SUMMARY
Sorting of ubiquitinated membrane proteins into lumenal vesicles of multivesicular bodies is mediated by the ESCRT apparatus and accessory proteins such as Bro1, which recruits the deubiquitinating enzyme Doa4 to remove ubiquitin from cargo. Here we propose that Bro1 works as a receptor for the selective sorting of ubiquitinated cargos. We found synthetic genetic interactions between BRO1 and ESCRT-0, suggesting Bro1 functions similarly to ESCRT-0. Multiple structural approaches demonstrated that Bro1 binds ubiquitin via the N-terminal trihelical arm of its middle V domain. Mutants of Bro1 that lack the ability to bind Ub were dramatically impaired in their ability to sort Ub-cargo membrane proteins, but only when combined with hypomorphic alleles of ESCRT-0. These data suggest that Bro1 and other Bro1 family members function in parallel with ESCRT-0 to recognize and sort Ub-cargos.
INTRODUCTION
Ubiquitin (Ub) is a sorting determinant mediating the degradation of a wide variety of membrane proteins in lysosomes by sorting them into the intralumenal vesicles (ILVs) of multivesicular endosomes/bodies (MVB) (Hanson and Cashikar, 2012). Ub is recognized by the ESCRT (Endosomal Sorting Complex Required for Transport) apparatus, which couples cargo-recognition and sorting with the formation and scission of intralumenal vesicles (Henne et al., 2011). Recognition of ubiquitinated cargo (Ub-cargo) occurs early during ILV formation, and is mediated in large part by ESCRT-0 and ESCRT-I, which each have multiple Ub-binding domains (UBDs) (Clague et al., 2012; Shields and Piper, 2011). ESCRT-0, composed of Vps27 and Hse1 in yeast, and by Hrs and STAM1/2 in humans, binds Ub via multiple UBDs housed within its VHS and UIM domains. ESCRT-0 is thought to be the endosomal Ub-sorting receptor that initiates cargo capture, and is equipped with a variety protein interactions that equip it for this task. Direct binding to clathrin positions it within clathrin-enriched endosomal subdomains where cargo is segregated for recycling or degradation. ESCRT-0 also binds ESCRT-I to facilitate assembly of the ESCRT apparatus and the transfer of Ub-cargo to ESCRT-I and -II. ESCRT-0 also associates with both Ub-ligases and deubiquitinating enzymes that may alter cargo ubiquitination and regulate its sorting into the MVB pathway. Several other proteins are proposed to work in parallel to ESCRT-0 as alternative ESCRT-0-like Ub-sorting receptors (Clague et al., 2012; Shields and Piper, 2011). Among these are the Tom1:Tollip complex and GGA3. Like the two subunits of ESCRT-0, both Tom1 and GGA3 have VHS domains and bind clathrin, ESCRT-I, and Ub. Functional studies implicate these as endosomal Ub-sorting receptors, although their site(s) of action and the repertoire of cargo substrates have yet to be clarified.
Late in the sorting process, ILVs are separated from the limiting membrane of the endosome by ESCRT-III, a heteropolymeric assembly of subunits centered on Snf7/CHMP4 (Babst et al., 2011). Yeast Snf7 also binds to Bro1, which in turn recruits the deubiquitinating enzyme (DUb) Doa4 to remove Ub from cargo prior to its entry into ILVs (Kim et al., 2005; Luhtala and Odorizzi, 2004; Richter et al., 2007). Loss of Doa4 causes Ub to hyperaccumulate in the vacuole lumen and be depleted from the cytosol (Amerik et al., 2000; Ren et al., 2008). The Bro1:Doa4 complex is thought to work late in the sorting process since earlier removal of Ub would permit cargo to escape incorporation into ILVs. However, loss of Bro1 produces a phenotype similar to loss of ESCRTs, while loss of Doa4 does not, demonstrating that Bro1 provides functions beyond the recruitment of Doa4 (Odorizzi et al., 2003; Raymond et al., 1992; Springael et al., 2002). Bro1 belongs to a larger family of related proteins including mammalian Alix and HD-PTP, which share a common architecture, bind ESCRT-I, and have both an N-terminal Bro1 homology domain that binds the ESCRT-III subunit Snf7/CHMP4 and a middle V domain that, in the case of Alix binds YPxL peptides motifs (Fisher et al., 2007; Kim et al., 2005; Lee et al., 2007). YPxL-binding enables Alix to sort YPxL-bearing cargos into MVBs and to mediate ESCRT-dependent budding of viruses whose Gag proteins bear YPxL motifs (Baietti et al., 2012; Dores et al., 2012; Strack et al., 2003). These data suggest that Alix and other Bro1-family proteins might recruit cargo to the ESCRT apparatus, at least along a Ub-independent pathway. Although a clear role for these proteins in the canonical sorting of Ub-cargo has not been established, recent studies have demonstrated that Alix can bind Ub thus supporting this possibility (Joshi et al., 2008; Keren-Kaplan et al., 2013; Sangsuriya et al., 2010).
Here we propose that Bro1 works in parallel with ESCRT-0 and contributes to the recognition and sorting of Ub-cargo into the MVB pathway. We show that deficiencies in Bro1 and ESCRT-0 yield synthetic phenotypes and, that like ESCRT-0, Bro1 binds clathrin. We show that the V domains of multiple Bro1 family members bind Ub, and that mutations compromising Ub-binding result in defective sorting of Ub-cargo.
RESULTS
Bro1 and the ESCRT-0 Ub-sorting receptor interact genetically
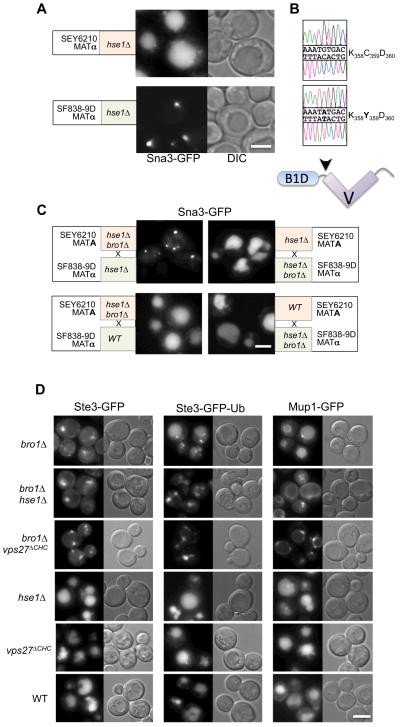
Deletion of either Vps27 or Hse1, two ESCRT-0 subunits (orthologous to Hrs and Stam1/2 in mammalian cells), causes a strong “class E” Vps phenotype, characterized by secretion of vacuolar proteases, accumulation of large late endosomal structures, and the inability to sort ubiquitinated membrane proteins into the vacuolar lumen via the MVB pathway (Bilodeau et al., 2002). This phenotype is observed when HSE1 is deleted from the SF838-9D parental strain, but not when deleted from the SEY6210 parental strain (Fig. 1A) as also noted in previous studies (Stringer and Piper, 2011). Diploid cells generated from these two hse1Δ strains also showed no sorting defects, providing us with a genetic tool for identifying genes that function in parallel with ESCRT-0 and whose depletion exacerbates the consequences of HSE1 loss. The homozygous hse1Δ diploid was sporulated and after one backcross to SEY6210 hse1Δ cells, we found that the phenotype of defective MVB sorting segregated 2:2 (Fig. 1SA). We determined the genomic sequence of the SEY6210 hse1Δ strain and compared it to that of hse1Δ segregants from a 2nd and 3rd backcross that showed MVB sorting defects to identify a single base-pair change in BRO1 (resulting in a C359Y change) that was present in defective progeny but not in the parental SEY6210 hse1Δ strain. Sanger sequencing confirmed this difference (Fig. 1B).
Figure 1. Synthetic interaction between Bro1 and ESCRT-0.
A. Localization of Ste3-GFP in hse1Δ null mutants generated from the SEY6210 and SF838-9D parental strains. Shown are DIC and GFP-fluorescence images
B. Chromatograms of BRO1 ORF sequence (Sanger sequencing of PCR-amplified DNA) from SEY6210 cells (top) or SF838-9D cells (bottom). Arrowhead in schematic below indicates location of residue 359 between the N-terminal Bro1 homology domain (B1D) and the middle V domain.
C. Demonstration that the bro1 Y359 allele causes a synthetic phenotype with hse1Δ. BRO1Δ was disrupted in MATA SEY6210 hse1Δ or MATα SF838 hse1Δ haploids, and these were subsequently mated to form diploids. Sorting of Sna3-GFP to the vacuole lumen was defective in hse1Δ homozygous diploids with only the bro1Y359 allele from the SF838-9D haplotype. However, Sna3-GFP was correctly sorted in hse1Δ homozygous diploids with only BRO1C359 from the SEY6210 haplotype. Diploids heterozygous for both hse1Δ bro1Δ also sorted Sna3-GFP properly regardless of from either haplotype.
D. Localization of Ste3-GFP, Ste3-GFP-Ub, and Mup1-GFP in SEY6210 cells of the indicated genotypes: wild-type (WT); bro1Δ null alone; bro1Δ hse1Δ double null; Vps27 lacking its C-terminal clathrin-binding motif (vps27ΔChc1); hse1Δ alone or the vps27ΔChc1 mutation alone. Bar=5μM
Residue 359 of Bro1 lies within the linker region between the N-terminal Bro1 homology domain and the middle V domain. Importantly, the SF838-9D parental strain has normal MVB and vacuolar protease sorting pathways despite having the BRO1 Y359 allele. However, a bro1Δ deletion has a typical “class E” Vps phenotype (Raymond et al., 1992) indicating that a sorting defect results only when the Y359-encoding allele of BRO1 is combined with loss of HSE1. As confirmation we made homozygous hse1Δ diploids from SF838-9D and SEY6210, in which the BRO1 gene was knocked out of either haplotype. The hse1Δ diploids having only the Y359 Bro1 from SF838-9D showed a strong MVB sorting defect as assessed by the localization of Sna3-GFP (Fig. 1C), a well-characterized MVB cargo (Macdonald et al., 2011). In contrast, hse1Δ homozygotes with the SEY6210 allele of BRO1 (C359) sorted Sna3-GFP normally. Although the biochemical defects of the Y359 Bro1 mutant were not investigated, the value of this mutant was to help uncover the genetic interaction between Bro1 and ESCRT-0.
To confirm the synthetic genetic interaction between Bro1 and Hse1, we analyzed MVB sorting in SEY6210 bro1Δ cells that also contained defined mutations in ESCRT-0 (Fig. 1D). Loss of Bro1 blocked the delivery of Ste3-GFP to the vacuole lumen, causing these cells to accumulate large “class E” endosomal compartments. Interestingly, other cargoes could still be sorted to the vacuole of bro1Δ cells, albeit to a limited degree. Both Ste3-GFP-Ub (an in-fame fusion of Ub onto Ste3-GFP) and GFP-tagged Mup1 (a methionine transporter) were sorted moderately well in bro1Δ cells and normally in hse1Δ cells. Combined loss of HSE1 and BRO1 caused substantial sorting defects that were observed by microscopy (Fig. 1D) and by immunoblotting for a GFP (Fig. S1B), which is cleaved from cargo upon delivery into the vacuolar lumen (Hettema et al., 2004). A synthetic defect was also observed when the bro1Δ mutation was combined with a hypomorphic mutation in Vps27 that blocks the ability of ESCRT-0 to bind to clathrin (vps27Δchc1) (Bilodeau et al., 2003; Shields et al., 2009). Finally, we found that levels of Bro1 were unperturbed by alterations in ESCRT-0, and that levels of ESCRT-0 subunit Vps27 were unperturbed by changes in BRO1 or HSE1, indicating the synthetic interaction between Bro1 and ESCRT-0 functions is specific (Fig. S1C).
Bro1 acts early in cargo-sorting and binds clathrin
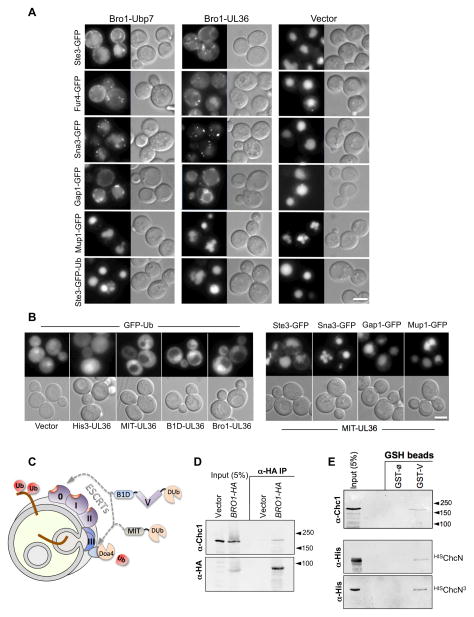
The synthetic genetic interaction between Bro1 and ESCRT-0 suggested that Bro1 may act early in the MVB sorting process in parallel with ESCRT-0. This function would be distinct from that previously established for Bro1, which is to recruit the Doa4 DUb to ESCRT-III to remove Ub from MVB cargo and rescue Ub from excessive vacuolar degradation and which is mediated by Bro1 binding to Snf7 via its N-terminal Bro1 domain and Doa4 via its proline-rich C-terminal domain (Amerik et al., 2006; Nikko and Andre, 2007; Richter et al., 2007). Previous studies show that fusing the catalytic domain of a deubiquitinating enzyme (DUb) onto ESCRT-0 effectively deubiquitinates cargo “early” in the sorting process, thereby blocking delivery to the vacuolar lumen (MacDonald et al., 2012; Stringer and Piper, 2011). This block can be circumvented by translationally fusing Ub to the C-termini of cargo making it resistant to the effects of ESCRT-DUb fusions. We reasoned that if Bro1 also could intervene early in the sorting process in parallel to ESCRT-0, then a Bro1-DUb fusion protein should also block cargo sorting to the vacuole and not merely replace the Bro1:Doa4 complex thought to work post-sorting to simply recycle Ub. Fusions of Bro1 to the catalytic domain of Ubp7 or UL36 (a yeast cysteine-based Ub-specific protease or a DUb found within the tegument protein of Herpes Simplex Virus-1, respectively) were expressed in wild-type cells (Fig. 2A). In addition to causing defective sorting of Ste3 and Sna3, Bro1-Ubp7 and Bro1-UL36 also perturbed sorting of Gap1 and Fur4 (the General Amino acid Permease and Uracil permease). In contrast, Ste3-GFP-Ub, containing an in-frame fusion of Ub that cannot be removed by DUbs, sorted normally demonstrating that Bro1-Ubp7 and Bro1-UL36 exert their inhibitory effect at the level of deubiquitinating cargo rather than altering the function of ESCRT apparatus itself. Mup1-GFP was only modestly affected by the Bro1-DUb fusions (Fig. 2A) and also underwent some level of MVB sorting even in the absence of Bro1 (Fig. 1D) suggesting that Bro1 may normally operate on a subset of MVB cargos.
Figure 2. Effect of Bro1-DUb fusion protein on the sorting of MVB cargo.
A. Bro1 fusion proteins containing the catalytic domain of either Ubp7 (Bro1-Ubp7) or UL36 (Bro1-UL36) were expressed from the copper-inducible CUP1 promoter in wild-type cells, in combination with the indicated GFP-tagged MVB cargo proteins. Shown are DIC and GFP fluorescence images.
B. Sorting of GFP-Ub in wild-type cells or in cells expressing the UL36 fused to His3 (His3-UL36), the N-terminal MIT domain of Vps4 (MIT-UL36), the N-terminal Bro1 domain (B1D-UL36), or full-length Bro1 (Bro1-UL36) (left). Wild-type cells or cells expressing a His3-UL36 fusion protein accumulate some GFP-Ub within vacuoles. Expression of Bro1-UL36 or MIT-UL36 excludes GFP-Ub from vacuoles. MIT-UL36 was also co-expressed in WT cells with the indicated GFP-tagged MVB cargo (right). Bar=5μM
C. Model for how Bro1 might work in two places: early in the process of cargo sorting in conjunction with ESCRT-0, such that the Bro1-DUb fusion proteins deubiquitinate cargo and block subsequent sorting into the MVB pathway; and late in the sorting process in conjunction with ESCRT-III, to recycle Ub from the MVB pathway and thus prevent its accumulation in the vacuole. This latter function can be mimicked by MIT-DUb.
D. Immunopreciptiation of HA-tagged Bro1 from spheroplasts prepared from cells transformed with vector alone or plasmid expressing Bro1-HA. Samples were immunoblotted with monoclonal antibodies against Chc1 (top) or HA (bottom).
E. Pulldown of GST alone (∅) or GST fused to the Bro1 V domain (GST-V). Bead-bound fractions were immunoblotted as was a 5% equivalent of input. Top shows pulldown of Chc1 from yeast lysates. Bottom shows pulldown of a monomeric 6xHis-tagged recombinant N-terminal clathrin β-propeller (HISChcN), or an oligomeric version containing a trimerization domain (HISChcN3).
The effect of Bro1-DUb in blocking cargo sorting is not due simply to recruiting DUb activity to ESCRT-III. We found that expressing a fusion of UL36 to the MIT domain of Vps4, which interacts with MIT-Interaction Motifs (MIM) within ESCRT-III subunits (Obita et al., 2007; Stuchell-Brereton et al., 2007), did not affect MVB cargo sorting (Fig. 2B). However, MIT-UL36 as well as UL36 fusion to full-length Bro1 and just the N-terminal Bro1 domain prevented the delivery of GFP-Ub into the vacuole, demonstrating that these proteins could stimulate recycling of Ub from cargo before it was consumed in the MVB sorting process. Thus, the dominant effect of Bro1-DUb proteins on the sorting of particular cargos likely reflects a function of Bro1 that works early in the cargo sorting process (Fig. 2C).
One of the biochemical features of ESCRT-0 and other proposed ESCRT-0-like Ub-sorting receptors such as Tom1 and GGAs is that they bind clathrin, which is found in distinct endosomal subdomains that concentrate Ub-cargo and help localize ESCRT-0 (Shields and Piper, 2011). Coimmunoprecipitation experiments showed that Bro1 shares the ability of ESCRT-0 to associate with clathrin in vivo (Fig. 2D). Clathrin-binding activity was housed within the middle Bro1 V domain since a GST-V domain fusion protein was sufficient to pull-down clathrin from yeast lysates (Fig. 2E). Also, the recombinant N-terminal β-propeller domain of clathrin heavy chain could specifically bind GST-V, indicating that the Bro1 V domain binds directly to clathrin (Fig. 2E). Interestingly, Bro1 has a conserved clathrin binding box motif (ter Haar et al., 2000) within an unstructured loop at the vertex of its V domain (Fig. S2A,B). However, mutations in this region did not block coimmunoprecipitation of Bro1 with clathrin from cell lysates suggesting other motifs may also be sufficient for clathrin-association (data not shown).
The V domains of Bro1 family proteins bind Ubiquitin
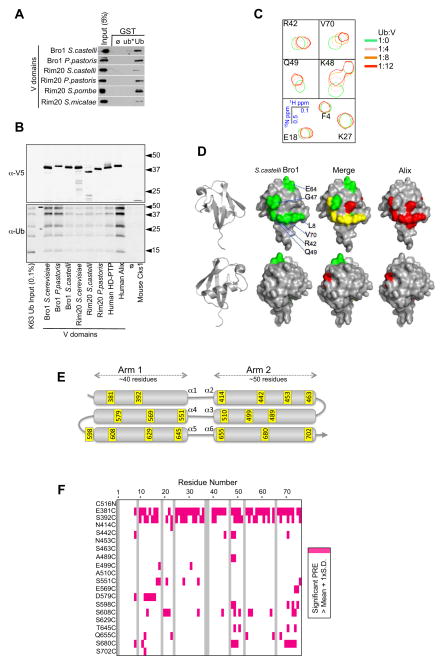
One of the key features of ESCRT-0 that allows it to act as a sorting receptor for Ub-cargo is its ability to bind Ub. Recently, the Bro1 homolog Alix was found to bind Ub via its middle V domain (Joshi et al., 2008). The Alix V domain is organized into 2 trihelical bundles adopting the shape of a “V” with a short and long arm (Fisher et al., 2007; Lee et al., 2007). We found the V domains of Bro1, Rim20, human Alix and HD-PTP bind directly to Ub even though they share only a moderate level of sequence identity. This was demonstrated by the ability of recombinant V domains to bind a Ub-GST fusion protein (Fig 3A), and to bind K63-linked polyubiquitin chains (Fig. 3B).
Figure 3. V domains of Bro1 and other family members bind Ub.
A. GST pull-down experiments of recombinant Bro1 and Rim20 V domains S. castelli, P. pastoris, S. pombe, and S. micatae using GST alone (∅), GST fused to Ub, or a mutant Ub with mutations in L8, I44, R42, and V70 (ub*)
B. Recombinant V domains were immobilized on α-V5 polyclonal antibody-coated beads, washed, and incubated with K63-linked polyubiquitin chains. Beads, alone or bound to an irrelevant V5-epitope-tagged protein were also included. Beads were washed and immunoblotted with α-V5 or α-Ub monoclonal antibodies.
C. HSQC NMR spectra of indicated backbone amides of 30μM 15N-Ub in the absence and presence of the S. castelli V domain at the designated ratios.
D. Significant chemical shift perturbations (>1 StDev of (0.2N2 + H2)1/2) caused by binding of the Bro1 or Alix V domain to 15N-Ub in HSQC experiments were mapped onto the molecular surface of Ub. Shown are front and back views of Ub, with the binding surface for Bro1 in green and Alix in red. Overlap in the two chemical shift perturbation profiles is shown as yellow in the merge.
E. Schematic of the S. castelli V domain, which forms two trihelical arms. The amino acid positions in yellow designate cysteine substitutions used to conjugate site-specific spin labels using MTSL.
F. Summary of paramagnetic relaxation experiments using the series of Bro1 cysteine mutants that bear spin-label at the indicated positions. The NMR 15N/1H HSQC spectra of 30μM 15N-Ub with 100μM of the indicated MTSL-labelled Bro1 V domains were collected in the presence (oxidized) and absence (reduced) of 2 mM ascorbate. Peak intensity ratios of these two spectra were calculated for each backbone amide. Plot of Ub residues that were subject to significant PRE, as reflected by a significant reduction in peak intensity ratio, defined as > 1 SD above the mean for that residue across the whole dataset.
Chemical shift perturbations measured by NMR HSQC experiments with 15N-Ub showed that the Bro1 V domain bound mono-Ub (Fig. 3C and S3A,B) and used a binding surface centered on the hydrophobic patch of Ub comprised of I44, V70, L8, and R42 (Fig. 3D), which serves as a common surface engaged by the vast majority of Ub-binding proteins (Husnjak and Dikic, 2012). These experiments utilized the V domain from S. castelli Bro1, which shares 56% identity with S. cerevisiae Bro1 V but had better stability in vitro. The Alix V domain also bound mono-Ub through the same general surface patch, although the profile of chemical shift perturbations in 15N-Ub induced by Alix V binding was slightly different indicating a different binding mode (Fig. 3D, Fig. S3A,B). Significant line broadening was evident for all peaks of 15N-Ub with increasing concentrations of V domains (Fig. S3C). This was most severe for HD-PTP V domain, which precluded the ability to use chemical shift changes to map the interface on Ub. The NMR peaks from Ub bound to V domain were likely broadened because the Ub-V complex is much larger than Ub alone and in range of intermediate exchange on the NMR timescale. Using loss of peak intensity as a measure of binding estimates the Kd of the V domains of Alix and HD-PTP in the range of ~50 μM and the S. castelli Bro1 V domain ~5 times higher.
To map the major Ub-binding site on Bro1 V in solution, we performed a series of paramagnetic relaxation enhancement (PRE) experiments using a set of Bro1 V mutants containing a nitroxide spin-label (MTSL:methanethiosulfonate) attached to cysteine residues substituted at different positions (Fig. 3E). MTSL-labeled Bro1 V proteins were used in HSQC experiments with 15N-Ub in the presence and absence of ascorbate. The oxidized form of the attached MTSL enhances the relaxation rate of nearby spin systems, resulting in a loss of peak intensity in HSQC spectra as compared to the ascorbate-reduced form that has lost its unpaired electron (Iwahara and Clore, 2006). Significant PRE effects on any of the residues of 15N-Ub were plotted for each of the labeled Bro1 V variants (Fig. 3F) revealing dramatic effects for spin labels incorporated in the N-terminal helix of first trihelical arm of the Bro1 V domain at residues 381 and 392. As confirmation, we generated a “half-V” protein comprised of only the first trihelical arm and found that it bound Ub in GST-pull-down assays and induced a profile of chemical shift perturbations on the surface 15N-Ub similar to full-length Bro1 V (Fig. S3D-I).
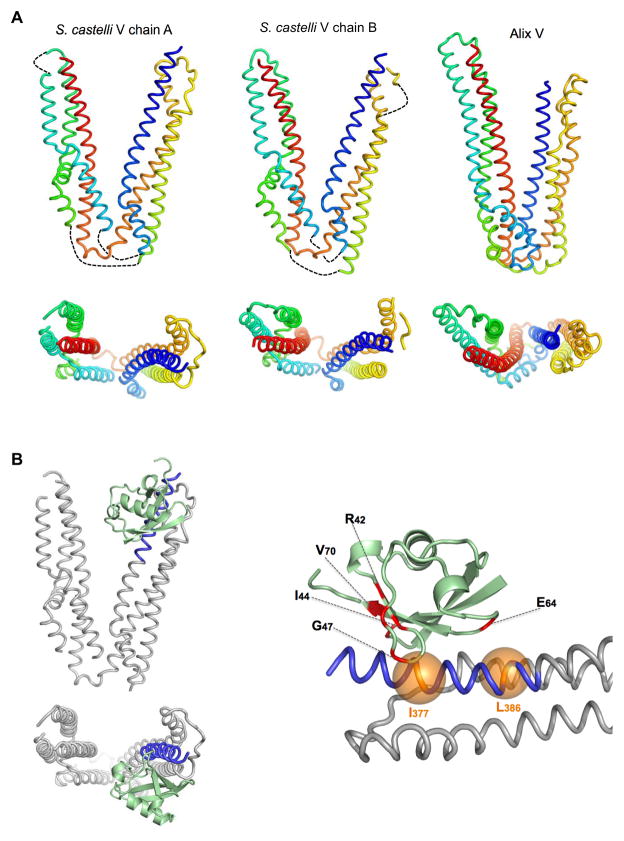
Further structural information about Ub-binding was obtained through protein crystallography experiments yielding a structure of Ub bound to the first trihelical arm the S. castelli Bro1 V domain. This structure was solved from crystals containing a selenomethionine-labelled Bro1V:Ub A28M complex diffracting to 3.6Å that allowed us to use the single-wavelength anomalous dispersion method to obtain experimental phases (Table I). Two V domains were found in the asymmetric unit with five Selenium sites per molecule of V domain. Despite the low level of sequence homology, the Bro1 V domain was remarkably similar to the Alix V domain, each with two trihelical arms with the protein sequence crisscrossing between the two arms. Unlike Alix V, the two arms of Bro1 V were of similar size (Fig. 4A) and the relative orientation of the trihelical arms and the helices within the distal C-terminal arm as they twist through the arm were different.
Table 1.
Crystallography Statistics
| SeMet Bro1VSEM:UbA28M | SeMet AlixV:Ub | |
|---|---|---|
| DATA COLLECTION | ||
| Space group | P212121 | I23 |
| Unit cell parameters (Å) | a=63.43, b=92.32, c=229.45 | a=b=c=223.77 |
| Resolution (Å) | 34.49–3.50 (3.62–3.50) | 39.56–6.50 (6.73–6.50) |
| Rmerge | 8.4 (61.1) | 6.1 (74.8) |
| Unique Reflections | 17664 (1700) | 3774 (370) |
| <I/σ(I)> | 6.6 (1.7) | 23.3 (2.1) |
| Completeness (%) | 100.0 (99.9) | 100.0 (100.0) |
| Multiplicity | 7.1 (7.3) | 21.4 (22.3) |
| Anomalous <I/σ(I)> | 5.3 (1.0) | 18.6 (1.2) |
| Anomalous completeness (%) | 99.9 (99.9) | 100.0 (100.0) |
| Anomalous multiplicity | 3.85 (3.83) | 11.4 (11.6) |
| REFINEMENT | ||
| Resolution (Å) | 34.49–3.60 | 39.56–6.50 |
| No. reflections | 16053 | 3760 |
| Rwork/Rfree | 36.6/42.1 | 20.3/28.3 |
| No. protein atoms | 3536 | 5398 |
| B FACTORS | ||
| Wilson (Å2) | 131.3 | 468.9 |
| Average (Å2) | 158.2 | 150.3 |
| RMS DEVIATIONS | ||
| Bond lengths (Å) | 0.002 | 0.002 |
| Bond angles (°) | 0.665 | 0.550 |
| MOLPROBITY STATISTICS | ||
| Ramachandran favored (%) | 91.2 | 90.6 |
| Allowed (%) | 7.4 | 8.2 |
| Outliers (%) | 1.31 | 1.2 |
| All-atom clashscore | 4.27 | 4.79 |
| Overall score | 1.72 | 2.01 |
| Solvent content (%) | 68.5 | 78.5 |
| Molecules of V domain/asymmetric unit | 2 | 2 |
Crystallography statistics for an open conformation of the human Alix V domain (PDB ID: 4JJY) and a crystal structure of the yeast Bro1 V domain in a complex with ubiquitin (PDB ID: 4JIO)
Figure 4. Crystal structure of the Bro1 V domain in complex with Ub.
A. 3.6Å structures of the Bro1 V domains found within the asymmetric unit (PDB ID: 4JIO). A cartoon model of the Alix V domain (PDB ID:2OEX) is shown at right. Below is alternate view from the top of the arms, looking into the vertex of the two Bro1 V domain structures together with Alix V. The backbone is colored blue-to-red according to amino acid order (N- to C-terminus). Loops not resolved in electron density maps are represented by dotted lines.
B. Structure of the Ub:Bro1 V domain complex, showing Ub (green) bound to the N-terminus of the Bro1 V domain within the first trihelical bundle (residues 370–392: blue). Model at right is closer view of Ub:V interaction. Ub residues that undergo significant NMR chemical perturbation upon Bro1 V binding are colored red. Positions of the Bro1 V I377 and L386 Cα atoms are shown in orange.
Ub was bound to one of the V domains (Chain A) within the asymmetric unit (Fig. 4B and Fig. S4A,B), where it was positioned along residues 375–386 of the exposed inner surface of the N-terminal trihelical bundle, in direct agreement with the PRE experiments placing residue 381 near the major site of Ub-binding in solution. Ub was orientated along the N-terminal V domain helix such that the C-terminus of Ub was near the beginning of the V domain and the N-terminus of Ub pointing towards the vertex of the V domain. The binding surface in Ub that was identified by chemical shift perturbations in NMR HSQC experiments (including residues L8, I44, and V70) was orientated towards the Bro1 V N-terminal helix (Fig 4B). To confirm the low-resolution crystal structure, we made an additional spin-labeled Bro1 V domain with MTSL attached to first residue of the V domain (residue 369) and compared its PRE profile on 15N-Ub to a Bro1 V domain spin-labelled at residue 392. The differential PRE effect between these two site-specific spin labels on the backbone amides of Ub showed that residue 369 of Bro1 V was nearer to the C-terminus of Ub while 381 was nearer to the N-terminal region of Ub (Fig. S4C). Together, these data demonstrate that Ub lies along the first alpha helix of the Bro1 V domain in an anti-parallel orientation and indicate that I377 likely participates in hydrophobic interactions with L8, I44, and V70 of Ub.
V domains adopt an open conformation in solution
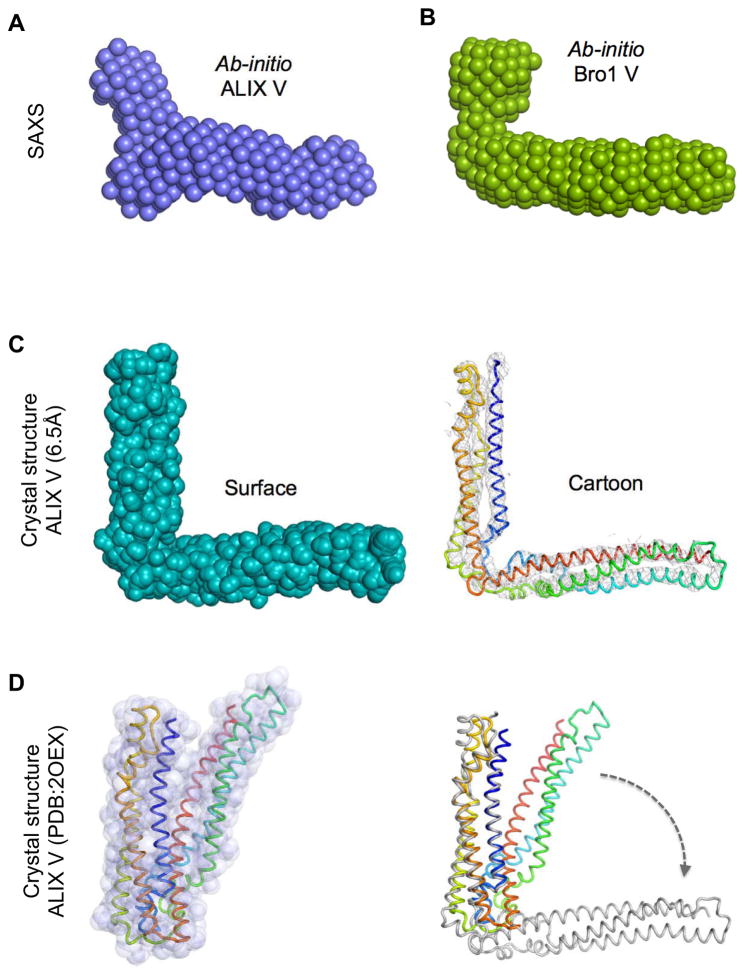
The structure of Bro1 V (Fig. 4) and previous structures of Alix V domains show a relatively “closed” conformation that might restrict access of some YPxL-containing proteins or ubiquitinated proteins to their binding sites on the inner surface (Fisher et al., 2007; Lee et al., 2007). Indeed, full-length Alix forms an intramolecular interaction (between its C-terminal proline-rich domain and its N-terminal Bro1 homology domain), which diminishes binding to YPxL-containing proteins. This interaction may function as a clasp, holding the V domain in a “closed” conformation that upon release, allows the V domain to spring “open” to expose protein interaction sites (Pires et al., 2009; Zhai et al., 2011; Zhou et al., 2010). We obtained several lines of evidence demonstrating that in the absence of such a clasp, the V domain adopts an “open” conformation in solution. Small Angle X-ray Scattering (SAXS) data from monodisperse Bro1 and Alix V domains were used to generate ab initio envelopes and revealed elongated V domains that were splayed open relative to their crystal structures (Fig. 5A and S5A). These elongated shapes were consistent with gel filtration data and dynamic light scattering showing that the monomeric 40 kDa Alix V and the 36 kDa of Bro1 V domains behaved as larger forms (Fig. S5C-E).
Figure 5. Alix and Bro1 V domains in the open conformation.
A.,B. ab initio envelopes of (A) human Alix V domain and (B) the S. castelli Bro1 V domain, as determined by SAXS.
C. Left: Space-filling and cartoon rendering of the 6.5Å crystal structure of human Alix V domain in an alternative “open” conformation (PDB ID: 4JJY). Right: Experimental electron density contoured at 1σ in left-hand panel is shown as a grey mesh at right.
D. Left: Molecular surface and cartoon overlay of Alix V (PDB:2OEX) in the closed conformation. Right: Overlay of closed Alix V (colored) onto the open conformation of Alix V (grey) (right).
Attempts to crystallize Alix V in complex with Ub yielded a 6.5Å crystal structure of Alix V in an “open” conformation without clear electron density for Ub (Table I, Fig. 5C). The single wavelength anomalous dispersion technique with isomorphous crystals of seleniomethionine-labelled Alix V allowed us to locate the Selenium sites, determine phases, and obtain traceable electron density maps. The features in the electron density maps clearly matched details other Alix V structures, however, the angle between the two arms of Alix V was far greater (e.g. an “open” conformation) than that found for the previously determined “closed” conformations (Fig. 5D, Supplemental movie). Having both the open and closed crystal structures of Alix V and the closed structure of Bro1 V allowed us to calculate experimental fit to the SAXS data of isolated V domains in solution. This analysis (Fig. S5B) showed that the SAXS data best describe a mix of V domains in both open and closed forms at a ratio of roughly 50:50.
The large shape changes displayed by V domains prompted to test whether they might undergo more dramatic changes in solution involving the swapping of N and C terminal helices between the trihelical bundles. We engineered a TEV protease site into in the middle hinge region of the V domain such that if a swap did occur, the protein could separate into 2 roughly equal fragments. However, cleaved Bro1 V stayed intact assessed by gel-filtration even after extended incubation at 25°C (Fig. S5F).
Together, these data show that while the overall topology of V domains is retained in solution, they can undergo a variety of conformational changes with small-scale changes evident in the packing of the helices within Bro1 V crystals (Fig. S4A) and large-scale changes in mix of “open” and “closed” forms in solution (Fig. 5).
Ub-binding by Bro1 is required for MVB sorting of ubiquitinated cargo
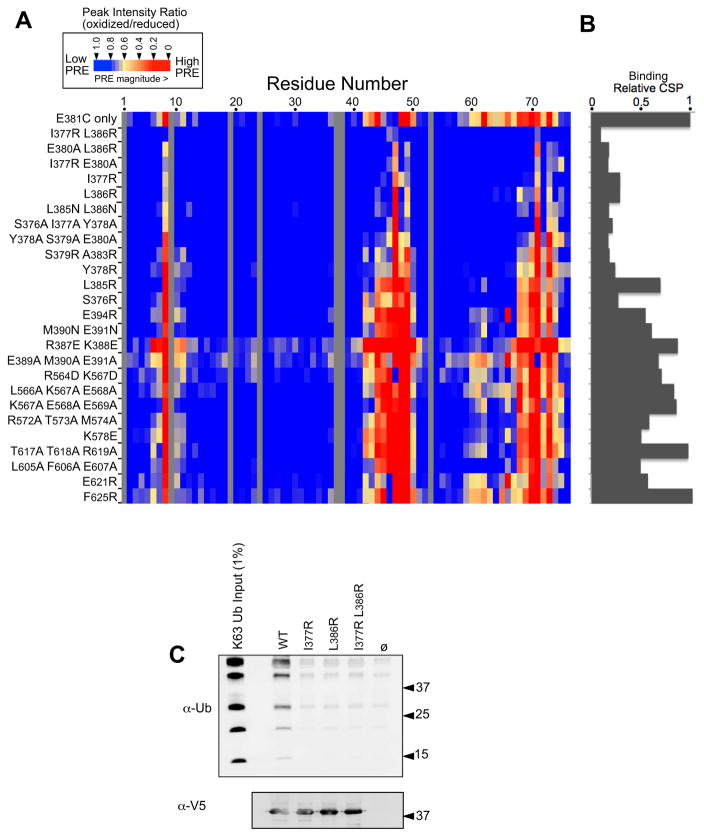
To test the functional relevance of Ub-binding by Bro1, we generated mutants of Bro1 V domain specifically defective in binding Ub. A series of mutations within the first trihelical arm were tested for their ability to disrupt Ub-binding using PRE experiments with 15N-Ub and Bro1 V mutants spin-labelled at position 381. Although side chains cannot be resolved with the diffraction data in the crystal structure, I377 is predicted from our structural data to be centered under the hydrophobic patch of Ub, whereas L386 lies under a loop region between α2 and β5 of Ub containing residues Q62, K63, E64 and S65. We found that mutation of I377 and/or L386 blocked Ub-binding as observed by the loss of PRE effect in 15N Ub (Fig. 6A). Loss of Ub-binding was also indicated by a loss in chemical shift perturbations in the backbone amides of Ub residues that mediate binding to Bro1 V (Fig. 6B) and by loss of the ability of Bro1 V domains immobilized on beads to bind K63 polyubiquitin chains (Fig. 6C). Compared to the WT Bro1 V protein, the mutant V domains were produced at similar levels, were just as soluble, and had identical NMR proton spectra (Fig. S6), indicating that the mutant V domains retained their overall structure.
Figure 6. Mutagenesis of the Ub-binding region of Bro1 V domain.
A. The indicated mutations were made in the context of the S. castelli Bro1 domain, which contains a cysteine residue at position 381. These proteins were labeled with MTSL and used in PRE experiments with 15N-Ub at a Brol V:Ub ratio of 3:1. Peak intensity ratios (oxidized vs. reduced spin label) for each Ub residue were calculated from HSQC spectra collected in the absence and presence of 2 mM ascorbate. The degree of PRE effect experienced by each Ub backbone amide is color-coded as indicated.
B. Index of chemical-shift perturbations in 15N-Ub upon binding to mutant Bro1 V domains. CSP index was calculated by summing the (0.2ΔN2 + ΔH2)1/2 values for Ub residues 8,42,44,48,49,69,70 and 71 and setting that value equal to 1 for the WT Bro1 V domain.
C. The WT and mutant recombinant V domains were immobilized on α-V5 polyclonal antibody-coated beads, washed, and incubated with K63-linked polyubiquitin chains. Beads alone or bound to an irrelevant V5-epitoped protein were also included. Beads were washed and immunoblotted with α-V5 or α-Ub monoclonal antibodies.
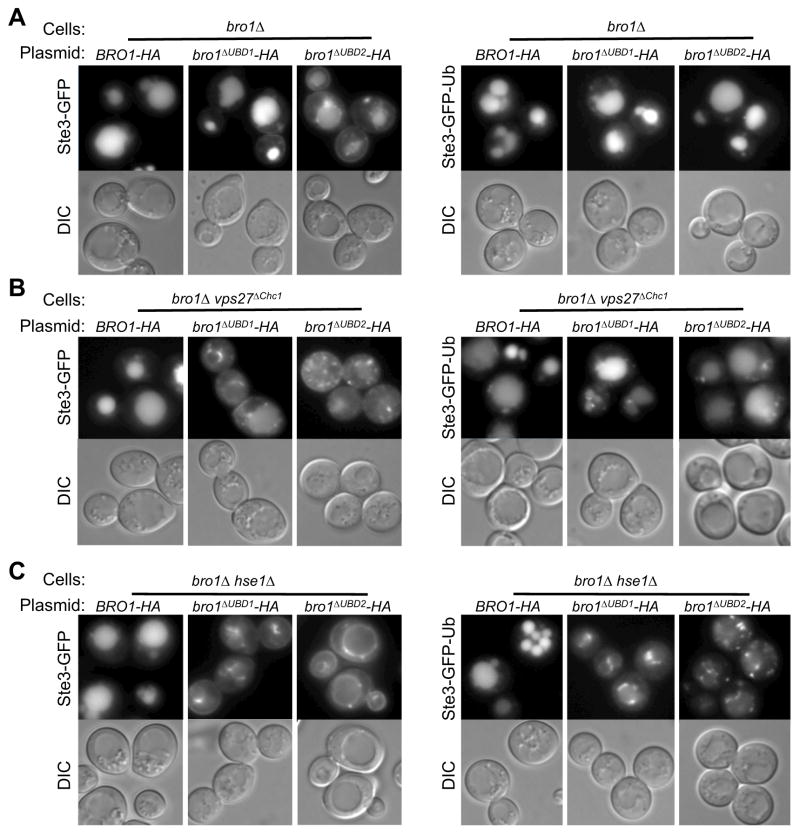
We next assessed the ability of Bro1 containing Ub-binding defective V domains to sort cargo into the MVB pathway (Fig. 7). Here we analyzed chimeric Bro1 proteins in which the V domain of an HA-epitope tagged Bro1 from S. cerevisiae was substituted with the mutant V domains of S. castelli characterized above. The wild-type chimeric Bro1-HA was produced at levels identical to those of full-length S. cerevisiae Bro1-HA and complemented the MVB sorting defects (Fig. 7A, S7). Two Ub-binding defective mutants of Bro1 (Bro1ΔUBD1, I377R; and Bro1ΔUBD2, L386R) also complemented bro1Δ mutants and were able to sort Ste3-GFP and Ste3-GFP-Ub into the vacuole, although modest defects were observed for Ste3-GFP sorting by the Bro1ΔUBD2 mutant. Their level of expression was identical to wild-type Bro1-HA and not affected by hse1Δ or vps27ΔChc1 mutations (Fig. S7). These data demonstrate that the Bro1 mutants retain their general folding and function. We reasoned that if Bro1, and in particular the Ub-binding capacity of Bro1, functions in parallel with ESCRT-0 as a Ub-sorting receptor then the functional defects of Bro1ΔUBD1 and Bro1ΔUBD2 would be compensated for by ESCRT-0. Thus, we analyzed the ability of Bro1ΔUBD1 and Bro1ΔUBD2 to mediate MVB sorting in cells where ESCRT-0 was compromised by either loss of Hse1 or loss of clathrin-binding to Vps27. Notably, hse1Δ mutants and vps27ΔChc1 mutants have no phenotype on their own; also, double mutants with bro1 (bro1Δ hse1Δ and bro1Δ vps27ΔChc1) were complemented for MVB sorting by Bro1-HA with the wild-type V domain. In contrast, sorting of Ste3-GFP was markedly defective when either of the Bro1ΔUBD1 and Bro1ΔUBD2 alleles was combined with the vps27ΔChc1 allele or loss of Hse1 (Fig. 7B,C). Severe sorting defects were also observed for Ste3-GFP-Ub. More moderate defects were observed when Bro1ΔUBD1 and Bro1ΔUBD2 combined with the vps27ΔChc1 allele, which may reflect that loss of clathrin-binding compromises ESCRT-0 function less than loss of Hse1. Both the Bro1ΔUBD1 and Bro1ΔUBD2 proteins were able to restore CPY sorting to not only bro1 mutants but also the bro1Δ hse1Δ and bro1Δ vps27ΔChc1 mutants, indicating that the deficiency caused by loss of Ub-binding was specific to MVB sorting and not to all other Bro1 functions. In addition, we found that Bro1ΔUBD1 and Bro1ΔUBD2 cells had a normal distribution of GFP-tagged Ub, indicating that the Doa4 deubiquiting enzyme was functioning normally to remove Ub from MVB cargo late in the process of sorting.
Figure 7. MVB sorting requires Bro1 to bind Ub.
A. Null mutant bro1Δ cells were transformed with low copy-number plasmids expressing a wild-type chimeric Bro1 with the V domain from S. castelli Bro1, with or without mutations in the Ub-binding region of the V domain. The bro1ΔUBD1 allele carries the I377R mutation; bro1ΔUBD2 carries the L386R mutation. Cells also expressed Ste3-GFP (left) or Ste3-GFP-Ub (right). Shown are GFP fluorescence and DIC images.
B. Same as in A, but bro1Δ cells also carried the vps27ΔChc1 allele that blocks the ability of Vps27 to bind directly to clathrin.
C. Same as in A but bro1Δ cells also lack the ESCRT-0 subunit Hse1. Bar=5μm
DISCUSSION
Based on the data presented here and elsewhere, we propose that Bro1 works early in the MVB biogenesis pathway, as a Ub-sorting receptor that functions in parallel with ESCRT-0. This role would be distinct from its later function as a recruitment factor for Doa4, the DUb that works late in the process of MVB biogenesis to recycle Ub from cargo post-sorting (Amerik et al., 2006; Richter et al., 2007). This model is supported by the genetic interactions we discovered between ESCRT-0 and Bro1, wherein hypomorphic mutations in both show dramatic synthetic phenotypes. Further support comes from the observation that expressing Bro1-DUb fusion proteins blocks the sorting of cargo into MVBs, and from the finding that Bro1 shares several binding partners with ESCRT-0, including Ub, clathrin, ESCRT-I, Ub-ligases, and deubiquitinating enzymes (Nikko and Andre, 2007). These interactions provide a biochemical rationale for how Bro1 can execute an ESCRT-0-like function as a Ub-sorting receptor. We found the interaction of the Bro1 V domain with Ub was critical for sorting Ub-cargo into MVBs only when ESCRT-0 function was weakened, an observation supporting the idea that Bro1 and ESCRT-0 provide similar overlapping functions. Interestingly, even when Ub-cargo sorting was blocked other functions such as sorting of the soluble vacuolar hydrolase CPY were normal, demonstrating that the MVB and vacuolar hydrolase sorting functions of Bro1 are separable, and that Ub-binding contributes to the former.
Together these data suggest that Bro1 belongs to an expanding coterie of endosomal Ub-sorting receptors, thereby diversifying membership beyond ESCRT-0, GGA, and Tom1-related proteins, which have a very similar domain organization (Clague et al., 2012; Shields and Piper, 2011). It is plausible that each of these Ub-receptors directs its attention to specific sets of cargos with some level of overlap. This possibility is supported by our findings that loss of Bro1 or mutation of the Ub-binding site in the V domain leads to more dramatic defects in MVB sorting for cargos such as Ste3 than for others such as Mup1. If Bro1 does work as an upstream Ub-sorting receptor in the MVB biogenesis pathway, then the mammalian Bro1-family members Alix and HD-PTP may likewise execute such a function. Correspondingly, Alix was was found to bind membrane proteins bearing YPxL motifs and to usher them into endosomal ILVs (Baietti et al., 2012; Dores et al., 2012). For one of these cargos (Par1), ubiquitination of the cargo itself was not required (although ubiquitination of associated proteins might be), suggesting that Alix can work as a sorting receptor for certain Ub-independent cargos. Since the Alix V domain binds Ub, it is possible that this protein also serves as a receptor for Ub-cargos. Unlike the loss of ESCRT-0 or ESCRT-I, loss of Alix in mammalian cells does not dramatically alter endosome morphology, nor does it block sorting of the few ubiquitinated membrane protein cargos that have been examined (Odorizzi, 2006). Thus, Alix may handle only a subset of Ub-cargos, or ESCRT-0 could substitute for Alix’s absence in experimental contexts. Interestingly, Dictyostelium discoideum does not have a canonical ESCRT-0 (Hrs/STAM) but does express a Bro1 homolog (Dd-Alix) and a Tom1 homolog (Dd-Tom1), the latter of which participates in many of the same protein interactions as a canonical ESCRT-0. Nevertheless, eliminating Dd-Tom1 affects neither the sorting of endosomal ubiquitinated proteins nor the biogenesis of MVBs, suggesting that Dd-Alix instead may provide much of the ESCRT-0-like function in this organism (Blanc et al., 2009).
Like both Alix and Bro1, HD-PTP also directly binds Ub, ESCRT-I and ESCRT-III and localizes to endosomes (Doyotte et al., 2008; Miura et al., 2008; Nikko and Andre, 2007; Stefani et al., 2011; Strack et al., 2003). Loss of HD-PTP promotes cell migration and is associated with cancer progression (Cao et al., 1998; Castiglioni et al., 2007; Gilbert et al., 2011; Lin et al., 2011). At the cellular level, RNAi-mediated depletion of HD-PTP causes morphological changes in endosomes and an accumulation of ubiquitinated proteins on them, a cellular phenotype similar to that resulting from ESCRT-0 depletion (Doyotte et al., 2008). Although it is not clear what types of Ub-cargo HD-PTP might sort, candidates include EGFR, integrins, and E-cadherin (Castiglioni et al., 2007; Lin et al., 2011; Miura et al., 2008).
Although we have speculated that other Bro1 family members can behave as endosomal Ub-sorting receptors, their capacity to bind Ub could instead be used for different or additional purposes. For instance, many Ub-binding proteins including Alix can themselves undergo ubiquitination, and may act as a scaffold for a variety of other Ub-binding proteins involved in endocytosis and/or viral budding (Hoeller and Dikic, 2010; Sette et al., 2010). In the future, such alternative models can be tested using Ub-binding defective mutants of other Bro1-family proteins. Predicting how to generate such mutants will be somewhat challenging, however. Although the Ub-binding region we identified is conserved amongst Bro1 orthologs, it is not obvious how to accurately predict the Ub-binding site in the V domains of other proteins. The V domains of HD-PTP, Alix, and Bro1 are only 13–15% identical and recent mutagenesis experiments indicate that Alix houses a UBD somewhere within its distal trihelical arm along a sequence that is only conserved across Alix orthologs (Dowlatshahi et al., 2012; Keren-Kaplan et al., 2013). This theme of different Ub-binding modes between functional orthologs is found for components of the ESCRT apparatus as well; Vps27 (ESCRT-0), Mvb12 (ESCRT-I), and Vps36 (ESCRT-II) share the ability to bind Ub with their mammalian counterparts Hrs, Mvb12A, UBAP1, and Eap45, but use different binding motifs to do so (Clague et al., 2012; Shields and Piper, 2011). This is consistent with the fact that low-affinity binding motifs for Ub are often structurally simple, suggesting they may not be that challenging to evolve.
EXPERIMENTAL PROCEDURES
Plasmids and yeast strains used are listed in Tables S1 and S2. BRO1-expressing plasmids cells with various V-domains were made in low copy/CEN plasmids encoding the BRO1 promoter and the flanking N-terminal and C-terminal portions of S. cerevisiae Bro1 upstream of 2 HA epitopes.
V domains were in produced in E.coli BL21(DE3) cells. TALON-Co2+ purified V domains were further purified by size exclusion chromatography and cleaved from their 6XHis tag with TEV protease prior to crystallography. SeMet-labelled proteins were expressed using the methionine pathway inhibition procedure (Doublie and Carter, 1992).
Crystals containing Ub (A28M to allow labeling with Selenium) and Bro1 V (with K417A, K418A, K419A mutations made to reduce surface entropy (Goldschmidt et al., 2007) were grown at 18°C by mixing equal amount of the protein samples and solution containing 0.1M bis-Tris propane pH 7, 0.2–0.4M Na malonate, 15–20% PEG 3350. AlixV:Ub crystals were grown at 4°C by mixing equal amount of the protein sample and solution containing 0.1 M Na citrate pH 5, 6–8 % PEG 8000, 5–10% ethylene glycol. Data were collected at 100 K on the 4.2.2 beamline at the Advanced Light Source (Bekeley, CA).
Diffraction data for SeMet-Bro1V:Ub (A28M) were processed using d*TREK (Pflugrath, 1999) and phased with phenix.autosol (Terwilliger et al., 2009) using the SAD method. Phenix.autobuild and phenix.refine were used to iteratively build and improve the Bro1V Cα-only model. Ub (pdb id: 1UBQ) was docked into the clearly visible density near the Bro1V N-terminus and refined with all atoms. The diffraction data for SeMet-AlixV:Ub were integrated and scaled using d*TREK and prepared for SHELXD (Sheldrick, 2008) using xprep (Bruker Corporation). Phenix was used to improve the quality of the map sufficiently such that the two arms of AlixV from the high-resolution structures could be independently placed in the electron density and then refined with secondary structure restraints in place. Coot was used to manually fit the structure and PyMOL to generate the structural figures.
SAXS data (collected at the 12-ID-B beamline at the Advanced Photon Source, Argonne National Laboratory) from 20μl samples of Alix and Bro V domains at concentrations ranging from 1 to 10 mg/ml at 20°C and exposure times of 1, 2 and 4 s were used to establish concentration dependence on scattering and exposure based radiation damage. The final SAXS dataset was collected at the SIBYLS 12.3.1 beamline at the Advanced Light Source (Lawrence Berkeley National Laboratory) at 10°C on samples ranging in concentration from 1 to 6 mg/ml and exposure times of 0.5, 1, 2 and 4s. Data were processed with PRIMUS (Konarev et al., 2003) after correction with matching buffers. The radius of gyration (Rg) estimated from the linear region of the Guinier plots was 34.3 Å for Alix V and 35.4 Å for Bro1 V. A pair distance distribution P(r) function was calculated using GNOM to estimate of the maximum dimension (Dmax) of the V domains and yielded values of 116.0 Å and 111.1 Å for Alix and Bro1 V domain respectively. The ab initio models using a chain-like ensemble of dummy residues were generated with GASBOR (Svergun et al., 2001). Theoretical SAXS profiles of open and closed V ensembles were generated with FoXS and its Minimal Ensemble Search algorithm was used to calculate weighted fit of the open and closed forms of V domains (Schneidman-Duhovny et al., 2010).
15N-HSQC data were collected at 25°C on a Bruker Avance II 800 MHz spectrometer and analyzed with SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, UCSF, CA) and NMRView (One Moon Scientific, Westfield, NJ). Chemical shift perturbations were measured by comparing peak positions to 15N-Ub alone using (0.2* ΔppmN2 + ΔppmH2)1/2). Paramagnetic relaxation enhancement effects were measured using cysteine-containing Bro1 proteins labeled with MTSL [(1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) –methanethio- sulfonate] (TRC Inc., Toronto, Canada). Incorporation to >90% was validated by electron spin resonance analysis. Peak intensity ratios of 15N-Ub when bound to the oxidizied vs. reduced MTSL-labelled Bro1 proteins were calculated to quantify the degree of PRE effects.
GFP-fusion proteins were localized in cells grown to mid-log phase at 30°C and imaged as previously described (Bilodeau et al., 2003). Genes under control of the CUP1 promoter were induced with 50μM CuCl2. Yeast protein extracts for SDS-PAGE and immunoblotting were prepared by NaOH pre-treatment of cells (Kushnirov, 2000). GST pull-down experiments were done as described previously (Pashkova et al., 2010). Immunoprecipitation from spheroplasts lysatges and bacterial lysates was performed as previously described (Bilodeau et al., 2003; Pashkova et al., 2010). Binding to K63 polyubiquitin was done by attaching recombinant 6xHis-V5-tagged V-domains to 30μls of beads coated with polyclonal anti-V5 antibodies, which were subsequently incubated for 1 hr with 2 μg of K63-linked polyUb (Boston Biochemicals, Cambridge, MA) in 200μl PBS containing 0.02% of Trition X100, 0.1% BSA or casein. Beads were washed 5 times; bound complexes were eluted with Laemmli sample buffer and heating to 70°C for 5 min.
To compare yeast genomic DNA, 7–9 million 75 base-pair, single-end reads were generated for each sample on an Illumina HiSeq 2000 platform machine (San Diego, CA). Data were analyzed by the University of Washington Genome Sciences group as detailed in supplementary materials.
Supplementary Material
HIGHLIGHTS.
Bro1 genetically interacts with ESCRT-0
Like ESCRT-0, Bro1 binds clathrin
The V domain of Bro1 and a wide range of Bro1 family proteins binds ubiquitin
Crystallography and NMR reveal the structural basis for Bro1 V binding to ubiquitin
Ubiquitin-binding by the Bro1 V domain contributes to sorting of ubiquitinated membrane proteins into multivesicular endosomes.
Acknowledgments
This work was supported by NIH GM58202. We would like to thank Ernesto Fuentes for SAXS data collection and preliminary analysis. X-ray scattering beam time resource was provided at beamline 12-ID-B at the Advanced Photon Source. Analysis of yeast genomic sequences was made possible through the NIH P41 GM103533 grant funding the Yeast Resource Center. MJD is supported as a Rita Allen Scholar, ABS was supported by F30CA165440 and T32 AG000057.
References
- Amerik A, Sindhi N, Hochstrasser M. A conserved late endosome-targeting signal required for Doa4 deubiquitylating enzyme function. J Cell Biol. 2006;175:825–835. doi: 10.1083/jcb.200605134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Davies BA, Katzmann DJ. Regulation of Vps4 during MVB sorting and cytokinesis. Traffic. 2011;12:1298–1305. doi: 10.1111/j.1600-0854.2011.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol. 2002;4:534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- Bilodeau PS, Winistorfer SC, Kearney WR, Robertson AD, Piper RC. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J Cell Biol. 2003;163:237–243. doi: 10.1083/jcb.200305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc C, Charette SJ, Mattei S, Aubry L, Smith EW, Cosson P, Letourneur F. Dictyostelium Tom1 participates to an ancestral ESCRT-0 complex. Traffic. 2009;10:161–171. doi: 10.1111/j.1600-0854.2008.00855.x. [DOI] [PubMed] [Google Scholar]
- Cao L, Zhang L, Ruiz-Lozano P, Yang Q, Chien KR, Graham RM, Zhou M. A novel putative protein-tyrosine phosphatase contains a BRO1-like domain and suppresses Ha-ras-mediated transformation. J Biol Chem. 1998;273:21077–21083. doi: 10.1074/jbc.273.33.21077. [DOI] [PubMed] [Google Scholar]
- Castiglioni S, Maier JA, Mariotti M. The tyrosine phosphatase HD-PTP: A novel player in endothelial migration. Biochem Biophys Res Commun. 2007;364:534–539. doi: 10.1016/j.bbrc.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Clague MJ, Liu H, Urbe S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev Cell. 2012;23:457–467. doi: 10.1016/j.devcel.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Dores MR, Chen B, Lin H, Soh UJ, Paing MM, Montagne WA, Meerloo T, Trejo J. ALIX binds a YPX(3)L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J Cell Biol. 2012;197:407–419. doi: 10.1083/jcb.201110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S, Carter C. Crystallization of Nucleic Acids and Proteins: A Practical Approach. Oxford: IRL Press; 1992. [Google Scholar]
- Dowlatshahi D, Sandrin V, Vivona S, Shaler T, Kaiser S, Melandri F, Sundquist WI, Kopito R. ALIX Is a Lys63-Specific Polyubiquitin Binding Protein that Functions in Retrovirus Budding. Dev Cell. 2012 doi: 10.1016/j.devcel.2012.10.023. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyotte A, Mironov A, McKenzie E, Woodman P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci U S A. 2008;105:6308–6313. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Gilbert MM, Tipping M, Veraksa A, Moberg KH. A screen for conditional growth suppressor genes identifies the Drosophila homolog of HD-PTP as a regulator of the oncoprotein Yorkie. Dev Cell. 2011;20:700–712. doi: 10.1016/j.devcel.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt L, Cooper DR, Derewenda ZS, Eisenberg D. Toward rational protein crystallization: A Web server for the design of crystallizable protein variants. Protein Sci. 2007;16:1569–1576. doi: 10.1110/ps.072914007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Cashikar A. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol. 2012;28:337–362. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Hettema EH, Valdez-Taubas J, Pelham HR. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 2004;23:1279–1288. doi: 10.1038/sj.emboj.7600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D, Dikic I. Regulation of ubiquitin receptors by coupled monoubiquitination. Subcell Biochem. 2010;54:31–40. doi: 10.1007/978-1-4419-6676-6_3. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- Joshi A, Munshi U, Ablan SD, Nagashima K, Freed EO. Functional replacement of a retroviral late domain by ubiquitin fusion. Traffic. 2008;9:1972–1983. doi: 10.1111/j.1600-0854.2008.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Kaplan T, Attali I, Estrin M, Kuo LS, Farkash E, Jerabek-Willemsen M, Blutraich N, Artzi S, Peri A, Freed EO, et al. Structure-based in silico identification of ubiquitin-binding domains provides insights into the ALIX-V:ubiquitin complex and retrovirus budding. EMBO J. 2013 doi: 10.1038/emboj.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev P, Volkov V, Sokolova A, Kocj M, Svergun D. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–1282. [Google Scholar]
- Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Aranda V, Muthuswamy SK, Tonks NK. Identification of PTPN23 as a novel regulator of cell invasion in mammary epithelial cells from a loss-of-function screen of the ‘PTP-ome’. Genes Dev. 2011;25:1412–1425. doi: 10.1101/gad.2018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N, Odorizzi G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J Cell Biol. 2004;166:717–729. doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C, Buchkovich NJ, Stringer DK, Emr SD, Piper RC. Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep. 2012;13:331–338. doi: 10.1038/embor.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald C, Stringer DK, Piper RC. Sna3 Is an Rsp5 Adaptor Protein that Relies on Ubiquitination for Its MVB Sorting. Traffic. 2011 doi: 10.1111/j.1600-0854.2011.01326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Neil SJ. Host factors involved in retroviral budding and release. Nat Rev Microbiol. 2011;9:519–531. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- Miura GI, Roignant JY, Wassef M, Treisman JE. Myopic acts in the endocytic pathway to enhance signaling by the Drosophila EGF receptor. Development. 2008;135:1913–1922. doi: 10.1242/dev.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E, Andre B. Split-ubiquitin two-hybrid assay to analyze protein-protein interactions at the endosome: application to Saccharomyces cerevisiae Bro1 interacting with ESCRT complexes, the Doa4 ubiquitin hydrolase, and the Rsp5 ubiquitin ligase. Eukaryot Cell. 2007;6:1266–1277. doi: 10.1128/EC.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Odorizzi G. The multiple personalities of Alix. J Cell Sci. 2006;119:3025–3032. doi: 10.1242/jcs.03072. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci. 2003;116:1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Yu L, Ramaswamy S, Piper RC. WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Mol Cell. 2010;40:433–443. doi: 10.1016/j.molcel.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugrath JW. The finer things in X-ray diffraction data collection. Acta crystallographica Section D, Biological crystallography. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, Roessle M, Moriscot C, Popov S, Hinz A, Jamin M, et al. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 2009;17:843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Pashkova N, Winistorfer S, Piper RC. DOA1/UFD3 plays a role in sorting ubiquitinated membrane proteins into multivesicular bodies. J Biol Chem. 2008;283:21599–21611. doi: 10.1074/jbc.M802982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, West M, Odorizzi G. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J. 2007;26:2454–2464. doi: 10.1038/sj.emboj.7601692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoul R. Do Alix and ALG-2 really control endosomes for better or for worse? Biol Cell. 2006;98:69–77. doi: 10.1042/BC20050007. [DOI] [PubMed] [Google Scholar]
- Sangsuriya P, Rojtinnakorn J, Senapin S, Flegel TW. Identification and characterization of Alix/AIP1 interacting proteins from the black tiger shrimp, Penaeus monodon. J Fish Dis. 2010;33:571–581. doi: 10.1111/j.1365-2761.2010.01156.x. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette P, Jadwin JA, Dussupt V, Bello NF, Bouamr F. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J Virol. 2010;84:8181–8192. doi: 10.1128/JVI.00634-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta crystallographica Section A, Foundations of crystallography. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Shields SB, Oestreich AJ, Winistorfer S, Nguyen D, Payne JA, Katzmann DJ, Piper R. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J Cell Biol. 2009;185:213–224. doi: 10.1083/jcb.200811130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields SB, Piper RC. How ubiquitin functions with ESCRTs. Traffic. 2011;12:1306–1317. doi: 10.1111/j.1600-0854.2011.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springael JY, Nikko E, Andre B, Marini AM. Yeast Npi3/Bro1 is involved in ubiquitin-dependent control of permease trafficking. FEBS Lett. 2002;517:103–109. doi: 10.1016/s0014-5793(02)02586-3. [DOI] [PubMed] [Google Scholar]
- Stefani F, Zhang L, Taylor S, Donovan J, Rollinson S, Doyotte A, Brownhill K, Bennion J, Pickering-Brown S, Woodman P. UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr Biol. 2011;21:1245–1250. doi: 10.1016/j.cub.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Stringer DK, Piper RC. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J Cell Biol. 2011;192:229–242. doi: 10.1083/jcb.201008121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophysical journal. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc Natl Acad Sci U S A. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta crystallographica Section D, Biological crystallography. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Landesman MB, Chung HY, Dierkers A, Jeffries CM, Trewhella J, Hill CP, Sundquist WI. Activation of the retroviral budding factor ALIX. J Virol. 2011;85:9222–9226. doi: 10.1128/JVI.02653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Si J, Corvera J, Gallick GE, Kuang J. Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. The Biochemical journal. 2010;432:525–534. doi: 10.1042/BJ20100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.