Abstract
We report 1) the anchoring of preformed fluorescent DNA nanodevices (NDs) and 2) the in situ self-assembly of fluorescent DNA NDs on target living cell surfaces. Three types of aptamer-tethered DNA NDs were built and anchored on target cell surfaces by specific target-aptamer association. The in situ nanodevice self-assembly was further demonstrated on the surfaces of target living cells in cell mixtures. These DNA NDs exhibited fluorescence emission and underwent fluorescence energy transfer on living cell surfaces.
Keywords: aptamer, DNA nanodevices, fluorescent, self-assembly, cell-surface engineering
Cell membrane is the interface of the intracellular and extracellular environments, where intracellular biological activities are regulated via signal transduction between membrane-bound receptors and signaling molecules, including hormones, neurotransmitters, or therapeutics from complex extracellular environments.[1] In situ analysis and regulation of these key role players is necessary for both 1) a complete, comprehensive understanding of biological pathways with high spatial and temporal resolution and 2) specific biological and therapeutic applications. Previous tactics to accomplish these goals have involved cell-surface engineering through genetic approaches[2] and chemical modification.[3]
Cell-surface modification with proteins has typically been achieved through genetic engineering, in which cells are transformed with plasmids or transfected with viruses containing genes that encode proteins of interest. This results in protein expression and secretion, and, hence, modification of cell surfaces with recombinant proteins.[2] However, this approach suffers from intrinsic drawbacks, including complex manipulation, prolonged protein expression and secretion, limited range of protein targets, and difficult construction of nanostructures using these proteins.[3b]
As an alternative, chemical modification of cell surfaces with such moieties as proteins, DNA, or nanomaterials has provided a new direction for cell-surface engineering.[3a–g, 3i, 3j] The interface of chemistry, material sciences and biomedical sciences has provided many opportunities to modify and manipulate cells.[3b, 3g, 3i, 4] For example, using Staudinger ligation, Bertozzi et al. modified mammalian cell surfaces with DNA and imparted specific recognition capability to cells, enabling programmed assembly of 3-dimensional microtissues.[3e] Similarly, Peterson et al. modified cell surfaces with artificial human Fc receptor, which can recognize the Fc region of human IgG and internalize this antibody.[5] However, this approach is often associated with complicated chemical reactions, potential harmful effects on cells, and limited ability to modify specific target cells under complex environments.
Beyond these strategies, DNA combined with nucleic acid aptamers offers a simple method to engineer nanodevices in situ on target cell surfaces. Owing to unique Watson-Crick base pairing and sequence programmability, oligonucleotides have been extensively explored as building blocks for the construction of various DNA nanostructures.[6] Selected through a process known as Systematic Evolution of Ligands by EXponential enrichment (SELEX), aptamers, which are single-stranded DNA or RNA, can bind cell-specific biomarkers on target cell surfaces with high affinities.[7] Thus, aptamers make excellent molecular probes for selective recognition and can provide the basis for selective modification and manipulation of target cell surfaces under complex environments.
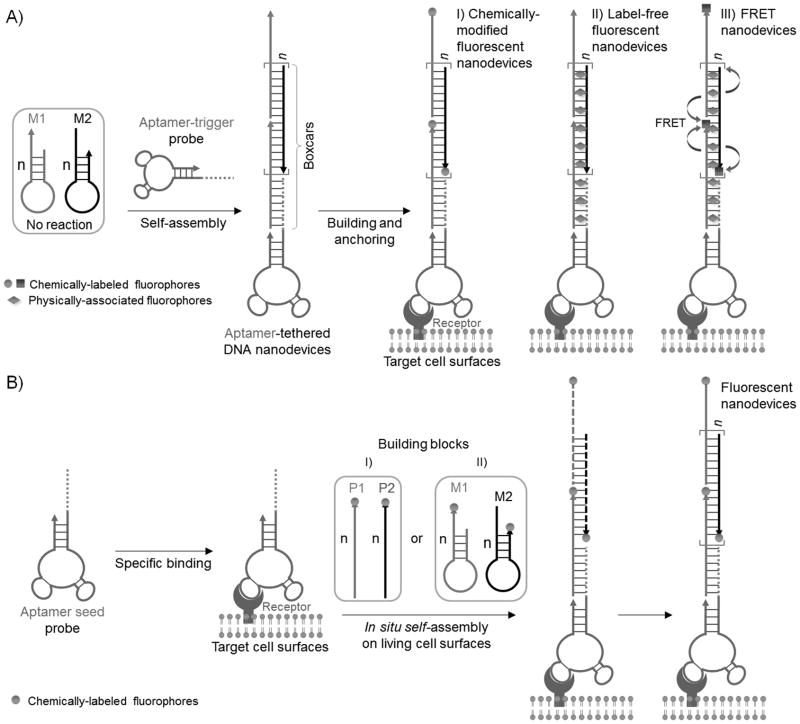
Based on aptamer-tethered DNA nanodevices (aptNDs), we report 1) the anchoring of preformed fluorescent aptNDs and 2) the in situ self assembly of fluorescent aptNDs, on target living cell surfaces. Fluorescence has been attractive for noninvasive sensing in living cells, and the ability of pin-point modification of DNA with fluorophores enables versatile application of fluorescence in DNA nanotechnology. The aptND is a long linear DNA nanostructure self-assembled via a hybridization chain reaction (HCR) (Scheme 1), by which many DNA nanoassemblies have been constructed.[8] The dsDNA portions in the preformed nanodevices work like a series of train boxcars, which can be loaded with fluorophores by either chemical modification on ssDNA monomers or physical association with dsDNA, while the aptamer moieties can guide the nanodevices to target cell surfaces. The features of multiple, repetitive, and alternating DNA building blocks in the resultant aptNDs provide an excellent platform for appropriate positioning of multi-chromophore arrays or multi-component nanofactories,[3g, 8b, 9] implicating the future in situ construction of nanofactories on target living cell surfaces for pinpoint biomolecular/pharmaceutical analysis or manipulation of biological activities.
Scheme 1.
illustration of the construction of fluorescent DNA nanodevices on target living cell surfaces based on an aptND platform, where A) three types of fluorescent DNA nanodevices, preformed via HCR-based self-assembly upon initiation by aptamer-tethered trigger probes, are anchored on target cell surfaces, or B) aptamer seed probes initiate in situ self-assembly of fluorescent DNA nanodevices on target cell surfaces by either I) cascading alternative hybridization of two partially complementary monomers or II) HCR.
We first designed two partially complementary hairpin monomers, M1 and M2, to construct aptNDs. These self-assembled nanodevices were anchored to the target cell surface as proof of concept. The stored energy in each loop of monomers was protected by the corresponding stem, preventing their hybridization and polymerization in the absence of an initiator trigger probe. Aptamers sgc8 and TDO5 were chosen to construct our model aptNDs. Sgc8 binds to target protein PTK7, which is overexpressed on target CEM cell surfaces, but not on nontarget Ramos cell surfaces.[10] TDO5 binds to the μ heavy chain of immunoglobulin M, which is overexpressed on Ramos cell surfaces, but not on CEM cells.[11]
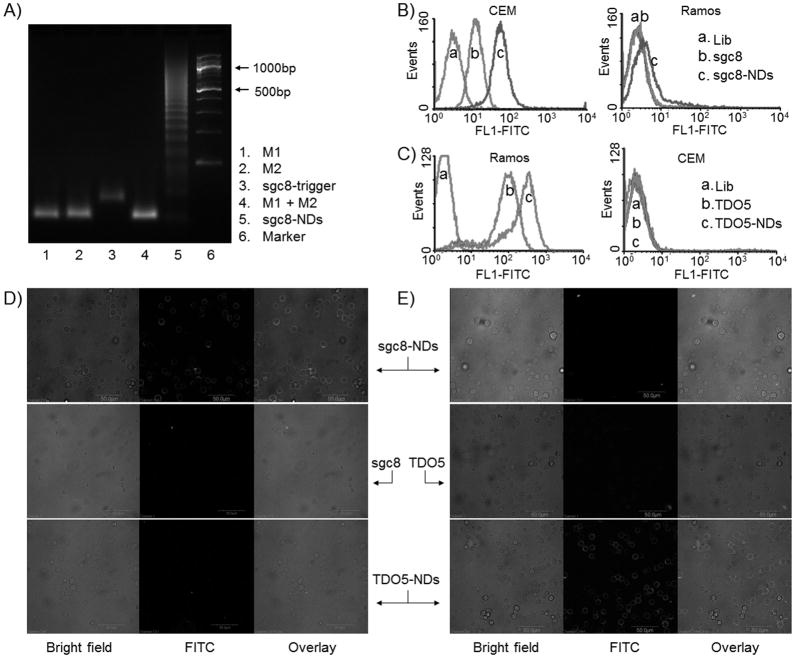
To engineer aptamer-tethered probes to trigger the self-assembly of nanodevices, a DNA initiator probe was modified on the 5′-ends of aptamers (aptamer-trigger; see sequences in Supporting Information, Table S1). The specific targeting abilities of the chimeric aptamer-trigger probes were verified with a binding assay (Supporting Information, Figure S1). Upon the initiation of aptamer-trigger probes, nanodevices were self-assembled in a cascading manner from M1 and M2 via HCR (Scheme 1A), as confirmed by agarose gel electrophoresis (Figure 1A). Nanodevices were observed only in mixed monomers in the presence of aptamer-trigger probe, which further verified the conditional nanodevice formation and assured the presence of aptamer on the ends to guide all nanodevices, providing the basis for specific recognition. The gel results also confirmed that one aptND carried multiple monomers, working like train boxcars, as indicated in Scheme 1A, which were implemented for chemical labeling (covalent) of multiple copies of fluorophores or physical association (noncovalent) with multiple fluorogenic dsDNA-intercalating fluorophores on each nanodevice.
Figure 1.
Selective anchoring of chemically-modified fluorescent DNA nanodevices on target cell surfaces. A, An agarose gel electrophoresis image verifying the self-assembly of sgc8-NDs upon the initiation of sgc8-trigger. M1 and M2 (Lane 4) did not react unless sgc8-trigger was present. B–C, Flow cytometric results indicating that fluorescent sgc8-and TDO5-nanodevices were selectively anchored on the surfaces of CEM cells (B) and Ramos cells (C), respectively, but not the corresponding nontarget cells. Fluorescent nanodevice-anchored target cells displayed enhanced fluorescence intensities, compared to cells bound by the corresponding aptamers only. D–E, Confocal microscopy images indicating that fluorescent sgc8- and TDO5-NDs were selectively anchored on the surfaces of CEM cells (D) and Ramos cells (E), respectively. (Scale bar: 50 μm; Lib: random sequences; lib, sgc8, TDO5, M1, M2: labeled with FITC.)
Corresponding to the different types of fluorescence signal transduction widely used in bioanalysis, three different types of fluorescent aptNDs were constructed and selectively anchored on target cell surfaces: 1) a chemically-modified fluorescent aptND, in which fluorophores were chemically modified on the ends of monomers; 2) a label-free fluorescent aptND, in which fluorogenic molecules were physically associated with dsDNA boxcars; and 3) a FRET aptND, in which two fluorophores were chemically modified on monomers and physically associated with boxcar, respectively, to enable energy transfer between the two (Scheme 1A).
To build chemically-modified fluorescent aptNDs, monomers were labeled with fluorescein isothiocyanate (FITC) as a model. Nanodevice formation was confirmed by agarose gel electrophoresis (Supporting Information, Figure S2A). To test whether these nanodevices, each carrying multiple copies of fluorophores, could be anchored on target cell surfaces, a binding assay was performed using flow cytometry. The sgc8- and TDO5-tethered DNA nanodevices were studied with CEM and Ramos cells. Compared with target cells incubated with FITC-labeled aptamers, only those cells incubated with the corresponding aptNDs displayed significant enhancement of fluorescence intensity, whereas nontarget cells did not (Figure 1B–C). This indicates that these aptNDs were anchored selectively on their corresponding target cell surfaces. In addition, since a single nanodevice was loaded with multiple copies of fluorophores, nanodevice-anchored target cells displayed enhanced fluorescence intensities compared to the corresponding cells labeled with aptamers only. The less fluorescence intensity enhancement (ca. 10 fold) shown in flow cytometric results than theoretical estimation (20 fold, based on 1:10:10 of aptamer-trigger: M1: M2 molar ratio), is presumably due to the self-quenching of FITC on nanodevices or some dissociation of nanodevices from cell surfaces prior to measurement. Consistently, in confocal microscopy study, high fluorescence intensities were observed on surfaces of aptND-labeled target cells (Figure 1D–E), again demonstrating that aptNDs were selectively immobilized on target cell surfaces. The higher fluorescence intensity of Ramos cells labeled with TDO5-NDs, compared to CEM cells labeled with sgc8-NDs, presumably results from the higher receptor density of TDO5.[11a, 12] When another fluorophore, Quasar 570, was utilized to construct chemically-modified fluorescent sgc8-NDs, they were also successfully anchored on the target CEM cell surfaces (Supporting Information, Figure S3).
We next studied building label-free fluorescent DNA nanodevices on target cell surfaces. The development of label-free fluorescent devices avoids chemical modification and allows for real-time signal monitoring.[9a, 13] Many of these devices were constructed based on the noncovalent interaction of dsDNA and fluorogenic dyes, including YOYO-1, TOTO-1, and EvaGreen, which are capable of intercalating into dsDNA with relatively high affinities compared to ssDNA, resulting in a dramatic enhancement of fluorescence intensity.[14] As a result of its low cell permeability and low background signal, EvaGreen (EG), a dsDNA-bisintercalator with high binding affinity and no apparent sequence dependence, was utilized in this study. AptNDs were prepared using non-labeled M1 and M2. The fluorogenic property of EG was demonstrated using the resultant aptNDs (Supporting Information, Figure S4). Label-free fluorescent nanodevices were constructed by mixing EG and aptNDs to allow EG intercalation into aptNDs. The signal-to-noise ratio (S/N) of this nanodevice was optimized (Supporting Information, Figure S5), and an EG: aptND molar ratio of 40: 1 was used in the subsequent study. The EG-intercalated aptNDs were then evaluated for selective anchoring on cell surfaces. As shown by flow cytometric data in Figure S6, EG-intercalated sgc8-NDs were selectively anchored on target CEM cell surfaces, but not on nontarget Ramos cells. Similarly, EG-intercalated TDO5-NDs were selectively anchored on target Ramos cell surfaces, but not on nontarget CEM cells. This demonstrates the selective anchoring of label-free fluorescent nanodevices on cell surfaces.
We then studied building FRET nanodevices on living cell surfaces. In nature, FRET is utilized by photosynthesis systems to improve efficiency and wavelength sensitivity.[15] For instance, phycobiliproteins can capture light energy, which is then passed on to chlorophylls.[16] The nature of the dipole-dipole mechanism constrains the length scales in FRET-based platforms to distances on the order of <10 nm.[17] The ability of FRET to shift emission wavelengths enables the excitation of a wider range of fluorophores with limited laser sources in bioanalysis. Indeed, based on FRET, a panel of multifluorophore conjugates (e.g., phycoerythrin-Cy5.5) has already been developed.[18] Moreover, FRET has been widely used to construct activatable biosensors.
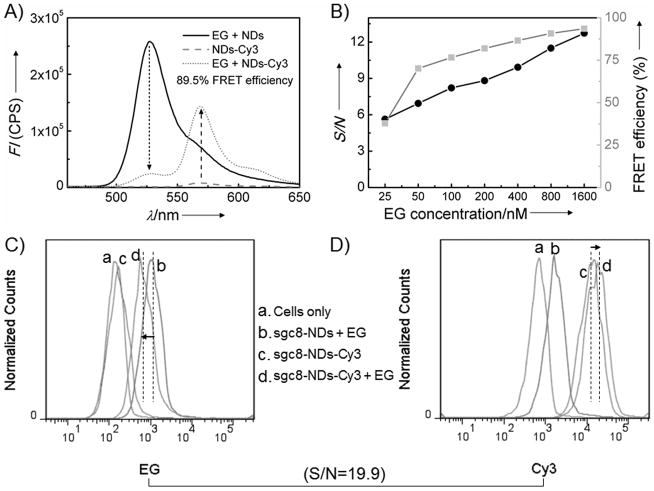
In a similar manner, our FRET nanodevices were constructed to capture one form of energy and pass it on. Structurally, this was made possible by the programmability of aptNDs and the tandem positioning of multiple FRET components by both covalent and noncovalent means (Supporting Information, Figure S2B). In this nanodevice, EG (energy donor) was intercalated into dsDNA boxcars in aptNDs, and Cy3 (energy acceptor) was chemically modified on the 3′-ends of M1 and M2. The intercalated EG was designed to work as an antenna to absorb short-wavelength light and then transfer the energy to nearby densely positioned and evenly distributed Cy3 on the aptNDs. Since each monomer contains 48 bases, the distance between EG and Cy3 molecules in the boxcars of aptNDs would theoretically be no longer than one-fourth of the length of one monomer, which is about 4 nm. This close proximity between energy donors and acceptors is essential to high-efficiency energy transfer.[17, 19] FRET efficiency was calculated based on donor quenching. Figure 2A shows efficient (ca. 89.5%) FRET from intercalated EG to chemically-attached Cy3 for 800 nM EG and 20 nM aptNDs. The FRET efficiency for a series of different EG concentrations was also determined (Figure 2B, gray line). We further determined the S/N by ratiometric measurements, which provide a built-in correction for environmental effects.[20] Figure 2B also displays the S/N with a series of different EG concentrations and a constant Cy3-labeled aptND concentration. Overall, these results confirmed the utility of our FRET-based fluorescent nanodevices.
Figure 2.
Development and determination of the energy transfer efficiency of FRET DNA nanodevices built on target cell surfaces. A, Fluorescence spectrometric results indicating energy transfer on aptNDs with chemically-labeled Cy3 (acceptor) on M1 and M2 and physically-associated EG (donor) in the duplex. Arrows denote the fluorescence changes (aptNDs: 20 nM; EG: 800 nM; Ex: 440 nm). B, FRET efficiency (gray) and S/N (black) of these aptNDs with a series of EG concentrations (aptNDs: 20 nM; Ex: 440 nm). C–D, Flow cytometric results indicating the anchoring of FRET aptNDs on target CEM cell surfaces by monitoring EG (C) and Cy3 (D) signals. Dotted lines denote the geometric mean fluorescence signal intensities, and arrows indicate signal changes. S/N was calculated as 19.9 (EG: 1 μM; aptNDs: 50 nM; Ex: 488 nm).
For bioanalysis or bioregulation, nanodevices anchored on the cell surfaces depend on the flawless operation of FRET. Therefore, we next tested FRET on living cells. Using flow cytometry, the fluorescence intensities of EG and Cy3 were monitored on target CEM cells. The high FRET efficiency of this nanodevice was verified with 488-nm excitation in buffer solution, as shown in Figure S7. Compared to cells modified with label-free fluorescent aptNDs and Cy3-labeled aptNDs, the cells modified with the FRET aptNDs displayed reduced fluorescence signal intensity from EG (Figure 2C), but enhanced intensity from Cy3 (Figure 2D). Based on the geometric mean of fluorescence intensities obtained from flow cytometry, the S/N was ca. 19.9. Overall, these data demonstrate that aptNDs successfully transferred energy when anchored on living cell surfaces and the principle of excitation of multi-chromophore arrays with a single laser source, providing the basis for future engineering and operation of related nanofactories on cell surfaces to signal and regulate biological activities.
The ability to build preformed nanodevices on cell surfaces would allow a wide variety of sophisticated nanodevices to be constructed before being anchoring on cell surface. However, situations arise making it difficult to transport preformed devices to, or anchor them on, target cells. Construction of such devices may also be hindered by the absence of local stimuli. Under these situations, in situ nanodevice assembly would be highly demanded. Thus, we further exploited the in situ self-assembly of fluorescent DNA nanodevices on target living cell surfaces. These aptNDs were built by 1) cascading alternative hybridization of two partially complementary non-hairpin monomers, and 2) HCR, a relatively difficult reaction (Scheme 1B).
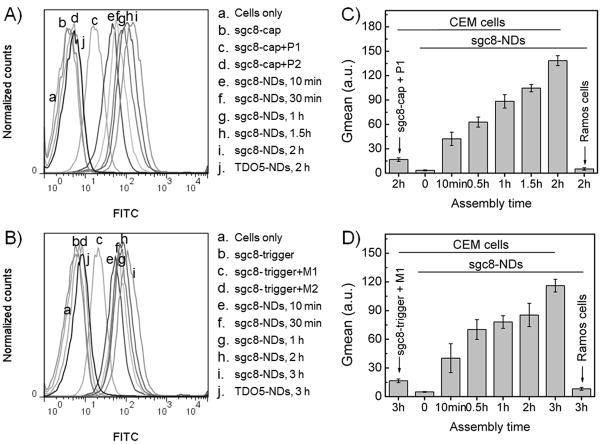
In approach 1), the reaction is relatively simple, and it was used first to demonstrate the principle of in situ nanodevice assembly on target cell surfaces. Aptamer sgc8 was modified with a capture sequence (sgc8-cap) to work as the seed probe for nanodevice assembly. Sgc8-cap was first incubated with target CEM cells to allow aptamer binding, and excess sgc8-cap was removed prior to the introduction of FITC-labeled building blocks, P1 and P2 (see sequences in Table S1). Subsequent incubation for different time periods (10 min, 30 min, 1 h, 1.5 h and 2 h) allowed in situ self-assembly of sgc8-NDs starting from the initiator aptamer seed probes bound on the cell surfaces. The resultant cells were washed and subjected to flow cytometric analysis. As shown in Figure 3A and 3C, after the introduction of building blocks, the fluorescence intensities of CEM cells increased with increasing incubation time, indicating the cascading self-assembly of fluorescent DNA nanodevices on cell surfaces. In contrast, the control aptamer, TDO5, did not allow nanodevice assembly on nontarget CEM cells, suggesting the selectivity of in situ self-assembly.
Figure 3.
Flow cytometric analysis (A, B) and statistical analysis of the geometric mean fluorescence intensities (Gmean) (C, D) of target CEM cells with sgc8-NDs assembled in situ on cell surfaces by a time series. Nanodevices were self-assembled via cascading alternative hybridization of two FITC-labeled partially complementary DNA strands (A, C; approach I), or HCR of two FITC-labeled monomers (B, D; approach II). The increase of fluorescence intensities indicates progressive nanodevice assembly (P1, P2, M1, and M2: labeled with FITC). Also see color graph of A and B in Supporting Information Figure S8.
In approach 2), in situ nanodevice assembly was carried out via HCR. Compared with the approach 1), HCR is a slow reaction which requires strand displacement in each elongation step, and previous studies have usually performed HCR in a nonliving cell environment without the requirement for rapid kinetics. To study such reaction in a living cell environment, sgc8-trigger seed probes were incubated with target CEM cells to allow aptamer binding, followed by the removal of excess sgc8-trigger and introduction of FITC-labeled M1 and M2. The continuous enhancement of fluorescence intensities indicated the progressive in situ self-assembly of fluorescent DNA nanodevices on target cell surfaces (Figure 3B,D). In contrast, no nanodevice assembly took place on nontarget CEM cells by TDO5 initiator probe, indicating selectivity. Compared to the approach 1), the relatively slow enhancement of fluorescence intensities in flow cytometric results also suggests that in situ nanodevice assembly by HCR is a relatively slow process.
We further investigated in situ assembly of sgc8-NDs on target cell surfaces in cell mixtures containing both target cells (CEM) and nontarget cells (Ramos). A biotinylated aptamer TDO5, combined with streptavidin-conjugated PE-Cy5.5, was utilized to label Ramos cells. The specific binding of TDO5 to Ramos cells at room temperature was confirmed by flow cytometry (Supporting Information, Figure S9). Again, sgc8-trigger was first incubated with cell mixtures, followed by removal of excess probes in solution and then introduction of FITC-labeled M1 and M2. The resultant solutions were incubated at room temperature for a series of time lengths to allow in situ nanodevice assembly by HCR, which was terminated by washing away free probes. TDO5 was introduced 30 min before the end of assembly, followed by removal of free aptamers and introduction of streptavidin-conjugated PE-Cy5.5. The resultant cell solutions were observed using confocal microscopy. As shown in Figure S10 (Supporting Information), with increasing assembly time, increasing FITC fluorescence intensities were observed on target CEM cell surfaces (not labeled with TDO5), while Ramos cells (labeled with TDO5) did not display FITC signal. This clearly demonstrated selective fluorescent nanodevice assembly in situ on living target cell surfaces in a complex mixture. The ability of in situ self-assembly of nanodevices on target cells in cell mixtures using a slow reaction is anticipated to be useful in localized engineering of complex biological nanofactories for biomolecule detection and regulation of biological pathways.
In conclusion, to achieve the long-term goal of pinpoint bioanalysis or manipulation of biological activities on target living cell membranes in complex extracellular environments, we have successfully built fluorescent DNA nanodevices on target living cell surfaces by anchoring preformed model nanodevices and by in situ self-assembly of nanodevices. These fluorescent DNA nanodevices consisted of aptamer-tethered nanodevices formed by cascading polymerization of monomeric building blocks. The concept of tethered aptamer moieties on nanodevices (aptNDs) provides the basis for selectively building nanodevices on target cell surfaces through specific aptamer-target interaction. The features of multiple, repetitive and alternating DNA building blocks in nanodevices provide an excellent platform for appropriate loading and positioning of multi-component molecular arrays through either chemical modification or physical association. Based on this aptND platform, three types of fluorescent DNA nanodevices were self-assembled as models and successfully anchored on target cell surfaces: chemically-labeled fluorescent nanodevices, label-free fluorescent nanodevices, and FRET nanodevices. We further demonstrated the principle of in situ self-assembly of nanodevices on target cell surfaces in heterogeneous cell mixtures. The ability of nanodevices to be self-assembled in situ on target living cells under a complicated environment could be useful in localized engineering of complex biological nanofactories on cell membranes for bioanalysis and the regulation of biological activities. Both fluorescence signaling and fluorescence activity of energy transfer were shown to be functional in these nanodevices. In the future, we envision that building nanodevices on target living cell surfaces could be useful for real-time tracking of target cells or sensing analytes in extracellular environments, cell-surface engineering, targeted drug delivery and manipulation of biological pathways.
Supplementary Material
Acknowledgments
The authors sincerely thank Dr. K. R. Williams for critical manuscript review. We acknowledge the Interdisciplinary Center for Biotechnology Research (ICBR) at the University of Florida for instrument assistance. This work is supported by grants awarded by the National Institutes of Health (GM066137, GM079359, and CA133086) and by the National Key Scientific Program of China (2011CB911000) and China National Grand Program (2009ZX10004-312).
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Guizhi Zhu, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Shands Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida Gainesville, FL 32611-7200 (USA).
Shengfeng Zhang, Molecular Science and Biomedicine Laboratory, State Key Laboratory of Chemo/Bio-Sensing and Chemometrics, College of Biology and College of Chemistry and Chemical Engineering, Collaborative Innovation Center for Chemistry and Molecular Medicine, Hunan University, Changsha, 410082, China.
Prof. Dr. Erqun Song, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Shands Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida Gainesville, FL 32611-7200 (USA). Key Laboratory of Luminescence and Real-Time Analysis of the Ministry of Education, College of Pharmaceutical Sciences, Southwest University, Chongqing 400715, China
Jing Zheng, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Shands Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida Gainesville, FL 32611-7200 (USA).
Rong Hu, Molecular Science and Biomedicine Laboratory, State Key Laboratory of Chemo/Bio-Sensing and Chemometrics, College of Biology and College of Chemistry and Chemical Engineering, Collaborative Innovation Center for Chemistry and Molecular Medicine, Hunan University, Changsha, 410082, China.
Prof. Dr. Weihong Tan, Email: tan@chem.ufl.edu, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Shands Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida Gainesville, FL 32611-7200 (USA). Molecular Science and Biomedicine Laboratory, State Key Laboratory of Chemo/Bio-Sensing and Chemometrics, College of Biology and College of Chemistry and Chemical Engineering, Collaborative Innovation Center for Chemistry and Molecular Medicine, Hunan University, Changsha, 410082, China
References
- 1.Geiger B, Bershadsky A, Pankov R, Yamada KM. Nat Rev Mol Cell Biol. 2001;2:793. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 2.a) Kulwichit W, Edwards RH, Davenport EM, Baskar JF, Godfrey V, Raab-Traub N. Proc Natl Acad Sci USA. 1998;95:11963. doi: 10.1073/pnas.95.20.11963. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) López-Casillas F, Cheifetz S, Doody J, Andres JL, Lane WS, Massague J. Cell. 1991;67:785. doi: 10.1016/0092-8674(91)90073-8. [DOI] [PubMed] [Google Scholar]
- 3.a) Lee K, Drachev VP, Irudayaraj J. ACS Nano. 2011;5:2109. doi: 10.1021/nn1030862. [DOI] [PubMed] [Google Scholar]; b) Mahal LK, Bertozzi CR. Chem & Biol. 1997;4:415. doi: 10.1016/s1074-5521(97)90193-9. [DOI] [PubMed] [Google Scholar]; c) Kellam B, De Bank PA, Shakesheff KM. Chem Soc Rev. 2003;32:327. doi: 10.1039/b211643j. [DOI] [PubMed] [Google Scholar]; d) Stephan MT, Moon JJ, Um SH, Bershteyn A, Irvine DJ. Nat Med. 2010;16:1035. doi: 10.1038/nm.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Gartner ZJ, Bertozzi CR. Proc Natl Acad Sci USA. 2009;106:4606. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Hochbaum AI, Aizenberg J. Nano Lett. 2010;10:3717. doi: 10.1021/nl102290k. [DOI] [PubMed] [Google Scholar]; g) Fernandes R, Roy V, Wu HC, Bentley WE. Nat Nanotechnol. 2009;5:213. doi: 10.1038/nnano.2009.457. [DOI] [PubMed] [Google Scholar]; h) Chittasupho C, Shannon L, Siahaan TJ, Vines CM, Berkland C. ACS Nano. 2011;5:1693. doi: 10.1021/nn102159g. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Koyfman AY, Braun GB, Reich NO. J Am Chem Soc. 2009;131:14237. doi: 10.1021/ja9015638. [DOI] [PubMed] [Google Scholar]; j) Wilson JT, Cui W, Kozlovskaya V, Kharlampieva E, Pan D, Qu Z, Krishnamurthy VR, Mets J, Kumar V, Wen J, Song Y, Tsukruk VV, Chaikof EL. J Am Chem Soc. 2011;133:7054. doi: 10.1021/ja110926s. [DOI] [PubMed] [Google Scholar]
- 4.a) Stevens MM, George JH. Science. 2005;310:1135. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]; b) Zhao WA, Teo GSL, Kumar N, Karp JM. Mater Today. 2010;13:14. doi: 10.1016/S1369-7021(10)70056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boonyarattanakalin S, Martin SE, Sun Q, Peterson BR. J Am Chem Soc. 2006;128:11463. doi: 10.1021/ja062377w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinheiro AV, Han D, Shih WM, Yan H. Nat Nanotechnol. 2011;6:763. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Ellington AD, Szostak JW. Nature. 1990;346:818. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; b) Tuerk C, Gold L. Science. 1990;249:505. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]; c) Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc Natl Acad Sci USA. 2006;103:11838. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Dirks RM, Pierce NA. Proc Natl Acad Sci USA. 2004;101:15275. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Choi HMT, Chang JY, Trinh LA, Padilla JE, Fraser SE, Pierce NA. Nat Biotechnol. 2010;28:1208. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Benvin AL, Creeger Y, Fisher GW, Ballou B, Waggoner AS, Armitage BA. J Am Chem Soc. 2007;129:2025. doi: 10.1021/ja066354t. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Teo YN, Kool ET. Chem Rev. 2012;112:4221. doi: 10.1021/cr100351g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. J Prot Res. 2008;7:2133. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikaratchy P, Chen HW, Li Y, Tan W. Anal Chem. 2007;79:4900. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]; b) Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Mol & Cell Prot. 2007;6:2230. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.a) Chen Y, Munteanu AC, Huang YF, Phillips J, Zhu Z, Mavros M, Tan W. Chem Eur J. 2009;15:5327. doi: 10.1002/chem.200802305. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shangguan D, Li Y, Tang Z, Cao Z, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc Natl Acad Sci USA. 2006;103:11838. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt HK, Armani AM. Nanoscale. 2010;2:1544. doi: 10.1039/c0nr00201a. [DOI] [PubMed] [Google Scholar]
- 14.a) Rye HS, Yue S, Wemmer DE, Quesada MA, Haugland RP, Mathies RA, Glazer AN. Nucleic Acids Res. 1992;20:2803. doi: 10.1093/nar/20.11.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mao F, Leung W-Y, Xin X. Bmc Biotechnol. 2007:7. doi: 10.1186/1472-6750-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Larsson A, Carlsson C, Jonsson M, Albinsson B. J Am Chem Soc. 1994;116:8459. [Google Scholar]
- 15.Melkozernov AN, Barber J, Blankenship RE. Biochemistry. 2005;45:331. doi: 10.1021/bi051932o. [DOI] [PubMed] [Google Scholar]
- 16.Glazer AN. J Biol Chem. 1989;264:1. [PubMed] [Google Scholar]
- 17.Chance RR, Prock A, Silbey R. Adv Chem Phy. 2007:1. [Google Scholar]
- 18.Hulspas R, Dombkowski D, Preffer F, Douglas D, Kildew-Shah B, Gilbert J. Cytometry Part A. 2009;75A:966. doi: 10.1002/cyto.a.20799. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, O’Donoghue MB, Huang YF, Kang H, Phillips JA, Chen X, Estevez MC, Yang CJ, Tan W. J Am Chem Soc. 2010;132:16559. doi: 10.1021/ja106360v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang P, Beck T, Tan W. Angew Chem. 2001;113:416. [Google Scholar]; Angew Chem Int Ed. 2001;40:402. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.