Abstract
Background
Two most commonly used animal models for studying breast cancer lung metastasis are: lung metastasis after orthotopic implantation of cells into the mammary gland, and lung implantations produced after tail vein (TV) injection of cells. Tail vein injection can produce lung lesions faster, but little has been studied regarding the differences between these tumors, thus, we examined their morphology and gene expression profiles.
Methods
Syngeneic murine mammary adenocarcinoma, 4T1-luc2 cells, were implanted either subcutaneously (Sq), orthotopically (OS), or injected via TV in Balb/c mice. Genome-wide microarray analyses of cultured 4T1 cells, Sq tumor, OS tumor, lung metastases after OS (LMet), and lung tumors after TV (TVt) were performed 10 days after implantation.
Results
Bioluminescence analysis demonstrated different morphology of metastases between LMet and TVt, confirmed by histology. Gene expression profile of cells were significantly different from tumors, OS, Sq, TVt or LMet (10,350 probe sets; FDR≤1%; P<0.0001). Sq tumors were significantly different than OS tumors (700 probe sets; FDR≤15%; P<0.01), and both tumor types (Sq and OS) were significantly different than LMet (1,247 probe sets; >1.5-fold-change; P<0.01), with no significant difference between TVt and LMet.
Conclusions
There were significant differences between the gene profiles of cells in culture and OS versus LMet, but there were no differences between LMet versus TVt. Therefore, the lung tumor generated by TVt can be considered genetically similar to those produced after OS, and thus TVt is a relevant model for breast cancer lung metastasis.
KEY WORDS : Breast cancer, lung metastasis, animal model, microarray, metastasis model
Introduction
Despite the fact that overall five years survival of breast cancer has recently improved to 98% in the United States (1), the survival of patients with metastatic disease remains low at only 23%, which accounts for approximately 40,000 deaths annually (2,3). It has been estimated that 20-30% of women with early stage breast cancer will eventually develop metastatic disease. The lung is the second most common anatomic site of first exclusive distant metastasis of breast cancer (4), thus advances in the understanding and management of lung metastases are expected to have a large impact on breast cancer survival (5). Despite the central role of mouse models in breast cancer research, these models have not been critically evaluated for their appropriateness for the study of lung metastasis (2-15,16-31).
A commonly employed murine model to study breast cancer lung metastasis entails injection of cancer cells via the tail vein (TV) to implant cells in the lung, and thus produce lung tumors (TVt) (32-34). Advocates of TV argue that it is an easy and quick method to form metastatic TVt lesions, especially with cell lines that take long periods of time to metastasize, if at all. However, others have argued that this model may not adequately mimic human metastatic breast cancer because it does not follow the biological steps that a primary tumor must take to produce a distant metastatic tumor (33,34), and it ignores the cross-talk between primary and metastatic lesions (32,35-38). Indeed, it has been reported that lung metastatic tumors (LMet), which progressed biologically from a primary tumor generated by orthotopic implantation (OS) of breast cancer cells have a different morphology compared to TVt tumors. Although there has been debate in the literature regarding advantages and disadvantages of TV implantations versus orthotopic implantation, evaluation by gene expression profiling of tumors from these models or of the metastatic lesions which they produce has not previously been examined (39). Using genome-wide gene expression microarrays, we have now found that primary OS tumors and their lung metastatic lesions have differentially expressed genetic profiles, but lung metastatic lesions produced by TV or OS have similar profiles.
Materials and methods
Virginia Commonwealth University Institutional Animal Care and Use Committee (IACUC) approval was obtained for all experiments. Female Balb/c mice, 12 weeks of age, weighing approximately 20 g were obtained from Harlan Laboratories (Frederick, MD). The 4T1-luc2, adenocarcinoma cell line derived from the mammary glands of Balb/c mice and genetically manipulated to overexpress the firefly luciferase gene was obtained from Caliper Life Sciences (Hopkinton, MA). The cells were cultured in RPMI media, suspended at a concentration of 1×106 cells/100 µL, and 10 µL of this solution were then injected, unless otherwise stated.
All cell implantations were performed under isoflurane anesthesia using sterile technique. Orthotopic implantation under direct vision (OS; Figure 1A): a 5 mm incision was made medial to the nipple, and a cotton swab was used to expose the mammary gland. The cells were implanted directly into the mammary gland under direct vision, using ×10 microscopic magnification, and the wound was closed. Subcutaneous implantation (Sq): under anesthesia the skin was tented up and 10 µL of the 4T1-luc2 cells were implanted into the subcutaneous space. Tail vein injection (TV; Figure 1B): 100 µL of 1×105 cells/100 µL were injected into the median tail vein.
Figure 1.
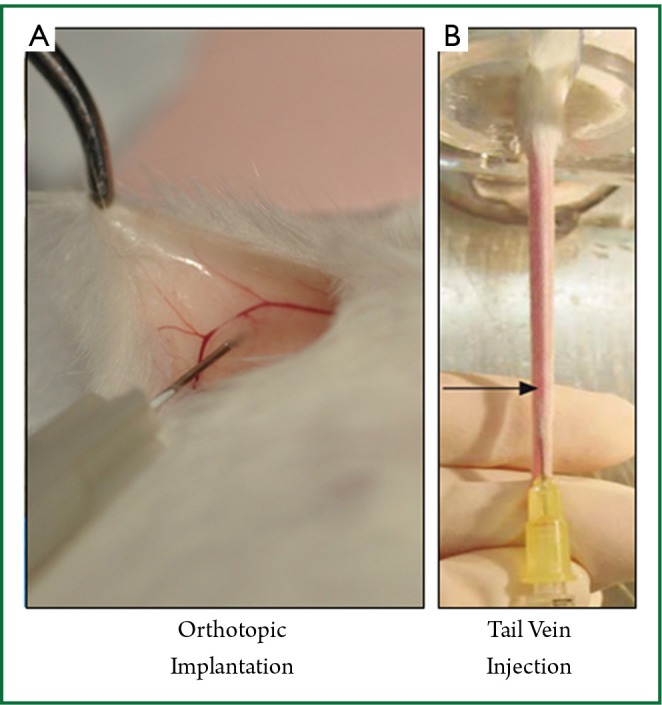
A. Demonstration of orthotopic cell implantation into the chest mammary gland under direct vision (OS). The right chest mammary gland is exposed through a small incision where 27G needle is used to inject the cells; B. Demonstration of tail vein injection of the cells percutaneously into the middle tail vein (TV).
Xenogen’s IVIS® 200 and Living Image® software (Caliper Life Sciences, Hopkinton, MA) were used to quantify the photon/sec emitted by 4T1-luc2 cells which reflects tumor burden after 200 µL of luciferin (Fisher Scientific, Inc.) was injected intraperitoneally. The quantification of photons emitted allowed for the quantification of tumor burden and cancer progression in vivo. To compare tumor growth and survival, 16 Balb/c mice in 2 experimental groups were used: OS and TV.
Gene expression profiling of 4T1 cells in vitro, OS and Sq tumors, and metastatic tumors
Ten days after OS or Sq (8 Balb/c mice per group, one implantation site each), the tumors were harvested and snap-frozen at –80 °C. Day ten after implantation was chosen based upon our previous study (36). RNA Extraction: Snap-frozen tissues were used for histopathological scoring after hematoxylin and eosin (H&E) staining of standard features performed on frozen sections adjacent, above and below the tissue used for RNA isolation. All samples contained more than 70% tumor. Total RNA was extracted and the quality evaluated using a method of sample processing established previously in our laboratory (40). Total RNA was extracted from multiple 10-µm thick frozen tissue sections using the MagMAXTM-96 for Microarrays Total RNA Isolation Kit (InvitrogenTM Life Technologies, Carlsbad, CA), in an automated fashion using the magnetic particle processor MagMAXTM Express. RNA purity was judged by spectrophotometry at 260, 270, and 280 nm. RNA integrity was assessed by running 1 µL of every sample in RNA 6000 Nano LabChips® on the 2100 Bioanalyzer (Agilent Technologies, Foster City, CA). Gene expression microarray analyses: The Affymetrix® protocol (Affymetrix, Santa Clara, CA) has been previously described (40). Briefly, starting from 500 ng of total RNA, cDNA synthesis and cRNA labelling were performed using the GeneChip® 3' IVT Express Kit (Affymetrix). Ten µg of fragmented cRNA were hybridized on the GeneChip® Mouse Genome 430A 2.0 array for 16 hrs at 60 rpm in a 45 °C hybridization oven. This array provides a comprehensive coverage of the transcribed murine genome by including over 22,600 probe sets that analyze the expression level of over 14,000 well-characterized mouse transcripts. The arrays were washed and stained with streptavidin phycoerythrin (SAPE; Molecular Probes, Eugene, OR) in the Affymetrix fluidics workstation. Every chip was scanned at a high resolution, with pixellations ranging from 2.5 µm down to 0.51 µm, by the Affymetrix GeneChip® Scanner 3000 according to the GeneChip® Expression Analysis Technical Manual procedures (Affymetrix). After scanning, the raw intensities for every probe were stored in electronic files (in .DAT and .CEL formats) by the GeneChip® Operating Software (GCOS v1.4) (Affymetrix). The overall quality of each array was assessed by monitoring the 3'/5′ ratios for a housekeeping gene (GAPDH) and the percentage of “Present” genes (%P) where arrays exhibiting GAPDH 3′/5′<3.0 and %P>40% were considered good quality arrays.
Statistical methods
For photon emission, Student’s t-test statistical analysis was utilized. For the microarray data analysis, standard statistical methods were utilized, including Robust Multi-array Analysis (RMA) method (41), hierarchical cluster analyses, and false discovery rates (FDR) as previously reported (42).
Results
Implantation methods
To evaluate how appropriate the commonly used mouse models are for studying breast cancer lung metastasis, we standardized our methodology based on the most commonly used implantation methods in the literature. OS is implantation of 4T1-luc2 cells into the mammary gland after exposing it via a small incision (Figure 1A), and TV is injection of the cells directly into the middle TV (Figure 1B).
TV obviates the biologic sequence of lung metastasis arising from a primary tumor and can produce tail injection site tumors
Bioluminescent technology allows detection of 4T1-luc2 cells in any part of the body due to the whole body scan. Figure 2 demonstrates bioluminescence imaging of lung metastasis 10 days after OS implantation (LMet). LMet tumors were obscured by OS tumor in the chest mammary gland (Figure 2A), which can be visualized in situ after removal of OS tumor (Figure 2B) or ex vivo (Figure 2C). Note that lung metastasis arising from OS (LMet) produced discrete tumors (Figure 2B,C). The cancer cells metastasized to the lung 7 to 10 days after OS implantation, which is consistent with our previous results (36,38). In contrast, lung implantation of 4T1-luc2 cells (TVt) was immediately confirmed after TV injection in the tail (Figure 2D). Note that TVt produced disseminated 4T1-luc2 cell implants throughout the lungs without discrete tumors. Additionally, we occasionally observed development of “tail tumor” at the injection site, which was easily detected with bioluminescent whole body scan.
Figure 2.
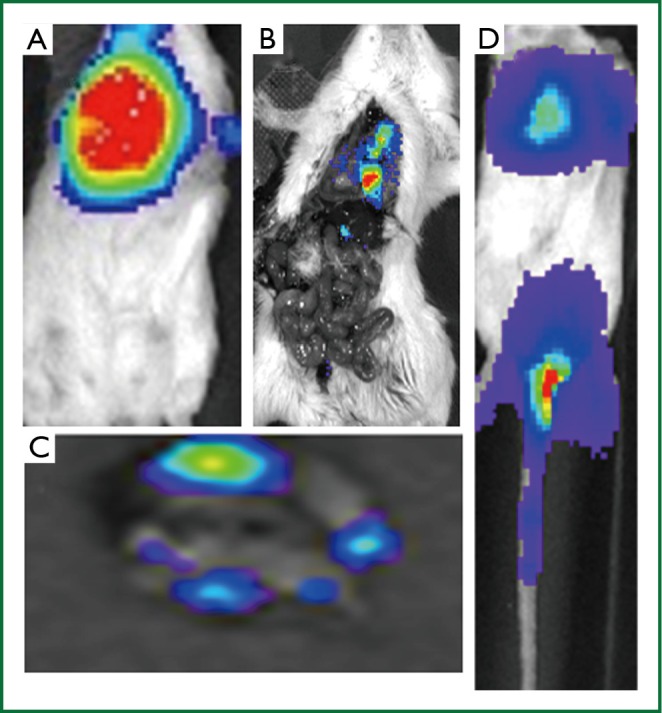
A. Bioluminescence technology allows detection of 4T1-luc2 cells. Lung metastases are not clearly visualized since they are obscured by the high signal from the primary tumor; B. In situ bioluminescence image of the lung metastasis. Note that LMet produced after OS demonstrates a nodular tumor in the lung; C. Ex vivo bioluminescence image of the lung demonstrates a nodular LMet tumor; D. TV injection immediately produced diffusely disseminated cell implantation throughout the lungs as well as tail tumor.
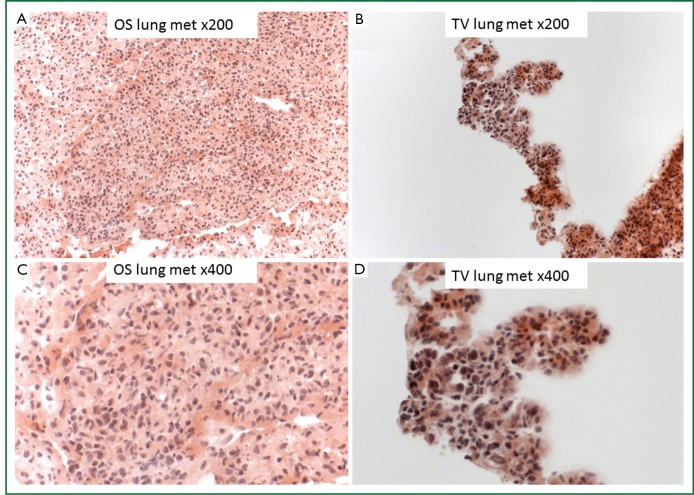
Histological analysis using H&E stain showed that OS lung metastases produced isolated LMet tumors (Figure 3A,B), in contrast to the TVt lung tumors in which the cancer cells colonized the lungs along the normal parenchymal architecture of the lung without forming isolated tumors (Figure 3C,D).
Figure 3.
H&E stain histological analysis of lung tumors. Low magnification (×200) (A) and high magnification (×400) (B) of LMet metastasis after OS implantation show an isolated lung tumor. Low magnification (×200) (C) and high magnification (×400) (D) of TVt tumor after TV implantation demonstrate disseminated colonization of 4T1-luc2 cells throughout the lungs along the parenchymal architecture without forming isolated tumors.
Genomic expression profiles were different between 4T1-luc2 cells in culture dish, Sq and OS primary tumors, and OS lung metastases (LMet), but not different between lung metastases after TV (TVt) and OS (LMet)
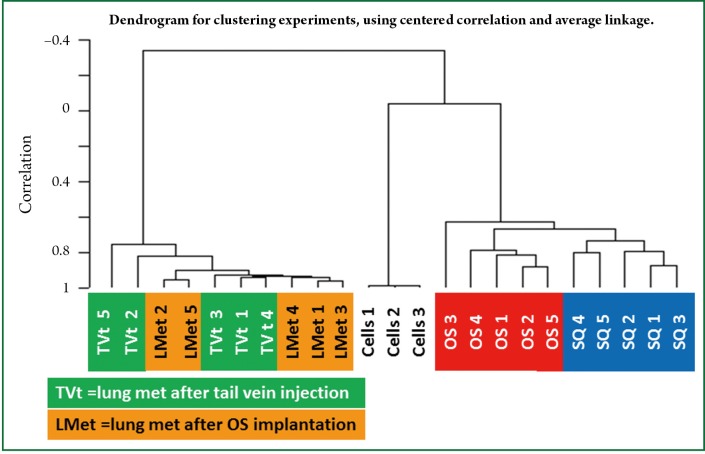
To evaluate the genomic profiles of the tumors produced by these methods, genome wide microarray analysis was performed of the 4T1 cells in culture, Sq and OS tumors as well as of the TVt and LMet metastases. Ten thousand three hundred fifty probe sets (45.7% of transcriptome) were significantly differentially expressed between 4T1 cells in culture compared to OS and Sq tumors, TVt and LMet metastases (FDR≤1%; P<0.0001). In addition, between Sq and OS tumors, 700 probe sets were differentially expressed (FDR≤15%; P<0.01). Furthermore, 1,247 probe sets were differentially expressed (1.5-fold; P<0.01) between OS primary tumors and LMet metastases, without any difference within each group. We also found that the same 1,247 probe sets were differentially expressed (1.5-fold; P<0.01) between OS primary tumors and TVt lung metastases without any difference within each group. Finally, there was no significant difference in gene expression between TVt and LMet metastases (Figure 4).
Figure 4.
Unsupervised hierarchical cluster analysis based on 22,690 probe sets on the GeneChip® Mouse Genome 430A 2.0 array. Gene Expression microarray analysis demonstrates that 4T1 cells in culture dish (Cells) are significantly different than tumors, OS, Sq, TVt and LMet (10,350 probe sets; FDR≤1%; P<0.0001). Moreover, Sq tumors were significantly different than OS tumors (700 probe sets; FDR≤15%; P<0.01), and both tumor types (Sq and OS) were significantly different than LMet (1,247 probe sets; >1.5-fold-change; P<0.01), with no significant difference between TVt and LMet.
Discussion
Breast cancer drug development is an expensive and inefficient process, and use of animal models for the screening of novel agents is a major component of it. However, the literature has not critically examined how appropriate these models are for the study of breast cancer metastasis (6-16,17-31). In translational research it is important to consider the degree to which breast cancer metastasis models provide clinically relevant endpoints (6,8-10,30,31,43-52). The most commonly used animal models for screening for anti-cancer drug development are Sq for local breast cancer and TV for breast cancer lung metastasis. Regardless of the model employed to evaluate the efficacy of novel therapeutics, it is critical to understand the limitations of each method.
It is important to appreciate to what extent the animal model produces clinically relevant endpoints that are translatable to human breast cancer. As we have demonstrated, TV does indeed produce TVt lung metastasis quickly after injection (Figure 2), but there are important limitations to consider. First, TVt obviates the biologic progression from primary tumor to distant lung metastasis and it will not evaluate the efficacy of the therapeutic agents that target the process of cancer progression to metastasis. Second, TV produces TVt lung metastasis without the presence of a primary tumor in the mammary gland. Human metastatic breast cancer patients are commonly treated with systemic therapy before the primary breast cancer is removed, which will not be modeled by TV which lacks a primary tumor. Situations where patients will be treated for metastatic breast cancer without a primary tumor include cases after urgent palliative mastectomy due to bleeding or ulceration of the primary tumor in the setting of metastatic disease, as well as cases of recurrence in the lung after mastectomy. Both situations present biological systems which are very different than the mere intravenous injection of cancer cells resulting in lung implantation. By producing lung metastasis without a primary tumor, TV ignores the cross-talk between primary and metastatic lesions (32,35-38). This element of breast cancer biology is not only important from a biological or basic science perspective, but it also has important implications in the clinical management of human breast cancer. In fact, there are ongoing clinical trials to evaluate the effect on metastatic progression and overall survival of mastectomy in patients with metastatic breast cancer (38). The third factor is the morphology of the TVt lung metastases (Figures 2,3). TVt lung metastases are diffusely disseminated throughout the lung because the cancer cells colonize the lung via hematogenous embolization. In contrast, OS produce discrete LMet metastatic tumors which progressed to the lung along the primary-tumor-to-distant-metastasis progression pathway. In humans, breast cancer forms lung metastases as discrete lung nodules which progress by pathways more analogous to LMet than TVt. Understanding this difference may have implications in terms of the bioavailability and drug delivery of therapeutics in the solid tumor versus the disseminated cells, the relationships of the nodules to the blood and lymphatic vessels in the lung, and the effects on mortality. In fact, mortality in TV typically follows a sudden death due to a thromboembolic phenomenon, rather than mortality via a more gradual process due to cancer progression and overall tumor burden, which occurs in LMet and human breast cancer (6). This factor cannot be underscored enough because the final translatable clinical endpoint, i.e. survival, to screen for the efficacy of novel therapeutics before entering clinical trials varies significantly between these methods.
In the era of targeted therapeutics, it is important to consider the genomic profiles of the tumors and lung metastases produced. The differential expression of over 10,000 genes between cells in culture and the tumors and lung metastases of the same cell line reinforces the importance of in vivo screening of the efficacy of novel therapeutics (Figure 4). In addition, the differential expression of so many genes between OS primary tumors and LMet metastases also reinforces the importance of not just evaluating the efficacy in primary tumors. Furthermore, the differential expression of so many genes between Sq and OS tumors implies that investigators should understand the differences in the potential target expressions of the tumors their model produces. Finally, there was no significant difference in the genomic expression profiles of TVt versus LMet metastases, which is an important strength of the TV model, especially in light of the limitations stated above. Based upon our findings, we cannot help but speculate that tumor microenvironments have a significant role in the gene expression profiles.
The choice of the appropriate model for breast cancer research relies upon the underlying hypothesis being tested. Understanding the limitations of these models, beyond the differences in genetic profile, is therefore of great importance. The benefit of Sq is that it produces tumors which can be followed locally for therapeutic effect, but they do not metastasize to the lung. The strength of the OS model is that it utilizes the biologic progression from primary tumor to distant metastasis, and it allows for testing hypotheses on primary-metastatic tumor interactions. It is also beneficial to evaluate the effect of tumor microenvironment especially when syngeneic cells are used. The weakness of this model is that some cell lines, especially utilizing xenograft models where human cell lines are implanted into mice, do not readily metastasize to the lung. TV has the strength in that it will implant cell lines immediately into the lungs without relying on metastasis from primary lesions that will significantly shorten the duration of the experiment, but it has no primary lesion for questions related to primary tumor-metastatic lesion interactions. In addition, TV colonizes the lung with cancer cells, rather than producing isolated lung “tumors”. This is an advantage for researchers since it allows them to quantify the amount of lung metastasis utilizing colony formation assay, which is an established commonly used method for 4T1 cells. On the other hand, TV often causes sudden death via thromboembolism, instead of by cancer progression, which makes it an unstable model to assess survival. As is the case for any animal model, TV should be utilized with its advantages and limitations in mind in order to translate the findings to human breast cancer lung metastasis. Strengths and weaknesses of these models are summarized in Table 1.
Table 1. Advantages and disadvantages of different implantation methods of mouse metastatic breast cancer models.
| Advantage | Disadvantage | |
|---|---|---|
| Orthotopic implantation (OS) | Progression from primary tumor to lung metastatic tumor similar to human | Some cells (especially xenografts) are difficult to metastasize |
| Possible to investigate primary tumor-metastasis interaction | Commonly takes time to metastasize | |
| Cancer progression produces mortality (translational endpoint) | ||
| Subcutaneous implantation (Sq) | Easy to perform | Does not metastasize readily |
| Commonly used | Heterotopic tumor microenvironment | |
| Superficial growth allows caliper measurement | Does not progress similar to human cancer | |
| Different genetic profile than OS primary tumors | ||
| Tail vein injection (TV) | Easy to perform | No primary tumor |
| Quick and easy implanation of cells in lung | No lung tumors, instead cell colonization | |
| Possible quantification using colony formation assay (4T1 cells) | Mortality by thromboembolic phenomena | |
| No progression from primary tumor to metastasis |
In summary, our study suggests that primary tumors and their lung metastatic lesions have differentially expressed genetic profiles, although there are no differences between metastatic lesions produced by TV injection versus orthotopic implantation of the same cancer cells. Although the TV injection method is limited because it evaluates lung metastasis without a primary tumor, ignores the biologic progression from primary tumor to metastatic lesions, and produces lung metastasis with a different morphology and mechanism of mortality than the orthotopic implantation method and human cancer, it does produce lung metastases with similar genomic profiles as lung metastases after orthotopic implantation. Utilizing these animal metastatic breast cancer models with an understanding of their limitations is expected to improve the efficiency of breast cancer drug development and the advancement of breast cancer research.
Acknowledgements
Kazuaki Takabe is supported by United States National Institute of Health (R01CA160688) and Susan G. Komen for the Cure [Investigator Initiated Research Grant (12222224)]. Masayuki Nagahashi is a Japan Society for the Promotion of Science Postdoctoral Fellow.
Disclosure: The authors declare no conflict of interest.
References
- 1.Edwards BK, Ward E, Kohler BA, et al. Annual report to the nation on the status of cancer, 1975-2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer 2010;116:544-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roth BJ, Krilov L, Adams S, et al. Clinical cancer advances 2012: annual report on progress against cancer from the american society of clinical oncology. J Clin Oncol 2013;31:131-61 [DOI] [PubMed] [Google Scholar]
- 3.DeSantis C, Siegel R, Bandi P, et al. Breast cancer statistics, 2011. CA Cancer J Clin 2011;61:409-18 [DOI] [PubMed] [Google Scholar]
- 4.Berman AT, Thukral AD, Hwang WT, et al. Incidence and patterns of distant metastases for patients with early-stage breast cancer after breast conservation treatment. Clin Breast Cancer 2013;13:88-94 [DOI] [PubMed] [Google Scholar]
- 5.O’Shaughnessy J.Extending survival with chemotherapy in metastatic breast cancer. Oncologist. 2005;10Suppl 3:20-9 [DOI] [PubMed] [Google Scholar]
- 6.Schuh JC. Trials, tribulations, and trends in tumor modeling in mice. Toxicol Pathol 2004;32:53-66 [DOI] [PubMed] [Google Scholar]
- 7.Burger MM. UICC study group on basic and clinical cancer research: Animal models for the natural history of cancer. Meeting held at Woods Hole, MA (USA), June 21-23, 1999. International Union Against Cancer. Int J Cancer 2000;85:303-5 [DOI] [PubMed] [Google Scholar]
- 8.Naito S, Giavazzi R, Walker SM, et al. Growth and metastatic behavior of human tumor cells implanted into nude and beige nude mice. Clin Exp Metastasis 1987;5:135-46 [DOI] [PubMed] [Google Scholar]
- 9.Naito S, von Eschenbach AC, Fidler IJ. Different growth pattern and biologic behavior of human renal cell carcinoma implanted into different organs of nude mice. J Natl Cancer Inst 1987;78:377-85 [PubMed] [Google Scholar]
- 10.Teicher BA. Tumor models for efficacy determination. Mol Cancer Ther 2006;5:2435-43 [DOI] [PubMed] [Google Scholar]
- 11.Fu X, Guadagni F, Hoffman RM. A metastatic nude-mouse model of human pancreatic cancer constructed orthotopically with histologically intact patient specimens. Proc Natl Acad Sci U S A 1992;89:5645-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furukawa T, Kubota T, Watanabe M, et al. A novel “patient-like” treatment model of human pancreatic cancer constructed using orthotopic transplantation of histologically intact human tumor tissue in nude mice. Cancer Res 1993;53:3070-2 [PubMed] [Google Scholar]
- 13.Furukawa T, Kubota T, Watanabe M, et al. Differential chemosensitivity of local and metastatic human gastric cancer after orthotopic transplantation of histologically intact tumor tissue in nude mice. Int J Cancer 1993;54:397-401 [DOI] [PubMed] [Google Scholar]
- 14.An Z, Jiang P, Wang X, et al. Development of a high metastatic orthotopic model of human renal cell carcinoma in nude mice: benefits of fragment implantation compared to cell-suspension injection. Clin Exp Metastasis 1999;17:265-70 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Yin L, Zhang Y, et al. In situ transduction of cytosine deaminase gene followed by systemic use of 5-fluorocytosine inhibits tumor growth and metastasis in orthotopic prostate cancer mouse models. Chin Med J (Engl) 2002;115:227-31 [PubMed] [Google Scholar]
- 16.Boyd DD, Kim SJ, Wang H, et al. A urokinase-derived peptide (A6) increases survival of mice bearing orthotopically grown prostate cancer and reduces lymph node metastasis. Am J Pathol 2003;162:619-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd M. eds. Status of the NCI preclinical antitumor drug discovery screen. Philadelphia: Lippincott, 1989. [Google Scholar]
- 18.Kuo TH, Kubota T, Watanabe M, et al. Site-specific chemosensitivity of human small-cell lung carcinoma growing orthotopically compared to subcutaneously in SCID mice: the importance of orthotopic models to obtain relevant drug evaluation data. Anticancer Res 1993;13:627-30 [PubMed] [Google Scholar]
- 19.Manzotti C, Audisio RA, Pratesi G. Importance of orthotopic implantation for human tumors as model systems: relevance to metastasis and invasion. Clin Exp Metastasis 1993;11:5-14 [DOI] [PubMed] [Google Scholar]
- 20.Hoffman RM. Orthotopic is orthodox: why are orthotopic-transplant metastatic models different from all other models? J Cell Biochem 1994;56:1-3 [DOI] [PubMed] [Google Scholar]
- 21.Hoffman RM. Orthotopic metastatic mouse models for anticancer drug discovery and evaluation: a bridge to the clinic. Invest New Drugs 1999;17:343-59 [DOI] [PubMed] [Google Scholar]
- 22.Bloomston M, Zervos EE, Rosemurgy AS., 2nd Matrix metalloproteinases and their role in pancreatic cancer: a review of preclinical studies and clinical trials. Ann Surg Oncol 2002;9:668-74 [DOI] [PubMed] [Google Scholar]
- 23.Hann B, Balmain A.Building ‘validated’ mouse models of human cancer. Curr Opin Cell Biol 2001;13:778-84 [DOI] [PubMed] [Google Scholar]
- 24.Bibby MC, Sleigh NR, Loadman PM, et al. Potentiation of EO9 anti-tumour activity by hydralazine. Eur J Cancer 1993;29A:1033-5 [DOI] [PubMed] [Google Scholar]
- 25.Fidler IJ, Wilmanns C, Staroselsky A, et al. Modulation of tumor cell response to chemotherapy by the organ environment. Cancer Metastasis Rev 1994;13:209-22 [DOI] [PubMed] [Google Scholar]
- 26.Dong Z, Radinsky R, Fan D, et al. Organ-specific modulation of steady-state mdr gene expression and drug resistance in murine colon cancer cells. J Natl Cancer Inst 1994;86:913-20 [DOI] [PubMed] [Google Scholar]
- 27.Killion JJ, Radinsky R, Fidler IJ. Orthotopic models are necessary to predict therapy of transplantable tumors in mice. Cancer Metastasis Rev 1998-1999;17:279-84 [DOI] [PubMed] [Google Scholar]
- 28.Yang M, Jiang P, Sun FX, et al. A fluorescent orthotopic bone metastasis model of human prostate cancer. Cancer Res 1999;59:781-6 [PubMed] [Google Scholar]
- 29.Rosol TJ, Tannehill-Gregg SH, LeRoy BE, et al. Animal models of bone metastasis. Cancer 2003;97:748-57 [DOI] [PubMed] [Google Scholar]
- 30.Le Dévédec SE, van Roosmalen W, Maria N, et al. An improved model to study tumor cell autonomous metastasis programs using MTLn3 cells and the Rag2(-/-) gammac (-/-) mouse. Clin Exp Metastasis 2009;26:673-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonkers J, Derksen PW. Modeling metastatic breast cancer in mice. J Mammary Gland Biol Neoplasia 2007;12:191-203 [DOI] [PubMed] [Google Scholar]
- 32.O’Reilly MS, Holmgren L, Shing Y, et al. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell 1994;79:315-28 [DOI] [PubMed] [Google Scholar]
- 33.Talmadge JE, Singh RK, Fidler IJ, et al. Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am J Pathol 2007;170:793-804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ottewell PD, Coleman RE, Holen I. From genetic abnormality to metastases: murine models of breast cancer and their use in the development of anticancer therapies. Breast Cancer Res Treat 2006;96:101-13 [DOI] [PubMed] [Google Scholar]
- 35.Kim MY, Oskarsson T, Acharyya S, et al. Tumor self-seeding by circulating cancer cells. Cell 2009;139:1315-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagahashi M, Ramachandran S, Kim EY, et al. Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res 2012;72:726-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagahashi M, Ramachandran S, Rashid OM, et al. Lymphangiogenesis: a new player in cancer progression. World J Gastroenterol 2010;16:4003-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rashid OM, Nagahashi M, Ramachandran S, et al. Resection of the primary tumor improves survival in metastatic breast cancer by reducing overall tumor burden. Surgery 2013;153:771-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding L, Ellis MJ, Li S, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010;464:999-1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dumur CI, Nasim S, Best AM, et al. Evaluation of quality-control criteria for microarray gene expression analysis. Clin Chem 2004;50:1994-2002 [DOI] [PubMed] [Google Scholar]
- 41.Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003;31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Storey JD. A direct approach to false discovery rates. J R Statist Soc B 2002;64:479-98 [Google Scholar]
- 43.Cabioglu N, Summy J, Miller C, et al. CXCL-12/stromal cell-derived factor-1alpha transactivates HER2-neu in breast cancer cells by a novel pathway involving Src kinase activation. Cancer Res 2005;65:6493-7 [DOI] [PubMed] [Google Scholar]
- 44.Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005;121:335-48 [DOI] [PubMed] [Google Scholar]
- 45.Viola RJ, Provenzale JM, Li F, et al. In vivo bioluminescence imaging monitoring of hypoxia-inducible factor 1alpha, a promoter that protects cells, in response to chemotherapy. AJR Am J Roentgenol 2008;191:1779-84 [DOI] [PubMed] [Google Scholar]
- 46.Tao K, Fang M, Alroy J, et al. Imagable 4T1 model for the study of late stage breast cancer. BMC Cancer 2008;8:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747-52 [DOI] [PubMed] [Google Scholar]
- 48.van ’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;415:530-6 [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 2005;365:671-9 [DOI] [PubMed] [Google Scholar]
- 50.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature 2005;436:518-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004;117:927-39 [DOI] [PubMed] [Google Scholar]
- 52.Gershwin ME, Ikeda RM, Kawakami TG, et al. Immunobiology of heterotransplanted human tumors in nude mice. J Natl Cancer Inst 1977;58:1455-61 [DOI] [PubMed] [Google Scholar]