Abstract
Purpose
Everolimus is a mammalian target of rapamycin (mTOR) inhibitor approved as an immunosuppressant and for second-line therapy of hepatocellular carcinoma (HCC) and renal cell carcinoma (RCC). Sorafenib is a multikinase inhibitor used as first-line therapy in HCC and RCC. This study assessed the pharmacokinetics (PK) of everolimus and sorafenib alone and in combination in plasma and tissues, developed physiologically based pharmacokinetic (PBPK) models in mice, and assessed the possibility of PK drug interactions.
Methods
Single and multiple oral doses of everolimus and sorafenib were administered alone and in combination in immunocompetent male mice and to severe combined immune-deficient (SCID) mice bearing low-passage, patient-derived pancreatic adenocarcinoma in seven different studies. Plasma and tissue samples including tumor were collected over a 24-h period and analyzed by liquid chromatography-tandem mass spectrometry (LC–MS/MS). Distribution of everolimus and sorafenib to the brain, muscle, adipose, lungs, kidneys, pancreas, spleen, liver, GI, and tumor was modeled as perfusion rate-limited, and all data from the diverse studies were fitted simultaneously using a population approach.
Results
PBPK models were developed for everolimus and sorafenib. PBPK analysis showed that the two drugs in combination had the same PK as each drug given alone. A twofold increase in sorafenib dose increased tumor exposure tenfold, thus suggesting involvement of transporters in tumor deposition of sorafenib.
Conclusions
The developed PBPK models suggested the absence of PK interaction between the two drugs in mice. These studies provide the basis for pharmacodynamic evaluation of these drugs in patient-derived primary pancreatic adenocarcinomas explants.
Keywords: Everolimus, Sorafenib, Pancreatic cancer, PBPK, Patient-derived pancreatic adenocarcinoma
Introduction
Everolimus (Afinitor®, Novartis) is a small molecule inhibitor of mTOR, approved by the FDA for treatment of renal cell carcinoma (RCC) [1]. Everolimus is a rapamycin analog having improved oral bioavailability [2] but similar pharmacodynamics. This class of agents target mTORC1 in the PI3 k-Akt-mTOR pathway, which acts as nutrient sensor and is involved in cell proliferation, angiogenesis, and survival [3, 4]. Major adverse events associated with everolimus treatment include hypercholesterolemia, hypertriglyceridemia, anemia, fatigue, rash, infections, and gastrointestinal (GI) effects such as diarrhea and nausea [5]. Sorafenib (Nexavar®, Bayer) is a small molecule multikinase inhibitor used for treatment of hepatocellular carcinoma (HCC) and RCC [6, 7]. The most common side effects associated with sorafenib treatment include dermatological events, such as hand-foot syndrome and edema of palms and soles, and GI events, such as diarrhea and anorexia. Hypertension is also a prevalent adverse event associated with the VEGF inhibitory action of sorafenib [8]. Everolimus and sorafenib are molecularly targeted agents that have inhibitory effects on compensatory cellular signaling mechanisms, which suggests the possibility that a combination of the two drugs may be more efficacious as anti-cancer therapy [9]. They have been tested in combination with xenograft models and in patients for cancers such as melanoma, renal carcinoma, pancreatic adenocarcinoma, hepatic carcinoma, thyroid cancer, and other solid tumors [10–12]. According to the clinicaltrials.gov website (accessed 01/25/2012), multiple trials using these two drugs in combination are underway. However, studies to date have not investigated the potential for PK interactions of sorafenib and everolimus.
Everolimus is metabolized primarily in liver and undergoes oxidative metabolism mediated by CYP 3A4 [13]. In mice, the drug is 12 % absorbed and the bioavailability is 5 % [13, 14]. The PK properties of everolimus are different in mice and rats in terms of red blood cell partitioning, plasma protein binding, plasma half-life, oral bioavailability, volume of distribution, tissue kinetics, and elimination [13, 14]. Therefore, the published PBPK model in rats [15] is not suitable to characterize plasma, tissue, and tumor concentrations in mice.
Sorafenib is metabolized primarily in the liver and also undergoes oxidative metabolism mediated by CYP 3A4. It can also undergo glucuronidation, which is mediated by UGT1A9 [16]. In mice, the drug is 92 % absorbed and the bioavailability is 80 %, thus indicating rapid first-pass metabolism [16]. The PK analysis showed rapid absorption with a Tmax of 1–1.6 h, terminal half-life of 3.2–4.2 h in CD-1 mice, and dose proportionality in Cmax and AUC at the doses tested [8]. Most preclinical information about sorafenib has been presented in FDA submissions and posters at conferences, and little has been published. However, a recent population PK model that took into account the delayed solubility of sorafenib, its limited GI absorption, and enterohepatic circulation was able to describe the high PK variability observed in patients [17].
Everolimus is known to exhibit pharmacokinetic interactions with co-administered drugs that increase its own exposure [15, 18]. Such interactions can lead to augmentation of toxicity owing to high organ exposure, as seen in the kidney during co-administration of rapamycin (everolimus analog) with cyclosporine [14]. Furthermore, because both drugs undergo oxidative metabolism via CYP 3A4, an evaluation of the pharmacokinetics of the drugs alone and in combination in plasma and tissues of interest can lend insights into their potential for toxicities when combined. To that end, a PBPK model was developed to assess data acquired in immunocompetent mice and tumor-bearing SCID mice. The effect of the combination on PK and drug distribution into tumors and multiple tissues was evaluated. A population approach was employed that enabled simultaneous analysis of data from the multiple studies performed.
Materials and methods
Preparation of sorafenib and everolimus
A microemulsion of 2 % w/v everolimus (obtained from Novartis Pharma AG, Basel, Switzerland) was diluted with pyrogen-free water to the required dose. The sorafenib dose was prepared by dissolving its p-toluenesulfonate salt (purchased from LC Laboratories, Wolburn, MA) in Cremophor EL/ethanol 50:50 at fourfold (4×) the dose, foil wrapped in amber colored vials, and stored at room temperature. This stock solution was prepared fresh every week. The solution for administration was prepared each day 1 h before dosing by diluting the stock solution with water to provide the required dose in a volume of 10 mL/kg. Both drugs were administered orally using a bead-tipped curved gavage needle.
Animals
Male BALB/c mice (7–8 weeks old, average weight 25–30 g) were obtained from Harlan (Indianapolis, IN). CB17 SCID mice (7–8 weeks old, average weight 20–25 g) were obtained from Roswell Park Cancer Institute (RPCI) (Buffalo, NY). Male SCID mice were implanted with low-passage, patient-derived, histopathologically verified, pancreatic adenocarcinomas as described earlier [19]. Pieces (2 × 2 × 2 mm) of donor tumors were subcutaneously implanted under anesthesia in the abdominal wall of 4–6 week-old mice. All procedures were approved by the Institutional Animal Care and Use Committees of the University at Buffalo and RPCI.
Study design
Dosing
The animals used and treatment design are summarized in Table 1. For experiments with tumor-bearing mice, two different patient-derived pancreatic adenocarcinomas that successfully engrafted into SCID mice [19] were selected.
Table 1. Study designs for PK studies.
| Study number | Animal model | Dosing | Dosing frequency | Sampling (h after last dose) |
|---|---|---|---|---|
| 1 | BALB/c (n = 18) |
|
Single dose | 0.5, 1, 4, 6, 10, 24 |
| 2 | SCID (n = 18) |
|
Single dose | 0.5, 1, 4, 6, 10, 24 |
| 3 | SCID (n = 18) |
|
5 days ON, 2 days OFF for 3 weeks | 0.5, 1, 4, 6, 10, 24 |
| 4 | SCID (n = 9) |
|
5 days ON, 2 days OFF for 3 weeks | 1, 6, 24 |
| 5 | SCID (tumored) (n = 18) |
|
5 days ON, 2 days OFF for 5 weeks | 1, 6, 24 |
| 6 | SCID (tumored) (n = 18) |
|
5 days ON, 2 days OFF for 5 weeks | 1, 6, 24 |
| 7 | SCID (tumored) (n = 18) |
|
5 days ON, 2 days OFF for 5 weeks | 1, 24 |
n indicates number of samples available for each drug
Treatment was initiated ∼6–8 weeks after tumor implantation, when tumors reached a volume of 100–200 mm3. In all studies, animals were randomized into treatment groups having similar tumor size distributions.
Sample collection and tissue sampling
At the appropriate time within the dosing regimen, groups of three animals were sacrificed at intervals over 24 h. At sacrifice, animals were weighed, anesthetized by ketamine/xylazine, and sacrificed by aortic exsanguination. Blood, lungs, muscle, brain, adipose tissue, kidneys, liver, pancreas, skin, spleen, gut, and tumor were collected. Blood was drawn from the abdominal aortic artery into syringes using ethylenediaminetetraacetic acid (EDTA) as an anticoagulant. Plasma was prepared by centrifugation (2,000 g for 15 min at 4 °C) and frozen at −20 °C until further analysis. The excised tissues were weighed and frozen in liquid nitrogen immediately after sacrifice and stored at −80 °C until homogenization. Liver samples were dissected carefully to avoid contamination with bile. The entire spleen, liver, brain, tumor, and pancreas were used for drug quantification. Partial samples of skin, skeletal muscle, and adipose tissue were analyzed. Both lungs and kidneys were analyzed. Tissue samples of small intestine were obtained after irrigation to remove gut contents. Skeletal muscle, adipose tissue, and brain were analyzed in Study# 1 (Table 1) and subsequently were omitted because of the minimal drug uptake by these organs.
Determination of drug concentrations in plasma and tissues
Everolimus
Everolimus concentrations in plasma and tissues were measured using a previously published LC–MS/MS assay [20], using a Thermo TSQ Quantum Ultra (Thermo Scientific, Waltham, MA) in ESI positive mode. The mobile phase gradient was composed of (A) 0.2 M ammonium formate, (B) methanol, and (C) water. The standard curve was validated for concentrations of 2.5–100 ng/mL. Plasma samples from high-concentration time points were diluted with blank plasma when necessary, and 200 μl was mixed with 500 μl of 1:1 methanol/water (vol/vol) and analyzed along with standards (prepared fresh for each run) and QC samples. One ng/μl of the internal standard ascomycin was added to each sample. Weighed tissue samples were homogenized with 0.75 mL methanol/water (1:1 vol/vol), and the total volume was made up to 1.5 mL and processed as described for plasma samples. The recovery from tissue samples was more than 98 %. Within-and between-day CV % were less than 15 %, and the assay was linear over a concentration range of 2.5–100 ng/mL.
Sorafenib
Sorafenib concentrations in plasma and tissues were measured using a previously published LC–MS/MS assay [21] in ESI positive mode. The mobile phase consisted of 65/35 (vol/vol) acetonitrile/water with 0.1 % formic acid. The standard curve was validated for concentrations of 0.5–100 ng/mL. Plasma samples were prepared as described for everolimus except the internal standard was 1 ng/μl sorafenib D3, and it was added to plasma samples in acetonitrile rather than methanol/water. Weighed tissue samples were prepared by homogenization in methanol/water (1:1 vol/vol) as described above for everolimus. The lower limit of quantitation was 0.5 ng/mL, and recovery from tissue samples was more than 99 %. Within- and between-day CV % were less than 15 %, and the assay was linear over the concentration range of 0.5–100 ng/mL.
Data transformation
Drug concentrations obtained in each tissue were converted from ng/mL to ng/g tissue by assuming a tissue density of 1 g/mL. Tissue concentrations were corrected for residual trapped blood using
| (1) |
where Ct and Ct(meas) are the corrected and measured tissue concentrations, Vmeas is the measured volume of collected tissue, and Vvasc/Vt is the fractional vascular volume of blood trapped in tissues as obtained from the literature [22]. The corrected tissue concentrations were used for further analysis.
Non-compartmental analysis
All samples were obtained via terminal sampling. Non-compartmental analysis (NCA) of data was carried out using the sparse sampling computational option in WinNonlin (version 5.0, Pharsight Corporation, Palo Alto, CA). Maximum concentrations (Cmax) and time to reach Cmax (Tmax) were observed values. NCA was used initially to characterize the PK profiles after single doses, and the estimates obtained were used as initial conditions during subsequent modeling.
Model development
Plasma and tissue data from the studies listed in Table 1 were used in constructing PBPK models. Local models were initially tested with blood- and tissue concentration– time data. The choice of final structural models was based upon evaluation of goodness of fits and the Akaike Information Criterion (AIC). The parameter values obtained with local models were used as initial estimates for the whole body model.
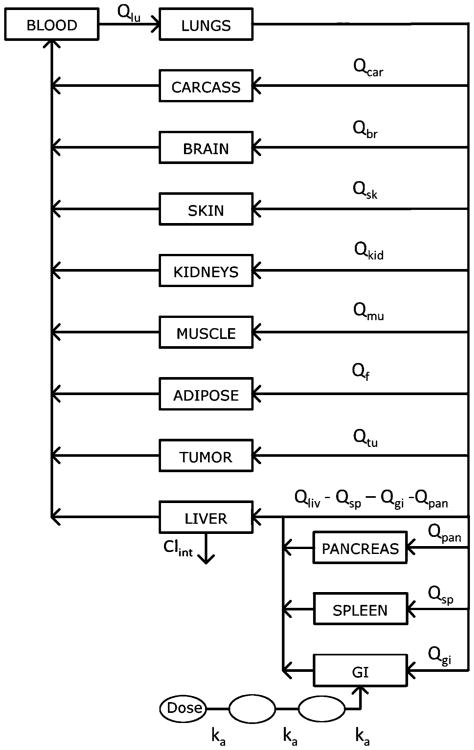
The schematic of the PBPK model used for everolimus and sorafenib is presented in Fig 1. The model includes 9 compartments: lungs, brain, kidneys, pancreas, spleen, liver, adipose, muscle, gut, tumor, and the carcass remainder. These compartments are linked by blood flow (Q). The oral absorption model includes a lumen compartment to describe absorption of drug from the GI tract, which was described using transit compartments owing to the delayed absorption of sorafenib [11]. Drug distribution in all tissues is assumed to occur instantaneously and homogenously, and therefore, perfusion-limited models were used to describe tissue concentrations. In tumor-bearing mice, an additional parallel vascular bed representing tumor was included. It was assumed that the animal physiology and drug transport kinetics in tumor-bearing mice were identical to non-tumor-bearing mice. Mass balance was maintained throughout the system. Plasma protein binding of everolimus and sorafenib was not considered. Elimination of the drug from the liver was modeled as a linear process given by Clint for both drugs.
Fig. 1. Whole body PBPK model for everolimus and sorafenib in mice. All organs are represented by a rectangular compartment and interconnected via blood flow Q.
The concentrations in plasma and tissues were described as: Plasma
| (2) |
Lungs:
| (3) |
Liver:
| (4) |
Small Intestine:
| (5) |
| (6) |
| (7) |
Other organs:
| (8) |
where Q is organ blood flow, V is organ volume, K is tissue-to-blood partition coefficient, F is fraction absorbed, ka is absorption rate constant, A1 is lumen concentration, An is amount in the nth transit compartment, and Clint is hepatic intrinsic clearance. The subscripts on Q, V, and K indicate specific tissues under consideration. The fraction absorbed in mice was fixed to 12 % for everolimus and to 92 % for sorafenib based on information from the FDA Summary Basis of Approval for both drugs [13, 16].
Tissue volumes and blood flow rates were obtained from the literature [10, 12, 22, 23] and scaled to the experimental animal weights. All physiological parameters used in the model are listed in Supplemental Information Table S1. An initial value for Qtu of 0.1 mL/min was obtained from the literature for a human colon carcinoma tumor [22]. However, because pancreatic cancers are poorly perfused [9], tumor blood flow Qtu was subsequently estimated in the model. The organ volumes were determined by weight (assuming a density of 1 g/mL) for lungs, brain, kidneys, pancreas, spleen, liver, and tumor. The organ volumes were maintained the same throughout the study for all tissues except tumor; for the latter, tumor volumes were measured throughout the study, and the observed volume for each individual animal was incorporated into the dataset as a continuous covariate. The hepatic clearance Clint, absorption rate constant ka, and tissue partition coefficients K were estimated for both drugs. Data obtained from each study were treated as replicate animals sampled as a single individual with averaged tissue volumes.
The equations for plasma and all tissues were solved simultaneously to estimate the model parameters using Maximum a Posteriori Bayesian estimation (MAP) implemented in ADAPT 5 [24] with an additive and proportional error variance model. Model selection was guided using the negative log likelihood and diagnostic plots.
Results
Sorafenib and everolimus disposition were evaluated after oral administration with extensive sampling in single- and multiple-dose studies in mice (Table 1). In multiple-dose studies, sampling was performed after the last dose. Several parameters obtained from initial data analysis were fixed for subsequent analysis (Table 2, Table 3). The oral absorption rate constant ka and the blood to tissue partition coefficient for lungs Klu, were fitted to the data obtained from Study#1, and then fixed in subsequent analyses. A value of 1.00 was obtained for Klu,, with a CV % < 10 %. The blood flow to the tumor Qtu was estimated to be 0.017 ml/min, with a CV % < 5 %. Because Qtu was assumed to be drug-independent, this value was used for both drugs. The population mean and inter-study variability were obtained for all parameters, but ADAPT 5 did not report their precision.
Table 2. Parameter estimates for everolimus pharmacokinetics.
| Parameter (Units) | Parameter description | Mean | CVa (%) |
|---|---|---|---|
| Clint (ml/h) | Hepatic clearance | 4.07 | 35.8 |
| Klu | Partition coefficient lungs | 1.00 | Fixed |
| Kbr | Partition coefficient brain | 0.018 | 6.36 |
| Kmu | Partition coefficient muscle | 0.105 | 9.36 |
| Kf | Partition coefficient adipose | 0.097 | 6.67 |
| Ksk | Partition coefficient skin | 0.232 | 63.1 |
| Kkid | Partition coefficient kidney | 0.435 | 20.6 |
| Kpan | Partition coefficient pancreas | 0.582 | 50.5 |
| Ksp | Partition coefficient spleen | 0.361 | 40.9 |
| Kliv | Partition coefficient liver | 0.452 | 36.9 |
| Kgi | Partition coefficient GI | 1.43 | 90.3 |
| ka (1/h) | Absorption rate constant | 9.45 | Fixed |
| Fa | Fraction absorbed | 0.12 | Fixed |
| Ktu | Partition coefficient tumor | 0.480 | 45.3 |
| Kcar | Partition coefficient carcass | 5.67 | 135 |
Coefficient of variation of the estimate; reflects inter-study variability
Table 3. Parameter estimates for sorafenib pharmacokinetics.
| Parameter (units) | Parameter description | Mean | CVa (%) |
|---|---|---|---|
| Clint (ml/h) | Hepatic clearance | 6.64 | 24.4 |
| Klu | Partition coefficient lungs | 1.00 | Fixed |
| Kbr | Partition coefficient brain | 0.06 | 4.11 |
| Kmu | Partition coefficient muscle | 0.356 | 5.13 |
| Kf | Partition coefficient adipose | 0.624 | 3.83 |
| Ksk | Partition coefficient skin | 0.361 | 55.4 |
| Kkid | Partition coefficient kidney | 3.08 | 20.0 |
| Kpan | Partition coefficient pancreas | 1.75 | 40.2 |
| Ksp | Partition coefficient spleen | 1.58 | 48.0 |
| Kliv | Partition coefficient liver | 4.37 | 29.8 |
| Kgi | Partition coefficient GI | 3.85 | 131 |
| ka (1/h) | Absorption rate constant | 3.83 | Fixed |
| Fa | Fraction absorbed | 0.920 | Fixed |
| Ktu_Low | Partition coefficient tumor-low dose | 0.741 | 3.63 |
| Ktu_high | Partition coefficient tumor-high dose | 2.12 | 3.63 |
| Kcar | Partition coefficient carcass | 3.71 | 113 |
Coefficient of variation of the estimate; reflects inter-study variability
Everolimus
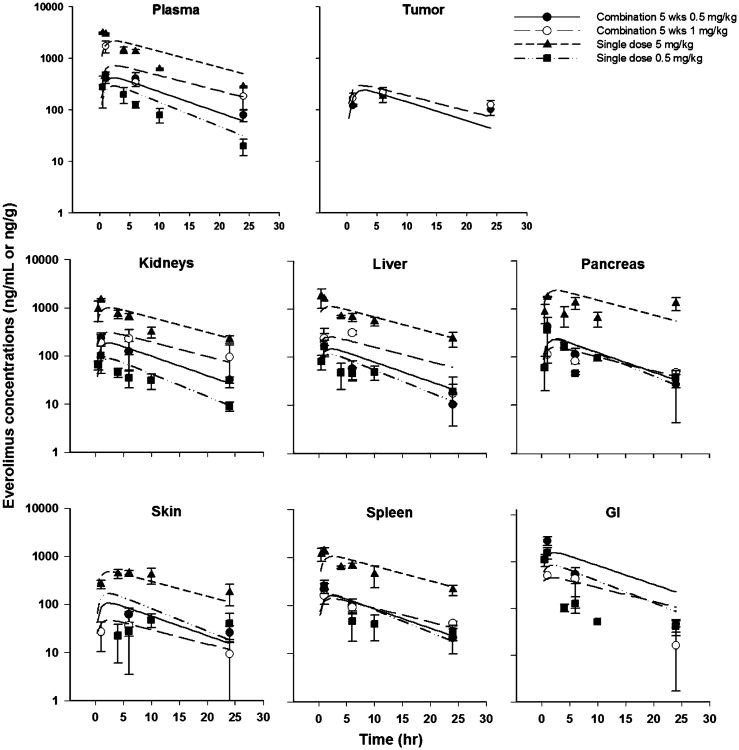
Plasma and tissue concentration versus time profiles were obtained for everolimus administered to BALB/c, CB17-SCID, and tumor-bearing CB17-SCID mice. After oral administration, the drug was rapidly absorbed (Fig. 2), with a Tmax of 1 h in plasma and most tissues, but with Tmax delayed up to 6 h in adipose and skin. Everolimus was detected in plasma and all tissues for up to 24 h after the last dose. The plasma and tissues showed parallel elimination phases, and the Cmax in plasma and tissues was dose-proportional for all mouse strains.
Fig. 2. Everolimus concentration–time profiles from studies 1, 2, 5, and 6 in mice (Table 1). Symbols represent mean (SD) of experimental data and lines represent predictions.
A PBPK model was employed to fit the concentration versus time data for plasma and tissues from various groups of animals and for different doses. There was no delay in the absorption of everolimus, and therefore, the number of transit compartments was 0, that is, the drug was absorbed directly from the GI tract. The parameter estimates, along with the inter-study variability for the population model, are given in Table 2. The overall Clint was estimated to be 4.07 mL/h, with 35.8 % variability between studies. The partition coefficients of everolimus into brain and adipose were low (0.02–0.1), and partition coefficients for muscle and skin were the next lowest (0.1–0.2). High variability was observed in the estimates of partition coefficients for skin, which can be attributed to difficulties in homogenization of the tissue. The partition coefficient for kidneys (0.44), pancreas (0.58), spleen (0.36), and liver (0.45) were close to 0.5, with a low variability between studies. Everolimus partitioned into the patient-derived primary pancreatic adenocarcinoma explants with a coefficient of 0.48. The gut partition coefficient Kgi was greater than 1, and the variability in the parameter was high, which was likely a result of the high GI concentrations owing to oral dosing.
Based upon a comparison of the everolimus AUC from each study in which the drug was given alone and in combination, there was no statistically significant difference in the PK of everolimus when it was given in combination with sorafenib (p < 0.05 as determined by student's t test). Parameter estimates for select PK parameters were also compared for everolimus administered alone or in combination. Sorafenib dosing did not cause significant variability in the hepatic clearance or tissue partition coefficient of everolimus.
The model provided reasonable fits and was able to describe well the everolimus concentrations in plasma and various tissues. The PBPK model was adequate in characterizing the concentrations as indicated by the good individual predictions for drug concentrations in plasma and tissues, except for the occasional overestimation of some profiles from the 0.5 mg/kg single dose. The concentrations were measured in tumor to permit assessment of pharmacodynamic effects [25]. The plots of standardized residuals versus predicted concentrations for all tissues show an unbiased distribution, indicative of suitable fittings (Supplemental information Fig S1). With the somewhat sparse sampling, oral dosing, and multiple studies, the present model could not be improved upon.
Sorafenib
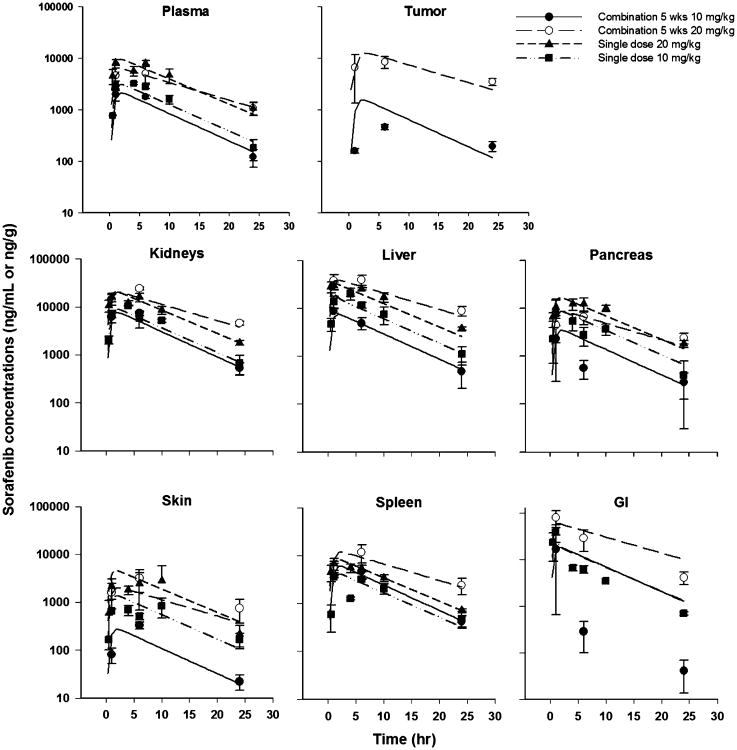
The plasma and tissue concentration versus time profiles are shown in Fig. 3 for sorafenib after oral administration to BALB/c, CB17-SCID, and tumor-bearing CB17-SCID mice. The Cmax in plasma and all tissues except the tumor was dose-proportional across the strains of mice. After oral dosing, sorafenib absorption was slow and delayed, with a Tmax of 6 h in plasma and tissues and 1 h in the GI tract. Sorafenib was detected in plasma and all tissues for up to 24 h after the last dose.
Fig. 3. Sorafenib concentration–time profiles from studies 1, 2, 5, and 6 in mice. Symbols represent mean (SD) of experimental data and lines represent predictions.
The PBPK model was used to fit the sorafenib concentration data for plasma and tissues from various groups of animals and different doses. The model includes two transit compartments and absorption rate constant ka to account for the delay in the appearance in plasma [17]. The fraction of dose absorbed was fixed to 92 %, as obtained from the literature [16]. The parameter estimates and the inter-study variability for the population model are given in Table 3. The Clint was estimated to be 6.64 ml/h, with modest variability of 24.4 %. The partition coefficient of sorafenib into the brain (0.06), skin (0.36), muscle (0.36), and adipose tissue (0.62) was low. The partition coefficient for kidneys (3.08), pancreas (1.75), spleen (1.58), and liver (4.37) were very high, and variability between studies was modest. The partition coefficient for the GI tract was high (3.85), with large variability, which is likely because the drug was dosed orally. Although the data were limited and variable, the concentrations in tumor were not dose-proportional as in plasma and other tissues. Thus, two different values for the tumor partition coefficient were obtained: 0.741 for the low dose and 2.12 for the high dose.
There was no statistically significant difference in the PK of everolimus when given alone or in combination with sorafenib, based upon student's t test (p > 0.05) of the AUCs from each study in which the drug was given alone and in combination. Alterations in everolimus PK when administered with sorafenib were also tested in the model, and selected individual parameter estimates were compared. No evidence of pharmacokinetic interaction was found, and dosing in combination did not cause significant variability in the hepatic clearance or everolimus tissue partition coefficients.
The PBPK model provided reasonable overall fits and was able to describe sorafenib concentrations well in plasma and other tissues, with the exception of some of the GI concentrations (Fig. 3). The model adequately characterized the concentrations as noted by the good individual predictions for concentrations of sorafenib in plasma and tissues. The plots of standardized residuals versus predicted concentrations for all tissues show an unbiased distribution, indicative of suitable fittings (Supplemental Information Fig. S2).
Discussion
Concomitant administration of drugs utilizing common pathways of absorption, distribution, metabolism, or elimination can lead to alterations in the PK of either drug. The present analysis characterized the concentration–time profiles for everolimus and sorafenib in plasma and various tissues after oral administration of the drugs alone and in combination. This information is essential to predict tissue toxicities due to drug accumulation or other interactions that could affect efficacy of the drug combination.
Tumor blood flow in the PBPK model was obtained initially from Baxter et al. [22] as used in other PBPK models [26]. That study employed a 86Rb uptake method [27] and measured blood flow to a human colon carcinoma xenograft in nude mice. Thus, the published value of 0.1 mL/min could be inaccurate for the poorly vascularized, patient-derived primary pancreatic adenocarcinoma explants. Therefore, Qtu was also estimated in our analysis of the everolimus data, and consequently fixed for subsequent analysis. The blood flow was estimated from the data was fivefold lower, 0.017 mL/min (Supplemental Information Table S1), underscoring the poorly perfused nature of patient-derived primary pancreatic adenocarcinoma explants [9].
The results indicate extensive distribution of everolimus and sorafenib into various tissues. Model-estimated partition coefficients (Kp) were close to values calculated by non-compartmental analysis. All tissue concentration–time data could be described well by the perfusion-limited model. It was assumed that everolimus and sorafenib are eliminated by metabolic biotransformation occurring primarily in the liver. This assumption was based on the fact that a negligible amount of either drug was detected in the urine in mice [13, 16]. Sorafenib is thought to undergo enterohepatic circulation (EHC) in humans based on the appearance of secondary peaks in the plasma concentration versus time profile [16]. Such EHC has been incorporated in a model to describe human PK of sorafenib [17] but was not included in our PBPK model for sorafenib owing to the limited sampling schedule.
Overall, sorafenib distributes more extensively into the tissues compared to everolimus, as expected based on the physicochemical properties of the drug such as lower molecular weight (637) compared to everolimus (958), lower log P (4.1) compared to everolimus (5.9), the weakly basic nature of the drug, and the comparatively lower polar surface area (sorafenib −92.4 vs. everolimus −205) (obtained from Pubchem Compound http://www.ncbi.nlm.nih.gov/pccompound/).
The sorafenib plasma concentrations observed here are consistent with the plasma PK in mice published in the FDA submission [16]. The Cmax and AUC after a single sorafenib dose which we obtained compared well with published results [14]. There are no published data available for PK of sorafenib in tissues, except for brain [4]. The K estimated here (0.06) agrees well with the calculated value of 0.02 for brain resulting from iv dosing [4]. The partition coefficients (K) estimated for brain, muscle, and adipose in our study were essentially identical to those calculated from published data for single doses [14], whereas the K estimated for kidneys, liver, spleen, lungs, and GI were two times higher than the tissue/plasma AUC ratio calculated from previous single-dose studies.
The partition coefficients of both drugs for the GI tract are high compared to other tissues. Such GI accumulation would be consistent with the occurrence of diarrhea and nausea in patients, which are the most common adverse clinical events associated with both drugs [5, 28]. The relatively higher partitioning of both drugs into liver and kidney might provide a PK basis for their effective use in hepatic and renal carcinomas.
Metabolism-related PK interactions were possible for sorafenib and everolimus but were not observed. The concomitant administration of everolimus with the strong CYP inhibitor ketoconazole increased AUC 15-fold in humans [29]. The moderate CYP inhibitors erythromycin, verapamil, and cyclosporine increased AUC 2.7–4.5 fold [15, 18, 30], and the weak inhibitor atorvastatin caused no change in everolimus AUC [31]. In vitro studies with human liver microsomes indicate that sorafenib is a moderate inhibitor of several CYP isoenzymes including CYP3A4, CYP2C19, and CYP2D6 [16]. In advanced melanoma patients, a PK interaction study with various CYP isoenzyme substrates such as midazolam, omeprazole, and dextromethorphan revealed no clinically significant changes in the exposures of these substrate probes or of sorafenib. There was also lack of clinically relevant adverse events or change in Cmax and exposure when administered concomitantly with a CYP3A4 inhibitor ketoconazole [32]. Induction of CYP3A protein in the liver of mice following extended dosing was investigated using immunoblot analysis, but no evidence of change in CYP enzymes was observed for either drug, alone or in combination (data not shown).
Extensive studies were conducted in both immunocompetent and SCID non-tumor-bearing mice as a baseline for comparison with tumor-bearing SCID mice (Table 1). Each study provided concentration versus time profiles for the various mouse models, duration of study, dosing regimen (single drug/combination; single- or multiple-dosing), drug doses, and presence of tumor. Population modeling enabled us to incorporate all these factors and evaluate potential variabilities arising from differing experimental conditions. The variability of the parameters reflects variability between studies. Due to limited sample sizes, we tested only the effect of single agent versus combination dosing on the estimated partition coefficients and clearances, as well as the effect of dose upon partition coefficient of sorafenib into tumor. Combining drugs did not have any significant impact on these parameters for either drug. An increase in dose increased the partition coefficient of sorafenib in tumors, thus altering tumor exposure. Tumor volumes in the study varied across treatment groups and also changed during the time course of the study. The population modeling approach enabled the inclusion of all these data jointly in the model.
The non-linearity in sorafenib partitioning into tumor may be the result of saturation of efflux transporters that are known to be involved in its distribution. Sorafenib is a weak P-glycoprotein (P-gp) substrate in vitro [33], and breast cancer resistant protein (BCRP) and P-gp at the blood brain barrier have been shown to play a dominant role in the efflux of sorafenib [4].
Involvement of P-gp in the disposition of everolimus also has been suggested [34, 35]. However, our data were unable to detect any changes in the disposition of everolimus at the doses tested.
In conclusion, population PBPK models were developed for everolimus and sorafenib in mice to account for the time course of drug concentrations in blood, an extensive variety of tissues, and in patient-derived pancreatic tumors. Concurrent administration of the two drugs did not cause a significant change in the PK of either agent. There was significant non-linearity in the tumor partition coefficient (i.e., tumor uptake) of sorafenib, with higher doses yielding greater-than-proportional drug concentrations in the tumor. This observation raises the possibility that anti-tumor pharmacodynamics may not be dose-proportional. The lack of PK interactions suggests feasibility for evaluating the therapeutic efficacy of this two-drug combination in pancreatic cancer, and the non-dose-proportionality of sorafenib tumor deposition may have therapeutic ramifications.
Supplementary Material
Acknowledgments
We thank Nancy Pyszczynski, Ninfa Straubinger, Rose Pitoniak, Kim Clark, and Joshua Prey for excellent technical assistance. We are grateful to The Novartis Institute for Biomedical Research Basel, Switzerland, for providing everolimus for animal studies. Grant Support-GM57980 and a Clinical Translational Science Award from the University at Buffalo (2011).
Footnotes
Electronic supplementary material: The online version of this article (doi:10.1007/s00280-013-2116-y) contains supplementary material, which is available to authorized users.
Conflict of interest: None.
Contributor Information
Dipti K. Pawaskar, Department of Pharmaceutical Sciences, School of Pharmacy and Pharmaceutical Sciences, State University of New York at Buffalo, Buffalo, NY 14260, USA
Robert M. Straubinger, Department of Pharmaceutical Sciences, School of Pharmacy and Pharmaceutical Sciences, State University of New York at Buffalo, Buffalo, NY 14260, USA; Department of Cancer Pharmacology and Therapeutics, Roswell Park Cancer Institute, Buffalo, NY 14263, USA
Gerald J. Fetterly, Department of Medicine, Roswell Park Cancer Institute, Buffalo, NY 14263, USA
Bonnie H. Hylander, Department of Immunology, Roswell Park Cancer Institute, Buffalo, NY 14263, USA
Elizabeth A. Repasky, Department of Immunology, Roswell Park Cancer Institute, Buffalo, NY 14263, USA
Wen W. Ma, Department of Medicine, Roswell Park Cancer Institute, Buffalo, NY 14263, USA
William J. Jusko, Email: wjjusko@buffalo.edu, Department of Pharmaceutical Sciences, School of Pharmacy and Pharmaceutical Sciences, State University of New York at Buffalo, Buffalo, NY 14260, USA.
References
- 1.Atkins MB, Yasothan U, Kirkpatrick P. Everolimus. Nat Rev Drug Discov. 2009;8:535–536. doi: 10.1038/nrd2924. [DOI] [PubMed] [Google Scholar]
- 2.Piguet AC, Saar B, Hlushchuk R, St-Pierre MV, McSheehy PM, Radojevic V, Afthinos M, Terracciano L, Djonov V, Dufour JF. Everolimus augments the effects of sorafenib in a syngeneic orthotopic model of hepatocellular carcinoma. Mol Cancer Ther. 2011;10:1007–1017. doi: 10.1158/1535-7163.MCT-10-0666. [DOI] [PubMed] [Google Scholar]
- 3.Rubio-Viqueira B, Hidalgo M. Targeting mTOR for cancer treatment. Curr Opin Investig Drugs. 2006;7:501–512. [PubMed] [Google Scholar]
- 4.Agarwal S, Sane R, Ohlfest JR, Elmquist WF. The role of the breast cancer resistance protein (ABCG2) in the distribution of sorafenib to the brain. J Pharmacol Exp Ther. 2011;336:223–233. doi: 10.1124/jpet.110.175034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harzstark AL, Small EJ, Weinberg VK, Sun J, Ryan CJ, Lin AM, Fong L, Brocks DR, Rosenberg JE. A phase 1 study of everolimus and sorafenib for metastatic clear cell renal cell carcinoma. Cancer. 2011;117:4194–4200. doi: 10.1002/cncr.25931. [DOI] [PubMed] [Google Scholar]
- 6.McKeage K, Wagstaff AJ. Sorafenib: in advanced renal cancer. Drugs. 2007;67:475–483. doi: 10.2165/00003495-200767030-00009. discussion 484–5. [DOI] [PubMed] [Google Scholar]
- 7.Nexavar Tablet prescribing information 2007 [Google Scholar]
- 8.Wilhelm S, Chien DS. BAY 43–9006: preclinical data. Curr Pharm Des. 2002;8:2255–2257. doi: 10.2174/1381612023393026. [DOI] [PubMed] [Google Scholar]
- 9.Komar G, Kauhanen S, Liukko K, Seppanen M, Kajander S, Ovaska J, Nuutila P, Minn H. Decreased blood flow with increased metabolic activity: a novel sign of pancreatic tumor aggressiveness. Clin Cancer Res. 2009;15:5511–5517. doi: 10.1158/1078-0432.CCR-09-0414. [DOI] [PubMed] [Google Scholar]
- 10.Gjedde SB, Gjedde A. Organ blood flow rates and cardiac output of the BALB/c mouse. Comp Biochem Physiol A Physiol. 1980;67:5. [Google Scholar]
- 11.Mager DE, Jusko WJ. Pharmacodynamic modeling of time-dependent transduction systems. Clin Pharmacol Ther. 2001;70:210–216. doi: 10.1067/mcp.2001.118244. [DOI] [PubMed] [Google Scholar]
- 12.Brown RP, Delp MD, Lindstedt SL, Rhomberg LR, Beliles RP. Physiological parameter values for physiologically based pharmacokinetic models. Toxicol Ind Health. 1997;13:407–484. doi: 10.1177/074823379701300401. [DOI] [PubMed] [Google Scholar]
- 13.Leighton JK, Saber H, Lee H. Pharmacology review-Everolimus. Center for Drug Evaluation and Research; Rockville: 2009. [Google Scholar]
- 14.O'Reilly T, McSheehy PM, Kawai R, Kretz O, McMahon L, Brueggen J, Bruelisauer A, Gschwind HP, Allegrini PR, Lane HA. Comparative pharmacokinetics of RAD001 (everolimus) in normal and tumor-bearing rodents. Cancer Chemother Pharmacol. 2010;65:625–639. doi: 10.1007/s00280-009-1068-8. [DOI] [PubMed] [Google Scholar]
- 15.Kovarik JM, Kalbag J, Figueiredo J, Rouilly M, Frazier OL, Rordorf C. Differential influence of two cyclosporine formulations on everolimus pharmacokinetics: a clinically relevant pharmacokinetic interaction. J Clin Pharmacol. 2002;42:95–99. doi: 10.1177/0091270002042001011. [DOI] [PubMed] [Google Scholar]
- 16.Saber-Mahloogi H, Morse DE. Pharmacology review-Sorafenib. Center for Drug Evaluation and Research; Rockville: 2005. [Google Scholar]
- 17.Jain L, Woo S, Gardner ER, Dahut WL, Kohn EC, Kummar S, Mould DR, Giaccone G, Yarchoan R, Venitz J, Figg WD. Population pharmacokinetic analysis of sorafenib in patients with solid tumours. Br J Clin Pharmacol. 2011;72:294–305. doi: 10.1111/j.1365-2125.2011.03963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovarik JM, Beyer D, Bizot MN, Jiang Q, Allison MJ, Schmouder RL. Pharmacokinetic interaction between verapamil and everolimus in healthy subjects. Br J Clin Pharmacol. 2005;60:434–437. doi: 10.1111/j.1365-2125.2005.02434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hylander BL, Pitoniak R, Penetrante RB, Gibbs JF, Oktay D, Cheng J, Repasky EA. The anti-tumor effect of Apo2L/TRAIL on patient pancreatic adenocarcinomas grown as xenografts in SCID mice. J Transl Med. 2005;3:22. doi: 10.1186/1479-5876-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh Y, Galviz G, Long BJ. Ultra-performance hydrophilic interaction liquid chromatography/tandem mass spectrometry for the determination of everolimus in mouse plasma. Rapid Commun Mass Spectrom. 2009;23:1461–1466. doi: 10.1002/rcm.4022. [DOI] [PubMed] [Google Scholar]
- 21.Jain L, Gardner ER, Venitz J, Dahut W, Figg WD. Development of a rapid and sensitive LC-MS/MS assay for the determination of sorafenib in human plasma. J Pharm Biomed Anal. 2008;46:362–367. doi: 10.1016/j.jpba.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baxter LT, Zhu H, Mackensen DG, Jain RK. Physiologically based pharmacokinetic model for specific and nonspecific monoclonal antibodies and fragments in normal tissues and human tumor xenografts in nude mice. Cancer Res. 1994;54:1517–1528. [PubMed] [Google Scholar]
- 23.Davies B, Morris T. Physiological parameters in laboratory animals and humans. Pharm Res. 1993;10:1093–1095. doi: 10.1023/a:1018943613122. [DOI] [PubMed] [Google Scholar]
- 24.D'Argenio DZ, Schumitzky A, Wang X. ADAPT 5 User's Guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomed Simul Res 2009 [Google Scholar]
- 25.Pawaskar DK, Straubinger RM, Fetterly GJ, Hylander BH, Repasky EA, Ma WW, Jusko WJ. Synergistic interactions between sorafenib and everolimus in pancreatic cancer xenografts in mice. Cancer Chemother Pharmacol. 2012 doi: 10.1007/s00280-013-2117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urva SR, Yang VC, Balthasar JP. Physiologically based pharmacokinetic model for T84.66: a monoclonal anti-CEA antibody. J Pharm Sci. 2010;99:1582–1600. doi: 10.1002/jps.21918. [DOI] [PubMed] [Google Scholar]
- 27.Friedman JJ. Muscle blood flow and 86Rb extraction: 86Rb as a capillary flow indicator. Am J Physiol. 1968;214:488–493. doi: 10.1152/ajplegacy.1968.214.3.488. [DOI] [PubMed] [Google Scholar]
- 28.Hotte SJ, Hirte HW. BAY 43–9006: early clinical data in patients with advanced solid malignancies. Curr Pharm Des. 2002;8:2249–2253. doi: 10.2174/1381612023393053. [DOI] [PubMed] [Google Scholar]
- 29.Kovarik JM, Beyer D, Bizot MN, Jiang Q, Shenouda M, Schmouder RL. Blood concentrations of everolimus are markedly increased byketoconazole. J Clin Pharmacol. 2005;45:514–518. doi: 10.1177/0091270005275368. [DOI] [PubMed] [Google Scholar]
- 30.Kovarik JM, Beyer D, Bizot MN, Jiang Q, Shenouda M, Schmouder RL. Effect of multiple-dose erythromycin on everolimus pharmacokinetics. Eur J Clin Pharmacol. 2005;61:35–38. doi: 10.1007/s00228-004-0866-5. [DOI] [PubMed] [Google Scholar]
- 31.Kovarik JM, Hartmann S, Hubert M, Berthier S, Schneider W, Rosenkranz B, Rordorf C. Pharmacokinetic and pharmacodynamic assessments of HMG-CoA reductase inhibitors when coadministered with everolimus. J Clin Pharmacol. 2002;42:222–228. doi: 10.1177/00912700222011148. [DOI] [PubMed] [Google Scholar]
- 32.Lathia C, Lettieri J, Cihon F, Gallentine M, Radtke M, Sundaresan P. Lack of effect of ketoconazole-mediated CYP3A inhibition on sorafenib clinical pharmacokinetics. Cancer Chemother Pharmacol. 2006;57:685–692. doi: 10.1007/s00280-005-0068-6. [DOI] [PubMed] [Google Scholar]
- 33.Gnoth MJ, Sandmann S, Engel K, Radtke M. In vitro to in vivo comparison of the substrate characteristics of sorafenib tosylate toward P-glycoprotein. Drug Metab Dispos. 2010;38:1341–1346. doi: 10.1124/dmd.110.032052. [DOI] [PubMed] [Google Scholar]
- 34.Chu C, Abbara C, Noel-Hudson MS, Thomas-Bourgneuf L, Gonin P, Farinotti R, Bonhomme-Faivre L. Disposition of everolimus in mdr1a-/1b- mice and after a pre-treatment of lapatinib in Swiss mice. Biochem Pharmacol. 2009;77:1629–1634. doi: 10.1016/j.bcp.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 35.Crowe A, Lemaire M. In vitro and in situ absorption of SDZ-RAD using a human intestinal cell line (Caco-2) and a single pass perfusion model in rats: comparison with rapamycin. Pharm Res. 1998;15:1666–1672. doi: 10.1023/a:1011940108365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.