Abstract
Contact with macroalgae often causes coral mortality, but the roles of abrasion versus shading versus allelopathy in these interactions are rarely clear and effects on gene expression are unknown. Identification of gene expression changes within corals in response to contact with macroalgae can provide insight into the mode of action of allelochemicals, as well as reveal transcriptional strategies of the coral that mitigate damage from this competitive interaction, enabling the coral to survive. Gene expression responses of the coral Acropora millepora after long-term (20 d) direct contact with macroalgae (Chlorodesmis fastigiata, Dictyota bartayresiana, Galaxaura filamentosa and Turbinaria conoides) and short-term (1 h and 24 h) exposure to C. fastigiata thalli and their hydrophobic extract were assessed. After 20 d of exposure, T. conoides thalli elicited no significant change in visual bleaching or zooxanthellae PSII quantum yield within A. millepora nubbins, but stimulated the greatest alteration in gene expression of all treatments. Chlorodesmis fastigiata, D. bartayresiana and G. filamentosa caused significant visual bleaching of coral nubbins and reduced the PSII quantum yield of associated zooxanthellae after 20 d, but elicited fewer changes in gene expression relative to T. conoides at day 20. To evaluate initial molecular processes leading to reduction of zooxanthella PSII quantum yield, visual bleaching, and coral death, short-term exposures to C. fastigiata thalli and hydrophobic extracts were conducted; these interactions revealed protein degradation and significant changes in catalytic and metabolic activity within 24 h of contact. These molecular responses are consistent with the hypothesis that allelopathic interactions lead to alteration of signal transduction and an imbalance between reactive oxidant species production and antioxidant capabilities within the coral holobiont. This oxidative imbalance results in rapid protein degradation and eventually to apoptosis and/or necrosis when compensatory transcriptional action by the coral holobiont insufficiently mitigates damage by the allelochemicals of C. fastigiata.
Keywords: Allelopathy, algal-coral competition, gene expression, Fiji
Introduction
Coral reefs are in global decline due to climate-induced bleaching, disease, overfishing, pollution, ocean acidification, and the interaction of these and other factors (Jackson et al. 2001; Bruno et al. 2007; Hoegh-Guldberg et al. 2007; Mumby and Steneck 2008; Maliao et al. 2008). Over the last 3-4 decades, coral cover in the Caribbean has declined by 80% (Gardner et al. 2003) and along the Great Barrier Reef by 50% (Bellwood et al. 2004), with 30% of corals worldwide being at elevated risk of extinction (Carpenter et al. 2008). Significant losses of coral cover have resulted in phase shifts from coral to macroalgal dominance on many reefs. As a consequence of increases in algal biomass, remaining corals on degraded reefs experience a higher frequency of contact with macroalgae. Contact between corals and algae can result in damage to corals through abrasion, shading, overgrowth and allelopathy by macroalgae, causing partial bleaching and/or mortality, as well as reductions in coral growth rate and fecundity (McCook et al. 2001; Jompa and McCook 2002, 2003; Titlyanov et al. 2007; Foster et al. 2008; Rasher and Hay 2010; Rasher et al. 2011). Some macroalgae also negatively impact survivorship and settlement of coral larvae and increase mortality of juvenile corals, thereby limiting the recovery of corals following disturbance (Kuffner et al. 2006; Box and Mumby 2007; Birrell et al. 2008a, 2008b; Diaz-Pulido et al. 2010; Paul et al. 2011a).
Despite the potential significance of coral-algal interactions with regard to resilience and recovery of coral populations, there is little understanding of the physiological, cellular, and molecular mechanisms involved in coral-algal interactions. Allelopathy is often suggested as a mechanism by which macroalgae outcompete coral (de Nys et al. 1991; Jompa and McCook 2003; Titlyanov et al. 2007; Foster et al. 2008; Diaz-Pulido et al. 2010), but effects of algal allelopathy on corals have been demonstrated only recently (Rasher and Hay 2010; Paul et al. 2011a; Rasher et al. 2011). Macroalgae produce a variety of biologically active secondary metabolites that function in herbivore defense and as anti-fouling agents (Hay 2009; Paul et al. 2011b); however, few studies have directly linked algal secondary metabolites to algal-mediated coral tissue damage under realistic field conditions (but see de Nys et al. 1991; Rasher and Hay 2010; Paul et al. 2011a; Rasher et al. 2011).
In response to stressors, including allelopathy, organisms elicit molecular mechanisms to mitigate physiological damage by altering expression of genes involved in processes to protect cellular structures, repair damage, maintain internal homeostasis and continue normal metabolic functioning. Molecular stress responses involve altered expression of a suite of generalized stress-responding genes (e.g., some heat shock proteins), as well as genes specific to particular stressors (Thorpe et al. 2004). The nature, intensity and duration of the stressor will influence whether normal metabolism and functions (i.e., growth and reproduction) are diminished during the stress response. Analysis of differential gene expression can thus be used to evaluate cellular pathways and physiological processes differentially regulated to respond to, and potentially compensate for, exposure to the stressor.
This study utilized gene expression analysis to investigate changes in gene expression of the coral Acropora millepora when in contact with four sympatric macroalgae (Chlorodesmis fastigiata, Dictyota bartayresiana, Galaxaura filamentosa and Turbinaria conoides) whose effects on visual coral bleaching and mortality were recently evaluated (Rasher and Hay 2010; Rasher et al. 2011). Chlorodesmis fastigiata, D. bartayresiana and G. filamentosa were previously demonstrated to be allelopathic to A. millepora, resulting in visual coral bleaching, the reduction of PSII quantum yield of symbiotic zooxanthellae, and variable mortality of the coral after 20 d of direct contact with macroalgae; in contrast, T. conoides demonstrated minimal allelopathic effects (Rasher et al. 2011).
In this study, differential gene expression analyses were used to assess molecular pathways in A. millepora that were altered due to contact with these macroalgae. Molecular effects of 20 d algal-coral exposures were evaluated using A. millepora when in contact with each of four macroalgae that differed in their ability to damage corals chemically. We subsequently investigated gene expression of A. millepora after 1 h and 24 h exposures to C. fastigiata thalli and hydrophobic extracts because this alga produced the greatest effect on visual coral bleaching, most strongly suppressed PSII quantum yield of symbiotic zooxanthellae, and caused considerable mortality in this coral (Rasher et al. 2011). This is the first investigation into differential gene expression of coral in response to chronic contact with macroalgae and, although changes in gene expression do not necessarily translate to alterations in protein activity and effects on fitness (Feder and Walser 2005), this analysis provides insight into both the transcriptional responses of corals to different algal allelochemicals and the changes in gene expression that may allow the coral holobiont to mitigate damage from this stress.
Materials and Methods
Coral-algal contact experiments
The experimental design of the coral-algal contact manipulation is described in Rasher et al. (2011). In short, C. fastigiata, D. bartayresiana, G. filamentosa and T. conoides individuals of sizes representative of the local habitat were each inserted between strands of rope and attached 1-2 cm from an A. millepora nubbin (6-8 cm height) mounted in a cement cone (n = 10-12 for each algal species; Fig. 1). These manipulations allowed for algal movement and coral-algal contact representative of natural interactions. Experimental coral nubbins were allowed to acclimate for seven weeks after fragmentation prior to the start of the experiment. Control corals (n=12) possessing a rope but lacking macroalgae were also deployed to control for effects of the rope or other environmental effects that could erroneously be attributed to treatments. Metal racks holding coral-algal pairings were caged to exclude large herbivores, and maintained on Votua Reef, Viti Levu, Fiji (18°13.049′S, 177°42.968′E) for 20 d. On day 20, visual bleaching of each treatment and control coral was assessed by taking a photograph of the coral in the plane of algal-coral contact (and 180° from this plane, 5-10mm away from contact) and evaluating % 2-D area of the coral bleached using image analysis software (ImageJ v. 1.40g, NIH). To complement assessments of visual bleaching, in situ pulse-amplitude-modulated (PAM) fluorometry was also used in situ to quantify the effects of macroalgal contact on PSII quantum yield of symbiotic zooxanthellae within A. millepora (see below for detail). After exposure to macroalgae, portions of coral nubbins in contact with algae were preserved in Trizol (Invitrogen) and frozen until RNA extraction. Because some macroalgae caused whole-nubbin mortality of some A. millepora replicates (see Fig. 2a), only corals with living tissue at the algal contact site were included in the genetic analysis.
Figure 1.
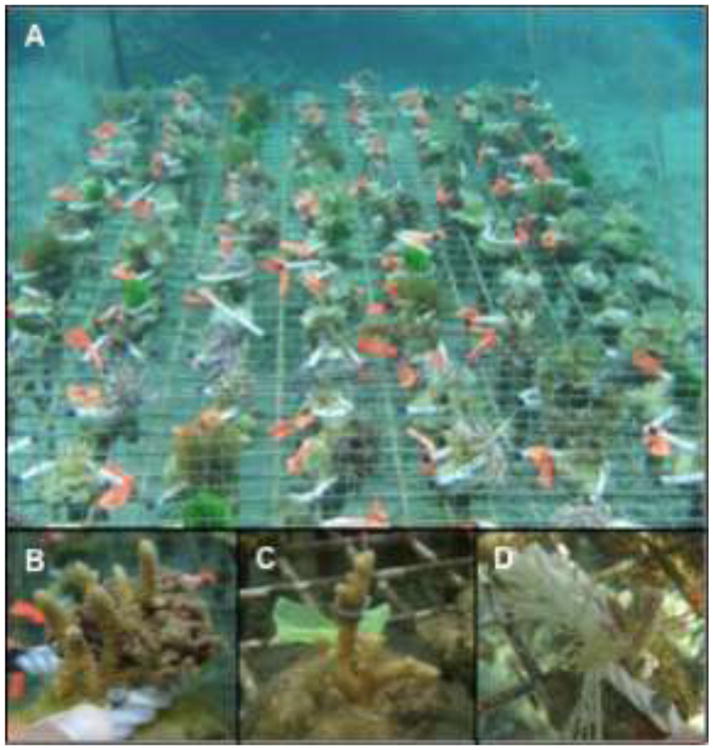
Experimental set-up in Votua village, Fiji. (a) A rack holding coral-algal pairings. (b-d) The coral A. millepora in contact with C. fastigiata (b), the hydrophobic extracts of C. fastigiata (c), or an algal mimic (d).
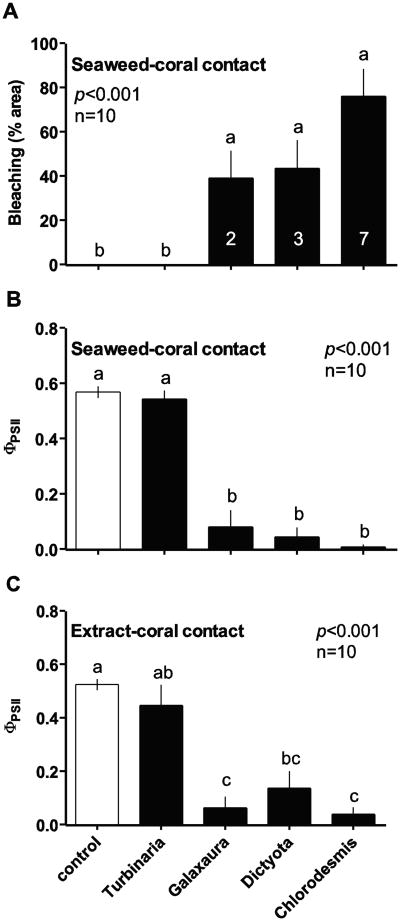
Figure 2.
Effects of contact with macroalgal thalli or hydrophobic extracts on Acropora millepora physiology. (a) Visual bleaching (2-D % area; mean ± SE) of A. millepora and (b-c) PSII quantum yield (ΦPSII; mean ± SE) of zooxanthellae within A. millepora when in contact with algal thalli for 20 d (a-b) or in contact with gel strips containing hydrophobic extracts from the same algae for 24 h (c), relative to controls. Evaluated by Kruskal-Wallis analyses of variance on ranks. Letters indicate significant groupings by Tukey's HSD post-hoc tests. Numbers inset within bars in (a) indicate number of replicates experiencing 100% mortality. Modified from Rasher et al. (2011).
This experimental design was subsequently repeated to assess initial gene expression responses of A. millepora following 1 h and 24 h exposures to C. fastigiata thalli (the most damaging of the four macroalgae in terms of bleaching and mortality). Supplemental experiments conducted in Rasher et al. (2011) demonstrated that inert algal mimics of C. fastigiata had undetectable effects on A. millepora, but we included identical C. fastigiata mimics in these short-term exposures to control for rapid physiological and gene expression responses of coral simply due to physical shading and/or abrasion. Thus, these short-term experiments included both rope controls (as above) and algal mimic controls (Fig. 1). In situ PAM fluorometry and tissue preservation were carried out using identical protocols as in the 20 d exposure experiment.
Coral-algal extract contact experiments
To assess whether allelochemicals from C. fastigiata, D. bartayresiana, G. filamentosa, and T. conoides might explain the effects of whole algal thalli on A. millepora after 20 d, we generated hydrophobic extracts of each alga and tested the allelopathic activities of these extracts on A. millepora, as described in Rasher et al. (2011). In short, each species of algae was extracted in 100% methanol and each extract was partitioned between water and ethyl acetate. The hydrophobic (ethyl acetate) fraction of each extract was retained and dried en vacuo. Hydrophobic extracts were then resuspended in methanol and embedded at natural volumetric concentration into a series of 1 cm2 Phytagel squares (Sigma-Aldrich) hardened on window-screen backing (n = 10 per extract). Control Phytagel squares contained solvent, but lacked algal extract (n = 10). Treatment and control gels were wrapped and cable-tied onto A. millepora nubbins (6-8 cm height) at mid-height on the nubbin and interspersed on an un-caged rack on Votua reef. After 24 h, Phytagel squares were removed and a single PAM fluorometry reading was taken under the center of each square (using identical protocol as for the 20d exposures using whole thalli, discussed below).
The preparation of C. fastigiata hydrophobic extracts and their application onto A. millepora for short-term extract exposure experiments were carried out as described above. Extracts embedded at natural concentration in Phytagel squares were applied to A. millepora nubbins (6-8 cm height) for 1 h or 24 h (n=10 per treatment). Gene expression responses of corals in contact with extracts were compared to control corals in contact with Phytagel squares containing solvent alone (n=10). The impacts of treatments and controls on coral photophysiology (i.e, PSII quantum yield) could not be assessed for these short-term extract experiments due to equipment failure at our remote field site. Coral tissue in contact with C. fastigiata extracts for 1 h and 24 h was preserved in Trizol and frozen until RNA extraction.
Pulse-Amplitude-Modulated (PAM) fluorometry
We used in situ PAM fluorometry (Diving PAM; Walz, Germany) to assess photosystem II (PSII) effective quantum yield (ΦPSII) of the symbiotic microalgae (zooxanthellae) living within A. millepora nubbins following 20 d, 24 h and 1 h exposure studies. PAM fluorometry is used to measure PSII quantum yield of in hospite zooxanthellae in an effort to understand the effects of biotic or abiotic stressors on the coral holobiont (Smith et al. 2006; Pawlik et al. 2007; Rasher et al. 2011), as well as the processes leading to coral bleaching (Warner et al. 1999, 2010; Fitt et al. 2001). While PAM fluorometry does not quantify coral bleaching per se, PAM readings are highly correlated with our assessments of visual bleaching for A. millepora and other co-occurring corals at this study site (Rasher and Hay 2010; Rasher et al. 2011). Single light-adapted measurements of ΦPSII were taken at the most damaged location on treatment corals in the plane of algal-coral contact (but excluding extremities), and at the same height on the opposite side (180°) of the coral, 5-10mm away from algal contact. Control corals were sampled using identical protocol, targeting the most damaged location but excluding extremities. All measurements were taken using the Walz Miniature Fiberoptic (DIVING-F1, diameter 2mm), held 4-5 mm from the sample and perpendicular to the coral surface. Care was taken to avoid self-shading.
Readings for all experiments were taken between 0900-1400 hrs and sampling of treatments and controls was interspersed through time so that measurements for a treatment would not be confounded by in situ diurnal changes in non-photochemical quenching (e.g., temperature and UV) throughout the sampling period. The sampling period for each experiment (exposure time) took no longer than 90 min. Although zooxanthellae and chl-a counts were not performed, the nubbins used in our studies were all fragmented from colonies in close proximity to our experimental rack, thus being collected from the same depth and environmental condition as that of our sampling area. Corals nubbins were (a) allowed to acclimate for 4-7 weeks prior to experiments, and (b) interspersed among controls and treatments in an effort to minimize and homogenize any initial variance in zooxanthellae density and diversity among nubbins at the beginning of our study.
Total RNA isolation
Coral tissue was scraped from the calcium carbonate skeleton where algal or extract contact had occurred and total RNA was extracted according to the manufacturer's protocol for RNA Isolation using Trizol (Invitrogen). Total RNA was purified using RNeasy MiniElute Clean-up kit (Qiagen) and RNA pellets were resuspended in nuclease-free water. RNA concentration and quality were assessed using a NanoDrop ND-1000 spectrophotometer (ThermoScientific). RNA integrity was confirmed using the RNA 6000 Nano kit and Bioanalyzer 2100 (Agilent Technologies).
Microarray platforms
The 20 d exposure study used custom microarrays (Combimatrix) specifically designed to detect expression of 148 anthozoan holobiont genes (host and zooxanthella symbiont; Electronic Supplementary Material, ESM Table 1) related to physiological stress, in addition to genes with functions involved in metabolism, development and regulation of programmed cell death (Morgan et al. 2001; Morgan and Snell 2002; Edge et al. 2005; Edge 2007). Anthozoan (primarily genes from Acropora and Montastraea species) and zooxanthella genes selected for this array have displayed differential expression under a variety of stressful conditions (Morgan et al. 2001; Morgan and Snell 2002; Edge et al. 2005) or were identified from public databases using a bioinformatics approach and categorized by function based on published literature and the Gene Ontology database (Ashburner et al. 2000; ESM Table 1). Multiple 35-40-mer probes (2-5 probes) for each gene were designed and replicated on the microarray three times in addition to positive (spike in) and negative controls. For each treatment and control, total RNA (5 ng) from 4-5 replicate corals with appropriate technical replicates (n=6) was reverse transcribed into cDNA, fluorescently labeled with amine-reactive dye using the ARES Alexa Fluor 546 DNA Labeling Kit (Invitrogen) and hybridized according to Edge (2007). Microarrays were scanned using a Microarray Express scanner (Perkin Elmer) to detect the intensity of the hybridized cDNA. Intensity data were extracted from the microarrays using Microarray Imager software (Combimatrix) and statistically analyzed using JMP Genomics 3 (SAS Institute).
For analysis of 1 h and 24 h C. fastigiata thallus and extract exposure experiments, an expanded coral holobiont array was used, which included 820 genes (769 coral host, 51 zoox; ESM Table 2) in order to survey a larger portion of the genomes including more functional groups. New genes were acquired from bioinformatic mining of recent transcriptome sequencing projects and new gene submissions into public databases (Genbank). Open reading frames from candidate genes were blasted in GenBank to confirm putative gene identity. Two 60-mer probes for each gene were designed from open reading frames using eArray (Agilent Technologies) and replicated eight times on the microarray in addition to positive (spike in) and negative controls. RNA (5 ng) from 5-6 biological replicates and appropriate technical replicates (n=6) was prepared and labeled using the One-Color Microarray-Based Gene Expression Analysis kit (Agilent Technologies). Microarray hybridization followed manufacturer's protocol and arrays were scanned using Agilent G2505C Microarray Scanner and Feature Extraction Software 10.7.1.1 (Morehouse School of Medicine, Atlanta, GA). Intensity data were statistically analyzed using JMP Genomics 3 (SAS Institute).
Statistical analysis
Visual coral bleaching and PAM fluorometry data from our long- and short-term algal-coral contact experiments were each evaluated with Kruskal-Wallis ANOVA on ranks, followed by Tukey's HSD or Dunn's post-hoc tests. Binomial coral mortality data were each evaluated with Fisher's exact tests. Raw genomic data were log2 transformed and loess normalized, background intensity was subtracted from each feature and replicate probes for each gene were averaged. Hierarchical clustering determined relationships among gene expression patterns of treatments (algal/extract contact) and control (rope contact or no-extract Phytagel square). Multivariate analysis of variance (MANOVA) was used to detect highly significant expression differences between control and treatment corals for all pairwise comparisons (p<0.01). Significance levels were adjusted using a false discovery rate correction to control for multiple testing (Benjamini and Hochberg 1995). Gene ontology (GO) terms were identified and analyzed using Blast2Go v2 (Conesa et al. 2005). Differential distribution of gene ontologies of differentially expressed genes for each treatment, compared to all genes represented on the arrays, were determined using Fisher's exact test in Blast2Go.
Results
Long-term exposure
Visual bleaching, reduction of zooxanthellae PSII quantum yield, and mortality of A. millepora nubbins varied as a function of contact with common macroalgae (Fig. 1; Rasher et al. 2011). When in direct contact for 20 d, C. fastigiata, D. bartayresiana and G. filamentosa caused significant visual bleaching (Fig. 2a) and a reduction of zooxanthellae PSII quantum yield (ΦPSII) within A. millepora nubbins (Fig. 2b) relative to controls. Negative effects of macroalgae on A. millepora were limited to sites of direct contact, with the exception of C. fastigiata, which caused significant visual bleaching of A. millepora 5-10 mm away from direct contact (Rasher et al. 2011) and significant whole-nubbin mortality (Fisher's exact test: p=0.003, Rasher et al. 2011). In contrast T. conoides contact did not elicit visual bleaching or a reduction of zooxanthellae ΦPSII within A. millepora nubbins (Figure 2a, b). After 24 hr, effects of algal hydrophobic extracts on zooxanthellae ΦPSII within A. millepora mirrored the effects of whole thalli after 20 d (Fig. 2c), demonstrating a role of allelopathy in these interactions.
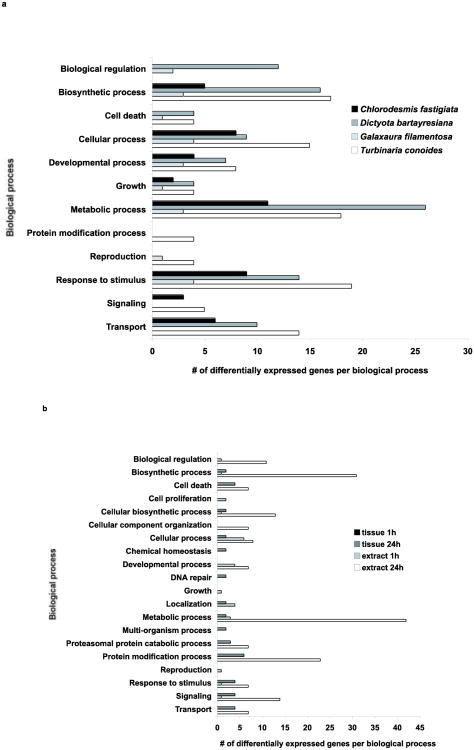
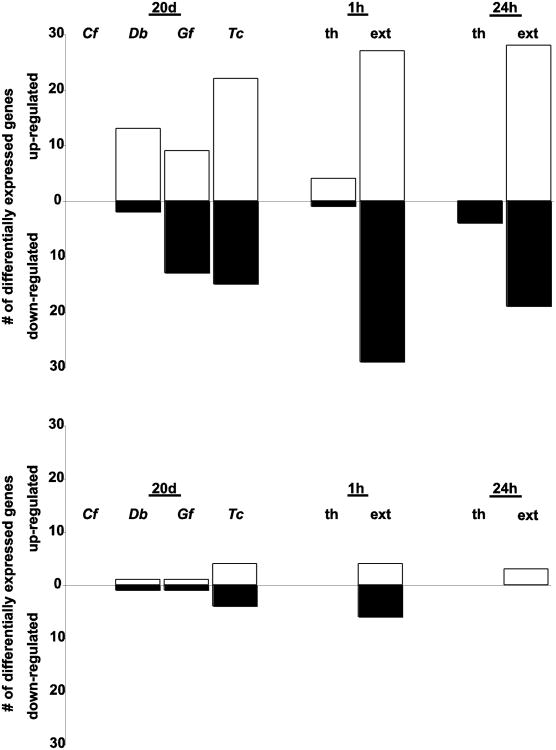
Relative to control corals with ropes but lacking macroalgae, 86 of the 148 coral host and zooxanthella genes were differentially expressed (at p<0.01) after direct contact with thalli of at least one of the four macroalgae (Table 1). Most affected genes were related to biosynthesis, cellular processes, metabolism and responses to stimuli (Fig. 3a). After 20 days of contact, T. conoides elicited the highest number of responsive genes in the A. millepora host (57 genes) and zooxanthellae (10 genes), while nine genes (all host genes) were differentially expressed in corals exposed to G. filamentosa relative to control expression levels at p<0.01 (Table 1; Fig. 4). D. bartayresiana and C. fastigiata also elicited differential expression in both coral and zooxanthella genes. Thirteen coral genes demonstrated consistent responses in at least three of the four algal treatments (Table 1), including multiple up-regulated genes involved in responses to damaged proteins (heat shock proteins and protein modification and transport genes). There was no significant differential distribution of specific GO terms represented after exposure to any macroalga for 20 d (Fisher's exact test, p>0.05).
Table 1.
Acropora millepora holobiont transcripts significantly (p=0.01) affected by allelopathic interaction with Chlorodesmis fastigiata (Chl), Dictyota bartayresiana (Dict), Galaxaura filamentosa (Gal) or Turbinaria conoides (Turb) thalli for 20 d (A) or with Chlorodesmis fastigiata thalli or extract for 1 h or 24 h (B). Positive expression fold changes indicate up-regulation after exposure and negative fold changes indicate down-regulation of mRNA transcripts relative to controls.
| Gene Name | Genbank Accession # | Chlorodesmis fastigiata | Dictyota bartayresian | |
|---|---|---|---|---|
| 28 kDa heat- and acid-stable phosphoprotein | EZ036134 | |||
| acidic glucanase | XM_002165050 | |||
| actin | AY360081 | -1.58 | ||
| actin (zooxanthella) | AB086828 | -0.45 | ||
| alpha-tubulin | L13999 | |||
| ATP-binding cassette, sub-family B, member 1 | DY447310 | |||
| bcl-2 apoptosis regulator | CV181079 | |||
| beta-tubulin | U60604 | |||
| beta-tubulin (zooxanthellae) | K03281 | -1.03 | ||
| calmodulin | AF507185 | -2.12 | -1.90 | |
| calreticulin | EZ022703 | |||
| caspase | DQ218058 | 1.98 | ||
| catalase | DQ104435 | -1.34 | ||
| cathepsin B | DT608217 | |||
| charged multivesicular body protein 2a | EZ009246 | 1.42 | 1.90 | |
| cluster of differentiation 36 scavenger receptor | DQ309525 | |||
| collagen alpha-1 chain, type XII-like | DQ309546 | |||
| cytochrome b | AB117374 | -1.08 | 2.40 | |
| cytochrome oxidase subunit I | AF013738 | -1.75 | ||
| DNA-3-methyladenine glycosylase | EZ033943 | -1.54 | ||
| F16P protein | AJ251054 | |||
| ferredoxin (zooxanthella) | EF134003 | 1.89 | ||
| ferritin heavy chain polypeptide 1 | AY130389 | 1.55 | ||
| ferrochelatase | NM_128592 | -1.50 | ||
| frizzled-8 | EZ019620 | |||
| glutamate carboxypeptidase | DQ309521 | |||
| glutaredoxin | EZ030451 | |||
| glutathione-s-transferase | EZ024210 | |||
| glyceraldehyde-3-phosphate dehydrogenase (zooxanthella) | AB106689 | 3.07 | ||
| glycerol 3-phosphate dehydrogenase | AM902265 | |||
| glycine amidinotransferase | EZ002467 | -2.29 | ||
| green fluorescent protein | AY181557 | |||
| heat shock 70kDa protein 8 | AY422994 | 1.20 | ||
| heat shock protein 16.2 | EZ020922 | 1.79 | 1.63 | |
| heat shock protein 27 | EF384631 | 3.26 | 3.34 | |
| heat shock protein 70 | AF152004 | 5.97 | ||
| heat shock protein 90 | Y17848 | 1.49 | 1.60 | |
| heat shock protein 90-alpha 1 | EZ011865 | |||
| hephaestin | EZ004840 | -3.44 | ||
| histones H3, H2B, H2A and H4 | L11067 | 1.36 | 2.07 | |
| homeodomain protein cnox-2 | AF245689 | -1.35 | -1.44 | |
| importin-5 | EZ001642 | |||
| integrin alpha-X | EZ024062 | |||
| integrin subunit | AF005356 | -1.54 | ||
| medium chain S-acyl fatty acid synthetase thio ester | DQ309537 | 1.56 | 1.96 | |
| hydrolase-like protein | ||||
| membrane-bound transcription factor site-1 protease | EZ004852 | 1.01 | ||
| metallothionein homolog | DR681654 | 1.96 | 1.44 | |
| methionine aminopeptidase 2 | EZ041485 | |||
| mini-collagen | D30747 | -1.09 | ||
| mitochondrial ATP synthase | DQ309536 | 1.65 | ||
| multidrug resistance efflux pump | DT622365 | -1.48 | ||
| NADH dehydrogenase | EZ011273 | |||
| NADH dehydrogenase subunit 1 gene | DQ351254 | -1.83 | ||
| peptidyl-prolyl cis-trans isomerase, cyclophilin-type | XM_001698484 | -2.09 | ||
| peridinin chlorophyll-a binding protein (zooxanthella) | AY149139 | 2.21 | ||
| peridinin chlorophyll-a binding protein apoprotein precursor (zooxanthella) | AY149170 | 1.92 | ||
| photosystem II protein D, psbA (zooxanthella) | AJ884906 | |||
| polyubiquitin (zooxanthella) | NM 001064913 | -1.33 | ||
| profilin | EZ023730 | 1.36 | 1.81 | |
| protein disulfide-isomerase A3 | EZ045810 | 2.01 | ||
| protein-tyrosine kinase (FAK) | AY841903 | |||
| ribophorin | CN629960 | -1.63 | ||
| ribosome-binding protein 1 | EZ012511 | |||
| ribulose 1,5-bisphosphate carboxylase oxygenase large subunit precursor (zooxanthella) | AF298221 | |||
| ribulose 1;5-bisphophate carboxylase oxygenase (zooxanthella) | AF299359 | -0.57 | -0.61 | |
| serine/threonine-protein kinase RIO3 | EZ015988 | |||
| spec1 | J01207 | |||
| spondin1 | DT620213 | |||
| superoxide dismutase (copper/zinc) | DQ309550 | -5.21 | 2.91 | |
| tartrate-resistant acid phosphatase type 5 | EZ020317 | -1.75 | -1.43 | |
| thioredoxin | Y17147 | 3.42 | 3.75 | |
| thymosin beta-10 | EZ010287 | 2.72 | ||
| translocon-associated protein subunit delta | EZ039782 | -1.39 | ||
| type I inositol-3,4-bisphosphate 4-phosphatase | EZ003000 | 2.49 | 3.44 | |
| Ubiquitin | DN252355 | 2.91 | ||
| unknown gene | DQ213995 | -0.65 | ||
| unknown gene | EU747061 | -1.25 | -1.27 | |
| unknown gene | EZ041133 | |||
| unknown gene | FE039580 | 3.40 | 3.58 | |
| unknown gene | XM_001922339 | |||
| unknown gene (zooxanthella) | EU153233 | |||
| unknown gene | DY585902 | |||
| unknown gene in response to copper exposure | ||||
| unknown gene in response to copper exposure | ||||
| unknown gene in response to dark exposure | ||||
| unknown gene in response to permethrin exposure | -0.84 | |||
| unknown gene in response to xenobiotic exposure | BI534459 | |||
| unknown gene in response to xenobiotic exposure | EZ016042 | |||
| UPF2 regulator of nonsense transcripts | EZ001321 | |||
| vasa-related protein | AB048853 | -0.91 | ||
| vitellogenin II precursor | CO539736 | -1.79 | ||
Figure 3.
Distribution of differentially expressed genes across biological process gene ontology according to Blast2Go after exposure to macroalgae for 20 d (a) or to Chlorodesmis fastigiata thalli or extracts for 1 h or 24 h (b). Different biological processes are included on each figure since two different array platforms were used for the long-term and short-term experiments.
Figure 4.
Transcriptional regulation of Acropora millepora (a) and associated zooxanthella (b) genes for all treatments. White bars indicate up-regulation of genes in treated corals relative to controls. Black bars indicate down-regulation of genes in treated corals relative to controls. Chlorodesmis fastigiata (Cf), Dictyota bartayresiana (Db), Galaxaura filamentosa (Gf), Turbinaria conoides (Tc).
Short-term Chlorodesmis fastigiata thalli exposures
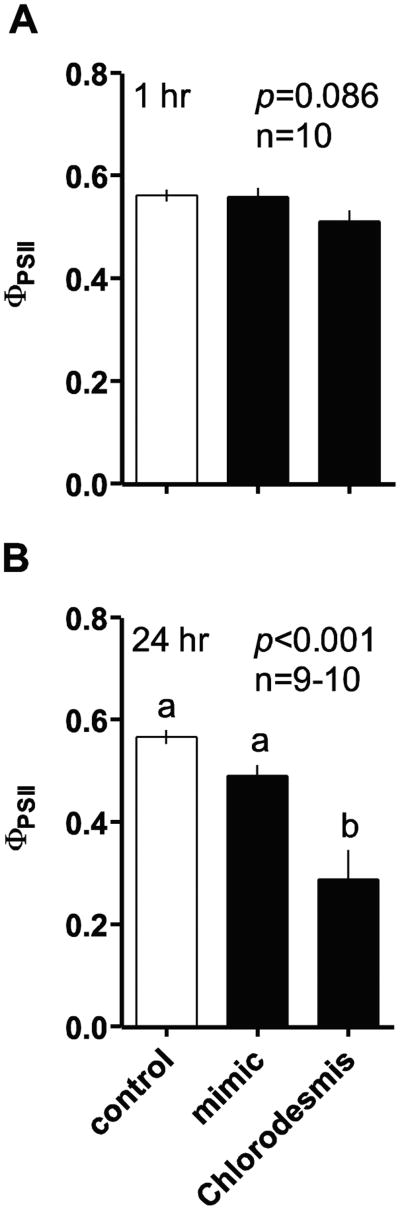
When placed in contact with A. millepora for 1 h, thalli of C. fastigiata produced no detectable effect on zooxanthellae ΦPSII within coral nubbins relative to controls (Fig. 5a); however, after 24 h of exposure to C. fastigiata, zooxanthellae ΦPSII was significantly reduced within A. millepora at the area of direct contact, relative to both controls with ropes and control algal mimics (Fig. 5b; p<0.001).
Figure 5.
Short-term effects of allelopathic macroalgal contact on Acropora millepora physiology. PSII quantum yield (ΦPSII; mean ± SE) of zooxanthellae within A. millepora when in contact with the allelopathic alga Chlorodesmis fastigiata for 1 h (a) or 24 h (b), relative to controls. Analyzed by Kruskal-Wallis Analyses of Variance on ranks. Letters in (b) indicate significant groupings by a Dunn's post-hoc test.
Corals contacting inert algal mimics (controls for shading and abrasion) did not demonstrate differential gene expression relative to rope-only controls after 1h or 24h of exposure, indicating that shading and abrasion did not elicit significant gene expression responses over this time scale; therefore, algal mimic controls were used as controls for gene expression comparisons in our short-term assays involving C. fastigiata. After 1 h of exposure to C. fastigiata thalli, expression of the 820 A. millepora host and zooxanthella genes were unchanged relative to algal mimic controls (Table 1). After 24 h exposures, 15 coral and two zooxanthella genes had altered expression levels (Table 1; Fig. 4), with the majority of the up-regulated genes involved in processes of protein modification (Fig. 3b), the 26S proteasome intracellular degradation system in particular (Table 1). In addition, expression was also altered in genes involved in cell death, responses to stimuli, signaling and transport. There was no significant differential distribution of specific GO terms represented after exposure to this macroalga for 24 h (Fisher's exact test, p>0.05).
Short-term Chlorodesmis fastigiata extract exposures
Measurements of zooxanthellae ΦPSII within A. millepora nubbins could not be made following 1 h and 24 h exposures to C. fastigiata hydrophobic extracts due to equipment failure at this remote field site. It has been previously demonstrated that exposure to C. fastigiata extracts for 24 h causes a reduction of zooxanthellae ΦPSII within A. millepora similar in magnitude to exposure to C. fastigiata thalli for 20 d (Fig. 2c, Rasher et al. 2011). Although not quantified in this study, visual coral bleaching occurred in areas of contact with C. fastigiata extracts, but not Phytagel controls, after 24 h (TLS, pers. obs.). This visual bleaching was not obvious after 1 h of exposure to C. fastigiata extracts.
After 24 h of exposure to hydrophobic extracts from C. fastigiata, A. millepora significantly increased expression of genes involved in catalytic activity and cellular metabolic processes, including pathways involving proteins and macromolecules (Fisher's exact tests, p<0.0001; Fig. 5). Although transcriptional changes of these pathways were not significantly over-represented after 1 h exposure to these extracts, four coral host genes were down-regulated, including three signal transduction genes (Table 1). As in the 24 h C. fastigiata thalli exposure, coral host genes activated by 24 h extract exposure were involved in processes of protein degradation via the 26S proteasome intracellular degradation system. Down-regulated genes included several involved in regulation of the cell cycle, DNA repair, as well as apoptosis regulation and protein modification and transport. Zooxanthella genes responsive to C. fastigiata extracts were involved in protein degradation, mRNA processing and ribosomal synthesis.
Discussion
Direct contact between A. millepora nubbins and some macroalgae (C. fastigiata, D. bartayresiana, G. filamentosa) resulted in visual coral bleaching, reductions of zooxanthellae ΦPSII, and apparent tissue mortality at the point of contact after 20 days. Bleaching is a reduction in coral colony pigmentation due to loss of symbiotic dinoflagellates (zooxanthellae) from coral cells and/or to loss of photosynthetic pigments from the zooxanthellae (Hoegh-Guldberg and Smith 1989; Kleppel et al. 1989). Coral bleaching most commonly results from thermal stress, high intensity light or ultraviolet radiation exposure, salinity fluctuations, increases in sedimentation or pollution, or microbial infection (Steen and Muscatine 1987; Hoegh-Guldberg and Smith 1989; Glynn and D'Croz 1990; Lesser et al. 1990; Glynn 1991; Kushmaro et al. 1996, 1997, 1998, 2001; Ben-Haim et al. 2002, 2003; Lesser 2006). Although the events and processes leading to the breakdown of the coral-zooxanthella symbiosis remain under investigation, mounting evidence indicates that thermal stress and high solar radiation damage the photosynthetic apparatus of the zooxanthella (Venn et al. 2008) and mitochondria of the host (Dykens et al. 1992; Nii and Muscatine 1997), resulting in increased reactive oxygen species (ROS) within host cells (Lesser 2006; Weis 2008). Both partners of the coral-zooxanthella symbiosis have adaptive mechanisms to tolerate a range of environmental stresses (Douglas 2003; Venn et al. 2008), however increasingly high concentrations of ROS due to photochemical or respiratory damage overwhelm the antioxidant defense systems of the symbiont and host (Lesser 1996; Franklin et al. 2004; Lesser and Farrell 2004; Mydlarz and Jacobs 2004) and destabilize the symbiosis resulting in bleaching via apoptosis (Dunn et al. 2007; Ainsworth and Hoegh-Guldberg 2008) or another mechanism (Weis 2008).
Increased ROS production, however, is also an important evolutionarily conserved innate immune response to acute stress that mobilizes intracellular defenses, and does not necessarily indicate dysfunction of mitochondrial or photosynthetic machinery. Rapid and transient increases in ROS production, which may peak within several minutes after exposure to the stressor with levels returning to baseline within 30 minutes (Finkel 1999), act as stress signaling mechanisms (Hensley et al. 2000) that activate additional signal transduction pathways and transcription factors, potentially triggering an apoptotic cascade. Typically, intracellular stress signaling pathways, including ROS bursts, are activated in response to a stressor, such as exposure to allelochemicals, followed by alteration of transcription factors, which regulate the cellular responses to the stress (Martindale and Holbrook 2002).
In this study, differential expression of genes involved in coral signaling pathways (bone morphogenic protein receptor 1, interferon-related developmental regulator) suggest the primary initial response (1 h) of A. millepora to exposure to C. fastigiata extracts was an alteration of signal transduction pathways, the process in which extracellular signals are intracellularly communicated to elicit cellular responses. By 24 h of exposure to extracts, different signaling genes were down-regulated, as were several genes involved in regulation of the cell cycle (cell division protein kinase 6, cyclin-dependent kinase 4, src tyrosine kinase 3, structural maintenance of chromosomes 3). Cell cycle suppression or arrest is a common response to stress, and may be a direct target of allelochemicals or may be an indirect effect after a cascade of signaling processes have elicited a physiological response (Peres et al. 2007; Zhang et al 2010).
Within 24 h of exposure to C. fastigiata, protein degradation was enhanced in A. millepora via the ATP-dependent 26S proteasome proteolytic pathway, which degrades and removes damaged or mis-folded proteins from cells. If accumulation of damaged proteins exceeds the cell's proteolytic capacity, protein aggregates will interfere with normal cellular functions leading to induction of apoptosis (Meriin et al. 1999; Sherman and Goldberg 2001). The up-regulation of molecular chaperones involved in refolding damaged proteins (heat shock protein 90) further supports the evidence for protein damage. Although protein degradation is a normal process in eukaryote cells, the rapid up-regulation of the protein degradation system within 24 h of exposure to apparently toxic compounds, indicates increased damage to proteins, possibly due to errors in transcription, translation or post-synthetic alterations of proteins such as those due to ROS exposure (Berlett and Stadtman 1997).
Up-regulation of antioxidant activity (copper/zinc superoxide dismutase, oxidoreductase NAD-binding domain, selenium binding protein) was initiated within 24 h of exposure to C. fastigiata thalli and extracts. This relatively rapid response is unlike oxidative stress responses due to high solar radiation in which gene regulation of genes involved in antioxidant activity were not transcriptionally altered by short-term (36 h) exposures to light and thermal stress (Michelmore et al. 2002; Rodriguez-Lanetty et al. 2009; Starcevic et al. 2010), but several days of exposure resulted in transcriptional activation of these enzymes (Csaszar et al. 2009; Fitt et al. 2009). In this study, all four macroalgal species triggered antioxidant defense gene expression (copper/zinc superoxide dismutase, ferredoxin, ferritin, metallothionein, thioredoxin) with 20 d exposures.
Expression of symbiont genes were also altered in response to C. fastigiata extracts by 24 h as demonstrated by increased expression of genes involved in GTP binding, protein degradation and RNA splicing. Although photosynthesis-associated genes were not significantly altered after 24 h exposure to C. fastigiata thalli, a significant reduction (p<0.001) in zooxanthellae ΦPSII within A. millepora was observed.
Chronic contact (20 d exposure) with three (C. fastigiata, D. bartayresiana and G. filamentosa) of the four macroalgae tested also resulted in differential expression in coral host genes associated with cytoskeletal organization, cell adhesion and cell signaling. Expression of zooxanthella genes involved in cytoskeletal organization, photosynthesis and antioxidant activity was altered by C. fastigiata and D. bartayresiana. These responses are consistent with an imbalance between cellular oxidant species production and antioxidant capabilities of the coral holobiont either due to disruption of the photosynthetic machinery of the zooxanthella, the mitochondria of the host, or both. Increases in ROS elicit modifications in a variety of cellular processes and may generate functional and structural damage to proteins, DNA and lipids sufficient to induce apoptosis, necrosis or other mechanisms (Weis 2008) in the coral host that resulted in bleaching observed after 20 d of chronic exposure to C. fastigiata, D. bartayresiana and G. filamentosa.
Gene expression patterns in A. millepora in contact with T. conoides for 20 d were consistent with ROS-induced apoptosis. In addition to similar patterns of transcriptional changes in coral and zooxanthella genes observed in the C. fastigiata, D. bartayresiana and G. filamentosa experiments, gene expression analysis after exposure to T. conoides thalli indicated that free oxygen radicals reduced expression of a cell death suppressor gene (Bcl-2 apoptotic repressor) enhancing the apoptotic response (caspase activation; Mates et al. 2008) of A. millepora, enabling damaged cells to be quickly removed and regenerated, as are damaged cells in Hydra (Augustin and Bosch 2011). Unlike exposures to C. fastigiata, D. bartayresiana and G. filamentosa, A. millepora was able to compensate for damage due to contact with T. conoides as evidenced by the lack of visual bleaching and impacts on symbiont photophysiology, perhaps as a consequence of the induction of the apoptotic pathway in response to contact with T. conoides.
Some naturally produced compounds trigger a cascade of oxidative stress responses leading to cell death. Plant triterpenes, oleanolic acid and maslinic acid are apparent stressors to some human cancer cells causing increased ROS accumulation, rearrangement of cytoskeleton organization and other signatures of oxidative stress leading to apoptosis (Martin et al. 2007). An allelochemical ((-)-catechin) of an invasive plant inhibits growth and germination of susceptible competitors by inducing a rapid increase in ROS, induction of oxidative stress-related genes, changes in cell morphology and death of root cells (Bais et al. 2003). Similarly, chemicals (gallic acid) from root exudates of an invasive marsh plant exhibit allelopathic activity by inducing elevated levels of ROS in root cells of competitors leading to microtubule disruption and root death (Rudrappa et al. 2007). These examples demonstrate cellular processes similar to those inferred by gene expression analysis in this study, and support the hypothesis that allelochemicals produced by some macroalgae (such as the terpenoids identified in Rasher et al. (2011) as allelopathic agents against corals) may trigger a cascade of oxidative stress responses leading to cell death if the coral holobiont is not capable of compensating at a molecular level for the effects of these allelochemicals.
Different macroalgae elicited different responses in coral holobiont gene expression. Variability in allelochemical presence, composition and concentration likely account for differences observed in molecular and physiological responses in A. millepora nubbins, because physiological responses to contact with macroalgae could be replicated by responses to their hydrophobic extracts alone (Fig. 2c; Rasher et al. 2011). In addition, cells vary in resistance to oxidative stress depending on the nature of the ROS (e.g., hydrogen peroxide, hydroxyl radicals or superoxide anions) generated due to the stress (Thorpe et al. 2004). Depending on the target of the allelochemicals, different ROS may be produced after exposure to different algal species resulting in variation in molecular and cellular responses of the coral. Coral exposed to T. conoides demonstrated no evidence of bleaching or damage to coral tissue after 20 d of exposure, and simultaneously demonstrated the greatest levels of differential gene expression in stress-responsive and structural genes. It appears that changes in gene expression within the A. millepora holobiont were capable of compensating for exposure to T. conoides thalli for at least 20 d. This compensation results in coral survival and normal zooxanthellae PSII functioning within the coral; however, the molecular responses could be energetically costly and require resource allocation away from coral growth and reproduction. In contrast molecular responses to the other three macroalgae did not adequately compensate for the effects of the alga's allelopathic metabolites, and so corals in contact with C. fastigiata, G. filamentosa, and D. bartayresiana suffered direct damage as evidenced by visual bleaching, reduced zooxanthellae PSII function and mortality (Rasher et al. 2011).
Despite exhibiting a significant effect on visual bleaching and symbiont ΦPSII, G. filamentosa elicited a modest genetic response in A. millepora. If our conservative threshold of including only highly significant differences (p<0.01) in expression between treatments and controls was relaxed to p<0.05, A. millepora exposed to G. filamentosa thalli exhibited differential expression in 20 coral genes (Table 1), 14 of which were similarly expressed in other coral-algal interactions. Additional genes involved in protein modification and transport, cytoskeletal structure and responses to stimuli were differentially expressed at p<0.05. Allelopathic effects of G. filamentosa extracts on A. millepora may have resulted in A. millepora tissue death (rather than bleaching) at the point of contact, but tissue integrity was not verified in these experiments. Since tissue removal from the skeleton during RNA extraction included the point of extract contact and its border with apparently healthy tissue, the proportion of RNA from the healthy boundary area would be higher than the RNA from the area of direct contact if that tissue was dead. The resulting gene expression analysis would reflect similar expression as healthy controls, which possibly masked, almost entirely, genetic expression signals from cells affected by the extracts.
Although quantitative analysis of specific protein activities will be necessary to verify the relationship between gene expression and effects on individual fitness, our results from transcriptome analysis suggest that allelopathic compounds in C. fastigiata, and potentially those from other damaging macroalgae, initially alter signal transduction pathways through induction of innate immune responses in corals and cause protein damage, possibly due to oxidization, with subsequent accumulation and removal of non-functioning proteins within 24 h of exposure. Chronic exposure to these allelopathic compounds can result in cytoskeletal damage and disruption of the coral/zooxanthella symbiosis leading to visual tissue bleaching, necrosis and death if the coral cannot mitigate this damage, perhaps through apoptosis.
Supplementary Material
Figure 6.
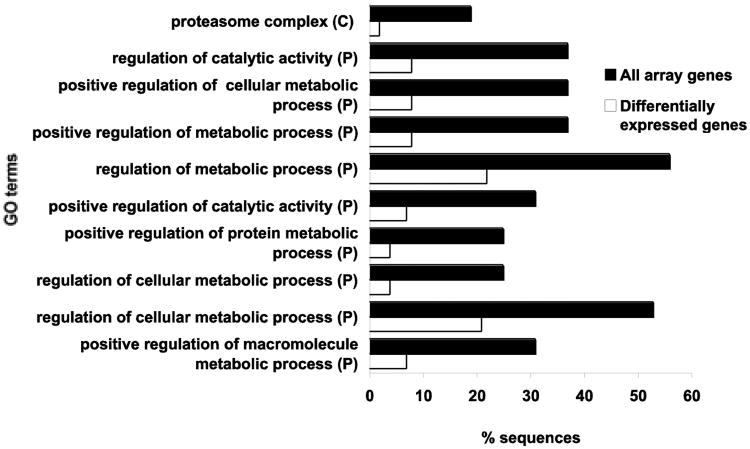
Significantly over-represented cellular components (C) and biological processes (P) for differentially expressed genes in Acropora millepora fragments exposed to Chlorodesmis fastigiata hydrophobic extracts for 24 h, based on Fisher's exact tests with multiple testing corrections for all Gene Ontology terms.
Acknowledgments
We thank the Fijian government for collection and research permits, the Votua Village elders for local research permissions and the University of South Pacific for logistical support. V Bonito, C Carter, S Engel and I Markham provided field assistance. SE Edge provided assistance with gene expression analysis and comments on the manuscript. Support was provided by the National Science Foundation (DGE 0114400 and OCE 0929119), the National Institutes of Health (U01 TW007401-01), and the Teasley Endowment to the Georgia Institute of Technology. The Morehouse School of Medicine microarray facility is supported by the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) (G12-RR03034).
References
- Ainsworth TD, Hoegh-Guldberg O. Cellular processes of bleaching in the Mediterranean coral Oculina patagonica. Coral Reefs. 2008;27:593–597. doi: 10.1038/ismej.2007.88. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin R, Bosch TCG. Cnidarian immunity: A tale of two barriers. In: Söderhäll K, editor. Invertebrate immunity. Landes Bioscience and Springer Science Business Media; 2011. pp. 1–16. [Google Scholar]
- Bais HP, Vepachedu R, Gilroy S, Callaway RM, Vivanco JM. Allelopathy and exotic plant invasion: From molecules and genes to species interactions. Science. 2003;301:1377–1380. doi: 10.1126/science.1083245. [DOI] [PubMed] [Google Scholar]
- Bellwood DR, Hughes TP, Folke C, Nystrom M. Confronting the coral reef crisis. Nature. 2004;429:827–833. doi: 10.1038/nature02691. [DOI] [PubMed] [Google Scholar]
- Ben-Haim Y, Rosenberg E. A novel Vibrio sp pathogen of the coral Pocillopora damicornis. Mar Biol. 2002;141:47–55. [Google Scholar]
- Ben-Haim Y, Zicherman-Keren M, Rosenberg E. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl Environ Microbiol. 2003;69:4236–4242. doi: 10.1128/AEM.69.7.4236-4242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- Birrell CL, McCook LJ, Willis BL, Diaz-Pulido GA. Effects of benthic algae on the replenishment of corals and the implications for the resilience of coral reefs. Oceanogr Mar Biol Annu Rev. 2008a;46:25–63. [Google Scholar]
- Birrell CL, McCook LJ, Willis BL, Harrington L. Chemical effects of macroalgae on larval settlement of the broadcast spawning coral Acropora millepora. Mar Ecol Prog Ser. 2008b;362:129–137. [Google Scholar]
- Box SJ, Mumby PJ. Effect of macroalgal competition on growth and survival of juvenile Caribbean corals. Mar Ecol Prog Ser. 2007;342:139–149. [Google Scholar]
- Bruno JF, Selig ER, Casey KS, Page CA, Willis BL, Harvell CD, Sweatman H, Melendy AM. Thermal stress and coral cover as drivers of coral disease outbreaks. PLoS Biol. 2007;5:1220–1227. doi: 10.1371/journal.pbio.0050124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter KE, Abrar M, Aeby G, Aronson RB, Banks S, Bruckner A, Chiriboga A, Cortes J, Delbeek JC, DeVantier L, Edgar GJ, Edwards AJ, Fenner D, Guzman HM, Hoeksema BW, Hodgson G, Johan O, Licuanan WY, Livingstone SR, Lovell ER, Moore JA, Obura DO, Ochavillo D, Polidoro BA, Precht WF, Quibilan MC, Reboton C, Richards ZT, Rogers AD, Sanciangco J, Sheppard A, Sheppard C, Smith J, Stuart S, Turak E, Veron JEN, Wallace C, Weil E, Wood E. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science. 2008;321:560–563. doi: 10.1126/science.1159196. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol T, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Csaszar NBM, Seneca FO, van Oppen MJH. Variation in antioxidant gene expression in the scleractinian coral Acropora millepora under laboratory thermal stress. Mar Ecol Prog Ser. 2009;392:93–102. [Google Scholar]
- de Nys R, Coll JC, Price IR. Chemically mediated interactions between the red alga Plocamium hamatum (Rhodophyta) and the octocoral Sinularia cruciata (Alcyonacea) Mar Biol. 1991;108:315–320. [Google Scholar]
- Diaz-Pulido G, Harii S, McCook LJ, Hoegh-Guldberg O. The impact of benthic algae on the settlement of a reef building coral. Coral Reefs. 2010;29:203–208. [Google Scholar]
- Douglas AE. Coral bleaching - how and why? Mar Pollut Bull. 2003;46:385–392. doi: 10.1016/S0025-326X(03)00037-7. [DOI] [PubMed] [Google Scholar]
- Dunn SR, Schnitzler CE, Weis VM. Apoptosis and autophagy as mechanisms of dinoflagellate symbiont release during cnidarian bleaching: Every which way you lose. Proc R Soc Lond Biol. 2007;274:3079–3085. doi: 10.1098/rspb.2007.0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykens JA, Shick JM, Benoit C, Buettner GR, Winston GW. Oxygen radical production in the sea anemone Anthopleura elegantissima and its endosymbiotic algae. J Exp Biol. 1992;168:219–241. [Google Scholar]
- Edge SE. Ph D dissertation. Georgia Institute of Technology; 2007. Using microarrays to quantify stress responses in natural populations of coral; p. 164. [Google Scholar]
- Edge SE, Morgan MB, Gleason DF, Snell TW. Development of a coral cDNA array to examine gene expression profiles in Montastraea faveolata exposed to environmental stress. Mar Pollut Bull. 2005;51:507–523. doi: 10.1016/j.marpolbul.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Feder ME, Walser JC. The biological limitations of transcriptomics in elucidating stress and stress responses. J Evol Biol. 2005;18:901–910. doi: 10.1111/j.1420-9101.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- Finkel T. Signal transduction by reactive oxygen species in non-phagocytic cells. J Leukocyte Biol. 1999;65:337–340. doi: 10.1002/jlb.65.3.337. [DOI] [PubMed] [Google Scholar]
- Fitt WK, Brown BE, Warner ME, Dunne RP. Coral bleaching: interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs. 2001;20:51–65. [Google Scholar]
- Fitt WK, Gates RD, Hoegh-Guldberg O, Bythell JC, Jatkar A, Grottoli AG, Gomez M, Fisher P, Lajuenesse TC, Pantos O, Iglesias-Prieto R, Franklin DJ, Rodrigues LJ, Torregiani JM, van Woesik R, Lesser MP. Response of two species of Indo-Pacific corals, Porites cylindrica and Stylophora pistillata, to short-term thermal stress: The host does matter in determining the tolerance of corals to bleaching. J Exp Mar Biol Ecol. 2009;373:102–110. [Google Scholar]
- Foster NL, Box SJ, Mumby PJ. Competitive effects of macroalgae on the fecundity of the reef-building coral Montastraea annularis. Mar Ecol Prog Ser. 2008;367:143–152. [Google Scholar]
- Franklin DJ, Hoegh-Guldberg P, Jones RJ, Berges JA. Cell death and degeneration in the symbiotic dinoflagellates of the coral Stylophora pistillata during bleaching. Mar Ecol Prog Ser. 2004;272:117–130. [Google Scholar]
- Gardner TA, Cote IM, Gill JA, Grant A, Watkinson AR. Long-term region-wide declines in Caribbean corals. Science. 2003;301:958–960. doi: 10.1126/science.1086050. [DOI] [PubMed] [Google Scholar]
- Glynn PW. Coral reef bleaching in the 1980s and possible connections with global warming. TREE. 1991;6:175–177. doi: 10.1016/0169-5347(91)90208-F. [DOI] [PubMed] [Google Scholar]
- Glynn PW, D'Croz L. Experimental evidence for high temperature stress as the cause of El Nino-coincident coral mortality. Coral Reefs. 1990;8:181–191. [Google Scholar]
- Hay ME. Marine chemical ecology: Chemical signals and cues structure marine populations, communities, and ecosystems. Annu Rev Mar Sci. 2009;1:193–212. doi: 10.1146/annurev.marine.010908.163708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley K, Robinson KA, Gabbita SP, Salsman S, Floyd RA. Reactive oxygen species, cell signaling, and cell injury. Free Radical Biol Med. 2000;28:1456–1462. doi: 10.1016/s0891-5849(00)00252-5. [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg O, Smith GJ. Influence of the population density of zooxanthellae and supply of ammonium on the biomass and metabolic characteristics of the reef corals Seriatopora hystrix and Stylophora pistillata. Mar Ecol Prog Ser. 1989;57:173–186. [Google Scholar]
- Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, Harvell CD, Sale PF, Edwards AJ, Caldeira K, Knowlton N, Eakin CM, Iglesias-Prieto R, Muthiga N, Bradbury RH, Dubi A, Hatziolos ME. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318:1737–1742. doi: 10.1126/science.1152509. [DOI] [PubMed] [Google Scholar]
- Jackson JBC, Kirby MX, Berger WH, Bjorndal KA, Botsford LW, Bourque BJ, Bradbury RH, Cooke R, Erlandson J, Estes JA, Hughes TP, Kidwell S, Lange CB, Lenihan HS, Pandolfi JM, Peterson CH, Steneck RS, Tegner MJ, Warner RR. Historical overfishing and the recent collapse of coastal ecosystems. Science. 2001;293:629–638. doi: 10.1126/science.1059199. [DOI] [PubMed] [Google Scholar]
- Jompa J, McCook LJ. The effects of nutrients and herbivory on competition between a hard coral (Porites cylindrica) and a brown alga (Lobophora variegata) Limnol Oceanogr. 2002;47:527–534. [Google Scholar]
- Jompa J, McCook LJ. Coral-algal competition: macroalgae with different properties have different effects on corals. Mar Ecol Prog Ser. 2003;258:87–95. [Google Scholar]
- Kleppel GS, Dodge RE, Reese CJ. Changes in pigmentation associated with the bleaching of stony corals. Limnol Oceanogr. 1989;34:1331–1335. [Google Scholar]
- Kuffner IB, Walters LJ, Becerro MA, Paul VJ, Ritson-Williams R, Beach KS. Inhibition of coral recruitment by macroalgae and cyanobacteria. Mar Ecol Prog Ser. 2006;323:107–117. [Google Scholar]
- Kushmaro A, Loya Y, Fine M, Rosenberg E. Bacterial infection and coral bleaching. Nature. 1996;380:396–396. [Google Scholar]
- Kushmaro A, Rosenberg E, Fine M, Loya Y. Bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar Ecol Prog Ser. 1997;147:159–165. [Google Scholar]
- Kushmaro A, Rosenberg E, Fine M, Ben Haim Y, Loya Y. Effect of temperature on bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar Ecol Prog Ser. 1998;171:131–137. [Google Scholar]
- Kushmaro A, Banin E, Loya Y, Stackebrandt E, Rosenberg E. Vibrio shiloi sp nov., the causative agent of bleaching of the coral Oculina patagonica. Int J Syst Evol Microbiol. 2001;51:1383–1388. doi: 10.1099/00207713-51-4-1383. [DOI] [PubMed] [Google Scholar]
- Lesser MP. Elevated temperatures and ultraviolet radiation cause oxidative stress and inhibit photosynthesis in symbiotic dinoflagellates. Limnol Oceanogr. 1996;43:271–283. [Google Scholar]
- Lesser MP. Oxidative stress in marine environments: Biochemistry and physiological ecology. Annu Rev Physiol. 2006;68:253–278. doi: 10.1146/annurev.physiol.68.040104.110001. [DOI] [PubMed] [Google Scholar]
- Lesser MP, Farrell JH. Exposure to solar radiation increases damage to both host tissues and algal symbionts of corals during thermal stress. Coral Reefs. 2004;23:367–377. [Google Scholar]
- Lesser MP, Stochaj WR, Tapley DW, Shick JM. Bleaching in coral reef anthozoans: Effects of irradiance, ultraviolet radiation, and temperature on the activities of protective enzymens against active oxygen. Coral Reefs. 1990;8:225–232. [Google Scholar]
- Maliao RJ, Turingan RG, Lin J. Phase-shift in coral reef communities in the Florida Keys National Marine Sanctuary (FKNMS), USA. Mar Biol. 2008;154:841–853. [Google Scholar]
- Martin R, Carvalho J, Ilbeas E, Hernandez M, Ruiz-Gutierrez V, Nieto ML. Acidic triterpenes compromise growth and survival of astrocytoma cell lines by regulating reactive oxygen species accumulation. Cancer Res. 2007;67:3741–3751. doi: 10.1158/0008-5472.CAN-06-4759. [DOI] [PubMed] [Google Scholar]
- Martindale JL, Holbrook NJ. Cellular response to oxidative stress: Signaling for suicide and survival. J Cell Physiol. 2002;192:1–15. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- Mates JM, Segura JA, Alonso FJ, Marquez J. Intracellular redox status and oxidative stress: implications for cell proliferation, apoptosis, and carcinogenesis. Arch Toxicol. 2008;82:273–299. doi: 10.1007/s00204-008-0304-z. [DOI] [PubMed] [Google Scholar]
- McCook LJ, Jompa J, Diaz-Pulido G. Competition between corals and algae on coral reefs: a review of evidence and mechanisms. Coral Reefs. 2001;19:400–417. [Google Scholar]
- Meriin AB, Yaglom JA, Gabai VL, Mosser DD, Zon L, Sherman MY. Protein-damaging stresses activate c-Jun N-terminal kinase via inhibition of its dephosphorylation: A novel pathway controlled by HSP72. Mol Cell Biol. 1999;19:2547–2555. doi: 10.1128/mcb.19.4.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchelmore CL, Schwarz JA, Weis VM. Development of symbiosis-specific genes as biomarkers for the early detection of cnidarian-algal symbiosis breakdown. Mar Environ Res. 2002;54:345–349. doi: 10.1016/s0141-1136(02)00201-5. [DOI] [PubMed] [Google Scholar]
- Morgan MB, Snell TW. Characterizing stress gene expression in reef-building corals exposed to the mosquitoside dibrom. Mar Pollut Bull. 2002;44:1206–1218. doi: 10.1016/s0025-326x(02)00177-7. [DOI] [PubMed] [Google Scholar]
- Morgan MB, Vogelien DL, Snell TW. Assessing coral stress responses using molecular biomarkers of gene transcription. Environ Toxicol Chem. 2001;20:537–543. [PubMed] [Google Scholar]
- Mumby PJ, Steneck RS. Coral reef management and conservation in light of rapidly evolving ecological paradigms. Trends Ecol Evol. 2008;23:555–563. doi: 10.1016/j.tree.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Mydlarz LD, Jacobs RS. Comparison of an inducible oxidative burst in free-living and symbiotic dinoflagellates reveals properties of the pseudopterosins. Phytochemistry. 2004;65:3231–3241. doi: 10.1016/j.phytochem.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Nii CM, Muscatine L. Oxidative stress in the symbiotic sea anemone Aiptasia pulchella (Carlgren, 1943): Contribution of the animal to superoxide ion production at elevated temperature. Biol Bull. 1997;192:444–456. doi: 10.2307/1542753. [DOI] [PubMed] [Google Scholar]
- Paul VJ, Riston-Williams R, Sharp K. Marine chemical ecology in benthic environments. Nat Prod Rep. 2011b;28:345–387. doi: 10.1039/c0np00040j. [DOI] [PubMed] [Google Scholar]
- Paul VJ, Kuffner IB, Walters LJ, Ritson-Williams R, Beach KS, Becerro MA. Chemically mediated interactions between macroalgae Dictyota spp. and multiple life-history stages of the coral Porites astreoides. Mar Ecol Prog Ser. 2011a;426:161–170. [Google Scholar]
- Pawlik JR, Steindler L, Henkel TP, Beer S, Ilan M. Chemical warfare on coral reefs: Sponge metabolites differentially affect coral symbiosis in situ. Limnol Oceanogr. 2007;52:907–911. [Google Scholar]
- Peres A, Churchman ML, Hariharan S, Himanen K, Verkest A, Vandepoele K, Magyar Z, Hatzfeld Y, Van Der Schueren E, Beemster GTS, Frankard V, Larkin JC, Inze D, De Veylder L. Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. J Biol Chem. 2007;282:25588–25596. doi: 10.1074/jbc.M703326200. [DOI] [PubMed] [Google Scholar]
- Rasher DB, Hay ME. Chemically rich seaweeds poison corals when not controlled by herbivores. Proc Natl Acad Sci U S A. 2010;107:9683–9688. doi: 10.1073/pnas.0912095107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasher DB, Stout EP, Engel MS, Kubanek J, Hay ME. Macroalgal terpenes function as allelopathic agents against reef corals. Proc Natl Acad Sci U S A. 2011;108:17726–17731. doi: 10.1073/pnas.1108628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lanetty M, Harii S, Hoegh-Guldberg O. Early molecular responses of coral larvae to hyperthermal stress. Mol Ecol. 2009;18:5101–5114. doi: 10.1111/j.1365-294X.2009.04419.x. [DOI] [PubMed] [Google Scholar]
- Rudrappa T, Bonsall J, Bais HP. Root-secreted allelochemical in the noxious weed Phragmites australis deploys a reactive oxygen species response and microtubule assembly disruption to execute rhizotoxicity. J Chem Ecol. 2007;33:1898–1918. doi: 10.1007/s10886-007-9353-7. [DOI] [PubMed] [Google Scholar]
- Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: A cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- Smith JE, Shaw M, Edwards RA, Obura D, Pantos O, Sala E, Sandin SA, Smriga S, Hatay M, Rohwer FL. Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol Lett. 2006;9:835–845. doi: 10.1111/j.1461-0248.2006.00937.x. [DOI] [PubMed] [Google Scholar]
- Starcevic A, Dunlap WC, Cullum J, Shick JM, Hranueli D, Long PF. Gene expression in the scleractinian Acropora microphthalma exposed to high solar irradiance reveals elements of photoprotection and coral bleaching. PLoS ONE. 2010;5:1–10. doi: 10.1371/journal.pone.0013975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen RG, Muscatine L. Low temperature evokes rapid exocytosis of symbiotic algae by a sea anemone. Biol Bull. 1987;172:246–263. [Google Scholar]
- Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: Oxidative-stress-response genes. Proc Natl Acad Sci U S A. 2004;101:6564–6569. doi: 10.1073/pnas.0305888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titlyanov EA, Yakovleva IM, Titlyanova TV. Interaction between benthic algae (Lyngbya bouillonii, Dictyota dichotoma) and scleractinian coral Porites lutea in direct contact. J Exp Mar Biol Ecol. 2007;342:282–291. [Google Scholar]
- Venn AA, Loram JE, Douglas AE. Photosynthetic symbioses in animals. J Exp Bot. 2008;59:1069–1080. doi: 10.1093/jxb/erm328. [DOI] [PubMed] [Google Scholar]
- Warner ME, Fitt WK, Schmidt GW. Damage to photosystem II in symbiotic dinoflagellates: A determinant of coral bleaching. Proc Natl Acad Sci USA. 1999;96:8007–8012. doi: 10.1073/pnas.96.14.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner ME, Lesser MP, Ralph P. Chlorophyll fluorescence in reef building corals. In: Suggett D, Prasil O, Borowitzka M, editors. Chlorophyll a fluorescence in aquatic sciences: Methods and applications. Springer; 2010. pp. 209–222. [Google Scholar]
- Weis VM. Cellular mechanisms of cnidarian bleaching: Stress causes the collapse of symbiosis. J Exp Biol. 2008;211:3059–3066. doi: 10.1242/jeb.009597. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gu M, Shi K, Zhou YH, Yu JQ. Effects of aqueous root extracts and hydrophobic root exudates of cucumber (Cucumis sativus L.) on nuclei DNA content and expression of cell cycle-related genes in cucumber radicles. Plant Soil. 2010;327:455–463. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.