Summary
Periodontitis is a disease that affects and destroys the tissues that support teeth. Tissues damage results from a prolonged inflammatory response to an ecological shift in the composition of subgingival biofilms. Three bacterial species that constitute the red complex group, Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola, are considered the main pathogens involved in periodontitis.
In the present study a real-time PCR based assay was designed to detect and quantify red complex species, then used to investigate 146 periodontal pocket samples from 66 periodontitis patients and 80 controls. Results demonstrated a significant higher prevalence of red complex species and increased amount of P. gingivalis and T. denticola in periodontal pocket of periodontitis patients.
Keywords: tooth, ligament, periodontal, diseases, inflammation, bone, resorption
Introduction
Periodontitis represents a destructive chronic inflammatory disease with a bacterial infection resulting from the complex actions of a small subset of periodontal pathogens (1).
From a pathological point of view, periodontitis can be defined as the presence of gingival inflammation at sites where there has been a pathological detachment of collagen fibres from the cementum and the junctional epithelium has migrated apically (2). The inflammatory response of the periodontal tissues to infection is influenced by environmental factors as well as by genetic factors (3). The primary microbial factor contributing to periodontitis is a shift in the content of the oral microflora, while the primary immunological factor is the destructive host inflammatory response (4).
The microbiota associated with periodontal health and disease has been intensely studied for well over a century by several generations of skilled scientists and clinicians (5, 6). Oral microbiota is an enormously complex and dynamic entity that is profoundly affected by perpetually changing local environments and host-mediated selective pressures (7). The presence of a commensal microbiota, including potential pathogens, is essential for the proper development of mucosal immunity (8).
The normal oral flora is hence in a balance between pathogens and commensals that requires regular cleaning to be maintained. A decrease in oral hygiene is quickly followed by the build-up of oral biofilms on tooth surfaces and, if left untreated, will progress to gingival inflammation and possibly periodontitis, alveolar bone loss and loss of teeth. It is likely that differences in host-defence mechanisms, including antimicrobial protein profiles, determine whether bacterial colonization progresses to overt disease (9).
Recent data estimate that the oral cavity may contain up to 19000 bacterial phylo-types (10), but each individual will only have a rate of the total numbers of pathogens. Indeed, there is a substantial diversity in the content of the microflora between individuals (11) and between different oral sites within the same individual (12, 13). Research has indicated that dietary changes combined with poor hygiene can cause a shift in the composition of the oral bacteria (13, 14). Moreover, some evidence in recent studies suggests that the oral microbiome changes as humans age and the dysbiosis in the oral cavity can lead to periodontitis (4).
Several methods have been used for microbiological testing in periodontitis (15). However, many techniques have not been fully accepted due to low sensitivity or specificity, moreover sometimes they are slow, expensive and laborious. In our laboratory (LAB srl, Ferrara, Italy), we developed a rapid and sensitive test to detect and quantify the three bacterial species more involved in periodontitis that constituted the red complex group: Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola. Both P. gingivalis and T. denticola occur concomitantly with the clinical signs of periodontal destruction. They appear closely ‘linked’ topologically in the developing biofilm, shown an in vitro ability to produce a number of outer membrane-associated proteinases and are considered the first pathogens involved in the clinical destruction of periodontal tissues. Moreover both them and T. forsythia, show an higher prevalence in disease than in health suggesting that these bacterial are associated with the local development of periodontitis (16).
The presence and the level of these pathogens can be effectively revealed by real time polymerase chain reaction analysis using bacterial species-specific primers and probes.
Our findings support the hypothesis that detection and quantification of red complex bacteria in crevicular fluid could be an appropriate tool for diagnosis and prognosis of periodontitis.
Material and methods
A total of 146 individuals participated to the study, 66 were affected by chronic periodontitis, while 80 constituted the control group. Controls include 46 healthy individuals and 34 affected by a moderate gingivitis. Table 1 summarizes principal characteristics of the groups.
Table 1.
Principal characteristics of the groups.
| Total | Healthy | Gingivitis | Periodontitis | |
|---|---|---|---|---|
| Subjects | 146 | 46 | 34 | 66 |
| Males | 63 | 12 | 14 | 37 |
| Females | 83 | 34 | 20 | 29 |
| Age (means years ± SD) | 39.8±18.9 | 31.6±18.6 | 34.3±15.4 | 48.9±18.2 |
| Sampling depht (mm±SD) | 3.9±1.7 | 2.6±0.6 | 3.2±1.0 | 5.0±1.7 |
A sample of the periodontal pocket microbiota was obtained from a single site by a paper probe. DNA was extracted and purified using standard protocols that include two consecutive incubation with lysozyme and proteinase K, followed by spin-column purification.
Real-Time Polymerase Chain Reaction
Primers and probes oligonucleotides were designed basing on 16S rRNA gene sequences of the Human Oral Microbiome Database (HOMD 16S rRNA RefSeq Version 10.1) counting 845 entries. All the sequences were aligned in order to find either consensus sequence or less conservate spots. Two real-time polymerase chain reaction (PCR) runs were performed for each sample. The first reaction quantified the total amount of bacteria using two degenerate primers and a single probe matching an highly conservated sequence of the 16S ribosomal RNA gene. The second reaction detected and quantified the three red complex bacteria, i.e. P. gingivalis, T. forsythia and T. denticola, in a multiplex PCR. This reaction included a total of six primers and three probes that were highly specific for each specie. Oligonucleotide concentrations and PCR conditions were optimized to ensure sensitivity, specificity and no inhibitions in case of unbalanced target amounts. Absolute quantification assays were performed using the Applied Biosystems 7500 Sequence Detection System. The amplification profile was initiated by a 10 min incubation period at 95°C to activate polymerase, followed by a two-step amplification of 15 s at 95°C and 60 s at 57°C for 40 cycles. All these experiments were performed including nontemplate controls to exclude reagents contamination.
Plasmids containing synthetic DNA target sequences (Eurofin MWG Operon, Ebersberg Germany) were used as standard for the quantitative analysis. Standard curves for each target were constructed in a triplex reaction, by using a mix of the same amount of plasmids, in serial dilutions ranging from 101 to 107 copies. There was a linear relationship between the threshold cycle values plotted against the log of the copy number over the entire range of dilutions (data not shown). The copy numbers for individual plasmid preparations were estimated using the Thermo NanoDrop spectrophotometer.
The absolute quantification of total bacterial genome copies in samples allowed for the calculation of relative amount of red complex species. To prevent samples and polymerase chain reaction contamination, plasmid purification and handling was performed in a separate laboratory with dedicated pipettes.
Statistical analysis
Descriptive statistics was performed using Microsoft Excel spreadsheets. The Freeman-Halton extension of Fisher’s exact test to compute the (two-tailed) probability of obtaining a distribution of values in a 2×3 contingency table, given the number of observations in each cell. Odds ratio calculation was performed online at the OpenEpi web site (www.openepi.com).
Absolute bacteria amount were normalized against the total bacterial load, obtaining the relative bacteria amount (RBA). The one-way analysis of variance (ANOVA) was used to determine whether there was any significant differences between the mean RBA value of three patients group, i.e. healthy, gingivitis and periodontitis.
Results
Occurrence and amount of red complex bacteria from crevicular fluid were evaluated in 146 individuals. A single specimen from each patient was analyzed by quantitative real time PCR (LAB test LAB® s.r.l, Ferrara, Italy) (Figs. 1, 2), obtaining measures of total bacteria load and of three species involved in periodontitis, i.e. P. gingivalis, T. forsythia, and T. denticola. Here we report a preliminary study focused mainly on prevalence of these three species among groups of patients with different diagnosis - regardless of different clinical aspects that may describe severity of the disease – in order to understand whether the presence of the red complex species and their relative amount may be considered predictive factors of periodontitis.
Figure 1.
LAB®-test kit.
Figures 2.
Method of sampling bacterial DNA.
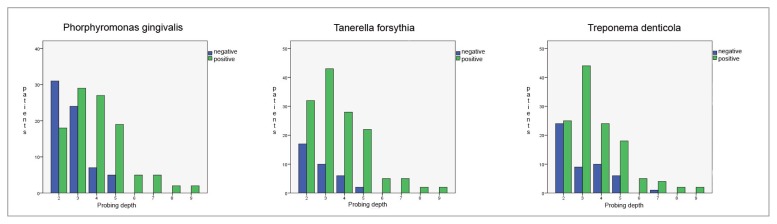
Prevalence of the three investigated species among health, gingivitis and periodontitis patients was shown in Table 2. Each specie was common among healthy patients, however the prevalence was roughly double in periodontitis group. Intermediate values, but closer to healthy individuals were observes among patients affected by gingivitis.
Table 2.
Association analysis between red complex bacteria and periodontitis.
| OR (95% C.I.) | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Healthy [1] | Gingivitis [2] | Periodontitis [3] | [1] vs. [3] | [1]+[2] vs. [3] | ||
| P. gingivalis | negative | 32 | 48 | 32 | 5.1(1.8–4.3) | 4.2(2.6–6.8) |
| positive | 38 | 32 | 34 | |||
|
| ||||||
| T. forsythia | negative | 49 | 31 | 22 | 6.1(3.1–11.9) | 4.6(2.6–7.9) |
| positive | 21 | 39 | 44 | |||
|
| ||||||
| T. denticola | negative | 42 | 38 | 43 | 3.4(1.8–6.4) | 2.4(1.5–3.9) |
| positive | 24 | 46 | 23 | |||
The Freeman-Halton extension of Fisher’s exact test indicated that the prevalence of each red complex specie is different among groups of patients with high degree of statistical significance, P. gingivalis P value = 2×10−8, T. forsythia P value = 1×10−8, and T. denticola P value = 2×10−4. The higher level of association with periodontitis was observed for T. forsythia, indeed the observed odds ratio was 6.1 (95% C.I. 3.1–11.9) when healthy individuals were compared to periodontitis patients, and 4.6 (95% C.I. 2.6–7.9) when healthy and gingivitis groups where combined and compared to periodontitis patients.
Results of quantitative data indicated that the normalized amount of P. gingivalis significantly differs among patient groups F(2, 304) = 7.77, P value = 0.001; as well as for T. denticola F(2, 304) = 7.47, P value = 0.001. On the contrary did not vary for T. forsythia F (2, 304) = 1.41, P value = 0.25. The calculated mean values were plotted in Figure 3.
Figure 3.
Plots representing the relative amount of each red complex bacterial specie in the different groups of patients.
Discussion
The polymerase chain reaction (PCR) is the most sensitive and rapid method to detect microbial pathogens in clinical specimens. In particular, the diagnostic value of PCR is significantly higher when specific pathogens that are difficult to culture in vitro or require a long cultivation period such as for anaerobic bacteria species involved in periodontitis onset. A recent improvement of this technique is the real-time PCR that allow for quantitatation of DNA target using fluorogenic probes in a close setup. Beside the opportunity to quantify target, the advantage to perform the assay is a closed system, in which the reaction tube is never opened after amplification, is of great value to prevent laboratory contamination and false positive results. In addition the need of a probe in addition to the two PCR primers, further increase the specificity of the reaction.
In the present investigation we designed and tested the performance of a real-time PCR based assay to detect and quantify the red complex bacteria involved in periodontal disease. In particular we found that P. gingivalis, T. forsythia, and T. denticola were strongly related to periodontitis because their prevalence was higher among periodontitis patient. The presence of these bacterial species can significantly increase the risk to develop periodontitis, being the OR comprised between 6.1 (T. forsythia) and 3.4 (T. denticola). The results of quantitative data analysis indicated that the relative amount of P. gingivalis and T. denticola in periodontal pocket was sensibly higher in affected patients. This indicated that both the presence and relative amount of red complex bacteria is relevant data in periodontal disease diagnosis.
Conclusion
Molecular analysis of periodontal pocket microflora by real-time PCR represent an effective inexpensive method to rapidly detect and quantify red complex bacterial species. This test was performed in a large patient sample and results demonstrated that the test is a valuable tool to improve diagnosis of periodontal disease.
Acknowledgenment
This work was supported by LAB® s.r.l, Ferrara, Italy.
References
- 1.Kuboniwa M, Inaba H, Amano A. Genotyping to distinguish microbial pathogenicity in periodontitis. Periodontology 2000. 2010;54:136–159. doi: 10.1111/j.1600-0757.2010.00352.x. [DOI] [PubMed] [Google Scholar]
- 2.Savage A, Eaton KA, Moles DR, Needleman I. A systematic review of definitions of periodontitis and methods that have been used to identify this disease. J Clin Periodontol. 2009;36:458–467. doi: 10.1111/j.1600-051X.2009.01408.x. [DOI] [PubMed] [Google Scholar]
- 3.Kinane DF, Peterson M, Stathopoulou PG. Environmental and other modifying factors of the periodontal diseases. Periodontology 2000. 2006;40:107–119. doi: 10.1111/j.1600-0757.2005.00136.x. [DOI] [PubMed] [Google Scholar]
- 4.Berezow AB, Darveau RP. Microbial shift and periodontitis. Periodontology 2000. 2011;55:36–347. doi: 10.1111/j.1600-0757.2010.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanner ACR IJ. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontology 2000. 2006;42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 6.Teles RP, Haffajee AD, Socransky SS. Microbial goals of periodontal therapy. Periodontol 2000. 2006;42:180–218. doi: 10.1111/j.1600-0757.2006.00192.x. [DOI] [PubMed] [Google Scholar]
- 7.Armitage GC. Comparison of the microbiological features of chronic and aggressive periodontitis. Periodontol 2000. 2010;53:70–88. doi: 10.1111/j.1600-0757.2010.00357.x. [DOI] [PubMed] [Google Scholar]
- 8.Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorr SU, Abdolhosseini M. Antimicrobial peptides and periodontal disease. J Clin Periodontol. 38(Suppl 11):126–141. doi: 10.1111/j.1600-051X.2010.01664.x. [DOI] [PubMed] [Google Scholar]
- 10.Slots J. Update on Actinobacillus Actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease. J Int Acad Periodontol. 1999;1:121–126. [PubMed] [Google Scholar]
- 11.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avila M, Ojcius DM, Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 2009;28:405–411. doi: 10.1089/dna.2009.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Ahmad A, Roth D, Wolkewitz M, Wiedmann-Al-Ahmad M, Follo M, Ratka-Kruger P, et al. Change in diet and oral hygiene over an 8-week period: effects on oral health and oral biofilm. Clin Oral Investig. 14:391–396. doi: 10.1007/s00784-009-0318-9. [DOI] [PubMed] [Google Scholar]
- 15.Loomer PM. Microbiological diagnostic testing in the treatment of periodontal diseases. Periodontology 2000. 2004;34:49–56. doi: 10.1046/j.0906-6713.2002.003424.x. [DOI] [PubMed] [Google Scholar]
- 16.Mineoka T, Awano S, Rikimaru T, Kurata H, Yoshida A, Ansai T, et al. Site-specific development of periodontal disease is associated with increased levels of Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia in subgingival plaque. J Periodontol. 2008;79:670–676. doi: 10.1902/jop.2008.070398. [DOI] [PubMed] [Google Scholar]