Sassone-Corsi. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Esquirol Y, Bongard V, Mabile L, Jonnier B, Soulat J-M, Perret B. Shift work and metabolic syndrome: respective impacts of job strain, physical activity, and dietary rhythms. Chronobiol Int. 2009;26:544–559. doi: 10.1080/07420520902821176. [DOI] [PubMed] [Google Scholar]
- Etchegaray J-P, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci USA. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hems DA, Rath EA, Verrinder TR. Fatty acid synthesis in liver and adipose tissue of normal and genetically obese (ob/ob) mice during the 24-hour cycle. Biochem J. 1975;150:167–173. doi: 10.1042/bj1500167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121:2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Maji T, Ashida K. Periodicity of food intake and lipogenesis in rats subjected to two different feeding plans. J Nutr. 1970;100:691–697. doi: 10.1093/jn/100.6.691. [DOI] [PubMed] [Google Scholar]
- Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia Katja A, Storch K-F, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-S, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Liu XS, Lazar MA. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes & Development. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur S, Li X-Y, Jingyi Li, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keränen SV, Knowles DW, Stapleton M, Bickel P, Biggin MD, Eisen MB. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse Y, Oh-hashi K, Iijima N, Naruse M, Yoshioka H, Tanaka M. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol Cell Biol. 2004;24:6278–6287. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TÅ, Hagenbeek D, Moulos P, Siersbæk R, Megens E, Denissov S, Børgesen M, Francoijs K-J, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes & Development. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Perreau-Lenz S, Zghoul T, Spanagel R. Clock genes running amok. Clock genes and their role in drug addiction and depression. EMBO Rep. 2007;8(Spec No):S20–23. doi: 10.1038/sj.embor.7401016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietroiusti A, Neri A, Somma G, Coppeta L, Iavicoli I, Bergamaschi A, Magrini A. Incidence of metabolic syndrome among night-shift healthcare workers. Occup Environ Med. 2010;67:54–57. doi: 10.1136/oem.2009.046797. [DOI] [PubMed] [Google Scholar]
- Preitner N, Francesca Damiola, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Rekowski M, von W, Giannis A. Histone acetylation modulation by small molecules: a chemical approach. Biochim Biophys Acta. 2010;1799:760–767. doi: 10.1016/j.bbagrm.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Repouskou A, Sourlingas TG, Sekeri-Pataryas KE, Prombona A. The circadian expression of c-MYC is modulated by the histone deacetylase inhibitor trichostatin A in synchronized murine neuroblastoma cells. Chronobiol Int. 2010;27:722–741. doi: 10.3109/07420521003786800. [DOI] [PubMed] [Google Scholar]
- Salgado-Delgado R, Angeles-Castellanos M, Saderi N, Buijs RM, Escobar C. Food intake during the normal activity phase prevents obesity and circadian desynchrony in a rat model of night work. Endocrinology. 2010;151:1019–1029. doi: 10.1210/en.2009-0864. [DOI] [PubMed] [Google Scholar]
- Sanchis-Segura C, Lopez-Atalaya JP, Barco A. Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition. Neuropsychopharmacology. 2009;34:2642–2654. doi: 10.1038/npp.2009.125. [DOI] [PubMed] [Google Scholar]
- Scheer FAJL, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes (Lond) 2008;32:658–662. doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- Siersbæk R, Nielsen R, John S, Sung M-H, Baek S, Loft A, Hager GL, Mandrup S. Extensive chromatin remodelling and establishment of transcription factor “hotspots” during early adipogenesis. EMBO J. 2011;30:1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookoian S, Castaño G, Gemma C, Gianotti T-F, Pirola C-J. Common genetic variations in CLOCK transcription factor are associated with nonalcoholic fatty liver disease. World J Gastroenterol. 2007;13:4242–4248. doi: 10.3748/wjg.v13.i31.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci USA. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woon PY, Kaisaki PJ, Bragança J, Bihoreau M-T, Levy JC, Farrall M, Gauguier D. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci USA. 2007;104:14412–14417. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Liu A, Weidenhammer A, Cooksey RC, McClain D, Kim MK, Aguilera G, Abel ED, Chung JH. The role of mPer2 clock gene in glucocorticoid and feeding rhythms. Endocrinology. 2009;150:2153–2160. doi: 10.1210/en.2008-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Yao YL, Sun JM, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- Yang X, Lamia KA, Evans RM. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:387–394. doi: 10.1101/sqb.2007.72.058. [DOI] [PubMed] [Google Scholar]
- Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- Zardoya R, Diez A, Serradilla MC, Madrid JA, Bautista JM, Garrido-Pertierra A. Lipogenic activities in rat liver are subjected to circadian rhythms. Rev Esp Fisiol. 1994;50:239–244. [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, Brenner DA, Montminy M, Kay SA. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]