Abstract
Purpose
To determine the possible effects of 125I-to-DNA distance on the magnitude and mechanism of Auger-electron induced-double-strand break (DSB) production.
Materials and methods
We have synthesized a series of 125I-labeled Hoechst (H) derivatives (125IE–H, 125IB-H, 125I-C8-H and 125I-C12-H). While all four molecules share a common DNA minor groove binding bis-benzimidazole motif, they are designed to position 125I at varying distances from the DNA helix. Each Hoechst derivative was incubated at 4 °C in phosphate buffered saline (PBS) together with supercoiled (SC) 3H-pUC19 plasmid DNA (ratio 3:1) ± the •OH scavenger dimethyl sulfoxide (DMSO) (0.2 M). Aliquots were analyzed on agarose gels over time and DSB yields per decay of 125I atom were determined. Docking of the iodinated compounds on a DNA molecule was carried out to determine the distance between the iodine atom and the central axis of DNA.
Results
In the absence of DMSO, the results show that the DSB yields decrease monotonically as the 125I atom is distanced – by 10.5 Å to 13.9 Å – from the DNA helix (125IEH: 0.52 ± 0.01; 125IB– H: 0.24 ± 0.03; 125I-C8–H:0.18 ± 0.02; 125I-C12–H: 0.10 ± 0.00). In the presence of DMSO, DSB yields for 125IEH (0.49 ± 0.02) and 125IB-H (0.26 ± 0.04) remain largely unchanged indicating that DSB are entirely produced by direct effects. Strikingly, 125I-C8–H or 125I-C12 –H, did not produce detectable DSB in the presence of DMSO under similar conditions suggesting when 125I atom is positioned > 12 Å from the DNA, DSB are entirely produced by indirect effects.
Conclusion
These results suggest that at a critical distance between the 125I atom and the DNA helix, DSB production switches from an ‘all’ direct to an ‘all’ indirect mechanism, the latter situation being comparable to the decay of 125I free in solution. These experimental findings were correlated with theoretical expectations based on microdosimetry.
Keywords: Auger electron, 125I decays, double-strand break, scavenger
Introduction
More than half of radionuclides known so far decay by internal conversion and/or electron capture resulting in the emission of a cascade of extremely low energy electrons, a phenomenon known as the Auger effect (Auger 1925). These Auger, Coster-Kronig and super Coster-Kronig electrons typically have energies in the range of few eV to 1 keV (Cole 1969, Pomplun et al. 1987, Kassis 2004). The majority of these electrons travel short distances up to ∼ 500 nm from the decay site and deposit all their energy in a small spherical volume of few cubic nanometers (Sastry and Rao 1984, Kassis et al. 1987a, 1987b, Pomplun et al. 1987). Although the relative biological effectiveness of Auger electrons was initially ignored owing to their low energy, it later became evident, from the lack of shoulder in the survival curves (Bradley et al. 1975), that Auger electrons behave like high LET (linear energy transfer) radiation (2–25 keV/μm) imparting very high cytotoxicity to mammalian cells when the decay site is located in close proximity to the nuclear DNA (Hofer and Hughes 1971, Feinendegen 1975, Hofer et al. 1975). These phenomenal observations spurred much interest in exploring the cancer therapeutic potential of Auger electrons (Bloomer and Adelstein 1975, 1977).
Among a handful of prominent Auger emitters (125I, 123I, 77Br, 111In, 193mPt and 195mPt), 125I and 123I have been most extensively studied owing to their possible application to cancer therapy (Bloomer and Adelstein 1977, Kassis and Adelstein 1996, Reilly et al. 2000, Kassis et al. 2004, Rebi-schung et al. 2008, Costantini et al. 2010) and diagnostic imaging (Yaakob 1999). Theoretical dosimetric calculations estimated that each 125I decay deposits an energy of 106–109 cGy in a volume of 2 nm radius around the decay site (Sastry and Rao 1984, Kassis et al. 1987b). Furthermore, Monte Carlo simulations have shown that the energy deposited per unit volume drops precipitously with increasing distance from a decaying 125I atom (Sastry and Rao 1984, Charlton et al. 1987, Kassis et al. 1987b) implying that the relative biological effectiveness of the Auger electron cascade would depend heavily upon the proximity of the decaying 125I atom with respect to the DNA in the mammalian cell nucleus (Hofer et al. 1975, Kassis et al. 1987a).
The similarity in size between the iodine atom and a –CH3 group has made the in vivo incorporation of radioiodine into the nuclear DNA during cell division possible. 5-[125I/123I] iodo-2′-deoxyuridine (125IUdR, 123IUdR) readily replaces thymidine probably achieving the closest possible proximity of the decaying radioiodine atom to the DNA strands. T is agent produces efficient cell killing as predicted by theoretical dosimetry (Hofer and Hughes 1971, Feinendegen 1975, Chan et al. 1976, Makrigiorgos et al. 1989). Iodine-125-labeled DNA intercalators (Martin 1977, Kassis et al. 1989), 125I-labeled minor-groove binders, such as Hoechst 33342 and 33258 (Martin and Holmes 1983, Harapanhalli et al. 1996, Kassis et al. 1999a, 1999b, 2000, Balagurumoorthy et al. 2006, Balagurumoorthy et al. 2008a, 2008b, Yasui et al. 2007), 123I-labeled steroid hormones (DeSombre et al. 1992, 2000, Yasui et al. 1996, 2001), 125I-deoxycytidine in homopyrimidine triplex-forming oligonucleotides (Panyutin et al. 2000, 2001, Sedel-nikova et al. 2000) and more recently 111In conjugated to nuclear localization signal peptides (Costantini et al. 2007, 2008) have also been used to position Auger-electron emitters proximal to DNA in the nucleus. The common property of all these agents is their ability to position the decaying atom within few nanometers distance from DNA within the mammalian cell nucleus.
Our laboratory has been interested in understanding the biophysical mechanisms underlying the induction of DNA strand breaks by Auger electrons, particularly 125I (Kassis et al. 1988, 1999a, 1999b, 2000, Sastry et al. 1988, Balagurumoorthy et al. 2006, 2008a, 2008b, Kishikawa et al. 2006), 123I (Makrigiorgos et al. 1989, Kassis et al. 1996, Balagurumoorthy et al. 2008b) and 111In (Sahu et al. 1995). Experiments on supercoiled plasmid DNA model demonstrated that efficient induction of double-strand breaks (DSB) was observed when 125I is covalently attached to either DNA intercalators (Kassis et al. 1989) or groove binders (Harapanhalli et al. 1996, Lobachevsky et al. 2004, Balagurumoorthy et al. 2006), whereas 125I-antipyrene which does not bind DNA and is free in solution is relatively ineffective in inducing DSB (Kassis et al. 1999b). In the experiments reported here, we have attempted to delineate the role of an important parameter believed to affect Auger electron-induced DSB yield, namely the distance between a decaying 125I atom and DNA. We have synthesized a series of radioiodinated Hoechst derivatives that were designed to position 125I atom at various distances from the DNA in bound state. These 125I labeled minor groove binders were incubated with supercoiled form of 3H-pUC19 and analyzed as a function of time to determine the DSB yields, following the accumulation of 125I decays. DSB yields were correlated with the 125I-DNA distances estimated from molecular modeling and this 125I-DNA distance-DSB relationship was compared with the theoretical predictions.
Materials and methods
Synthesis of 125I–EH, 125I–BH, 125I–C8–H and 125I–C12–H
125I–EH was synthesized from its trimethylstannyl derivative as described previously (Harapanhalli et al. 1996, Kassis et al. 1999a, Balagurumoorthy et al. 2006). 125I–BH was synthesized by treating Hoechst33258 with 4-iodobenzoyl chloride (Sigma-Aldrich, St Louis, MO, USA) followed by conversion of the resulting compound into tributylstannyl derivative which was radiolabeled with Na 125I (Perkin Elmer Life and Analytical Sciences, Waltham MA, USA) in the presence of Iodogen. Hoechst33258 (Sigma-Aldrich) was refluxed with several molar excess malonyl dichloride (Sigma-Aldrich) (to add Cn linkers, where n = 8, 12) followed by treatment with (4-iodophenyl) methanamine (Sigma-Aldrich). The resulting products were converted to tributylstannyl derivatives which in turn were radioiodinated with Na 125I in the presence of Iodogen to obtain 125I-C8–H and 125I-C12–H. All the radioiodinated products were purified on a SepPak® Plus C18 cartridge (Waters, Milford MA, USA) using acetonitrile:water (1:1) as eluant. This process removed all ionic impurities including residual iodide. The radiochemical purity, checked by thin layer chromatography on reversed-phase silica (mobile phase, acetonitrile:methanol:water: tri-n -butylamine, 8:4:2:1) was found to be greater than 98%. The ability of these compounds to bind DNA was checked qualitatively by fluorescence spectroscopy. Non-radioactive iodinated analogs of the above four compounds were mixed with calf thymus DNA (Sigma-Aldrich) and the fluorescence spectra were recorded at 25°C in a PerkinElmer LS50B luminescence spectrometer (Perkin Elmer Life and Analytical Sciences) using a slit width of 5 nm, excitation wavelength of 335 nm and an emission wavelength of 450 nm. Fluorescence spectra were also recorded for these compounds without DNA under identical conditions.
Molecular modeling
Molecular docking was carried out with different algorithms to model the specific interactions between DNA and the minor groove binding ligands 125I–EH, 125I–BH, 125I-C8–H and 125I-C12–H. These models of the ligand-DNA complexes, were used to measure the distance between the iodine atom in the DNA bound Hoechst derivative and the central axis of the DNA molecule. All the docking processes were carried out blind with no specification of binding site. Two rounds of docking were performed. The first one was carried out using force field with GLIDE utility (a docking program in the Schrodinger Suite, designed to perform docking in high-throughput screening of potential ligands based on binding mode and affinity for a given receptor molecule), followed by an energy minimization step. The energetically favorable conformations for each DNA-ligand complex were subjected to a second quantum mechanical/ molecular mechanical (QM/MM) docking. The final poses were assigned docking scores using the GLIDE-XP docking function. The ligand-DNA conformations with top 20 docking scores for each compound were used to calculate the average distance between the iodine atom and central axis in the DNA molecule.
The X-ray co-crystal structure of m-iodo-p-methoxy-Hoechst (IMH) complexed with the oligonucleotide duplex d(CGCGAATTCGCG)2 is available in the protein data bank. Therefore to validate our approach, IMH was docked into the minor groove of d(CGCGAATTCGCG)2 following the above strategy. Strikingly, most of the IMH-d(CGCGAATTCGCG)2 conformations could be perfectly superimposed with the crystal structure with a root mean square deviation (RMSD) of < 0.5 Å, validating our docking methodology.
Preparation of 3HT-pUC19 plasmid DNA
pUC19 plasmid DNA (30 ng) (New England Biolabs, Incorporated, Beverly MA, USA) was transformed into Escherichia coli DH5α competent cells (Invitrogen Inc., GibcoBRL CA, USA). Individual colonies were inoculated into 3 ml Luria broth (LB) containing 50 μg/ml ampicillin and incubated overnight at 37 °C to prepare precultures. Precultures were inoculated into 100 ml LB-ampicillin to which 3H-thymidine deoxyribose ([3H]dThd, 37 MBq) (Perkin Elmer Life and Analytical Sciences) was added, and the bacteria were grown for 16 h at 37 °C. The medium was centrifuged at 6000 g in a GSA rotor, and the 3H-labeled plasmid DNA was isolated using standard procedures with the Qiagen Maxi preparation kit. The plasmid DNA was dissolved in phosphate buffered saline (PBS) (pH 7.4). The DNA concentration was determined spectrophotometrically by measuring absorbance at 260 nm (A260). The plasmid DNA was stored at −20 °C.
DNA – 125I–EH, 125IB–H, 125I-C8–H and 125I-C12–H incubations
Supercoiled (SC) form of 3HT-pUC19 plasmid DNA was incubated with the above compounds in 1 × PBS ± DMSO at 4°C, at a ligand to pUC19 DNA (2686 base pairs) ratio of ∼ 3:1, corresponding to a ligand/DNA bp ratio of 0.00112 (one ligand molecule is bound per ∼ 895 base pairs). Adhikary et al studied the binding of Hoechst 33258 to DNA at various ligand to DNA (bp) ratios. They concluded that at a Hoechst 33258/DNA bp ratio of 0.05 (one ligand molecule is bound per 20 bp), Hoechst 33258 binds quantitatively in the minor groove of DNA as inferred from the mono-exponential fluorescence decay characteristics (Adhikary et al. 2003). Additional modes of binding such as intercalation and stacking/adsorption (Buurma and Haq 2008) of Hoechst 33258 molecules on DNA are found only at higher ligand to DNA ratios above 0.05. Therefore, the much lower ratio of 125I- labeled Hoechst derivative to DNA used in the present study ensured that the ligand is bound to A:T sequences in the minor groove only. At various intervals, aliquots (6 μl) were removed from the incubation mixture, combined with loading-dye - glycerol mixture (3 μl) and PBS (3 μl), and loaded onto 1% agarose gels in 0.5 × TAE (Tris-Acetate-Ethylenediaminetetraaceticacid) buffer with ethidium bromide (0.5 μg/ml, Sigma-Aldrich). The gels were run at 200 V (7 V/cm) for 1 h and then photographed on a transilluminator (long wave) attached to a charge coupled device (CCD) camera. The DNA bands corresponding to the super-coiled (SC), nicked (N) and linear (L) forms were excised from the gel. The DNA present in each of these bands was quantified by dissolving the gel pieces in Opti-Fluor scintillation fluid (10 ml) and assaying the associated tritium in a liquid scintillation counter. Since the DNA bands also contained some undecayed 125IEH bound to DNA, correction was made for spillover of 125I into the tritium energy window using a 125I standard. Control incubation of DNA, without any 125I-labeled Hoechst derivative was also set up in parallel, under identical conditions. At various time intervals, aliquots were drawn and analyzed on 1% agarose gel. The DNA bands corresponding to SC, N and L forms were excised and DNA contents were quantified by assaying the associated tritium by liquid scintillation counting. The number of decays accumulated in any given time was obtained by measuring 125I activity in a γ counter (Perkin Elmer 1480 Wizard Automatic, Perkin Elmer Life and Analytical Sciences) prior to analysis of the aliquots on agarose gel.
Calculations and statistics
The DSB calculations were based on the assumption that the binding of 125I-labeled Hoechst derivatives (125I–EH, 125IB–H, 125I-C8–H and 125I-C12–H) to DNA and thus the strand breaks follow a Poisson distribution. The mean number of DSB (X DSB) per DNA molecule is calculated from the fraction of linear DNA formed at each time after exposure of super-coiled DNA to a given number of 125I disintegrations as described previously (Cowan et al. 1987, Balagurumoorthy et al. 2006, 2008a):
| (1) |
where FL is the fraction of linear DNA formed followed by 125I decays at any given time point. The rate of formation of DSB per DNA molecule per decay of 125I is obtained by plotting XDSB as a function of the number of accumulated 125I decays per ml and fitting the data to a linear regression. The standard errors for the slopes of the linear regressions reflect random errors that are associated with pipetting and loading. The straight lines are forced through zero, because at zero decay there are no DSB. The slopes of these linear regressions reflect the DSB yield (YDSB) expressed as the number of DSB generated in one DNA molecule per decay of 125I per ml, and the reciprocal of the slopes represents D0 (the number of decays per ml required to form one DSB in one pUC19 plasmid DNA molecule). The slope when multiplied by the concentration of plasmid DNA in total number of plasmid DNA molecules per ml, gives the yield of DSB per decay of 125I:
| (2) |
where the concentration of DNA ([DNA]) = 3.06 × 1013 molecules/ml.
Results
Detection of double-strand breaks produced by 125I–EH, 125IB–H, 125I-C8–H and 125I-C12–H in supercoiled 3H-pUC19 DNA
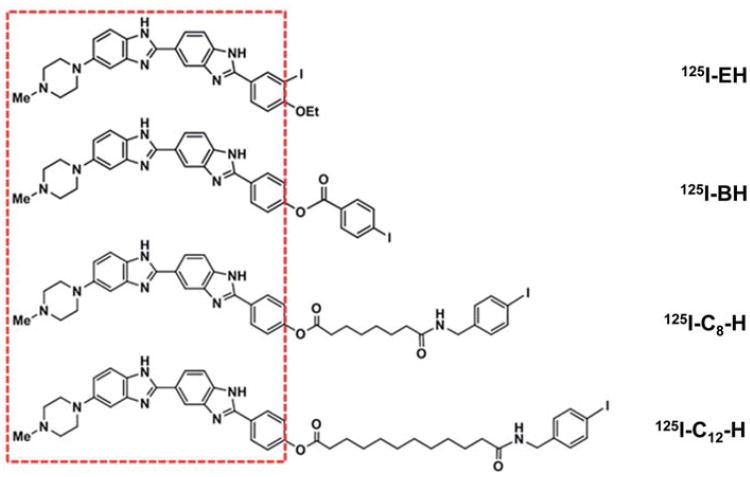
To study the possible effect of distance between the 125I atom and the central axis of DNA on the Auger electron induced DSB yield, a series of precursor Hoechst derivatives shown in Figure 1 were synthesized and radioiodinated with 125I. All the four precursor molecules share a common DNA binding bis-benzimidazole motif indicated by the red boundary (Figure 1) and therefore their DNA binding properties were anticipated to be similar. Preliminary data on the binding equilibrium of these four compounds to calf thymus DNA indicated that all of them bind to DNA with comparable strength; inferred from the enhanced fluorescence intensity of these compounds upon complexation with DNA (See Supplementary Table I, to be found online at http://www.informahealthcare.com/ doi/abs/10.3109/09553002.2012.706360) consistent with our earlier observations (Harapanhalli et al. 1996, Balagurumoorthy et al. 2008a). The differences among these four compounds (Figure 1) lie in the chemical moiety carrying the iodine atom outside the red boundary that is linked to the bis-benzimidazole skeleton. As a result, when these Hoechst derivatives bind to the DNA, the distance between the iodine atom and the DNA was expected to increase progressively going from 125I–EH to 125I-C12–H.
Figure 1.
Iodo-Hoechst derivatives synthesized and used in our strategy to study the effect of increasing distance between 125I and DNA on DNA DSB yields. The DNA binding bis-benzimidazole motif common to all the four compounds is shown within the red rectangular box.
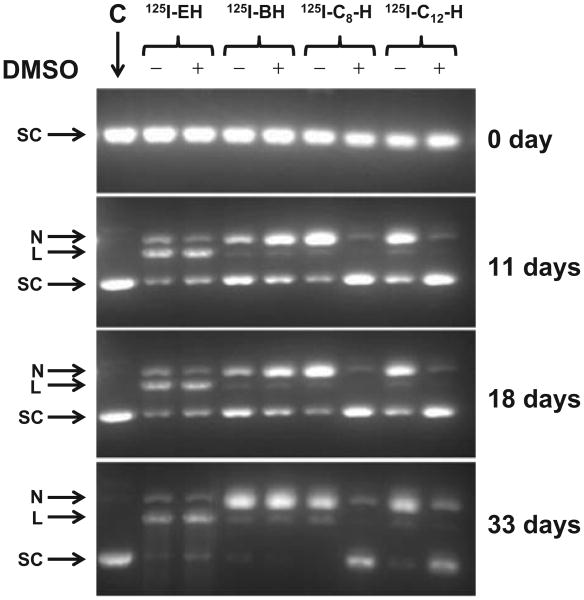
We have been using tritium-labeled pUC19 in all our studies as it allows us to quantify the fraction of supercoiled, nicked and linear forms after 125I decay. Supercoiled form of 3H-pUC19 was incubated with 125I–EH, 125IB–H, 125I-C8– H and 125I-C12– H in a DNA to ligand ratio of ∼1:3. Previously we had shown that at this ratio of 125I–EH to 3H-pUC19, the formation of linear form from nicked DNA is low, and the majority of linear DNA formed directly from supercoiled DNA through DSB (Balagurumoorthy et al. 2006) enables precise DSB quantification. A few representative agarose gels run on days 0, 11, 18 and 33 are shown in Figure 2. It can be seen that as a function of time, i.e., with an increase in 125I decays, a higher fraction of the supercoiled DNA progressively disappears resulting in the formation of nicked and linear DNA. The 3H–pUC19 DNA in the control incubation seen in lane C primarily remains intact throughout the course of the incubation at 4°C. At the beginning of the incubations (0 day), soon after the mixing of 3H-pUC19 with the 125I-labeled compounds (125I–EH, 125IB–H, 125I-C8–H and 125I-C12–H), when the number of accumulated 125I decays is insignificant, it can be seen that the supercoiled form of DNA remains intact in all the incubations, with and without 0.2 M DMSO (Figure 2). The induction of single and double-strand breaks was evident from the appearance of nicked open-circular and linear forms respectively with a concomitant disappearance of supercoiled form even on day 11 in the case of 125I–EH and 125I-B–H, ± DMSO. The intensities of the bands corresponding to the linear form (denoted by L) for the compounds 125I–EH and 125I-B–H in the presence and absence of DMSO are almost equal on day 11 and subsequently. This observation is consistent with our earlier finding for 125I–EH (Balagurumoorthy et al. 2006, 2008a) and suggests that in addition to 125I–EH, 125I-B-H also produces DSB predominantly by direct action, not through hydroxyl radical mediated indirect mechanisms. This trend is maintained throughout the course of the incubation.
Figure 2.
Representative agarose gels used to analyze the incubations of supercoiled 3HT-pUC19 plasmid DNA with 125I-labeled Hoechst (H) derivatives (125I–EH, 125I–BH, 125I-C8–H, and 125I-C12–H) at 4°C in PBS (pH 7.4): – and + indicate absence and presence of DMSO (0.2 M), respectively. Lane C is the control 3HT-pUC19 plasmid DNA incubation that does not contain 125I-labeled compound and remains intact throughout the course of the incubation period. SC, L and N represent supercoiled, linear and relaxed-circular forms of 3HT-pUC19 plasmid DNA. Gels (containing ethidium bromide) were visualized using ultraviolet (320 nm) transillumination.
On the other hand for 125I-C8 –H and 125I-C12 –H, the formation of linear and relaxed-circular DNA was observed on day 11 and subsequently, could be seen only in the absence of DMSO, indicating that both DSB and SSB (single-strand break) are produced by hydroxyl radical mediated indirect mechanisms (Figure 2). The intensities of the intact supercoiled DNA remaining in incubations containing 125I-C8–H and 125I-C12–H in the presence of DMSO are almost equal to the intensity of the supercoiled DNA in control incubation which does not have any radioiodinated compound indicating that any strand break produced, whether it be SSB or DSB, results from hydroxyl radical mediated indirect mechanisms for these two compounds, respectively containing eight and 12 carbon linkers between 125I and the bis-benzimidazole motif. Furthermore, in the absence of DMSO, these 125I-C8 –H and 125I-C12 –H are efficient only in predominantly producing SSB rather than DSB. Essentially, the efficiency of linearization produced by 125I-EH and 125I-B-H is independent of the presence of DMSO and it appears that 125I–B–H is less efficient than 125I–EH in inducing linearization in supercoiled form of 3H-pUC19 DNA. In contrast to this, 125I–C8 –H and 125I–C12–H produce little or no linear form in the presence of DMSO; even in the absence of DMSO, the faint band corresponding to linear form is slightly less intense for 125I–C12 –H than that of 125I– C8 –H suggesting that the former could be less efficient than the latter in inducing DSB. However, further quantification of the gel data corroborated these qualitative analyses and revealed novel insights into the variation of both magnitude and mechanism of DSB production among 125I–EH, 125IB–H, 125I-C8–H and 125I-C12–H, probably because of the difference in the distance between 125I atom and the supercoiled 3H-pUC19 DNA.
Quantification of DSB yields produced by 125I decays in 125I-EH, 125I-B-H, 125I-C8 -H and 125I-C12-H
Following the accumulation of 125I decays in the supercoiled 3HT-pUC19 DNA bound to 125I–EH, 125I-B–H, 125I-C8–H and 125I–C12–H, the incubation mixtures were analyzed to determine the fraction of DNA in each of these three topological states. Quantification of the fraction of DNA in various forms enabled us to calculate SSB and DSB yields per decay of 125I atom. In this paper, we report only the DSB yields obtained for each of these compounds in supercoiled plasmid DNA.
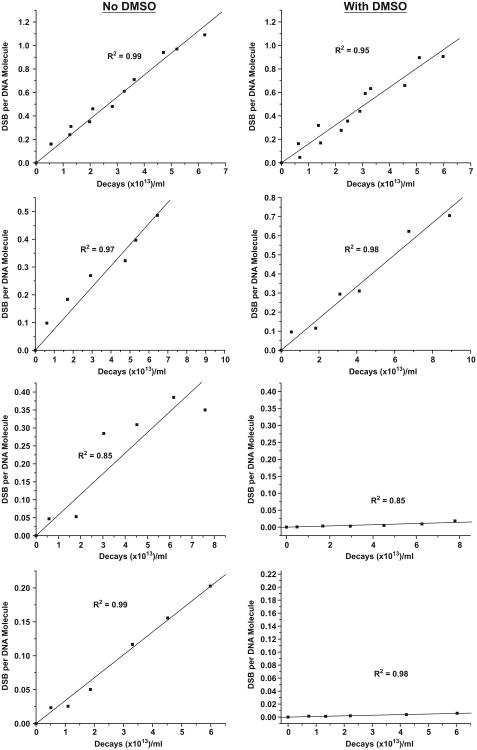
The appearance of linear DNA, an indicator of DSB in the incubations of supercoiled form of 3H-pUC19 with the 125I-labeled 125I–EH, 125I–B–H, 125I-C8–H and 125I-C12–H, was used to quantify the DSB yield. The rate of formation of DSB per DNA molecule as a function of accumulated 125I decays for each of these 125I-labeled compounds in the presence and absence of 0.2 M DMSO is shown in Figure 3. The D0, the number of decays required to produce a single DSB by 125I–EH [–DMSO, (5.57 ± 0.13) × 1013 decays/ml and + DMSO, (6.22 ± 0.24) × 1013] and 125I–B–H [–DMSO, (13.14 ± 0.67) × 1013 decays/ml and +DMSO (11.98 ± 0.50) × 1013 decays/ml] in the presence and absence of 0.2 M DMSO are similar in magnitude (Table I). This finding suggests that the entire DSB are produced by the direct ionization of DNA backbone resulting from the traversal of Auger electrons and OH radicals do not contribute to this process of DSB induction. Accordingly, similar DSB yields of 0.52 ± 0.01 and 0.49 ± 0.02 were obtained for 125I–EH in the presence and absence of 0.2 M DMSO, respectively (Table I). The trend for 125I–B–H was the same as that of 125I–EH with DSB yields of 0.24 ± 0.03 and 0.26 ± 0.04 (Table I) in absence and presence of DMSO respectively. Further, D0 values for 125I–B–H ± DMSO [(13.14 ± 0.67) × 1013 and 11.98 ± 0.50) × 1013 decays/ml] are roughly two times higher than those of 125I–EH [(5.57 ± 0.13) × 1013 and (6.22 ± 0.24) × 1013 decays/ml] indicating that the efficiency with which 125I–B–H could induce DSB in supercoiled 3H-pUC19 is only half of that of 125I–EH.
Figure 3.
Quantitative analysis of agarose gel electrophoresis to determine DSB yields in SC plasmid DNA induced by 125I–EH, 125I–BH, 125I-C8–H and 125I-C12–H (±DMSO) at 4°C in PBS (pH 7.4). DSB yields calculated from the appearance of linear form from supercoiled plasmid DNA were plotted as a function of accumulated 125I decays. Linear regressions were carried out and R2 values were obtained using Origin. Linear regression straight lines were forced through zero.
Table I.
Radiosensitivity of supercoiled form of pUC19 plasmid DNA as a function of distance between the decaying 125I atom and the DNA; Effect of 125I-DNA distance on the mechanism and magnitude of DSB induction.
| Compound | Mean Distance (Å) between 125l and DNA † | D0(×1013 Decays/ml) | DSB yield per Decay | DMF | Mechanism | |||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| −DMSO | +DMSO | −DMSO | +DMSO | Direct | •OH radical mediated Indirect | |||
| 125I-EH | 10.5 (20) | 5.57 ±0.13 | 6.22 ± 0.24 | 0.52 ± 0.01 | 0.49 ± 0.02 | 1.12 ± 0.05 | 100% | 0 |
| 125l-B-H | 12.0 (20) | 13.14 ± 0.67 | 11.98 ± 0.50 | 0.24 ± 0.03 | 0.26 ± 0.04 | 0.91 ± 0.06 | 100% | 0 |
| 125l-C8-H | 13.9 (14) 8.1 (6) | 17.41 ± 1.72 | 558.65 ± 68 | 0.18 ± 0.02 | 0.01 ± 0.00 | 32.09 ± 5.03 | 0% | 100% |
| 8 125I-C12 -H | 13.2 (11) 6.0 (9) | 29.62 ± 0.82 | 1059.61 ±49 | 0.10 ± 0.00 | 0.00 ± 0.00 | 35.77 ± 1.93 | 0% | 100% |
The numbers in parantheses indicate the number of conformational poses used to calculate the mean distances shown. Errors indicate standard errors in the slope of the linear regressions shown in figure 3, used to calculate the DSB yields.
In contrast to this, D 0 value for 125I-C8–H without DMSO [(17.41 ± 1.72) × 1013] is about ∼32-fold smaller than the D0 obtained in the presence of DMSO [(558.65 ± 68) × 1013 decays/ml] and strikingly the DSB yield was found to be proportionately much higher in the absence of DMSO (0.18 ± 0.02 per 125I decay) compared to that with DMSO (0.01 ± 0.00 per decay of 125I) as shown in Table I. These observations suggest that 125I-C8 –H induces DSB predominantly by hydroxyl radical mediated indirect mechanisms unlike 125I–EH and 125I-B–H, which produces the entire DSB by direct action of Auger electrons.
The behavior of 125I-C12–H is similar to that of 125I-C8–H; D0 values of (29.62 ± 0.82) × 1013 and (1059.61 ± 49.52) × 1013 decays/ml were obtained in the absence and presence of DMSO respectively. A DSB yield of 0.10 ± 0.00 per decay obtained in the absence of DMSO compared to almost no DSB observed in the presence of DMSO for this compound (Table I) which is expected to position 125I atom at a much greater distance than the other three compounds indicated the mechanism of DSB production by 125I-C12–H is similar to that of 125I-C8–H.
Results from the present experiments summarized in Table I indicate: (i) In the absence of DMSO, there is a progressive decrease in the Auger electron-induced DSB yields in the following order: 125I–EH>125I–B–H>125I–C8–H> 125I–C12–H, (ii) the trend is the same even under scavenging conditions with 0.2 DMSO, (iii) the magnitudes of DSB yields ± DMSO are about the same for compounds 125I–EH and 125I–B–H, and (iv) the magnitudes of DSB yields in the presence of 0.2 M DMSO for compounds 125I-C8–H and 125I-C12–H are always much smaller or insignificant and almost close to none compared to the DSB yield without DMSO.
Distance between the 125I atom and the central axis of DNA as measured by molecular modeling studies
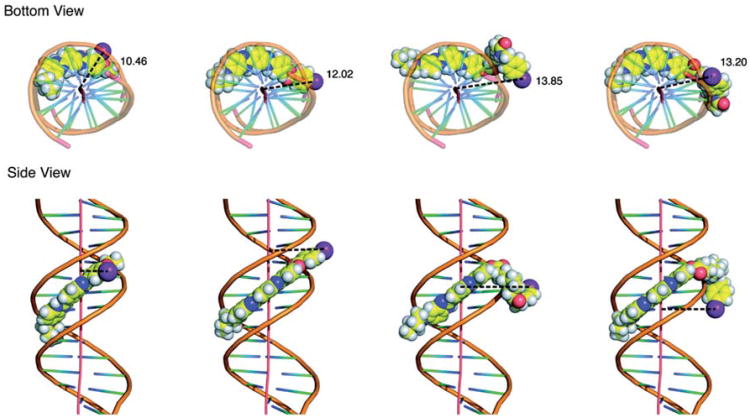
The ligand-DNA conformations with top 20 docking scores for each compound (Figure 1) were used to calculate the average distance between the iodine atom and central axis in the DNA molecule. Since the X-ray co-crystal structure of IMH complexed with the oligonucleotide duplex d(CGCGAATTCGCG)2 is available in the protein data bank (Squire et al. 2000), we validated our approach by first docking IMH into the minor groove of d(CGCGAATTCGCG)2. These simulations demonstrated that: (i) Most of the IMH-d(CGCGAATTCGCG)2 conformations could be perfectly superimposed with the crystal structure with a RMSD of < 0.5 Å, and (ii) all the interactions between DNA and IMH observed in the X-ray crystal structure were found to be conserved in the molecular models obtained from the docking procedure. These results validated the parameters and criteria used in our docking procedure and gave us confidence in using the same in building the models of other Hoechst derivatives (125I–B–H, 125I–C8–H and 125I–C12–H) to predict the distance between the 125I atom and the central axis of DNA. Therefore, the same docking procedures were used for each of the iodinated Hoechst derivatives and the distances between their iodine atoms and the DNA were measured. The average distances of 125I–EH and 125I–B–H to the DNA central axis are 10.4 Å and 12.5 Å, respectively, while for 125I–C8–H and 125I–C12H, the distance from docking is 8.2 Å and 8.4 Å . After examining each pose of the latter two compounds in the ligand-DNA complex structure, we noticed that the C8 and C12 linkers in several (six out of 20 for 125I–C8–H and nine out of 20 for 125I–C12–H) of the conformations of these two compounds were flipped towards the DNA and the phenyl ring containing the iodine atom was stacked onto the terminal base pair of the 12 base pair long DNA duplex. We rationalized that it could be due to the shorter length of the duplex DNA (12 base pair). To avoid this from happening in the energy minimization process, the four Hoechst derivatives shown in Figure 1 were docked onto a longer DNA template of 20 base pair (Protein Data Base code: 2V3L). The representative poses with their respective 125I-DNA distances are shown in Figure 4. In these simulations, the distances between the iodine atoms of 125I–EH and 125I–B–H and the DNA were found - as expected - to be similar to those obtained with the shorter DNA template (10.5 Å vs. 10.4 Å and 12.0 Å vs. 12.5 Å , respectively). These encouraging results also served as a validation to use the longer 20 base pair DNA in the docking to avoid or minimize the flipping back of the carbon linker on to the DNA terminus for 125I-C8–H and 125I-C12–H. Based on these expectations, when longer DNA is used to dock 125I-C8–H and 125I-C12–H derivatives, (i) no interactions of iodine atom were observed with the DNA termini, and (ii) the distances between the iodine atoms and the central DNA axis of 13.9 Å and 13.2 Å, respectively, were found to be more in reasonable agreement with our expectations from their chemical structures (Figure 1).
Figure 4.
(Panel A): Molecular model of m-iodo-p-ethoxyHoechst (I–EH) and its derivatives listed in Figure 1 docked into minor groove of DNA shown in Corey-Pauling-Koltun space-filling model as viewed down the helical axis. Distance between the iodine atom relative to the DNA helical axis (in Å ) indicated is the mean distance of several poses presented in Table I. (Panel B): Iodo-Hoechst derivative-DNA complexes as seen from the side. Bis-benzimidazole motif binds to d(AATT) tetranucleotide sequence through hydrogen bonds to N3 of adenine and O2 of thymine in oligonucleotide duplex d(CGCGAATTCGCG)2. The effect of the flexibility of the C-C linker in positioning the iodine atom could be seen clearly.
The top 20 energetically favorable conformations for 125I–EH or 125I–B–H docked on 12 bp and 20 bp DNA used for calculating mean 125I-DNA distance were highly similar and the ligand-DNA interactions were conserved among all of them and as a result the distances between iodine atom and the DNA are very close. On the other hand, for 125I-C8–H and 125I-C12 –H owing to the flexibility of the C8 and C12 linker region between bis-benzimidazole and the iodine atom, diverse conformations with a range of 125I-DNA distances were possible. As shown in Table I, the top 20 conformations could be segregated into two clusters, 14 and six based on mean 125I-DNA distances of 13.9 Å and 8.1 Å for 125I-C8–H and 11 and nine for 125I-C12–H with mean distances of 13.2 Å and 6.0 Å . For practical purposes pertinent to interpreting the experimentally observed DSB yields produced by these 125I-labeled compounds reported in this paper, the mean 125I-DNA distances of 13.9 Å and 13.2 Å (Table I) obtained for the majority of the conformations (Figure 4) are used.
Discussion
Several in vitro/in vivo studies with compounds labeled with the Auger-electron emitting radionuclide Iodine-125, e.g., intercalators (Kassis et al. 1989), groove binders (Martin and Haseltine 1981, Kassis et al. 1999a, 1999b), and steroid hormones (DeSombre et al. 1992) have demonstrated that a decaying iodine atom does not need to be covalently incorporated into the DNA of the target cell to produce DSB or effective cell kill, if it is placed in sufficient proximity to the DNA. Hofer et al. (1975) studied the effect of proximity of the decaying Auger emitter to the nuclear DNA on the cytotoxicity produced using 125I and 67Ga and concluded that Auger cascade produced in the cytoplasm is relatively ineffective in producing radiobiological effects compared to decays occurring in the nucleus of a mammalian cell. The conclusions of that study are phenomenological and are limited by the usage of two different Auger emitting radionuclides in nucleus and cytoplasm. On the other hand, later studies from our laboratory (Kassis et al. 1987a), using the same Auger emitter (125I), showed that decays occurring outside the nucleus are ineffective in killing the labeled cell. Even in cases where decays occurring within the nucleus of a mammalian cells, 125I covalently attached to the nuclear DNA (125IUdR) exhibited a higher RBE as compared to non-specifically bound 125I-dihydrorhodamine localized within the nucleus but not covalently attached to DNA (Kassis et al. 1987a). Followed by these early findings, several other recent in vitro and in vivo studies suggested that close proximity of the decaying iodine-125 atom to the DNA is crucial to the efficient induction of DSB in plasmid models (Kassis et al. 1999a, 1999b, Lobachevsky et al. 2008) as well as in mammalian cells (Walicka et al. 1999).
There is a ∼1000-fold lesser energy deposited at distances greater than few nanometers away from the 125I decay site predicted by the dosimetric calculations (Kassis et al. 1987b, Kassis 2004). In accordance with these expectations the probability of DSB yields were calculated to decrease as the DNA is moved away from the 125I decay site (Humm and Charlton 1989). Comparison of the DSB yield produced by a 125I-labeled groove binder which positions 125I atom within ∼ 1 nm from the center of the helix, with that of 125I-antipyrene, which does not bind DNA and hence decays far away from the helix, at distances > 500 nm within which > 98% of Auger electrons deposit their energy, also emphasized that proximity of 125I to the DNA is a critical factor in inducing DSB (Kassis et al. 1999a). Recent studies (Lobachevsky et al. 2008) have shown that there is a 25– 40% decrease in DSB yield when the distance between the decaying 125I atom and the DNA is increased from 9.2 Å to 11.2 Å . However, a comprehensive understanding underlying the exact relationship between DSB yield and the position of decaying iodine-125 is needed for the better design of Auger electron based therapeutic strategies.
The binding of bis-benzimidazole Hoechst dyes to naked DNA and chromatin in mammalian cells have been well documented (Arndt-Jovin and Jovin 1977). Hoechst 33342 and 33258 could be successfully radioiodinated with both iodine-125 and iodine-123 either directly (Martin and Pardee 1985) or through destannylation of tributyltin derivatives (Harapanhalli et al. 1996) and thereby could be exploited to position the decaying 125I atom within few nanometers from the target DNA. In the context of determining the exact relationship between DSB production efficiency and the distance between the radioiodine and the DNA, we previously proposed that substitutions on bis-benzimidazole skeleton of these compounds could lead to derivatives that may be useful in making radioiodinated derivatives to position iodine at different distances from the DNA in the bound state (Chen et al. 2004). High resolution X-ray crystal structure is available for iodinated Hoechst 33258 derivative (IMH)-DNA complex (Squire et al. 2000). In the absence of X-ray or nuclear magnetic resonance (NMR) structures for Hoechst derivatives complexed with the DNA listed in Figure 1 with linkers to position 125I at greater distances, molecular modeling is the only viable way to estimate the distance between the iodine atom and the DNA. A RMSD of 0.5 Å obtained for the energy minimized IMH-DNA model from the crystal structure reassures that the parameters and criteria used in modeling are appropriate and further if the same parameters are used to model other derivatives shown in Figure 1 containing linkers between iodine atom and bis-benzimidazole skeleton, the likelihood of obtaining reliable distances is very high. Nevertheless, we are aware of the fact that some degree of conformational flexibility associated with such (C-C)n linkers would: (i) Introduce certain degrees of ambiguity in precisely fixing the position of 125I atom with regard to the DNA in the ligand-DNA complex, and (ii) there could be more than one energetically favorable structure possible for the ligand-DNA complex arising from conformational freedom inherent in the C-C bonds. In spite of these possible caveats, we decided to proceed owing to the simple chemistry and ease of chemical synthesis. We have taken the influence of the flexibility of the (C-C) n linker on the position of the iodine atom with regard to DNA into consideration in predicting the distances between 125I and the DNA by averaging several energetically favorable conformations generated by docking. To minimize the possible uncertainty in the estimated Iodine-DNA distances, for each compound the mean of several Iodine-DNA distances from conformations with top 20 docking scores was calculated (Table I) and used in the DSB-distance correlation. Our contention is that the 125I decay is a random event that can take place when iodine atom is in any one of these positions in a dynamic equilibrium among multiple ligand-DNA conformations.
In the absence of DMSO, the results (Figure 3 and Table I) show that the DSB yields decrease monotonically as the 125I atom is distanced - a few Angstroms - from the DNA helix (125IH: 0.52 ± 0.01; 125IB- H: 0.24 ± 0.03; 125I-C8-H: 0.18 ±0.02; 125IC12- H: 0.10 ±0.00). In the presence of DMSO, DSB yields for 125IH (0.49 ± 0.02) and 125I–B–H (0.26 ± 0.04) remain the same (i.e., DSB are entirely produced by direct effects). However, no DSB were detected when SC plasmid DNA was incubated under similar conditions with 125I-C8 –H or 125I-C12–H (i.e., DSB are entirely produced by indirect effects). In addition to the formation of DNA radicals, these indirect effects may also be mediated by the scavenging of a hydroxyl radical by the linker region or the Hoechst skeleton itself to form a reactive radical which in turn could open up the sugar ring and cause strand scission (Adhikary et al. 1997, 2000).
These results suggest that at a critical distance between the 125I atom and the DNA helix, DSB production switches from an ‘all’ direct to an ‘all’ indirect mechanism, a situation that is reminiscent of the decay of 125I bound to DNA vs. 125I free in solution (Kassis et al. 1999b).
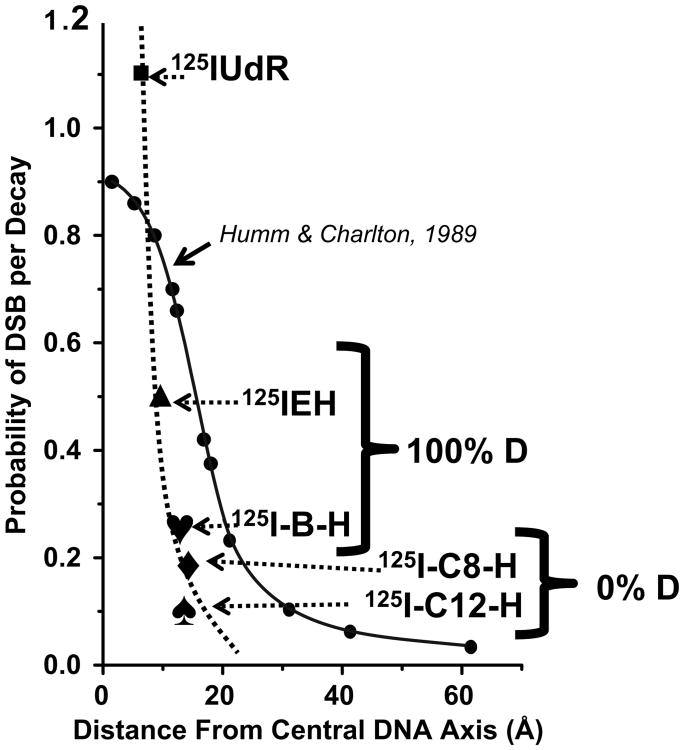
We have compared these DSB yields obtained when the decaying 125I is positioned at different distances in the complexes of supercoiled form of pUC19 DNA and 125I-labeled Hoechst derivatives with that of theoretical predictions of Humm and Charlton based on Monte Carlo simulations (Figure 5). Before we did so, we reminded ourselves of the assumptions used in these theoretical predictions most notably that DNA was considered as a rigid rod, which is not true for supercoiled DNA of size 2686 bps which, rather, is in a conformationally flexible and dynamic state (Ramachandran and Schlick 1997). In this context, perhaps it would be more appropriate to compare the DSB yields obtained using linear plasmid DNA template (closer to the rod like model) with the theory (a work currently underway in our laboratories), given the fact that 125IEH induced DSB yields very much depend on the DNA topology and DSB yield for linear pUC19 is ∼ 3-fold higher than that of its supercoiled form (Balagurumoorthy et al. 2008a).
Figure 5.
Comparison between the variation of experimental DSB yields with 125I-DNA distance reported in this work [dotted line connecting (■), 125IUdR; (▲), 125IEH; (♥), 125I-B-H; (♦), 125I-C8 –H; (♠), 125I-C12–H] and the theoretical prediction [solid line connecting (●)] of Humm and Charlton (1989).
In principle, major contributions of Auger electron-induced DSB for 125I can come from three sources: Direct ionizations, charge neutralization of the highly positive daughter tellurium ion and hydroxyl radicals mediated indirect mechanisms (Hofer 2000). For the first two compounds at distances < 12 A hydroxyl radical-mediated indirect mechanisms seem to have no role in DSB production and direct ionizations and charge neutralization contribute to the entire DSB produced. Our observation that all the DSB produced by 125I-C8–H and 125I-C12–H could be scavenged by DMSO rules out the role of direct ionization and charge neutralization followed by Coulombic explosion (Pomplun and Sutmann 2004) in DSB production by these two compounds. This makes sense particularly as: (i) The probability of direct ionizations in DNA by electron traversal is expected to decrease as the distance between the 125I-DNA is increased, and (ii) charge neutralization followed by Coulombic explosion could be effective in producing strand breaks only if the decaying 125I atom is either covalently attached or close enough to the helix so that the highly positive daughter tellurium ion can drain the ‘π’ electron density of the heterocyclic bases leading to opening of the sugar ring followed by scission in the phosphodiester bond. An additional important point to note is the experimental curve of the variation of DSB as a function of 125I-DNA distance (Figure 5) is more steeper than the theoretical curve and left-shifted suggesting that theory always overestimates the DSB yield at any given distance. This observation is in agreement with that of Lobachevsky et al. (2008) in the range of distances (7.4–11.2 Å ) that they examined. This could again be consequent to the assumptions made in the simulations to imply that energy deposited by the 125I decay process could lead to these three processes contributing to DSB with equal probability, but in reality it may be true that each of these three mechanisms leading to DSB varies as a function of distance as indicated by the difference in DSB yields in the presence and absence of DMSO. Another possibility is that the actual distances are higher than the ones measured by docking. In this case, either X-ray crystal structures or the NMR solution structures need to be solved for the ligand-DNA complexes using oligonucleotides with various sequences to average out the sequence-dependent microheterogeneity in the DNA structure. However, the overall correlation observed between the experimental DSB yields and the theoretical 125I-DNA distances itself is in reasonable agreement with our expectations based on Auger-electron microdosimetry.
Conclusions
The distances between the DNA and the decaying 125I atom were estimated from molecular modeling for various bis-benzimidazole ligands. Auger electron-induced DSB yields were determined experimentally. We do find a correlation between DSB yield and the distances consistent with the trend expected from the theoretical dosimetry. However, we have found that in our experimental system the magnitude of 125I-induced DSB drops much more sharply as a function of distance than in the original theoretical Monte Carlo predictions of Humm and Charlton (1989) and the experimentally observed DSB yield at a given distance is always smaller than the yields predicted from theoretical simulations. There appears to be a critical distance between the 125I atom and the DNA of 12 Å beyond which direct ionizations of Auger cascade and charge neutralizations of the highly positive daughter tellurium ion are ineffective in inducing double-strand breaks. We believe these findings may have important ramifications for the better design of Auger electron-emitting radiopharmaceuticals.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health 5R01 CA015523 to AIK. PB was a recipient of National Research Service Award under National Institutes of Health 5T32 CA009078.
Footnotes
Supplementary material available online Supplementary Table 1
Notice of correction: This paper published online on July 24, 2012 contained a small grammatical error in the title. The error has been corrected in this version of the paper. The authors and publisher would like to apologize for any inconvenience caused.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Adhikary A, Bothe E, Jain V, Von Sonntag C. Pulse radiolysis of the DNA-binding bisbenzimidazole derivatives Hoechst 33258 and 33342 in aqueous solutions. International Journal of Radiation Biology. 2000;76:1157–1166. doi: 10.1080/09553000050134393. [DOI] [PubMed] [Google Scholar]
- Adhikary A, Bothe E, von Sonntag C, Jain V. DNA radioprotection by the bisbenzimidazole derivative Hoechst 33258: Model studies at the nucleotide level. Radiation Research. 1997;148:493–494. [Google Scholar]
- Adhikary A, Buschmann V, Muller C, Sauer M. Ensemble and single-molecule fluorescence spectroscopic study of the binding modes of the bis-benzimidazole derivative Hoechst 33258 with DNA. Nucleic Acids Research. 2003;31:2178–2186. doi: 10.1093/nar/gkg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt-Jovin DJ, Jovin TM. Analysis and sorting of living cells according to deoxyribonucleic acid content. Journal of Histochemistry and Cytochemistry. 1977;25:585–589. doi: 10.1177/25.7.70450. [DOI] [PubMed] [Google Scholar]
- Auger P. Sur les rayons b secondaires produits dans un gaz par des rayons X. Comptes Rendus Hebdomadaires des Seances de l Academie des Sciences. 1925;180:65–68. [Google Scholar]
- Balagurumoorthy P, Chen K, Adelstein SJ, Kassis AI. Auger electron-induced double-strand breaks depend on DNA topology. Radiation Research. 2008a;170:70–82. doi: 10.1667/RR1072.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagurumoorthy P, Chen K, Bash RC, Adelstein SJ, Kassis AI. Mechanisms underlying production of double-strand breaks in plasmid DNA after decay of 125I-Hoechst. Radiation Research. 2006;166:333–344. doi: 10.1667/RR3591.1. [DOI] [PubMed] [Google Scholar]
- Balagurumoorthy P, Wang K, Adelstein SJ, Kassis AI. DNA double-strand breaks induced by decay of (123)I-labeled Hoechst 33342: Role of DNA topology. International Journal of Radiation Biology. 2008b;84:976–983. doi: 10.1080/09553000802512568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomer WD, Adelstein SJ. Antineoplastic effect of iodine- 125-labelled iododeoxyuridine. International Journal of Radiation Biology. 1975;27:509–511. doi: 10.1080/09553007514550521. [DOI] [PubMed] [Google Scholar]
- Bloomer WD, Adelstein SJ. 5-125I-iododeoxyuridine as prototype for radionuclide therapy with Auger emitters. Nature. 1977;265:620–621. doi: 10.1038/265620a0. [DOI] [PubMed] [Google Scholar]
- Bradley EW, Chan PC, Adelstein SJ. The radiotoxicity of iodine- 125 in mammalian cells I. Effects on the survival curve of radioiodine incorporated into DNA. Radiation Research. 1975;64:555–563. [PubMed] [Google Scholar]
- Buurma NJ, Haq I. Calorimetric and spectroscopic studies of Hoechst 33258: Self-association and binding to non-cognate DNA. Journal of Molecular Biology. 2008;381:607–621. doi: 10.1016/j.jmb.2008.05.073. [DOI] [PubMed] [Google Scholar]
- Chan PC, Lisco E, Lisco H, Adelstein SJ. The radiotoxicity of iodine-125 in mammalian cells. II. A comparative study on cell survival and cytogenetic responses to 125IUdR, 131IUdR, and 3HTdR. Radiation Research. 1976;67:332–343. [PubMed] [Google Scholar]
- Charlton DE, Pomplun E, Booz J. Some consequences of the Auger effect: Fluorescence yield, charge potential, and energy imparted. Radiation Research. 1987;111:553–564. [PubMed] [Google Scholar]
- Chen K, Adelstein SJ, Kassis AI. Molecular modeling of the interaction of iodinated Hoechst analogs with DNA: Implications for new radiopharmaceutical design. Journal of Molecular Structure (Theochem) 2004;711:49–56. [Google Scholar]
- Cole A. Absorption of 20-eV to 50,000-eV electron beams in air and plastic. Radiation Research. 1969;38:7–33. [PubMed] [Google Scholar]
- Costantini DL, Chan C, Cai Z, Vallis KA, Reilly RM. (111)In-labeled trastuzumab (Herceptin) modified with nuclear localization sequences (NLS): an Auger electron-emitting radiotherapeutic agent for HER2/neu-amplified breast cancer. Journal of Nuclear Medicine. 2007;48:1357–1368. doi: 10.2967/jnumed.106.037937. [DOI] [PubMed] [Google Scholar]
- Costantini DL, Hu M, Reilly RM. Peptide motifs for insertion of radiolabeled biomolecules into cells and routing to the nucleus for cancer imaging or radiotherapeutic applications. Cancer Biotherapy and Radiopharmaceuticals. 2008;23:3–24. doi: 10.1089/cbr.2007.0430. [DOI] [PubMed] [Google Scholar]
- Costantini DL, McLarty K, Lee H, Done SJ, Vallis KA, Reilly RM. Antitumor effects and normal-tissue toxicity of 111In-nuclear localization sequence-trastuzumab in athymic mice bearing HER-positive human breast cancer xenografts. Journal of Nuclear Medicine. 2010;51:1084–1091. doi: 10.2967/jnumed.109.072389. [DOI] [PubMed] [Google Scholar]
- Cowan R, Collis CM, Grigg GW. Breakage of double-stranded DNA due to single-stranded nicking. Journal of Theoretical Biology. 1987;127:229–245. doi: 10.1016/s0022-5193(87)80133-9. [DOI] [PubMed] [Google Scholar]
- DeSombre ER, Hughes A, Hanson RN, Kearney T. Therapy of estrogen receptor-positive micrometastases in the peritoneal cavity with Auger electron-emitting estrogens: Theoretical and practical considerations. Acta Oncologica. 2000;39:659–666. doi: 10.1080/028418600750063695. [DOI] [PubMed] [Google Scholar]
- DeSombre ER, Hughes A, Shafii B, Púy L, Kuivanen PC, Hanson RN, Harper PV. Estrogen receptor-directed radiotoxicity with Auger electron-emitting nuclides: E-17a-[123I] iodovinyl-11b-methoxyestradiol and CHO-ER cells. In: Howell RW, Narra VR, Sastry KSR, Rao DV, editors. Biophysical aspects of Auger processes American Association of Physicists in Medicine Symposium Series No 8. Woodbury, NY: American Institute of Physics; 1992. pp. 352–371. [Google Scholar]
- Feinendegen LE. Biological damage from the Auger effect, possible benefits. Radiation and Environmental Biophysics. 1975;12:85–99. doi: 10.1007/BF01328970. [DOI] [PubMed] [Google Scholar]
- Harapanhalli RS, McLaughlin LW, Howell RW, Rao DV, Adelstein SJ, Kassis AI. [125I/127I]iodoHoechst 33342: Synthesis, DNA binding, and biodistribution. Journal of Medicinal Chemistry. 1996;39:4804–4809. doi: 10.1021/jm9602672. [DOI] [PubMed] [Google Scholar]
- Hofer KG. Biophysical aspects of Auger processes. Acta Oncologica. 2000;39:651–657. doi: 10.1080/028418600750063686. [DOI] [PubMed] [Google Scholar]
- Hofer KG, Harris CR, Smith JM. Radiotoxicity of intracellular 67Ga, 125I and 3H: Nuclear versus cytoplasmic radiation effects in murine L1210 leukaemia. International Journal of Radiation Biology. 1975;28:225–241. doi: 10.1080/09553007514550991. [DOI] [PubMed] [Google Scholar]
- Hofer KG, Hughes WL. Radiotoxicity of intranuclear tritium, 125iodine and 131iodine. Radiation Research. 1971;47:94–109. [PubMed] [Google Scholar]
- Humm JL, Charlton DE. A new calculational method to assess the therapeutic potential of Auger electron emission. International Journal of Radiation Oncology *Biology* Physics. 1989;17:351–360. doi: 10.1016/0360-3016(89)90450-1. [DOI] [PubMed] [Google Scholar]
- Kassis AI. The amazing world of auger electrons. International Journal of Radiation Biology. 2004;80:789–803. doi: 10.1080/09553000400017663. [DOI] [PubMed] [Google Scholar]
- Kassis AI, Adelstein SJ. 5-[125I]iodo-2'-deoxyuridine in the radio-therapy of solid CNS tumors in rats. Acta Oncologica. 1996;35:935–939. doi: 10.3109/02841869609104048. [DOI] [PubMed] [Google Scholar]
- Kassis AI, Fayad F, Kinsey BM, Sastry KSR, Adelstein SJ. Radiotoxicity of an 125I-labeled DNA intercalator in mammalian cells. Radiation Research. 1989;118:283–294. [PubMed] [Google Scholar]
- Kassis AI, Fayad F, Kinsey BM, Sastry KSR, Taube RA, Adelstein SJ. Radiotoxicity of 125I in mammalian cells. Radiation Research. 1987a;111:305–318. [PubMed] [Google Scholar]
- Kassis AI, Harapanhalli RS, Adelstein SJ. Comparison of strand breaks in plasmid DNA after positional changes of Auger electron-emitting iodine-125. Radiation Research. 1999a;151:167–176. [PubMed] [Google Scholar]
- Kassis AI, Harapanhalli RS, Adelstein SJ. Strand breaks in plasmid DNA after positional changes of Auger electron-emitting iodine-125: Direct compared to indirect effects. Radiation Research. 1999b;152:530–538. [PubMed] [Google Scholar]
- Kassis AI, Howell RW, Sastry KSR, Adelstein SJ. Positional effects of Auger decays in mammalian cells in culture. In: Baverstock KF, Charlton DE, editors. DNA damage by Auger emitters. London: Taylor and Francis; 1988. pp. 1–13. [Google Scholar]
- Kassis AI, Kirichian AM, Wang K, Safaie Semnani E, Adelstein SJ. Therapeutic potential of 5-[ 125I]iodo-2′-deoxyuridine and methotrexate in the treatment of advanced neoplastic meningitis. International Journal of Radiation Biology. 2004;80:941–946. doi: 10.1080/09553000400017671. [DOI] [PubMed] [Google Scholar]
- Kassis AI, Sastry KS, Adelstein SJ. Kinetics of uptake, retention, and radiotoxicity of 125IUdR in mammalian cells: Implications of localized energy deposition by Auger processes. Radiation Research. 1987b;109:78–89. [PubMed] [Google Scholar]
- Kassis AI, Tumeh SS, Wen PYC, Baranowska-Kortylewicz J, Van den Abbeele AD, Zimmerman RE, Carvalho PA, Garada BM, DeSisto WC, Bailey NO, et al. Intratumoral administration of 5-[123I] iodo-2′-deoxyuridine in a patient with a brain tumor. Journal of Nuclear Medicine. 1996;37(4 suppl):19S–22S. [PubMed] [Google Scholar]
- Kassis AI, Walicka MA, Adelstein SJ. Double-strand break yield following 125I decay: Effects of DNA conformation. Acta Oncologica. 2000;39:721–726. doi: 10.1080/028418600750063785. [DOI] [PubMed] [Google Scholar]
- Kishikawa H, Wang K, Adelstein SJ, Kassis AI. Inhibitory and stimulatory bystander effects are differentially induced by Iodine-125 and Iodine-123. Radiation Research. 2006;165:688–694. doi: 10.1667/RR3567.1. [DOI] [PubMed] [Google Scholar]
- Lobachevsky PN, Karagiannis TC, Martin RF. Plasmid DNA break-age by decay of DNA-associated Auger electron emitters: Approaches to analysis of experimental data. Radiation Research. 2004;162:84–95. doi: 10.1667/rr3187. [DOI] [PubMed] [Google Scholar]
- Lobachevsky PN, White J, Leung M, Skene C, Martin RF. Plasmid breakage by (125)I-labelled DNA ligands: effect of DNA-iodine atom distance on breakage efficiency. International Journal of Radiation Biology. 2008;84:991–1000. doi: 10.1080/09553000802478091. [DOI] [PubMed] [Google Scholar]
- Makrigiorgos GM, Kassis AI, Baranowska-Kortylewicz J, McElvany KD, Welch MJ, Sastry KSR, Adelstein SJ. Radiotoxicity of 5-[123I] iodo-2′-deoxyuridine in V79 cells: A comparison with 5-[125I]iodo-2′-deoxyuridine. Radiation Research. 1989;118:532–544. [PubMed] [Google Scholar]
- Martin RF. Induction of double-stranded breaks in DNA by binding with an 125I-labelled acridine. International Journal of Radiation Biology. 1977;32:491–497. doi: 10.1080/09553007714551261. [DOI] [PubMed] [Google Scholar]
- Martin RF, Haseltine WA. Range of radiochemical damage to DNA with decay of iodine-125. Science. 1981;213:896–898. doi: 10.1126/science.7256283. [DOI] [PubMed] [Google Scholar]
- Martin RF, Holmes N. Use of an 125I-labelled DNA ligand to probe DNA structure. Nature. 1983;302:452–454. doi: 10.1038/302452a0. [DOI] [PubMed] [Google Scholar]
- Martin RF, Pardee M. Preparation of carrier free [ 125I]iodoHoechst 33258. International Journal of Applied Radiation and Isotopes. 1985;36:745–747. doi: 10.1016/0020-708x(85)90049-3. [DOI] [PubMed] [Google Scholar]
- Panyutin IG, Winters TA, Feinendegen LE, Neumann RD. Development of DNA-based radiopharmaceuticals carrying Auger-electron emitters for anti-gene radiotherapy. The Quaterly Journal of Nuclear Medicine and Molecular Imaging. 2000;44:256–267. [PubMed] [Google Scholar]
- Panyutin IV, Luu AN, Panyutin IG, Neumann RD. Strand breaks in whole plasmid dna produced by the decay of (125)I in a triplex-forming oligonucleotide. Radiation Research. 2001;156:158–166. doi: 10.1667/0033-7587(2001)156[0158:sbiwpd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Pomplun E, Booz J, Charlton DE. A Monte Carlo simulation of Auger cascades. Radiation Research. 1987;111:533–552. [PubMed] [Google Scholar]
- Pomplun E, Sutmann G. Is coulomb explosion a damaging mechanism for (125)IUdR? International Journal of Radiation Biology. 2004;80:855–860. doi: 10.1080/09553000400017614. [DOI] [PubMed] [Google Scholar]
- Ramachandran G, Schlick T. Buckling transitions in superhelical DNA: Dependence on the elastic constants and DNA size. Biopolymers. 1997;41:5–25. doi: 10.1002/(SICI)1097-0282(199701)41:1<5::AID-BIP2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Rebischung C, Hofmann D, Stefani L, Desruet MD, Wang K, Adelstein SJ, Artignan X, Vincent F, Gauchez AS, Zhang H, et al. First human treatment of resistant neoplastic meningitis by intrathecal administration of MTX plus (125)IUdR. International Journal of Radiation Biology. 2008;84:1123–1129. doi: 10.1080/09553000802395535. [DOI] [PubMed] [Google Scholar]
- Reilly RM, Kiarash R, Cameron RG, Porlier N, Sandhu J, Hill RP, Vallis K, Hendler A, Gariépy J. 111In-labeled EGF is selectively radiotoxic to human breast cancer cells overexpressing EGFR. Journal of Nuclear Medicine. 2000;41:429–438. [PubMed] [Google Scholar]
- Sahu SK, Kassis AI, Makrigiorgos GM, Baranowska-Kortylewicz J, Adelstein SJ. The effects of indium-111 decay on pBR322 DNA. Radiation Research. 1995;141:193–198. [PubMed] [Google Scholar]
- Sastry KSR, Howell RW, Rao DV, Mylavarapu VB, Kassis AI, Adelstein SJ, Wright HA, Hamm RN, Turner JE. Dosimetry of Auger emitters: Physical and phenomenological approaches. In: Baverstock KF, Charlton DE, editors. DNA damage by Auger emitters. London: Taylor and Francis; 1988. pp. 27–38. [Google Scholar]
- Sastry KSR, Rao DV. Dosimetry of low energy electrons. In: Rao DV, Chandra R, Graham MC, editors. Physics of nuclear medicine: Recent advances. Woodbury, NY: American Institute of Physics; 1984. pp. 169–208. [Google Scholar]
- Sedelnikova OA, Panyutin IG, Luu AN, Reed MW, Licht T, Gottesman MM, Neumann RD. Targeting the human mdr1 gene by 125I-labeled triplex-forming oligonucleotides. Antisense and Nucleic Acid Drug Development. 2000;10:443–452. doi: 10.1089/oli.1.2000.10.443. [DOI] [PubMed] [Google Scholar]
- Squire CJ, Baker LJ, Clark GR, Martin RF, White J. Structures of m -iodo Hoechst-DNA complexes in crystals with reduced solvent content: Implications for minor groove binder drug design. Nucleic Acids Research. 2000;28:1252–1258. doi: 10.1093/nar/28.5.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walicka MA, Ding Y, Roy AM, Harapanhalli RS, Adelstein SJ, Kassis AI. Cytotoxicity of [125I]iodoHoechst 33342: Contribution of scavengeable effects. International Journal of Radiation Biology. 1999;75:1579–1587. doi: 10.1080/095530099139188. [DOI] [PubMed] [Google Scholar]
- Yaakob W, Gordon L, Spicer KM, Nitke SJ. The usefulness of Iodine-123 whole body scans in evaluating thyroid carcinoma and metastases. Journal of Nuclear Medicine Technology. 1999;27:279–281. [PubMed] [Google Scholar]
- Yasui LS, Chen K, Wang K, Jones TP, Caldwell J, Guse D, Kassis AI. Using Hoechst 33342 to target radioactivity to the cell nucleus. Radiation Research. 2007;167:167–175. doi: 10.1667/rr0584.1. [DOI] [PubMed] [Google Scholar]
- Yasui LS, Hughes A, DeSombre ER. DNA damage induction by 125I-estrogen. Acta Oncologica. 1996;35:841–847. doi: 10.3109/02841869609104035. [DOI] [PubMed] [Google Scholar]
- Yasui LS, Hughes A, DeSombre ER. Cytotoxicity of 125I-oestrogen decay in non- oestrogen receptor-expressing human breast cancer cells, MDA-231 and oestrogen receptor-expressing MCF-7 cells. International Journal of Radiation Biology. 2001;77:955–962. doi: 10.1080/09553000110063377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.