Summary
The nuclear pore complex, composed of proteins termed nucleoporins (Nups), is responsible for nucleocytoplasmic transport in eukaryotes. Nuclear pore complexes (NPCs) form an annular structure composed of the nuclear ring, cytoplasmic ring, a membrane ring, and two inner rings. Nup192 is a major component of the NPC’s inner ring. We report the crystal structure of Saccharomyces cerevisiae Nup192 residues 2–960 [ScNup192(2–960)], which adopts an α-helical fold with three domains (i.e., D1, D2, and D3). Small angle X-ray scattering and electron microscopy (EM) studies reveal that ScNup192(2–960) could undergo long-range transition between “open” and “closed” conformations. We obtained a structural model of full-length ScNup192 based on EM, the structure of ScNup192(2–960), and homology modeling. Evolutionary analyses using the ScNup192(2–960) structure suggest that NPCs and vesicle-coating complexes are descended from a common membrane-coating ancestral complex. We show that suppression of Nup192 expression leads to compromised nuclear transport and hypothesize a role for Nup192 in modulating the permeability of the NPC central channel.
INTRODUCTION
Nuclear pore complexes (NPC) are large, octagonally symmetric, cylindrical macromolecular assemblies formed at the fusion of the inner and outer nuclear envelope membranes. The NPC is responsible for the rapid, active, and selective exchange of macromolecules between the nucleus and cytoplasm (Aitchison and Rout, 2012; Grossman et al., 2012). As both the gatekeepers of nucleocytoplasmic transport and a platform for the organization of numerous nuclear activities, NPCs contribute to the regulation of myriad physiological processes, including gene expression, cell cycle control, and spindle and kinetochore assembly (Wozniak et al., 2010). The typical diameter of the central channel of the NPC is ~35 nm, which allows for the passive diffusion of small molecules and proteins with molecular weights less than ~40 kDa. However, the passage of higher molecular weight cargo proteins requires that the cargo be recognized by cognate transport factors, most of which belong to the karyopherin family (Peters, 2009). This transport is active and mediated by the interaction of karyopherins with the “FG” repeat-containing nucleoporins, which line the central channel of the NPC (Terry et al., 2007).
Each yeast NPC is an ~50 MDa assembly made of at least 456 protein molecules represented by multiple copies of ~30 distinct protein types, termed nucleoporins (Nups; Rout et al., 2000). The core scaffold of the NPC is composed of almost half of the NPC’s Nups. It is an annular structure comprising two outer rings (the nuclear and cytoplasmic rings) and two inner rings and serves to anchor the peripheral Nups. (Alber et al., 2007a, b). Computational and crystallographic analyses have revealed that the core scaffold Nups are predominately composed of β-propeller and α-solenoid-like folds or consecutive arrangements of β-propeller domains followed by α-solenoid-like folds (Brohawn et al., 2009; DeGrasse et al., 2009; Devos et al., 2006). The “Y-shaped” Nup84 complex, composed of seven core scaffold Nups, is the major component of the outer rings of the yeast NPC and represents a structurally well-characterized subcomplex (Brohawn et al., 2009; Fernandez-Martinez et al., 2012; Hoelz et al., 2011; Lutzmann et al., 2002). High-resolution crystal structures (Jeudy and Schwartz, 2007; Schrader et al., 2008) are also available for Nic96, the S. cerevisiae homolog of vertebrate Nup93 believed to play a key role in linking together Nups within the core scaffold of the NPC to the FG Nups. The inner ring components are also highly conserved (DeGrasse et al., 2009; Kosova et al., 1999) and, in yeast, consist of four large Nups: Nup192, Nup188, Nup170, and Nup157. Nup192 (1,683 amino acids; 191,535 Da) is essential for cell growth, and studies have revealed extensive interaction networks between Nup192 (Nup205 in vertebrates), Nup170 (Nup155 in vertebrates), Nic96 (Nup93 in vertebrates), Nup53, and membrane-ring Nups, such as Pom152 and Ndc1, underscoring the importance of inner ring Nups in anchoring the NPC to the nuclear envelope and in maintaining its structural integrity (Amlacher et al., 2011; Fahrenkrog et al., 2000; Flemming et al., 2009; Gomez-Ospina et al., 2000; Hawryluk-Gara et al., 2005; Mansfeld et al., 2006). In yeast, Nup192 and Nup188 compete for a predicted α-helix near the N terminus of Nic96, supporting the existence of distinct Nup192:Nic96 and Nup188:Nic96 binary complexes (Flemming et al., 2012). Deletion of S. cerevisiae Nup170 and Nup188 in both S. cerevisiae and Xenopus has been shown to affect the permeability of the NPC (Shulga et al., 2000; Theerthagiri et al., 2010). Similarly, in the case of Caenorhabditis elegans, depletion of Nup205 and Nup93 (Nup192 and Nic96 homologs, respectively) resulted in nuclear accumulation of nonnuclear macromolecules of ~70 kDa, suggesting a role for inner ring Nups in modulating the permeability of the NPC’s central channel (Galy et al., 2003).
Recent studies have improved our biochemical and functional understanding of the interactions between the inner ring Nups and the rest of the NPC. However, high-resolution structures are not available for the core scaffold Nups of the NPC’s inner ring, except for the C-terminal domain of Nup170 (Whittle and Schwartz, 2009). Here, we report the crystal structure of the N-terminal half of S. cerevisiae Nup192, consisting of residues 2–960 [ScNup192(2–960)] at 3.25 Å resolution, revealing an α-helical fold with three structural domains, namely, D1, D2, and D3. Small angle X-ray scattering (SAXS), negative-stain electron microscopy (EM), and normal mode analyses revealed flexibility of the D1 and D3 domains relative to the D2 domain, resulting in “open” and “closed” conformations. A structural model of full-length ScNup192 (ScNup192FL) was constructed based on EM density maps and sequence-structure threading, illustrating the extended α-solenoid-like structure of the whole protein. Structural analyses showed similarity between ScNup192(2–960) and karyopherins, β-catenins, and adaptins, elaborating on our understanding of the potential evolutionary link between vesicle-coating complexes and the NPC (Alber et al., 2007b; Dacks and Field, 2007; Devos et al., 2004, 2006; Field et al., 2011). Finally, we show that suppression of Nup192 expression leads to compromised nuclear transport, suggesting a role for Nup192 in modulating the permeability of the NPC’s central channel.
RESULTS
Structure Determination of ScNup192(2–960)
The construct encompassing residues 2–960 [ScNup192(2–960)], corresponding to the N-terminal half of ScNup192, yielded crystals (resolution ~3.25 Å) that belong to the tetragonal space group P43212 with one molecule in the asymmetric unit. The structure was determined by Se-single-wavelength anomalous dispersion (SAD) with the high solvent content (~75%) of the crystals resulting in an excellent quality electron density map (Figure S1 available online). ScNup192(2–960) also crystallized in a hexagonal form, which diffracted only to 4.75 Å resolution. Structure solution obtained by molecular replacement showed that only one molecule is present in the asymmetric unit of the crystals belonging to the P3221 space group. Structural analyses revealed that, while the overall fold of the protein is similar between the two crystal forms, the molecular arrangement across the symmetry axes are different, even though some of the interfaces involved in the crystal contacts are retained (data not shown). The final model of ScNup192(2–960) in the tetragonal form contains 850 out of 959 residues and was refined to R and Rfree values of 18.8% and 24.3%, respectively (Table 1).
Table 1.
Crystallographic Statistics
| Data Collection | ScNup192(2–960) |
|---|---|
| PDB code | 4IFQ |
| space group | P43212 |
| unit-cell dimensions (Å) | a = 134.6, b = 134.6, c = 234.8 |
| Matthew’s coefficient (Å3/Da) | 4.79 |
| solvent content (%) | 74 |
| wavelength (Å) | 0.9791 |
| resolution (Å) | 50.00–3.25 (3.29–3.25)a |
| number of unique reflections | 64,698 (2,186)b |
| completeness (%) | 99.1 (98.8) |
| Rsymm (%) | 13.6 (96.9)b |
| multiplicity | 8.4 (8.4)b |
| <I/σ(I)> | 17.5 (2.6)b |
| Refinement | |
| number of reflections | 32,643 |
| number of reflections in test set | 1,930 |
| R-factor (%) | 18.8 |
| Rfree (%) | 24.3 |
| root-mean-square deviations from ideal values | |
| bond length (Å) | 0.0137 |
| bond angles (°) | 1.7551 |
| Ramachandran plot | |
| MolProbity (Chen et al., 2010) residues in | |
| favored region (%) | 92.5 |
| allowed region (%) | 99.9 |
Values in parentheses correspond to the highest resolution shell.
Bijvoet pairs are not merged.
The Overall Architecture of ScNup192(2–960)
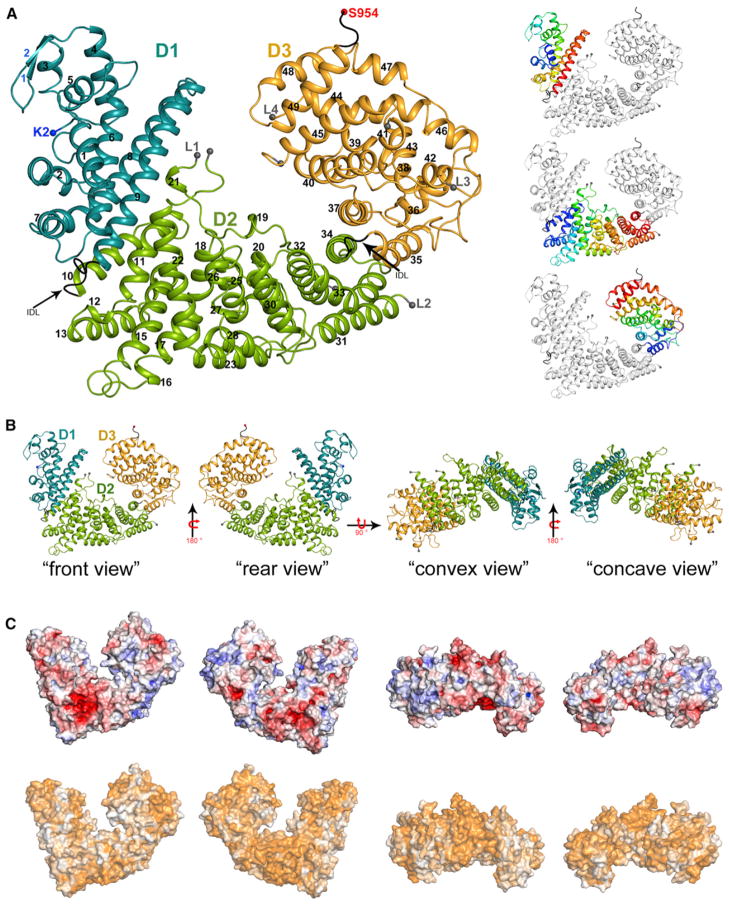
The overall structure of ScNup192(2–960) reveals an α-solenoid-like fold containing tandem repeats of 49 helices and a short β-hairpin near the N terminus (Figure 1A) with dimensions of 84 Å × 78 Å × 98 Å. The structural arrangement of helices suggests that ScNup192(2–960) can be described in terms of three distinct structural domains, namely, D1, composed of residues 2–202; D2, composed of residues 210–660; and D3, composed of residues 665–949 (Figure 1A). Secondary structure elements do not cross over between the domains and are arranged in a sequential manner from the N terminus to C terminus. The D1 and D3 domains appear slightly twisted toward each other in opposite directions relative to the central D2 domain, resulting in a bowl-like shape with distinct convex and concave surfaces (Figure 1B).
Figure 1. Structure and Surface Properties of ScNup192(2–960).
(A) Overall structure of ScNup192(2–960) is shown as a cartoon representation with the D1, D2, and D3 domains colored cyan, green, and gold, respectively. Interdomain loops comprising residues 204–211 and 663–670 are marked by arrows. Residues at the boundaries of the disordered loops are depicted as gray spheres, and the beginning of these loops are marked as L1, L2, L3, and L4. Side panels show individual D1, D2, and D3 domains as the blue to red rainbow from N to C terminus. Secondary structure elements are shown as defined by the DSSP (Kabsch and Sander, 1983) program. Strands of the β-hairpin are labeled in blue; additionally, helices are numbered consecutively with 310 helices treated as α helices for simplicity. See also Figure S1.
(B) Representations of the front, rear, convex, and concave views of the ScNup192(2–960) are illustrated with the D1, D2, and D3 domains colored cyan, green, and gold, respectively.
(C) The top row shows electrostatic potential of ScNup192(2–960) plotted onto its solvent accessible surface. Missing side chains and charges were assigned for ScNup192(2–960) structure using Protein Data Bank (PDB) 2PQR (Dolinsky et al., 2007), and electrostatic surface was calculated using APBS (Baker et al., 2001) within PyMOL. Negative (−7 kT/e) and positive (+7 kT/e) potentials are shown in red and blue, respectively. The bottom row shows conservation of 11 different fungal Nup192 sequences plotted onto the surface of ScNup192(2–960) structure with least to absolutely conserved residues colored as a gradient from white to orange. See also Figure S2.
The N-terminal D1 domain is made up of a β-hairpin formed by two short α strands and nine α helices (α1–α9). Two long helices, α8 and α9, form a helical bundle (Figure 1A). The central D2 domain of ScNup192(2–960) is composed of 25 helices, from α10 to α34, which pack against each other. The D2 domain contains two long loops: (L1) a long, ~100 amino acid (residues 328–424) loop that seems to adopt a mostly random coil conformation, within which residues 363–416 could not be modeled in the electron density maps and (L2) an ~30 amino acid loop (residues 575–601) that could not be modeled in the electron density maps. The D3 domain of ScNup192(2–960) has a globular shape composed of 15 helices (from α35 to α49) and includes two disordered loop regions (residues 799–813 between α41 and α42 [L3], and residues 850–856 between α44 and α45 [L4]). The D3 domain folds over the central D2 domain via interactions between helices α34 and α37 and α33 and α35 (Figure 1A).
Surface Properties and Sequence Conservation of ScNup192(2–960)
A multiple sequence alignment of ScNup192(2–960) with 11 other Nup192 sequences of fungal origin reveals segments of conserved residues across the entire length of the protein (Figure S2). A noticeable sequence divergence is seen within the loop L1 of the D2 domain. These divergences seem to be due to insertions and deletions between residues 363 and 416, which are disordered in the crystal structure. Aside from this, the D2 domain shows a higher degree of conservation compared to both the D1 and D3 domains. Mapping sequence conservation and electrostatic potential onto the surface of ScNup192(2–960) reveals the following: (1) the surface of ScNup192(2–960) is largely negatively charged and possesses several conserved grooves and invaginations (Figures 1B and 1C); (2) a negatively charged “funnel” contoured by residues from helices α11, α15, and α22 with domain D2, although distinct, is somewhat less conserved at its entrance (Figures 1B and 1C, front view); (3) residues from helices α23, α24, α28, and L2 loop connecting helices α23 and α24 are highly conserved, resulting in a large patch of conserved surface (Figures 1A and 1C, rear and convex views) that is predominantly negatively charged. It is noteworthy that this surface includes the conserved sequence AFLLTK-X-K-D/N-D/E-EEDSLLSGED-L/F-X-LD-D/E-I/V between residues 447 and 471, dominated by acidic side chains (Figure S2); and (4) the concave surface of ScNup192(2–960), which shows a uniform charge distribution, is only moderately conserved compared to the convex surface (Figures 1B and 1C, convex and concave views).
Structural Dynamics of ScNup192(2–960) Revealed by SAXS and EM
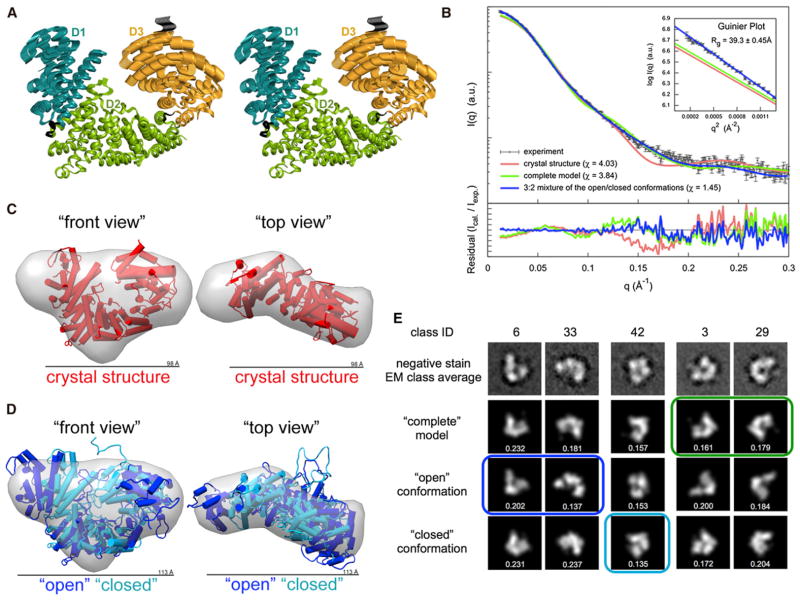
A noticeable feature during crystallographic model building is that the D1 domain of ScNup192(2–960) is apparently more mobile than the D2 and D3 domains, which indicates a possible conformational flexibility. Accordingly, the average main-chain B-factor for the D1 domain is 104 Å2 compared to that of 74 Å2 and 78 Å2 for the D2 and D3 domains, respectively. Elastic network model analysis using HingeProt (Emekli et al., 2008) suggested a long-range motion of D1 and D3 domains relative to the central D2 domain about the hinge residues of Ile196 and Leu675 (which are the near end of the D1 and D2 domains, respectively). This motion was also identified by elNémo (Suhre and Sanejouand, 2004) as one of the top normal modes (Figure 2A) of ScNup192(2–960). Such conformational dynamics might play a role in regulating the diameter of the central pore of the NPC. Therefore, we probed the dynamics of ScNup192(2–960) in solution using both SAXS and EM.
Figure 2. Conformational Flexibility of ScNup192(2–960).
(A) Stereoview of the top normal mode predicted by elNémo (Suhre and Sanejouand, 2004) is shown, depicting the motion of D1 and D3 domains relative to the D2 domain of ScNup192(2–960). Residues in the hinge regions 204–211 and 663–670, between the domains, are colored in black.
(B) Comparison of the merged experimental SAXS profile (black) of ScNup192(2–960) with the calculated SAXS profiles from the crystal structure (χ = 4.03, red), the “complete model” (χ = 3.84, green), and the 3:2 mixture of “open” and “closed” conformations (χ = 1.45, blue) are shown. The lower plot presents the residuals (calculated intensity/experimental intensity) of each calculated SAXS profile. The upper inset shows the SAXS profiles in the Guinier plot with an Rg fit of 39.30 ± 0.45 Å. The maximum particle size (Dmax) is 123 Å.
(C) The crystal structure shown as cylinders (in red, front and top views) was fitted to the ab initio shape (represented as a gray envelope) computed from the experimental SAXS profile. This comparison reveals extra volume in solution. See also Figure S3A.
(D) The “open” (in blue) and “closed” (in cyan) conformations of ScNup192(2–960) were fitted with the ab initio shape (represented as a gray envelope) computed from the experimental SAXS profile. The mixture of two conformation envelopes agrees well with the ab initio shape. See also Figure S3B and Movies S1, S2, S3, and S4.
(E) Selected negative stain EM class averages are shown along with the projections of the “complete model”, “open”, and “closed” conformations. The em2D scores are shown in the bottom of each projection. See also Figure S4, showing 44 EM class averages along with projection of the “complete model”, “open”, and “closed” conformations, and Figure S5, illustrating histogram of em2D scores for selected class averages.
See also Tables S1 and S2.
Indeed, the experimentally measured SAXS profile (Table S1) of ScNup192(2–960) in solution (black in Figure 2B) did not match the theoretical SAXS profiles computed from either the crystal structure (χ = 4.03, red in Figure 2B) or a “complete model” (χ = 3.84, green in Figure 2B), which fully complemented the crystal structure by MODELLER (Sali and Blundell, 1993). Further, the experimentally determined radius of gyration (Rg) of 39.30 ± 0.45 Å in the Guinier plot (subset in Figure 2B) was 11%–18% higher than the Rg values of 33.33 Å and 35.41 Å computed from the crystal structure and the “complete model”, respectively. Similarly, the maximum particle size (Dmax) of 123 Å was 25.5% larger than the maximum dimension of 98 Å from the crystal structure. Accordingly, both the crystal structure and the “complete model” did not match the more extended ab initio shape (Figures 2C and S3A) computed from the experimental SAXS profile.
To study the flexibility of the ScNup192(2–960) structure in solution, conformational sampling by molecular dynamics was carried out by BILBOMD (Pelikan et al., 2009; Movies S1, S2, S3, and S4). The analysis of the resulting 110,000 conformations using the minimal ensemble search (MES) indicated that a 3:2 mixture of only two conformations (“open” and “closed”) was sufficient to explain the experimental SAXS profile within its error bars (χ = 1.45, blue in Figures 2B and S3B). In addition, the mixture of the two conformation envelopes also agrees well with the ab initio shape computed from the SAXS profile (Figure 2D). The “open” and “closed” conformations of ScNup192(2–960) support the long-range motion indicated by HingeProt and elNémo (Figure 2A).
EM analysis of ScNup192(2–960), using the iterative stable alignment and clustering (ISAC) method (Yang et al., 2012) revealed 44 class averages (Figures S4A–S4D, top row) that represent 83% of the entire data set. The ScNup192(2–960) in these classes shows a bowl-like shape that agrees with the overall architecture of the crystal structure. However, numerous class averages may correspond to different conformational states of ScNup192(2–960). We thus explored if the conformations identified by SAXS are consistent with these EM class averages. In particular, we quantified the overlap of projections of the “complete model”, “open”, and “closed” conformations with each of 44 EM class averages. The best matching models were determined for each class average. As expected, the 44 class averages are not explained by a single conformation. Moreover, several class averages could not be explained by any of the conformations (Table S2). However, a number of class averages could be assigned to either the “complete model”, “open”, or “closed” conformations (Figures 2E, S4, and S5; Table S2). Taking these diverse data in aggregate, it appears that ScNup192(2–960) is a rather dynamic molecule with long-range flexibility in the range of 10 Å–20 Å in solution.
Structural Homologs of ScNup192(2–960)
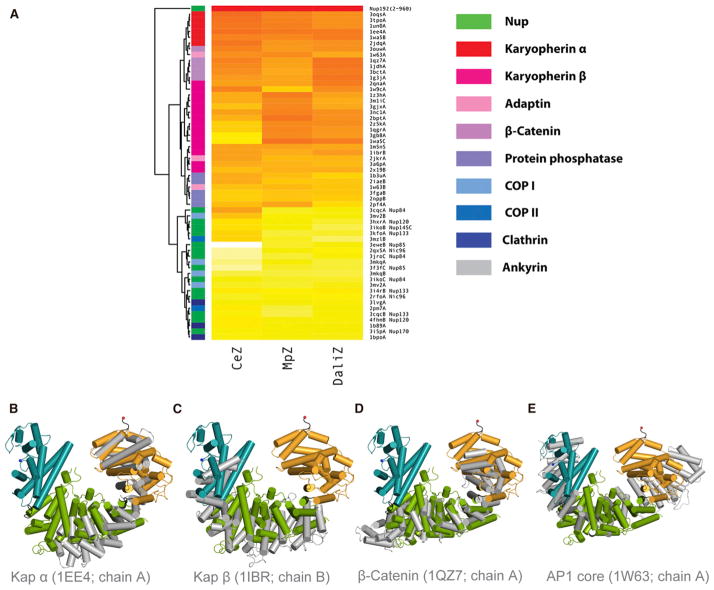
Nucleoporins share a common fold pattern with vesicle-coating complexes and likely share an evolutionary origin with clathrin, COPI, and COPII (Alber et al., 2007b; Dacks and Field, 2007; Devos et al., 2004, 2006). Although such distant relationships are not detectable by primary sequence comparisons, they can be revealed by sequence-structure threading and structural comparisons. To investigate these relationships, the ScNup192(2–960) structure was compared with 59 publicly available structures of similarly sized fragments from α-helical proteins representing ten functional groups. Structural alignments were carried out using Dali (Holm and Rosenström, 2010), CE (Shindyalov and Bourne, 1998), and Multiprot (Schneidman-Duhovny et al., 2010), and we combined standardized scores from these three methods into a composite Z score (Figure 3A; Table S3). The resulting scores and trees suggested that Nup192(2–960) is most closely related to karyopherins, β-catenins, and adaptins (Figures 3B–3E). Notably, the overall fold of ScNup192(2–960) does not show strong structural similarity to other α-solenoid-like Nups. Many of the structural similarities between ScNup192 and other α-solenoid-like Nups are limited only to relatively short segments (i.e., structural coverage of only ~10% of ScNup192), reducing confidence in these (Figure S6A). In addition, we also performed similar analyses on the structures of yeast Nup85 (Figure S6B; Table S4) and Nup170 (Figure S6C; Table S5). In contrast to Nup192, Nup85 appears more closely related to COPII and the COPII-like Nups, as expected (Brohawn et al., 2008; Devos et al., 2004; Hsia et al., 2007), while Nup170 shows closer similarity to clathrin (ter Haar et al., 1998; Whittle and Schwartz, 2009). This suggests that the different α-solenoid Nups might have evolved from several distinct coating protein architectures.
Figure 3. Structural Relatives of ScNup192(2–960).
(A) The heat map shown above illustrates the structural relationship of ScNup192(2–960) with selected α-helical proteins representing ten functional groups. Standardized scores for Dali, CE, and Multiprot alignments represented as a yellow to red gradient; red indicates stronger alignment scores (Table S2). The structure dendrogram is computed by hierarchical clustering using pairwise distances between alignment Z scores. The bar on the left shows the protein class for each structure, showing that karyopherins, β-catenin, and adaptin proteins are closest to Nup192. See also Figures S6B and S6C and Tables S4 and S5 for analyses on Nup85 and Nup170 structures.
(B–E) Structural superpositions of Nup192(2–960) with karyopherin 60 (PDB: 1EE4, chain A), karyopherin 95 (PDB: 1IBR, chain B), β-catenin (PDB: 1QZ7, chain A), and adaptor protein 1 (PDB: 1W63, chain A), respectively, are illustrated based on DALI alignments. These were the top structural alignment hits among the karyopherin alpha, karyopherin beta, β-catenin, and adaptin protein families. The D1, D2, and D3 domains of ScNup192(2–960) structure are shown in cyan, green, and gold, respectively, and the superposed molecules are in gray. See also Figure S6A.
See also Table S3.
Electron Microscopy and Structural Model of Full-Length ScNup192
To assess that the structural features shown by the recombinant ScNup192(2–960) are reflected in the native protein, ScNup192FL was isolated (Figure S7A; Fernandez-Martinez et al., 2012) and analyzed by EM. The random conical tilt method was used to reconstruct an initial three-dimensional structure (Radermacher, 1988). Analysis of ScNup192FL untilted particles by ISAC resulted in 23 class averages that accounted for 676 images (33% of the entire data set). The tilted counterparts of three class averages were used for reconstruction of an initial model for refinement. The resolution of the final structure of ScNup192FL was ~26 Å (Figure S7B). An angular coverage plot at the final angular interval of 10 degrees is nearly complete (Figure S7C). The three-dimensional structure of ScNup192FL exhibits a twisted S-like morphology (Figure S7D), similar to the Chaetomium thermophilum Nup192 homolog (Amlacher et al., 2011) or yeast Nup188 (Flemming et al., 2012). The overall structure of ScNup192FL, which is 18 nm in length and 10 nm in width, displays a pronounced semicircle followed by an extended arm. Comparison with the crystal structure of the ScNup192(2–960) (Figure 1A) clearly shows that the “semicircle” corresponds to the N terminus and the “extended arm” to the C terminus of the molecule (Figure 4). To generate a structural model, the C-terminal region of ScNup192FL, comprising residues 961–1683, was modeled using sequence/structural homology-based methods (Table S6). Indeed, both the crystal structure of ScNup192(2–960) and the homology model of the C-terminal region could be docked into the “semicircle” and the “extended arm” of the EM structure of ScNup192FL, respectively (Figure 4). The cross-correlation value for docking of ScNup192(2–960) crystal structure to the “semicircle” of ScNup192FL is 0.754. Thus, we obtained an atomic model for ScNup192FL that reveals the architecture of this key piece of the NPC inner ring complex.
Figure 4. Structural Model of the ScNup192FL.
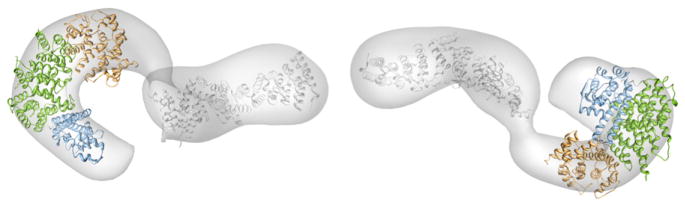
The EM reconstruction of ScNup192FL refined at 26 Å resolution, represented as envelope, is shown in two distinct views. The crystal structure of ScNup192(2–960) and a homology model of ScNup192 C-terminal region, residues 961–1,683, were fitted on the EM map using Chimera (Pettersen et al., 2004). The D1, D2, and D3 domains of ScNup192(2–960) are shown in cyan, green, and gold, respectively. The homology model of ScNup192 C-terminal region is shown in gray.
ScNup192 Plays a Role in Modulating Transport through the NPC
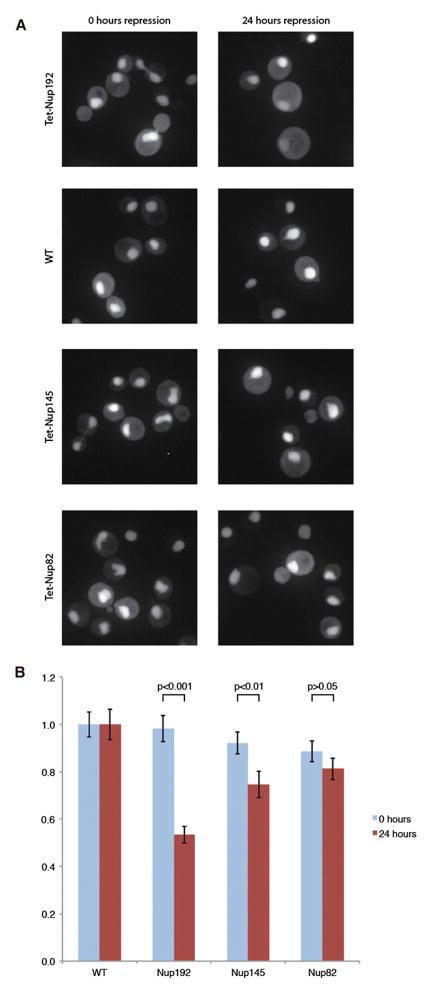
Flexibility in components of the structural core of the NPC may be necessary to allow the NPC channel to dilate during transport events. Removing such Nups may alter transport through the NPC. To address this, we generated a strain where chlorotetracyline can block the biogenesis of Nup192 and examined the consequences on basic NPC function. We measured the distribution of a fusion protein of the nuclear localization sequence (NLS) of Nab2 to mCherry. The nuclear to cytoplasmic ratio (N/C) of such a reporter represents the equilibrium of active import into the nucleus and passive permeability leaking outward (Shulga et al., 2000; Timney et al., 2006). Images were collected of Tet-Nup192 cells expressing the fluorescent reporter either immediately after addition of chlorotetracyline or after 24 hr of repression (Figure 5A). N/C ratios of the reporter protein were measured from the image data of these cells, normalized to that of identically treated wild-type (WT) control cells (Figure 5B). The N/C ratio of NLS-mCherry was reduced to a statistically significant ~50% of the wild-type distribution, indicating a partial loss of NPC function due to either a reduced import rate or increased permeability. Repression of Nup145 and Nup82, both also essential Nups, caused either a lesser effect on import/permeability or no measureable effect, respectively. This indicated that import/permeability defects, such as those observed upon Nup192 depletion, are not a general effect of the depletion of any nucleoporin.
Figure 5. Steady State Localization of Transport Reporter after Depletion of Nup192.
(A) The different panels show the localization of the Nab2NLS-mCherry-PrA NPC function reporter protein in strains where Nup192, Nup145, or Nup82 levels were repressed with chlortetracycline. Example images of these strains are shown after 0 or 24 hr of repression.
(B) The N/C ratios of the reporter were quantified from ~100 cells per condition and normalized to a WT N/C value of 1 at the respective time point. These mean values are plotted with error bars of the standard error of the normalized means. Reported p values were the result of a Mann-Whitney rank sum test, comparing the distributions of normalized N/C values with or without repression. See also Figure S8.
DISCUSSION
Nup192 (the metazoan homolog of vertebrate Nup205) is a major component of the NPC’s inner ring subcomplex, together with Nup188, Nup170, and Nup157 (Alber et al., 2007a, b). So far, only a few structural insights into the functional role of inner ring Nups have been gained (Amlacher et al., 2011; Flemming et al., 2012; Whittle and Schwartz, 2009). Our crystal structure of ScNup192(2–960) may eventually improve our understanding of the NPC at atomic-level resolution. Moreover, through complementary techniques, such as SAXS and EM, we have been able to reveal the conformational dynamics and overall architecture of ScNup192. The structure of ScNup192(2–960) does not closely resemble that of any other α-helical Nups solved to date (though it is expected that Nup188 will be similar; Devos et al., 2006; Flemming et al., 2012). Predictive algorithms suggest the concave and convex surfaces of Nup192 might mediate protein-protein interactions within the NPC (data not shown). Thus, they may resemble the situation in karyopherins, whose concave and convex surfaces interact with cargoes and nucleoporins, respectively (Chook and Blobel, 2001; Conti et al., 2006; Wente and Rout, 2010), and have also been shown to be flexible (Forwood et al., 2010; Fukuhara et al., 2004).
Our structural analyses reveal that ScNup192(2–960) achieves conformational flexibility through the rigid-body movements of D1 and D3 domains relative to the central D2 domain. This flexibility is imparted by two short inter-domain loops comprising residues 204–211 and 663–670, which serve as hinges, and may mediate several possible functions. First, it may insulate the structure of the NPC from morphological changes of the nuclear envelope during cell division and growth. Such flexibility in the NPC has been suggested by prior high-resolution EM studies (Akey, 1995; Yang et al., 1998). Second, this flexibility may be required during the biogenesis of NPC in order to interlock various nucleoporins (Melcák et al., 2007; Solmaz et al., 2011). Nup192 is known to form multiple interactions with other Nups (Alber et al., 2007a, 2007b; Amlacher et al., 2011), and it has been suggested that it can wrap around the linker-Nup Nic96 (Amlacher et al., 2011). A related possibility that should also be considered is that, once assembled into the mature NPC, Nup192 could be preferentially stabilized in one of the two conformations described in this work. Third, the integrity of the NPC permeability barrier is particularly sensitive to mutation or deletion of inner ring components (Galy et al., 2003; Shulga et al., 2000), and flexibility in the region may contribute to allowing bulky cargoes to pass through the restricted central channel in this region of the NPC. We tested a potential contribution of ScNup192 to maintaining the permeability barrier by depleting it and compared the results in a nuclear transport assay to the depletion of similarly essential components of the outer ring and cytoplasmic filaments. Though the assay cannot formally discriminate between effects on active import versus passive NPC permeability, previous studies indicated that gross NPC permeability, not import, was affected by depletion of inner ring components Nup170, Nup188, or Nup205 (Galy et al., 2003; Shulga et al., 2000); therefore, it seems likely that NPC permeability is also increased in cells depleted in Nup192. This permeability may be a direct result of alteration in the diameter of the inner ring due to Nup192 loss or indirectly through loss of interactions through Nup192 that maintain the core scaffold’s contribution to the NPC’s permeability barrier. Further research should address the detailed role of Nup192 flexibility on NPC function.
Strikingly, weak but potentially significant structural similarities are observed between ScNup192(2–960) and karyopherins, β-catenins, and adaptins, though our structural analyses did not reveal the canonical ARM and HEAT repeats considered characteristic of these proteins. In our original protocoatomer hypothesis, we noted that the NPC’s core scaffold is made of a set of cage-like structures containing Nups composed entirely of either a β-propeller fold, an α-solenoid-like fold, or a distinctive arrangement of both—a combination otherwise unique to the proteins that coat transport vesicles. These similarities suggested a common evolutionary origin for NPCs and vesicle-coating complexes, such as the COPI, COPII, and clathrin/adaptin complexes in an early membrane-curving module that led to the formation of the internal membrane systems in modern eukaryotes (Alber et al., 2007a, 2007b; Devos et al., 2004, 2006). Our discovery of an α-solenoid-like fold variant in the NPC, resembling that of both karyopherins and adaptins, provides additional information regarding the protocoatomer hypothesis. We now see three distinct coating protein architectures spread between the NPC and vesicle-coating complexes: clathrin/COPI-like, characterized by a β-propeller N terminus followed by an extended α-solenoid leg (Lee and Goldberg, 2010; ter Haar et al., 1998); COPII-like, in which a C-terminal α-solenoid-like polypeptide folds back on itself to form a compact rod, often with a β-propeller N terminus as well as a β-blade for binding to a separate β-propeller subunit (Brohawn et al., 2008; Fath et al., 2007; Hsia et al., 2007); and karyopherin/adaptin-like, with the whole protein length characterized by a serpentine, continuous α-solenoid-like fold (Conti et al., 2006; Owen et al., 2004). Presuming the veracity of the protocoatomer hypothesis, the first possibility for the evolutionary origin of these architectures is that an original β-propeller/α-solenoid protocoatomer molecule diverged to form three separate coating complexes, each characterized by a specific molecular architecture—namely COPI-like, COPII-like, and adaptin-like (Figure 6A). However, modern NPCs and coating complexes carry mixtures of these three architectures. This may have arisen through a complicated series of reintegration of these separate coating complexes into hybrid complexes (Figure 6A) or through an elaborate convergent evolution process. Alternatively, a more parsimonious scenario is one in which these three architectures evolved together within a single ancestral complex (Figure 6B), which then diverged and specialized into the separate NPC and vesicle-coating complexes of modern eukaryotes, each complex retaining various aspects of the original three architectures (Figure 6B). Further high-resolution structural work on membrane-coating complexes is needed to distinguish these possible scenarios.
Figure 6. Evolutionary Relationship between Membrane-Coating Complexes.
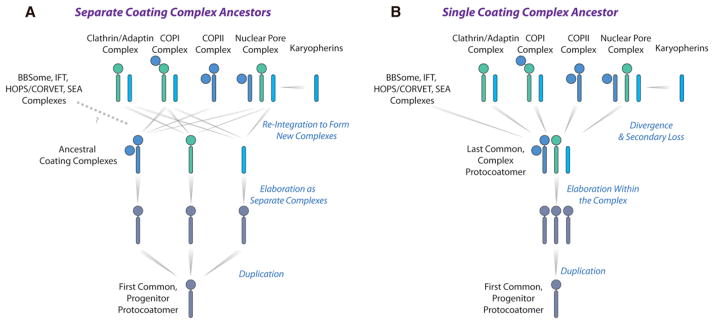
Two possible evolutionary scenarios based on the structural relationship of ScNup192(2–960) with the karyopherin, β-catenin, and adaptin protein families are shown in (A) and (B). The α-solenoid-like and β-propeller folds are represented as rods and circles, respectively. See Discussion for the explanation.
EXPERIMENTAL PROCEDURES
Purification, Crystallization, and Structure Determination of ScNup192(2–960)
The Se-Met-labeled ScNup192(2–960) with a C-terminal His-tag was expressed in BL21(DE3) Codon+RIL E. coli cells (see Supplemental Information). ScNup192(2–960) was purified using Ni-NTA pull-down followed by a gel filtration over Superdex 200 column equilibrated with 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid pH 8.0, 500 mM NaCl, 10% (v/v) glycerol, and 5 mM dithiothreitol, containing four complete protease inhibitor tablets in 400 ml (protein storage buffer). The sample (crystal number 1) used for phasing by SAD appeared in the presence of 14% polyethylene glycol 3350 (PEG3350) and 200 mM potassium sulfate, and the sample (crystal number 2) used for final refined structure at 3.25 Å resolution grew in the presence of 10% PEG3350 and 100 mM potassium iodide. Diffraction data sets were processed with HKL3000 (Minor et al., 2006), and structure solution was obtained using AutoSol (Terwilliger et al., 2009) in Phenix (Adams et al., 2010). An initial model assembled using AutoBuild (Terwilliger et al., 2008) and Buccaneer (Cowtan, 2008), as implemented in CCP4 (Winn et al., 2011), was refined against the data set from crystal number 2. Subsequent model completion involved several rounds of manual building in COOT (Emsley et al., 2010) and refine-ment using Refmac5 (Murshudov et al., 1997).
Small Angle X-Ray Scattering
SAXS profiles of ScNup192(2–960) were obtained at concentrations of 0.4, 1.0, 1.5, 2.0, and 2.5 mg/ml in the protein storage buffer at 15°C, and twenty-four 1 s exposures were made for each protein sample (Table S1). The buffer SAXS profile was obtained in the same manner and subtracted from a protein profile. The merged experimental SAXS profile was compared with SAXS profiles calculated using both the crystal structure of ScNup192(2–960) and for the “complete model”, in which disordered components and eight Se-Met residues were built using MODELLER (Sali and Blundell, 1993) with FoXS (Schneidman-Duhovny et al., 2010).
Conformational Sampling Analysis Using Molecular Dynamics
The “complete model” of ScNup192(2–960) was subjected to energy minimization and heated up to 1,500 K in the CHARMM (Brooks et al., 1983) force field. The D1 and D3 domains were allowed to move relative to the D2 domain about the inter-domain loop residues 204–211 and 663–670, subject to a Rg constraint of 30 Å to 50 Å. Disordered loops not present in the crystal structure were also allowed to move. The resulting 110,000 conformations of ScNup192(2–960) were analyzed to identify a minimal ensemble of conformations that reproduce the experimental SAXS profile, using the MES (Pelikan et al., 2009) and the FoXS (Schneidman-Duhovny et al., 2010) program. We tried larger ensembles of up to ten conformations to avoid oversimplification, and the MES results revealed that an ensemble of two conformations is sufficient to explain the experimental SAXS profile (Figure S3B).
EM Analyses of ScNup192(2–960)
Purified ScNup192(2–960) was applied to glow-discharged carbon-coated copper grids and stained with 0.75%–1% uranyl formate. Images were collected on a Tecnai F20 (FEI, USA) transmission electron microscope. Particles were selected using Boxer from EMAN (Ludtke et al., 1999). The contrast transfer function (ctf) of the images was determined using ctfit and the phases flipped accordingly. The particles were normalized and were then subjected to the ISAC (Yang et al., 2012) technique to produce stable class averages. We quantified, based on the em2D score, the overlap of projections of the “complete model” and the 110,000 molecular dynamics-generated conformations of ScNup192(2–960) with each of 44 EM class averages using the EMageFit application (Velázquez-Muriel et al., 2012) of the IMP software package (Russel et al., 2012) (Figure S4; Table S2). The em2D score is defined as one minus the cross-correlation coefficient between the image and the best-matching projection. The histograms of the em2D scores for all class averages were generated to determine the best matching conformation (or subset conformations), while selected histograms are shown (Figure S5).
Three-Dimensional EM Construction of Native ScNup192FL
Native ScNup192FL, isolated by affinity purification and sucrose gradient, as described previously (Fernandez-Martinez et al., 2012), was applied to glow-discharged carbon-coated copper grids and stained with 0.75%–1% uranyl formate. The random conical tilt reconstruction method was used to create an initial model of ScNup192FL (Frank and Radermacher, 1992). The untilted images were aligned and classified using the ISAC method (Yang et al., 2012). Three-dimensional (3D) reconstructions were made from three class averages, aligned, and averaged, and this combined reconstruction was used as an initial model for reference-based alignment in SPIDER (Frank et al., 1996).
Functional Analysis of ScNup192
Tetracycline-repressible mutants of Nup192, Nup145, and Nup82 were engineered by inserting three tetracycline-binding aptamers from pTDH3-tc3-3xHA upstream of the corresponding open reading frames. These conditional mutant strains and WT cells were transformed with a Nab2NLS-mCherry-PrA yeast constitutive expression plasmid (pBT054). These strains were grown, treated with chlortetracycline to inhibit Nup translation, and imaged as described in the Supplemental Information. Nuclear and cytoplasmic regions from all cells in these images were segmented using purpose-built MatLab scripts and the mean pixel intensities used to calculate each cell’s N/C reporter-protein ratio.
Supplementary Material
Acknowledgments
Authors are grateful to the scientists at Eli Lilly & Company, San Diego, CA who contributed towards cloning and expression of ScNup192(2–960) during PSI2. P.S. acknowledge the help of R.P. Kumar (Almo lab), W. Shi (BNL), L.L. Morisco (APS), S.T. Sojitra (APS), and S.R. Wasserman (APS) during crystallographic data collection. Funding for the NYSGXRC and NYSGRC were provided by NIH Grants U54 GM074945 (S.K.B.) and U54 GM094662 (S.C.A.), respectively. Additional funding for this work was provided by NIH grants R01 GM062427 (M.P.R.), R01 GM083960 (A.S.), U54 GM103511 and U01 GM098256 (A.S. and M.P.R.). This publication was in part made possible by the Center for Synchrotron Biosciences grant, P30-EB-009998, from the NIBIB. Use of the BNL was supported by the U.S. Department of Energy (DOE) under Contract Number DE-AC02-98CH10886. Use of the APS, ANL was supported by DOE. Access to the LRL-CAT beam line at the APS was provided by Eli Lilly, which operates the facility. Portions of this research were carried out at the SSRL, SLAC National Accelerator Laboratory operated for DOE by Stanford University. The SSRL SMBP is supported by the DOE Office of Biological and Environmental Research, by the NIH, NCRR, Biomedical Technology Program (P41RR001209), and by the NIGMS (P41GM103393). The investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant number C06 RR017528-01-CEM from the NCRR, NIH. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH.
Footnotes
ACCESSION NUMBERS
The Protein Data Bank accession code for the coordinate and structure factor of ScNup192(2–960) is 4IFQ. The Electron Microscopy Data Bank accession code for ScNup192FL is 5556. The NYSGRC target identifier for the PSI:Biology target S. cerevisiae Nup192 is NYSGRC-019330.
Supplemental Information includes eight figures, four movies, six tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.str.2013.02.005.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Rout MP. The yeast nuclear pore complex and transport through it. Genetics. 2012;190:855–883. doi: 10.1534/genetics.111.127803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW. Structural plasticity of the nuclear pore complex. J Mol Biol. 1995;248:273–293. doi: 10.1016/s0022-2836(95)80050-6. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- Conti E, Müller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Cowtan K. Fitting molecular fragments into electron density. Acta Crystallogr D Biol Crystallogr. 2008;64:83–89. doi: 10.1107/S0907444907033938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks JB, Field MC. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J Cell Sci. 2007;120:2977–2985. doi: 10.1242/jcs.013250. [DOI] [PubMed] [Google Scholar]
- DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35(Web Server issue):W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emekli U, Schneidman-Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. HingeProt: automated prediction of hinges in protein structures. Proteins. 2008;70:1219–1227. doi: 10.1002/prot.21613. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B, Hübner W, Mandinova A, Panté N, Keller W, Aebi U. The yeast nucleoporin Nup53p specifically interacts with Nic96p and is directly involved in nuclear protein import. Mol Biol Cell. 2000;11:3885–3896. doi: 10.1091/mbc.11.11.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath S, Mancias JD, Bi X, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, Rout MP. Structure-function mapping of a heptameric module in the nuclear pore complex. J Cell Biol. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Sali A, Rout MP. Evolution: On a bender—BARs, ESCRTs, COPs, and finally getting your coat. J Cell Biol. 2011;193:963–972. doi: 10.1083/jcb.201102042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming D, Sarges P, Stelter P, Hellwig A, Böttcher B, Hurt E. Two structurally distinct domains of the nucleoporin Nup170 cooperate to tether a subset of nucleoporins to nuclear pores. J Cell Biol. 2009;185:387–395. doi: 10.1083/jcb.200810016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming D, Devos DP, Schwarz J, Amlacher S, Lutzmann M, Hurt E. Analysis of the yeast nucleoporin Nup188 reveals a conserved S-like structure with similarity to karyopherins. J Struct Biol. 2012;177:99–105. doi: 10.1016/j.jsb.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Forwood JK, Lange A, Zachariae U, Marfori M, Preast C, Grubmüller H, Stewart M, Corbett AH, Kobe B. Quantitative structural analysis of importin-α flexibility: paradigm for solenoid protein structures. Structure. 2010;18:1171–1183. doi: 10.1016/j.str.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M. Three-dimensional reconstruction of single particles negatively stained or in vitreous ice. Ultramicroscopy. 1992;46:241–262. doi: 10.1016/0304-3991(92)90018-f. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Fernandez E, Ebert J, Conti E, Svergun D. Conformational variability of nucleo-cytoplasmic transport factors. J Biol Chem. 2004;279:2176–2181. doi: 10.1074/jbc.M309112200. [DOI] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ospina N, Morgan G, Giddings TH, Jr, Kosova B, Hurt E, Winey M. Yeast nuclear pore complex assembly defects determined by nuclear envelope reconstruction. J Struct Biol. 2000;132:1–5. doi: 10.1006/jsbi.2000.4305. [DOI] [PubMed] [Google Scholar]
- Grossman E, Medalia O, Zwerger M. Functional architecture of the nuclear pore complex. Annu Rev Biophys. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell. 2005;16:2382–2394. doi: 10.1091/mbc.E04-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeudy S, Schwartz TU. Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J Biol Chem. 2007;282:34904–34912. doi: 10.1074/jbc.M705479200. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kosova B, Panté N, Rollenhagen C, Hurt E. Nup192p is a conserved nucleoporin with a preferential location at the inner site of the nuclear membrane. J Biol Chem. 1999;274:22646–22651. doi: 10.1074/jbc.274.32.22646. [DOI] [PubMed] [Google Scholar]
- Lee C, Goldberg J. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell. 2010;142:123–132. doi: 10.1016/j.cell.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J, Güttinger S, Hawryluk-Gara LA, Panté N, Mall M, Galy V, Haselmann U, Mühlhäusser P, Wozniak RW, Mattaj IW, et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell. 2006;22:93–103. doi: 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Melcák I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315:1729–1732. doi: 10.1126/science.1135730. [DOI] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macro-molecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Collins BM, Evans PR. Adaptors for clathrin coats: structure and function. Annu Rev Cell Dev Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R. Translocation through the nuclear pore: Kaps pave the way. Bioessays. 2009;31:466–477. doi: 10.1002/bies.200800159. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J Electron Microsc Tech. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel D, Lasker K, Webb B, Velázquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, Sali A. Putting the pieces together: integrative modeling platform software for structure determination of macro-molecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38(Web Server issue):W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol Cell. 2008;29:46–55. doi: 10.1016/j.molcel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Shulga N, Mosammaparast N, Wozniak R, Goldfarb DS. Yeast nucleoporins involved in passive nuclear envelope permeability. J Cell Biol. 2000;149:1027–1038. doi: 10.1083/jcb.149.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solmaz SR, Chauhan R, Blobel G, Melčák I. Molecular architecture of the transport channel of the nuclear pore complex. Cell. 2011;147:590–602. doi: 10.1016/j.cell.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Sanejouand YH. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32(Web Server issue):W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Musacchio A, Harrison SC, Kirchhausen T. Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker. Cell. 1998;95:563–573. doi: 10.1016/s0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol. 2010;189:1129–1142. doi: 10.1083/jcb.200912045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timney BL, Tetenbaum-Novatt J, Agate DS, Williams R, Zhang W, Chait BT, Rout MP. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J Cell Biol. 2006;175:579–593. doi: 10.1083/jcb.200608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez-Muriel J, Lasker K, Russel D, Phillips J, Webb BM, Schneidman-Duhovny D, Sali A. Assembly of macromolecular complexes by satisfaction of spatial restraints from electron microscopy images. Proc Natl Acad Sci USA. 2012;109:18821–18826. doi: 10.1073/pnas.1216549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Schwartz TU. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J Biol Chem. 2009;284:28442–28452. doi: 10.1074/jbc.M109.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci. 2010;67:2215–2230. doi: 10.1007/s00018-010-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.