Abstract
Synaptophysin is an integral synaptic vesicle (SV) protein that accounts for ∼10% of total SV protein cargo. Deletion of synaptophysin results in the defective retrieval of synaptobrevin II (sybII) from the plasma membrane during endocytosis, coupled with a slowing in the speed of endocytosis. Synaptophysin has been implicated in X-linked intellectual disability, with a recent study identifying four separate synaptophysin gene mutations in families affected by the disorder. To determine how these mutations may affect synaptophysin function, we expressed them in cultured neurons derived from synaptophysin knock-out mice. Two distinct truncating mutants were mislocalized throughout the axon and phenocopied the arrest of sybII retrieval in synaptophysin knock-out cultures. The remaining two mutants displayed a nerve terminal localization but did not support efficient sybII retrieval. Interestingly, one mutant fully rescued SV endocytosis kinetics, suggesting that sybII retrieval and endocytosis speed are independent from each other. These studies suggest that the efficient retrieval of sybII by synaptophysin may be key to maintaining synaptic health and perturbation of this event may contribute to the pathogenesis underlying neurodevelopmental disorders such as X-linked intellectual disability.
Introduction
Intellectual disability has a prevalence of ∼2% in the general population (Ropers and Hamel, 2005) and X-linked intellectual disability (XLID; also known as X-linked mental retardation) accounts for 5–10% of intellectual disability in males (Lubs et al., 2012). XLID is a heterogeneous group of conditions and, accordingly, a large number of X-chromosome gene mutations have been linked to this disorder (Ropers and Hamel, 2005; Lubs et al., 2012). A large-scale systematic sequencing of families containing multiple individuals with intellectual disability identified four mutations in the gene encoding synaptophysin that segregated with the condition and were potentially causal for the individuals affected (Tarpey et al., 2009).
Synaptophysin is a four transmembrane integral synaptic vesicle (SV) protein comprising ∼10% of total SV protein content (Takamori et al., 2006). Despite its abundance, synaptophysin knock-out mice reproduce normally and have no gross ultrastructural defects (Eshkind and Leube, 1995; McMahon et al., 1996). Synaptophysin knock-out mice do, however, display mild behavioral alterations and defects in learning and memory (Schmitt et al., 2009), suggesting a defect in synaptic function.
Recently, two aspects of presynaptic dysfunction have been identified in synaptophysin knock-out mice. First, nerve terminals from these mice displayed a reduced speed of SV endocytosis (Gordon et al., 2011; Kwon and Chapman, 2011). Second, these mice exhibited a profound deficit in the retrieval of a specific SV cargo from the plasma membrane, the integral SV protein synaptobrevin II (sybII) (Gordon et al., 2011). SybII drives SV fusion via interactions with the plasma membrane SNARE (soluble NSF attachment protein receptor) proteins syntaxin and SNAP-25 (Südhof, 2004) via an α-helical SNARE motif proximal to its transmembrane domain (Sutton et al., 1998).
Because synaptophysin is essential for efficient sybII retrieval, we investigated whether identified XLID mutants display defects in this process. To this end, we replicated XLID mutations in human synaptophysin (hSyp) and determined whether they could rescue function in cultured neurons derived from synaptophysin knock-out mice. We found that all XLID hSyp variants displayed a reduced ability to retrieve sybII from the plasma membrane during endocytosis, with different effects on SV endocytosis kinetics. Therefore, inefficient sybII retrieval may underlie neuronal defects in XLID patients harboring synaptophysin mutations.
Materials and Methods
Materials.
hSyp clone cDNA BC064550 was obtained from Source Bioscience. Synaptobrevin II-pHlourin (sybII-pHluorin) and vesicular glutamate transporter 1 (vGLUT1)-pHluorin constructs were provided by Prof. G. Miesenbock (Oxford University) and Prof. R. Edwards (University of California), respectively. Neurobasal media, B-27 supplement, penicillin/streptomycin, MEM, and lipofectamine 2000 were obtained from Invitrogen. All other reagents were obtained from Sigma-Aldrich.
Plasmid preparation.
hSyp clone cDNA (full-length human synaptophysin gene plus ∼1500 bp of downstream intronic sequence) was obtained in a pCMV-SPORT6 vector. Primers were generated to encompass the entire hSyp coding sequence plus 1267 bases of downstream sequence (forward primer: TCTCGAGCTATGCTGCTGC TGGCGGACATGGACG, reverse primer: GGATCCGCCATATCACTCGACTCTTCTCAGG, restriction sites underlined). The 2 kb PCR transcript was digested with XhoI and BamHI and ligated into a pEGFP-C1 vector backbone (Clontech) in which the coding sequence for EGFP was replaced by mCerulean (mCer; Rizzo et al., 2004) using AgeI and BsrGI enzymes. The hSyp clone contained a point mutation (Pro196Leu), which was reverted back to its original form by mutagenesis (forward: GGAGCTGAGAGACCCTGTGACCTCGG, reverse: CCGAGGTCACAGGGTCTCTCAGCTCC). Human XLID synaptophysin mutations were then introduced into mCer-hSyp via mutagenesis (mutated bases underlined: 177–178CA>GT, forward: GGATTGTGCCAAGTAGACCGAGAGTGACCTCAGC, reverse: GCTGAGGTCACTCTCGGTCTACTTGGCACAATCC; 274–275insA, forward: CGAGGGGGCACCAACCAAGGTCTTCTTAG, reverse: CTAAGAAGACCTTGGTTGGTGCCCCCTCG; 649G>C, forward: GGTGCTCTGGGTCCGCAACCTGTGG, reverse: CCACAGGTTGCGGACCCAGAGCACC; 829–832delGACT, position of deleted bases highlighted, forward: GCTACCAGCCT- - - -ATGGTCAACCAGCCGGC, reverse: GCCGGCTGGTTGACCAT- - - -AGGCTGGTAGC) and confirmed by sequencing.
Cortical neuronal cultures.
Synaptophysin knock-out mice were maintained as heterozygous breeding pairs on a C56BL/6J background (Gordon et al., 2011). Dissociated primary cortical neuronal cultures were prepared from both embryonic day 17.5 (E17.5) knock-out and wild-type embryos of either sex and then cotransfected with both mCer vectors and pHluorin reporter using lipofectamine 2000 after 7 d in culture (Gordon et al., 2011). Cells were imaged after 13–16 d in culture.
Fluorescent imaging protocols.
Cortical cultures were mounted in a Warner imaging chamber with embedded parallel platinum wires (RC-21BRFS) and placed on the stage of Zeiss Axio Observer D1 epifluorescence microscope. Neurons transfected with mCer vectors were visualized with a Zeiss PlanNeofluar 40× oil-immersion objective (numerical aperture 1.3) at 430 nm excitation, whereas pHluorin reporters were visualized at 500 nm (both using a 525 nm dichroic and long-pass emission filter >535 nm). In all experiments, cultures were stimulated with a train of 300 action potentials delivered at 10 Hz (100 mA, 1 ms pulse width). Cultures were subjected to continuous perfusion with imaging buffer containing the following (in mm): 136 NaCl, 2.5 KCl, 2 CaCl2, 1.3 MgCl2, 10 glucose, and 10 HEPES, pH 7.4. Fluorescent images were captured at 4 s intervals (exposure times ranged between 0.9 and 1.3 s) using a Hamamatsu Orca-ER digital camera and processed offline using ImageJ version 1.43 software. Regions of interest of identical size were placed over nerve terminals and the total fluorescence intensity was monitored over time. Only regions that responded to action potential stimulation were selected for analysis. All statistical analyses were performed using Microsoft Excel and GraphPad Prism software. The pHluorin fluorescence change was calculated as ΔF/F0, and n refers to the number of individual coverslips examined.
The diffuseness of mCer-hSyp variants and sybII-pHluorin fluorescence along axons was determined by calculating the coefficient of variation (CV; Lyles et al., 2006), which was determined by drawing a >100 pixel line over an axonal segment and determining the SD/mean fluorescence signal. The CV for sybII-pHluorin was calculated after exposure of the cells to alkaline imaging buffer (50 mm NH4Cl substituted for 50 mm NaCl) to reveal total sybII-pHluorin fluorescence, whereas mCer-hSyp CV was calculated after exposure to acidic imaging buffer (20 mm MES substituted for 20 mm HEPES, pH 5.5) to quench contaminating pHluorin fluorescence (in both cases, n refers to the mean of 5 different axonal segments on an individual neuron on a single coverslip).
Results
Synaptophysin XLID mutants display different localization profiles
Mutations in hSyp segregate with XLID in a number of affected families, implicating hSyp in this condition (Tarpey et al., 2009). These mutations affect both the sequence and reading frame of hSyp (Fig. 1A). 177-178CA>GT introduces a premature stop codon that truncates hSyp after 59 aa (hSyp59). 274-275insA produces a frameshift that results in 45 new amino acids being expressed and truncates the protein after 136 aa (hSyp136). 649G>C introduces an amino acid substitution in the fourth transmembrane domain of the protein (hSypG217R). Finally, the 829-832delGACT mutation produces a frameshift in the cytoplasmic C-terminal tail of hSyp, resulting in the expression of 156 new amino acids. This mutant is therefore 469 aa long (hSyp469), whereas the wild-type hSyp is 313 aa long.
Figure 1.
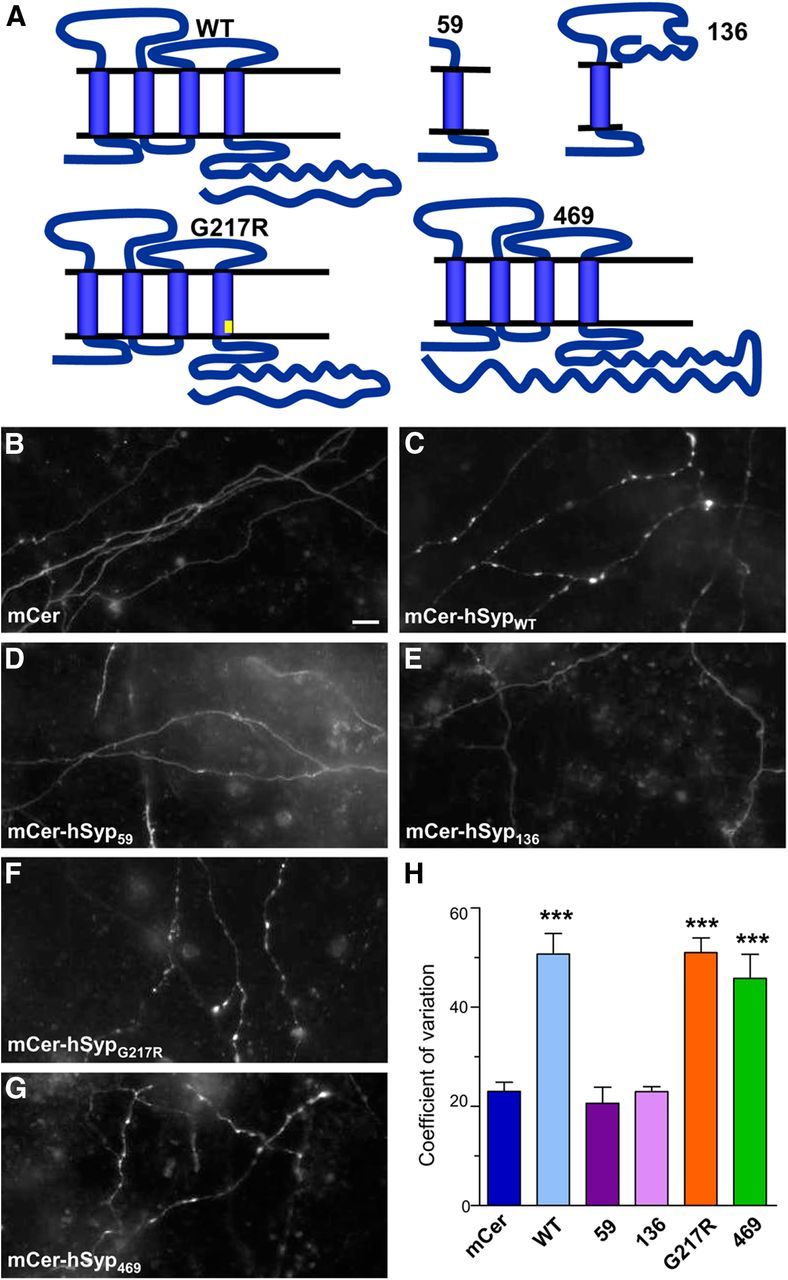
XLID mCer-hSyp mutants display different localization profiles. A, Illustration depicting putative mutant hSyp structures. Compared with the full-length wild-type (WT) protein, hSyp59 is a truncation after the first transmembrane domain; hSyp136 is a truncation and frame shift after the first transmembrane domain; hSypG217R is a point mutation in the fourth transmembrane domain (G217R); and hSyp469 is a frameshift resulting in an extension of the C terminus. Synaptophysin knock-out cortical neurons were cotransfected with sybII-pHluorin and either empty mCer vector or mCer-hSyp variants. Representative images show the differential distribution of the hSyp proteins along axonal segments after quenching pHluorin fluorescence with acidic imaging buffer (B, mCer; C, mCer-hSypWT; D, mCer-hSyp59; E, mCer-hSyp136; F, mCer-hSypG217R; and G, mCer-hSyp469). Scale bar, 10 μm. H, Bar graph displaying the CV of mCer fluorescence intensity along axonal segments. Data are presented as mean ± SEM, n = 5 for all. ***p < 0.001, one-way ANOVA with Bonferroni's multiple-comparison post test for mCer-hSypWT, mCer-hSypG217R, and mCer-hSyp469 against mCer, mCer-hSyp59, or mCer-hSyp136.
To determine how these XLID mutations might affect hSyp function, mCer-tagged mutants were coexpressed in cultured synaptophysin knock-out neurons with the genetic reporter sybII-pHluorin. We first determined whether XLID mutations could influence the targeting of hSyp to SVs by examining their neuronal localization. The mCer empty vector displays a diffuse localization throughout the axon, whereas wild-type (mCer-hSypWT) is distributed in a punctate fashion, indicating its localization to nerve terminals (Fig. 1B,C). The truncated hSyp mutants mCer-hSyp59 and mCer-hSyp136 displayed a similar distribution profile to the empty vector (Fig. 1D,E), whereas mCer-hSypG217R and mCer-hSyp469 mimicked the distribution of mCer-hSypWT (Fig 1F,G). To quantify the axonal distribution of hSyp variants, the CV was analyzed. CV quantifies the diffuseness of protein distribution, with a lower CV indicating a homogeneous localization along the axon and a higher CV signifying a punctate distribution (Lyles et al., 2006). When this analysis was performed, the CV of mCer-hSypWT, mCer-hSypG217R, and mCer-hSyp469 was significantly higher than that of mCer, mCer-hSyp59, and mCer-hSyp136 (Fig. 1H). Therefore, hSyp59 and hSyp136 mutations result in mislocalization of the protein, whereas hSypG217R and hSyp469 retained a distribution commensurate with a nerve terminal localization.
Synaptophysin XLID mutants differentially affect sybII-pHluorin distribution
The absence of synaptophysin causes a mislocalization of sybII from nerve terminals, with both endogenous and exogenous sybII displaying a diffuse axonal distribution (Gordon et al., 2011). Therefore, we next examined the ability of XLID mutants to rescue the punctate localization of the genetic reporter sybII-pHluorin in synaptophysin knock-out neurons. Expression of mCer-hSypWT results in a punctate localization of sybII-pHluorin, whereas in neurons expressing the empty mCer vector, sybII-pHluorin is diffuse (Fig. 2A,B). In neurons expressing either mCer-hSyp59 or mCer-hSyp136, sybII-pHluorin is localized diffusely along axons (Fig. 2C,D), closely mimicking the distribution of these hSyp mutants (Fig. 1). In knock-out neurons rescued with either mCer-hSypG217R (Fig. 2E) or mCer-hSyp469 (Fig. 2F), sybII-pHluorin displays a punctate distribution profile similar to that of mCer-hSypWT, suggesting that both mutants can effectively target sybII to nerve terminals. When CV analysis was performed, the value for sybII-pHluorin was significantly lower in neurons transfected with mCer, mCer-hSyp59, or mCer-hSyp136 compared with neurons expressing mCer-hSypWT, mCer-hSypG217R, or mCer-hSyp469 (Fig. 2G). Therefore, sybII cannot be efficiently targeted to nerve terminals by either mCer-hSyp59 or mCer-hSyp136.
Figure 2.
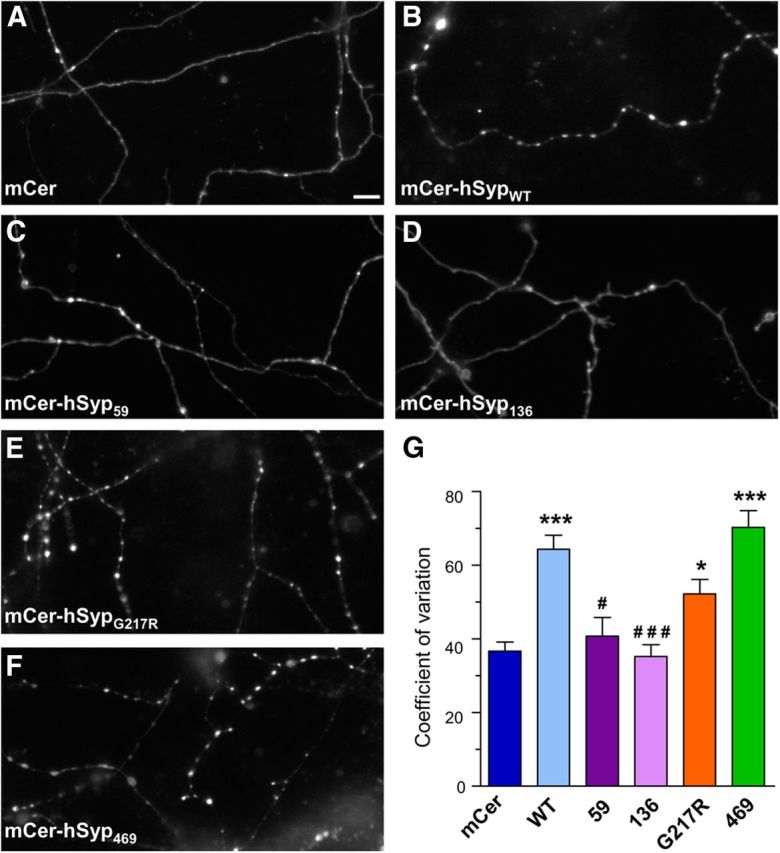
XLID mCer-hSYP mutants differentially affect the localization of sybII-pHluorin. Synaptophysin knock-out cortical neurons were cotransfected with sybII-pHluorin and either empty mCer vector or mCer-hSyp variants. Representative images show the differential distribution of sybII-pHluorin along axonal segments after exposure to alkaline imaging buffer (A, mCer; B, mCer-hSypWT; C, mCer-hSyp59; D, mCer-hSyp136; E, mCer-hSypG217R; F, mCer-hSyp469). Scale bars, 10 μm. G, Bar graph displaying the CV of sybII-pHluorin fluorescence intensity along axonal segments. Data are presented as mean ± SEM. n = 4 for mCer-hSyp59 and mCer-hSyp136; n = 5 for mCer-hSyp469; n = 8 for mCer and mCer-hSypWT; and n = 11 for mCer-hSypG217R. *p < 0.05 and ***p < 0.001, one-way ANOVA with Bonferroni's multiple-comparison post test for mCer-hSypWT, mCer-hSypG217R, and mCer-hSyp469 against mCer; #p < 0.05 and ###p < 0.001 for mCer-hSyp59 and mCer-hSyp136 against mCer-hSypWT.
XLID mutants do not rescue sybII-pHluorin retrieval
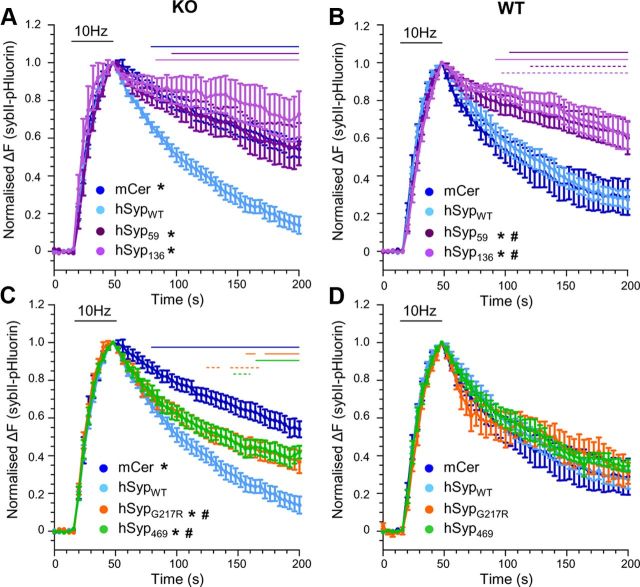
The altered axonal distribution of sybII-pHluorin in synaptophysin knock-out neurons is due to a defect in sybII retrieval from the plasma membrane during endocytosis (Gordon et al., 2011). Therefore we next investigated whether expression of hSyp XLID mutants also perturbed sybII-pHluorin retrieval. The pHluorin tag reports changes in the pH of its local environment, so the observed fluorescence increase on stimulation is due to SV exocytosis. After stimulation, a fluorescence decrease occurs, representing SV endocytosis followed by SV acidification. Changes in this fluorescent downstroke strictly reports sybII retrieval, because SV acidification is unaffected in synaptophysin knock-out mice (Kwon and Chapman, 2011). Synaptophysin knock-out neurons cotransfected with both mCer-hSyp mutants and sybII-pHluorin were stimulated with a train of 300 action potentials (10 Hz) to trigger SV recycling. SybII-pHluorin fluorescence decreased after stimulation in neurons rescued with mCer-hSypWT, in contrast to empty vector, in which the decrease in fluorescence was significantly slowed (Fig. 3A). When truncated variants of hSyp were examined, a pronounced defect in poststimulation sybII-pHluorin retrieval was observed for both mCer-hSyp59 and mCer-hSyp136 (Fig. 3A). Therefore, sybII-pHluorin retrieval is not supported by truncated XLID hSyp mutants.
Figure 3.
XLID-hSyp mutants differentially affect the retrieval of sybII-pHluorin. Synaptophysin knock-out (KO; A, C) or wild-type (WT; B, D) cortical neurons were cotransfected with sybII-pHluorin and either mCer or mCer-hSyp variants. Cultures were stimulated with a train of 300 action potentials (10 Hz, indicated by bar). Time course of mean ΔF/F0 of sybII-pHluorin ± SEM is displayed for mCer (n = 10 KO; n = 4 WT) and mCer-SypWT (n = 9 KO, n = 5 WT), mCer-hSyp59 (n = 5 KO, n = 9 WT), mCer-hSyp136 (n = 5 KO, n = 7 WT), mCer-hSypG217R (n = 9 KO, n = 4 WT), and mCer-hSyp469 (n = 8 KO, n = 6 WT). *p < 0.05, two-way ANOVA against mCer-hSypWT (over times indicated by solid lines); #p < 0.05, two-way ANOVA against mCer (over times indicated by dashed lines).
Both hSyp59 and hSyp136 mutants represent severe truncations of synaptophysin; therefore, we predicted that they may be dominant negative when expressed in wild-type neurons. To determine this, we examined the retrieval of sybII-pHluorin in wild-type neurons coexpressing either mCer-hSyp59 or mCer-hSyp136. Expression of either of these mutants produced significant dominant-negative effects on sybII-pHluorin retrieval compared with either mCer-hSypWT or mCer empty vectors (Fig. 3B). Therefore, expression of truncated XLID hSyp mutants in wild-type neurons perturbs the ability of endogenous synaptophysin to retrieve sybII successfully.
In contrast to the truncated hSyp mutants, hSypG217R and hSyp469 both displayed a subcellular localization indicative of targeting to SVs. We next investigated whether these mutants could rescue sybII-pHluorin retrieval in synaptophysin knock-out neurons. These mutants displayed a partial rescue of sybII-pHluorin retrieval compared with mCer alone; however, they did not fully rescue to mCer-hSypWT levels (Fig. 3C). Neither mutant affected sybII-pHluorin retrieval when expressed in wild-type neurons, indicating no interference with endogenous synaptophysin (Fig. 3D). Therefore, the full-length XLID mutants hSypG217R and hSyp469 display defects in sybII retrieval despite being correctly localized to nerve terminals.
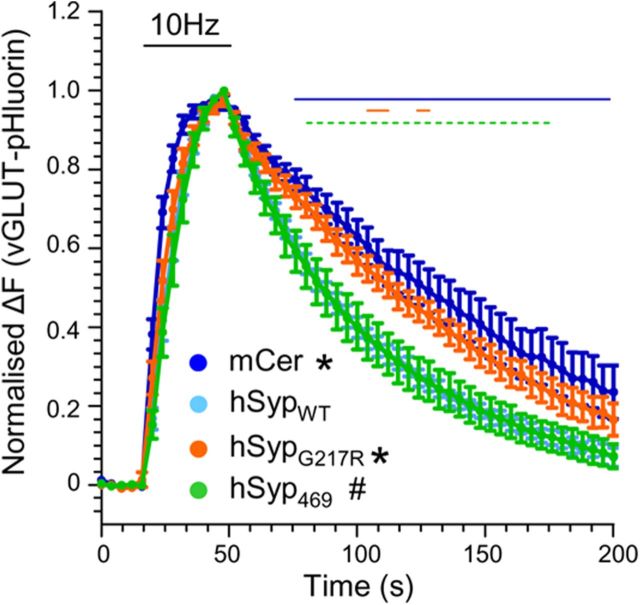
hSYPG217R and hSYP469 differentially regulate the rate of SV endocytosis
The absence of synaptophysin has no global effect on SV turnover; however, it does reduce the speed of SV endocytosis (Gordon et al., 2011; Kwon and Chapman, 2011). Therefore, we next determined whether XLID hSyp mutants could rescue SV endocytosis kinetics by coexpressing the genetic reporter vGLUT-pHluorin (Voglmaier et al., 2006). We were unable to examine the effect of either mCer-hSyp59 or mCer-hSyp136 because both were toxic when expressed in the absence of exogenous sybII (data not shown). Therefore, we limited our study to mCer-hSypG217R and mCer-hSyp469. After stimulation with 300 action potentials (10 Hz), vGLUT-pHluorin recovery was significantly slower in synaptophysin knock-out neurons expressing the empty mCer vector compared with mCer-hSypWT (Fig. 4; Gordon et al., 2011). There was no significant effect of vGLUT-pHluorin localization (data not shown), indicating this was purely a kinetic defect. Expression of mCer-hSypG217R failed to rescue vGLUT-pHluorin retrieval kinetics significantly, whereas mCer-hSyp469 fully rescued vGLUT-pHluorin retrieval in a manner identical to mCer-hSypWT (Fig. 4). Therefore, although both hSypG217R and hSyp469 display defects in sybII-pHluorin retrieval, only hSyp469 is able to fully rescue deficits in the speed of SV endocytosis.
Figure 4.
hSypG217R and hSyp469 differentially affect the retrieval of vGLUT-pHluorin. Synaptophysin knock-out cortical neurons were cotransfected with vGLUT-pHluorin and either mCer or mCer-hSyp variants and stimulated with a train of 300 action potentials (10 Hz, indicated by bar). Time course of mean ΔF/F0 of vGLUT-pHluorin ± SEM is displayed for mCer (n = 11), mCer-hSypWT (n = 10), mCer-hSypG217R (n = 9), and mCer-hSyp469 (n = 7). *p < 0.05, two-way ANOVA against mCer-hSypWT (over times indicated by solid line); #p < 0.05, two-way ANOVA against mCer (over times indicated by dashed lines).
Discussion
Synaptophysin is one of the most abundant proteins on SVs and is required for the efficient retrieval of sybII from the plasma membrane after exocytosis (Gordon et al., 2011). We show here that a range of mutant variants of hSyp identified in a screen of XLID families differentially affected both the localization and function of the protein. None of the XLID hSyp mutants examined could fully rescue sybII retrieval in synaptophysin knock-out neurons. Truncation of hSyp after its first transmembrane domain resulted in a dominant-negative form of the protein, which was mislocalized from nerve terminals and abrogated sybII-pHluorin retrieval, even in wild-type neurons. Full-length hSyp mutations displayed more subtle defects in sybII-pHluorin retrieval and a divergence in their ability to rescue the speed of SV endocytosis.
The hSyp59 and hSyp136 truncation mutants were dominant negative, with their expression in wild-type neurons inhibiting sybII-pHluorin retrieval to levels observed in synaptophysin knock-out neurons. An interesting observation was that these mutants were toxic to neurons unless they were coexpressed with exogenous sybII. In the presence of exogenous sybII, these hSyp mutants exit the Golgi and are diffusely localized along the length of the axon. This is in contrast to synaptophysin mutants lacking their N-glycosylation site, which are retained in the neuronal cell body (Kwon and Chapman, 2012). The fact that truncated hSyp mutants exit the secretory pathway when coexpressed with sybII suggests that their traffic is interdependent on each other. SybII interacts with synaptophysin in vivo (Calakos and Scheller, 1994; Edelmann et al., 1995) and synaptophysin has been proposed to facilitate the sorting of sybII to SVs (Pennuto et al., 2003; Bonanomi et al., 2007). The exact interaction site on synaptophysin for sybII has not been determined; however, both binding and trafficking assays suggest that the cytoplasmic C terminus is dispensable and that it is mediated via hydrophobic interactions through their transmembrane domains (Yelamanchili et al., 2005; Bonanomi et al., 2007; Felkl and Leube, 2008). The fact that exogenous sybII rescues survival in neurons expressing hSyp truncation mutants suggests that the first transmembrane domain of synaptophysin forms at least part of the sybII interaction site.
The dominant-negative nature of the truncated hSyp mutants may provide clues as to the mechanism of synaptophysin function in nerve terminals. Synaptophysin forms a hexameric structure on SVs (Arthur and Stowell, 2007), so these mutants may disrupt the function of endogenous synaptophysin by precluding association into correct multimeric structures that may be crucial for its role. It should be noted that in a disease context, mutant hSyp would not be coexpressed with hSypWT, because random silencing of the X-chromosome would prohibit this from occurring in females carrying XLID hSyp mutants.
Full-length XLID hSyp variants hSypG217R and hSyp469 are localized to nerve terminals but display defective sybII-pHluorin retrieval. Interestingly, hSyp469 completely rescues the speed of endocytosis in synaptophysin knock-out neurons, whereas hSypG217R does not. This was not due to differences in nerve terminal expression levels because both mutants displayed equal expression to mCer-hSypWT (% expression to WT; mCer-hSypG217R 121 ± 19; mCer-hSyp469 102 ± 12). We previously suggested that the slowing of SV endocytosis in synaptophysin knock-out mice was due to inefficient sybII retrieval, because a similar phenotype was observed in sybII knock-out synapses (Deák et al., 2004). These two hSyp mutants have demonstrated that control of sybII retrieval can be decoupled from SV endocytosis kinetics, suggesting that they are independently controlled by synaptophysin. The point mutation in the fourth transmembrane domain of hSypG217R affects both sybII-pHluorin retrieval and vGLUT-pHluorin speed, suggesting a general defect in synaptophysin function. Consistent with this, preliminary experiments to examine the trafficking of this mutant during neuronal activity suggest that its reentry back into nerve terminals is retarded, unlike hSyp469, which mimics wild-type hSyp (S. Gordon, unpublished observations). The selective deficit in sybII-pHluorin retrieval observed with hSyp469 suggests that its extended C-terminal cytosolic domain contains additional motifs that disrupt this event because all other aspects of its function correlate with wild-type hSyp.
The ability of hSyp469 to display a wild-type functional profile with the exception of inefficient sybII-pHluorin retrieval suggests that it is specifically this event that may be deleterious to individuals carrying hSyp mutations in XLID. Consistent with this, all hSyp mutants have an inability to fully rescue sybII retrieval. This effect should not be restricted to sybII-pHluorin because endogenous sybII is mislocalized to the same extent as the reporter in synaptophysin knock-out neurons (Gordon et al., 2011). Inefficient sybII retrieval still results in efficient neurotransmitter release and does not perturb global SV turnover in synaptophysin knock-out mice (McMahon et al., 1996; Gordon et al., 2011; Kwon and Chapman, 2011). The absence of an immediate consequence of perturbed sybII retrieval is potentially due to the excess reservoir of sybII present on SVs. Current estimates suggest that there are ∼60–70 copies of sybII on a SV (Takamori et al., 2006); however, <10 (possibly even only 1–3) copies of sybII are required for SV fusion (Mohrmann et al., 2010; van den Bogaart et al., 2010; Sinha et al., 2011). Therefore, rather than an immediate reduction in neurotransmission, it is likely that deficient sybII retrieval will have a subtle effect on synaptic efficiency over time. This may be exacerbated during high-intensity stimulation, resulting in small deficiencies in plastic changes at the synapse due to experience. Consistent with this, the only behavioral alteration identified in synaptophysin knock-out mice to date is a mild deficit in learning and memory (Schmitt et al., 2009).
The discovery of a series of mutations in hSyp during a large-scale study into familial XLID has linked synaptophysin to this disorder (Tarpey et al., 2009). Although they only account for a small percentage of the XLID cases, these mutations were highly penetrant, implicating synaptophysin in disease causation for these patients. We show in this study that all hSyp mutants examined were dysfunctional for the retrieval of sybII during endocytosis, providing a possible mechanistic basis for the condition. These XLID mutations have provided clues to the mechanism of action of synaptophysin in sybII retrieval and have illustrated that synaptophysin may control sybII retrieval independently of SV endocytosis kinetics. Finally, our results provide a valuable insight into how small perturbations in the efficiency of presynaptic function could translate into human disease.
Footnotes
This work was supported by the Wellcome Trust (Grant #088138). We thank R. Leube for providing the synaptophysin knock-out mice and Prof. G. Miesenbock (Oxford University) and Prof. R. Edwards (University of California) for the gift of sybII-pHluorin and vGLUT1-pHluorin, respectively.
The authors declare no competing financial interests.
References
- Arthur CP, Stowell MH. Structure of synaptophysin: a hexameric MARVEL-domain channel protein. Structure. 2007;15:707–714. doi: 10.1016/j.str.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanomi D, Rusconi L, Colombo CA, Benfenati F, Valtorta F. Synaptophysin I selectively specifies the exocytic pathway of synaptobrevin 2/VAMP2. Biochem J. 2007;404:525–534. doi: 10.1042/BJ20061907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calakos N, Scheller RH. Vesicle-associated membrane protein and synaptophysin are associated on the synaptic vesicle. J Biol Chem. 1994;269:24534–24537. [PubMed] [Google Scholar]
- Deák F, Schoch S, Liu X, Südhof TC, Kavalali ET. Synaptobrevin is essential for fast synaptic-vesicle endocytosis. Nat Cell Biol. 2004;6:1102–1108. doi: 10.1038/ncb1185. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Hanson PI, Chapman ER, Jahn R. Synaptobrevin binding to synaptophysin: A potential mechanism for controlling the exocytotic fusion machine. EMBO J. 1995;14:224–231. doi: 10.1002/j.1460-2075.1995.tb06995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshkind LG, Leube RE. Mice lacking synaptophysin reproduce and form typical synaptic vesicles. Cell Tissue Res. 1995;282:423–433. doi: 10.1007/BF00318874. [DOI] [PubMed] [Google Scholar]
- Felkl M, Leube RE. Interaction assays in yeast and cultured cells confirm known and identify novel partners of the synaptic vesicle protein synaptophysin. Neuroscience. 2008;156:344–352. doi: 10.1016/j.neuroscience.2008.07.033. [DOI] [PubMed] [Google Scholar]
- Gordon SL, Leube RE, Cousin MA. Synaptophysin Is Required for Synaptobrevin Retrieval during Synaptic Vesicle Endocytosis. J Neurosci. 2011;31:14032–14036. doi: 10.1523/JNEUROSCI.3162-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SE, Chapman ER. Synaptophysin regulates the kinetics of synaptic vesicle endocytosis in central neurons. Neuron. 2011;70:847–854. doi: 10.1016/j.neuron.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SE, Chapman ER. Glycosylation is dispensable for sorting of synaptotagmin 1 but is critical for targeting of SV2 and synaptophysin to recycling synaptic vesicles. J Biol Chem. 2012;287:35658–35668. doi: 10.1074/jbc.M112.398883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubs HA, Stevenson RE, Schwartz CE. Fragile X and X-linked intellectual disability: four decades of discovery. Am J Hum Genet. 2012;90:579–590. doi: 10.1016/j.ajhg.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles V, Zhao Y, Martin KC. Synapse formation and mRNA localization in cultured Aplysia neurons. Neuron. 2006;49:349–356. doi: 10.1016/j.neuron.2005.12.029. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Bolshakov VY, Janz R, Hammer RE, Siegelbaum SA, Südhof TC. Synaptophysin, a major synaptic vesicle protein, is not essential for neurotransmitter release. Proc Natl Acad Sci U S A. 1996;93:4760–4764. doi: 10.1073/pnas.93.10.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann R, de Wit H, Verhage M, Neher E, Sørensen JB. Fast vesicle fusion in living cells requires at least three SNARE complexes. Science. 2010;330:502–505. doi: 10.1126/science.1193134. [DOI] [PubMed] [Google Scholar]
- Pennuto M, Bonanomi D, Benfenati F, Valtorta F. Synaptophysin I controls the targeting of VAMP2/synaptobrevin II to synaptic vesicles. Mol Biol Cell. 2003;14:4909–4919. doi: 10.1091/mbc.E03-06-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Ropers HH, Hamel BC. X-linked mental retardation. Nat Rev Genet. 2005;6:46–57. doi: 10.1038/nrg1501. [DOI] [PubMed] [Google Scholar]
- Schmitt U, Tanimoto N, Seeliger M, Schaeffel F, Leube RE. Detection of behavioral alterations and learning deficits in mice lacking synaptophysin. Neuroscience. 2009;162:234–243. doi: 10.1016/j.neuroscience.2009.04.046. [DOI] [PubMed] [Google Scholar]
- Sinha R, Ahmed S, Jahn R, Klingauf J. Two synaptobrevin molecules are sufficient for vesicle fusion in central nervous system synapses. Proc Natl Acad Sci U S A. 2011;108:14318–14323. doi: 10.1073/pnas.1101818108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, Müller SA, Rammner B, Gräter F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmüller H, Heuser J, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, O'Meara S, Latimer C, Dicks E, Menzies A, Stephens P, Blow M, Greenman C, Xue Y, Tyler-Smith C, Thompson D, Gray K, Andrews J, Barthorpe S, Buck G, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogaart G, Holt MG, Bunt G, Riedel D, Wouters FS, Jahn R. One SNARE complex is sufficient for membrane fusion. Nat Struct Mol Biol. 2010;17:358–364. doi: 10.1038/nsmb.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmaier SM, Kam K, Yang H, Fortin DL, Hua Z, Nicoll RA, Edwards RH. Distinct endocytic pathways control the rate and extent of synaptic vesicle protein recycling. Neuron. 2006;51:71–84. doi: 10.1016/j.neuron.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Yelamanchili SV, Reisinger C, Becher A, Sikorra S, Bigalke H, Binz T, Ahnert-Hilger G. The C-terminal transmembrane region of synaptobrevin binds synaptophysin from adult synaptic vesicles. Eur J Cell Biol. 2005;84:467–475. doi: 10.1016/j.ejcb.2004.11.007. [DOI] [PubMed] [Google Scholar]